Abstract
Acute leukemias caused by translocations of the MLL gene at chromosome 11 band q23 (11q23) are characterized by a unique gene expression profile. More recently, data from several laboratories indicate that the most commonly encountered MLL fusion proteins, MLLT1, MLLT3, and AFF1 are found within a molecular complex that facilitates the elongation phase of mRNA transcription. Mutational analyses suggest that interaction between the MLLT1/3 proteins and AFF family proteins are required for experimental transformation of hematopoietic progenitor cells (HPCs). Here, we define a specific pairing of two amino acids that creates a salt bridge between MLLT1/3 and AFF proteins that is critically important for MLL-mediated transformation of HPCs. Our findings, coupled with the newly defined structure of MLLT3 in complex with AFF1, should facilitate the development of small molecules that block this amino acid interaction and interfere with the activity of the most common MLL oncoproteins.
Keywords: Oncoproteins, protein binding, mutation complementation, leukemic transformation
1. Introduction
A model is emerging that accounts for mechanisms by which several of the numerous MLL fusion partners, when fused to MLL, initiate acute leukemia. Findings from a number of laboratories indicate that the most common MLL fusion partners, MLLT1, MLLT3, and AFF1 (previously designated ENL, AF9, and AF4, respectively) are components of transcriptional elongation complexes variously termed elongation assisted proteins (EAPS) complex, super elongation complex (SEC), or AF4-ENL-P-TEFb (AEP) complex. MLL fusion proteins containing either MLLT1, MLLT3, or AFF1 are able to assemble these complexes at genes normally regulated by wild-type MLL. Presumably by facilitating transcriptional elongation, MLL target genes are expressed inappropriately by these fusion proteins [1-7]. In this context, the C-terminal ANC1 homology domains (AHD) of MLLT1 and MLLT3 directly bind AFF1 and/or one of its family members (AFF3/LAF4, AFF4/AF5q31) which in turn recruits P-TEFb. Accordingly, MLL-MLLT3 fusion genes with mutations of the MLLT3 AHD that prevent AFF1 binding fail to transform hematopoietic precursor cells (HPCs) [5, 6]. Experimentally, less is known about the requirements of MLL-AFF1 in leukemic transformation. This is due, in part, to the fact that MLL-AFF1 gene constructs fail to transform murine HPCs. However, Yokoyama and colleagues were able to experimentally transform murine HPCs with an MLL-AFF4 fusion gene (AFF4 was previously known as AF5q31) [5]. AFF4 is a close homolog of AFF1, and it has also been identified as an MLL fusion partner in human leukemia [8]. Mapping studies performed by this group showed that a C-terminal AFF1/AFF4 hetero/homodimerization domain is required for transformation. The MLLT1/3 binding domain of AFF4 contributes to the potency of transformation but is not absolutely required for this process. The N-terminal P-TEFb binding domain is not found within the truncated fusion protein and is thus dispensable as well. However, by way of either heterodimerization with AFF1 or by homodimerization, the MLL-AFF4 fusion protein complex is still able to recruit P-TEFb as well as an additional MLLT1/3 molecule.
In this report, we show that a point mutation within MLLT3 that impairs its binding to AFF1/4 significantly diminishes the activity of a MLL-MLLT3 fusion protein. Further, we use genetic complementation to extend the findings of others that recruitment of MLLT1/3 by an MLL-AFF4 fusion protein is critically important to transformation of mouse HPCs.
2. Materials and Methods
2.1. Recombinant DNA
The MLL-AFF4 viral expression vector was a gift of Dr. Michael Cleary (Stanford University). Other gene expression constructs were synthesized by PCR amplification of DNA followed by ligation of endonuclease restricted DNA into the corresponding expression vectors. MLLT3(D544R) and AFF4(K717D) mutations were obtained using the Agilent Quik Change Site Directed Mutagenesis kit according to the manufacturer’s protocol. The MLL-MLLT3(D544R) and MLL-AFF4(K717D) fusion genes were cloned by three-way ligation as previously described [9].
2.2. Cell Lines Used
HEK293T cells (Clontech) and Phoenix Ecotropic cell line (Orbigen, Inc., San Diego, CA) were cultured under standard aerobic conditions with 5% CO2 and in culture medium containing 10% fetal bovine serum.
2.3. Co-Immunoprecipitation and Western Blotting
HEK293T cells were transfected with the indicated gene expression vectors and cultured for 48 h under standard aerobic conditions. Cells were lysed and lysates were treated with anti-GFP (Life Technologies) or anti-IgG (Sigma-Aldrich) antibodies. Immune complexes were recovered with Protein A-agarose beads (Santa Cruz Technologies). Western blot was performed using anti-FLAG antibody (Sigma-Aldrich). One percent of the total cell lysate was used for input.
2.4. Confocal Microscopy
HEK293T cells were plated in 4-well chamber slides and transfected with plasmid DNA encoding mCherry fluorescent protein-tagged AFF1623-811 and GFP-tagged MLLT3. 48 h after transfection, cells were fixed using 4% paraformaldehyde in PBS. The slides were mounted with Prolong Goldantifade with DAPI before being examined using a Zeiss LSM-510 confocal microscope.
2.5. Murine Bone Marrow Colony Replating Assay
Murine bone marrow c-kit+ cells were transduced with MSCVneo, MSCVneo-MLL-MLLT3(WT), MSCVneo-MLL-MLLT3(D544R), MSCVneo-MLL-AFF4(WT) or MSCVneo-MLL-AFF4(K717D) retroviruses as previously described) [10]. Cells were plated in methylcellulose (M3234, Stem Cell Technologies) with cytokines IL-3, IL-6, SCF, GM-CSF and G418 for one week. Colony forming units were enumerated per 10,000 cells plated, and cells were serially replated after 7 days for each of four weeks. Colony assays were repeated three to ten times in duplicate, and statistical significance was calculated using Student’s t-test, p<0.05.
2.6. Genetic Complementation Assay
Murine bone marrow c-kit+ cells were transduced with MSCVyfpMLLT3(WT or D544R) and MSCVneoAFF4(WT or K717D). Cells were plated in methylcellulose with cytokines and G418 as described [10]. After one week, YFP+ cells were sorted to select for G418RYFP+ cells which were serially replated for two additional weeks. At the end of each week, colony number was enumerated per 10,000 cells.
2.7. Quantitative RT-PCR
RNA was isolated from bone marrow cells after one or two weeks in methylcellulose using TriReagent following the manufacturer’s protocol. cDNA was prepared and Real Time PCR using TaqMan probes for Meis1 (Mm00487664_m1) and Hprt (Mm01545399_m1, Applied Biosystems) was performed. Data were analyzed using the 2−ΔΔ Ct method. Expression was normalized to Hprt expression and was performed in triplicate. Results are reported as the mean ± standard error of the mean.
2.8. Binding Assays
Fluorescein AFF4 (WT) or AFF4 (K717D) peptides were titrated into MBP-AF9 (WT) or MBP-AF9 (D544R) as described [11].
3. Results and Discussion
3.1. A single electrostatic interaction is an important determinant for the stable association of AFF1 with MLLT3 as well as for MLL-MLLT3-induced transformation
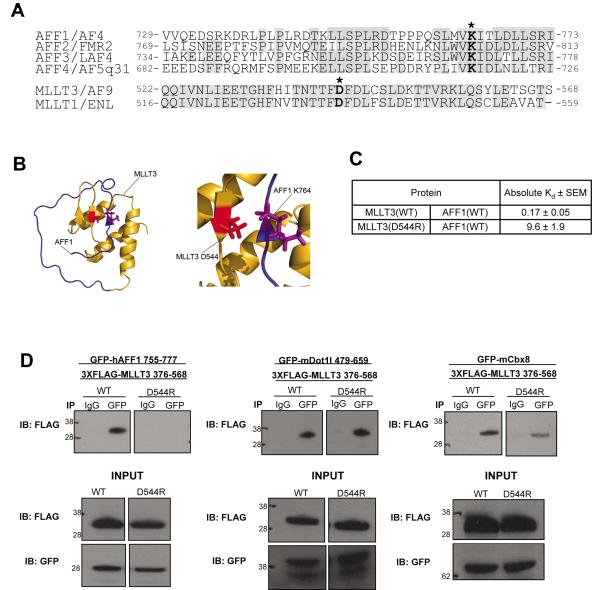
We previously identified a 14 amino acid sequence within AFF1 that binds MLLT3. This sequence is highly conserved in all AFF family members and includes a lysine residue at position 4 [12]. Moreover, the C-terminal AHDs of MLLT3 and MLLT1 are highly homologous (Figure 1A). When changed to aspartic or glutamic acid, the conserved lysine residue in AFF1 (residue 764) prevents its interaction with MLLT3 in a yeast two-hybrid assay. We used the two hybrid system to select a library of mutagenized MLLT3 cDNA molecules for mutations that restore AFF1 binding and found that substituting aspartic acid 544 for arginine has this effect (Supplementary Table 1). The structure of MLLT3 in complex with AFF1 has subsequently been reported, and it demonstrates that a salt bridge is formed between AFF1 K764 and MLLT3 D544 (Figure 1B) [11]. Using peptides and recombinant proteins in vitro, the Kd of wild type AFF1 and MLLT3 has been determined to be 0.17 nM[11] while that of AFF1 and MLLT3(D544R) is 9.6 nM (Figure 1C). This underscores the importance of this salt bridge to high affinity binding of the two proteins.
Figure 1. A single electrostatic interaction is a critical determinant for the stable association of AFF1 and MLLT3.
a) Protein sequence alignment of AFF family members shows a high degree of sequence conservation, including AFF1 lysine 764 and AFF4 lysine residue K717 which form a salt bridge with MLLT3 D544 (residues indicated by asterisks). MLLT3 is highly homologous to family member MLLT1. b) Ribbon representation of the structure of the MLLT3 – AFF1 complex (PDB code 2LMO [11]) with the side chains of MLLT3 D544 and AFF1 K764 shown. c) Binding assays were conducted as described in [11]. d) MLLT3(D544R) mutation disrupts the interaction with hAFF1, while maintaining an interaction with both Dot1l and Cbx8.
First, we wanted to determine specifically which protein interactions were affected by the MLLT3(D544R) mutation. We tested the ability of a FLAG-tagged C-terminal fragment of MLLT3 (residues 376-568) to precipitate hAFF1, as well as other known MLLT3 binding proteins mCbx8, and mDot1l. We compared these results to a similar FLAG-tagged MLLT3(D544R) protein. hAFF1 residues 755-777 (encompassing the MLLT3 interaction domain), mCbx8, and mDot1l residues 479-659 (previously shown to bind MLLT3 [13] were expressed as GFP fusion proteins in HEK293T cells. These cells co-expressed FLAG-MLLT3376-568 or FLAG-MLLT3376-568(D544R). Cell lysates were immunoprecipitated with an anti-GFP antibody and the immunoprecipitates were then analyzed by Western blot using an anti-FLAG antibody. As depicted in figure 1D, GFP-AFF1755-777, GFP-Cbx8, and GFP-Dot1l479-659 all efficiently co-precipitated FLAG-MLLT3376-568. In contrast, while GFP-Cbx8 and GFP-Dot1l479-659 were able to co-precipitate FLAG-MLLT3376-568(D544R), GFP-AFF1755-777 did not. These findings indicate that the MLLT3 D544R mutation specifically impairs binding to AFF1 to a much greater degree than it does to either Cbx8 or Dot1l. NMR HSQC spectra of MLLT3 in complex with CBX8 and DOT1L have been reported but do not immediately reveal the effects of a MLLT3 D544R mutation [11]. We await the complete NMR structures of these protein complexes.
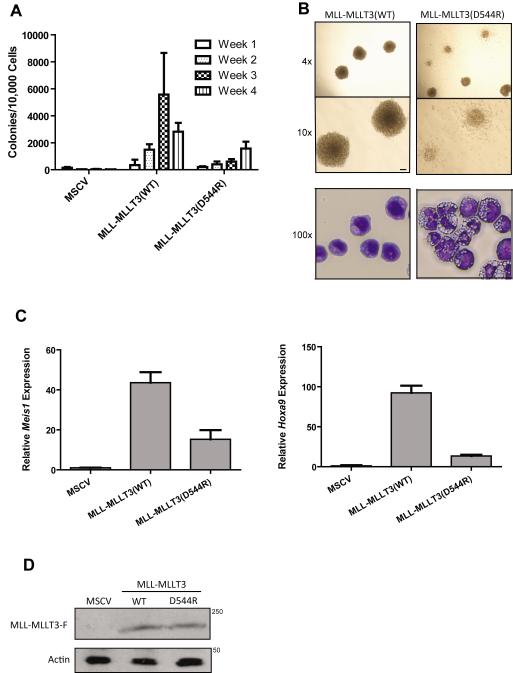
Next, we analyzed the effect of the MLLT3 D544R mutation on the ability of MLL-MLLT3 oncoprotein to transform mouse HPCs. Based on current models, we predicted that a MLL-MLLT3 molecule with a D544R substitution in MLLT3 would inefficiently transform HPCs due to its diminished affinity for AFF1/4. c-kit+ HPCs were isolated from the bone marrow of C57BL/6 mice and were transduced with retrovirus expressing either no oncogene, MLL-MLLT3, or MLL-MLLT3(D544R). Cells were then re-plated on a weekly basis in methylcellulose medium and colony numbers were enumerated. Figure 2A shows that while MLL-MLLT3 efficiently transformed HPCs, MLL-MLLT3(D544R) produced significantly fewer colonies at weeks 3 and 4. Additionally, the colonies formed by MLL-MLLT3(D544R)-transduced cells were more diffuse and the cells themselves were more differentiated when compared to cells transduced with the wild-type oncogene (Figure 2B). It is noteworthy that colony number was reduced but not abolished. We attribute this to the ability of MLLT3(D544R) to interact with AFF1/4, albeit weakly.
Figure 2. MLLT3-AFF1 interaction is required for MLL-MLLT3-induced transformation.
a) Cells expressing MLL-MLLT3(WT) efficiently re-plated, whereas cells expressing MLL-MLLT3(D544R) showed a statistically significant reduction in colony-forming ability (p<0.02). Colony assays were conducted in duplicate and repeated ten times. b) Top panel, MLL-MLLT3(D544R) expressing cells exhibited more diffuse colony morphology as compared to the dense, compact colonies formed by MLL-MLLT3(WT) cells; bottom panel, cytospin and staining with Wright-Giemsa indicate that MLL-MLLT3(WT)-expressing cells exhibit blast-like morphology, whereas cells expressing MLL-MLLT3(D544R) appear differentiated. c) Expression of MLL-MLLT3 target gene Meis1 and d) Hoxa9 was significantly reduced in c-kit+ HPCs cells harvested at week one expressing MLL-MLLT3(D544R) compared to MLL-MLLT3(WT) ( ** = p<0.02). e) MLL-MLLT3(WT) and MLL-MLLT3(D544R) proteins are stably expressed in bone marrow isolated at week one as shown by western blot.
We tested the effect of the mutant oncoprotein on activation of the MLL target genes Meis1 and Hoxa9. MLL fusion proteins are known to activate Meis1 and Hoxa9, and their expression is a requirement for transformation of HPCs [14,15]. One week following transduction of murine c-kit+ HPCs, gene expression was measured by quantitative reverse transcription polymerase chain reaction (QRT-PCR). As shown in figure 2C, Meis1 and Hoxa9 mRNA was found at significantly lower levels in cells transduced by MLL-MLLT3(D544R) in comparison to MLL-MLLT3-transduced cells. This effect was not due to instability of the mutant protein as it was detectable in mouse HPCs at levels similar to wild-type MLL-MLLT3 when analyzed one week following transduction (Figure 2D). These findings are consistent with those of Yokoyama and Biswas and their colleagues who studied the effects of other specific mutations within the MLLT3 AHD on MLL fusion protein-induced HPC transformation [5,6]. We conclude that the MLL-MLLT3 oncoprotein must interact with AFF1/4 in order to stimulate MLL target gene expression and to transform c-kit+ HPCs. Coupled with the findings of Tan and co-workers who demonstrated the importance of Cbx8[16], the data indicate that both AFF1/4 and CBX8 binding are required for MLL-MLLT3-induced HPC transformation. Additionally, structural data indicate that binding of AFF1/4 and CBX8 to MLLT3 is mutually exclusive, therefore distinct MLL-MLLT3 – AFF1/4 and MLL-MLLT3 – CBX8 complexes are likely to be necessary for transformation[11].
3.2. MLLT3(D544R) is able to complement a lysine to aspartic acid mutation within the MLLT3 binding domain of AFF1/4 both physically and functionally
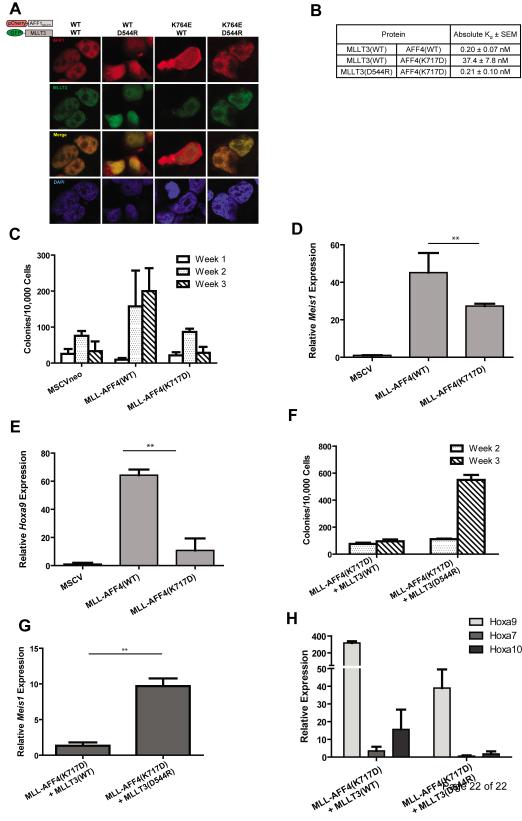
As indicated, the electrostatic interaction between AFF1 and MLLT3 D544 occurs at position K764 of AFF1. We tested the effects of a K764E mutation in AFF1 on MLLT3 binding in vivo. To do this, we took advantage of a fragment of AFF1 encompassing amino acids 623-811. This protein fragment contains the MLLT3 interaction domain but lacks a nuclear localization sequence. We previously showed that when tagged to a fluorescent protein, AFF1623-811 is found in the cytoplasm and to a lesser extent in the nucleus (some nuclear recruitment is due to the presence of endogenous MLLT3 in HEK293T cells). Co-expression of fluorescent protein-tagged MLLT3 recruits almost all AFF1623-811 into the nucleus where the two proteins co-localize[12]. Here, we expressed AFF1623-811 with a K764E mutation fused to a mCherry fluorescent tag. In contrast to wild-type AFF1623-811, co-expression of GFP-tagged MLLT3 does not recruit mCherry-AFF1623-811(K764E) into the nucleus. However, the combined expression of mCherry-AFF1623-811(K764E) with GFP-MLLT3(D544R) leads to co-localization of both proteins within the nucleus (Figure 3A). As expected, GFP-MLLT3(D544R) does not co-localize with mCherry-AFF1623-811 with a wild-type sequence. These results demonstrate the importance in vivo of the salt bridge that forms between AFF1 K764 and MLLT3 D544.
Figure 3. Restoration of the salt-bridge interaction between MLLT3 and AFF1/AFF4 is required for nuclear colocalization and rescue of in vitro leukemic transformation ability.
a) Co-expression of a fragment AFF1 encompassing residues 623-811and MLLT3(WT) in HEK293T cells leads to colocalization of both proteins in the nucleus. Disruption of the salt-bridge interaction by co-expression of AFF1623-811 with MLLT3(D544R), or AFF1623-811(K764E) with MLLT3(WT) results in a loss of this colocalization. Restoration of the salt-bridge interaction by expression of the two mutants AFF1(K764E) and MLLT3(D544R) restores nuclear colocalization. b) AFF4(K717D) reduces binding of AF9 by 187 fold. The AF9(D544R) mutant can restore binding of AFF4(K717D) to wild type levels. c) Cells expressing MLL-AFF4(WT) efficiently re-plated for the duration of the experiment, whereas cells expressing MLL-AFF4(K717D) showed reduced colony formation ability. d) Meis1 expression and e) Hoxa9 was reduced in MLL-AFF4(K717D)-expressing cells compared to MLL-AFF4(WT). ** = p<0.02. f) Cells co-expressing MLL-AFF4(K717D) and MLLT3(D544R) showed increased transformation capacity and g) relative Meis1 expression compared to cells co-expressing MLL-AFF4(K717D) and MLL-MLLT3(WT). h) Cells expressing mutant MLL-AFF4(K717D) in conjunction with wild-type MLLT3, and to a lesser degree mutant MLLT3(D544R), exhibit high-level Hoxa9 expression. Also shown is the expression of other MLL Hox gene targets, Hoxa7 and Hoxa10.
Our results led us to reason that a mutation in AFF1 at K764, by weakening its interaction with MLLT3, would attenuate the ability of MLL-AFF1 fusion proteins to transform HPCs. We relied on a MLL-AFF4 expression vector to test this prediction as MLL-AFF1 does not transform HPCs under standard experimental conditions [5]. A retroviral vector expressing MLL-AFF4 (gift of Dr. Michael Cleary) was modified to encode aspartic acid at residue 717 of AFF4. This residue functionally corresponds to AFF1 K764. Before conducting the HPC transformation assays, we first determined whether the dissociation constant for MLLT3 and AFF4 is similar to that of AFF1 and then tested the effects of the AFF4(K717D) mutation on AFF4 – MLLT3 binding in vitro. We found that the Kd of MLLT3 and AFF4 is 0.20 nM, a value that is nearly identical to that of MLLT3 and AFF1[11]. When the K717D mutation is introduced into AFF4, the Kd increases to 37.4 nM. As with AFF1(K764E), a D544R substitution in MLLT3 restores binding to AFF4(K717D). The Kd of the two mutant proteins is equivalent to that of the two wild-type proteins, 0.21 nM (Figure 3B).
As was previously demonstrated by Yokoyama et al., when introduced into c-kit+ selected HPCs by retroviral transduction, MLL-AFF4 efficiently transforms these cells in vitro. In contrast, in the same serial replating assay, MLL-AFF4(K717D) exhibits transforming activity that does not differ significantly from cells transduced with the empty retroviral expression vector (Figure 3C). Expression of Meis1 and Hoxa9 measured one week following retroviral transduction was also impaired by the MLL-AFF4(K717D) mutation but expression was nevertheless significantly greater than in cells transduced with the empty expression vector (Figures 3D and 3E). This result differs somewhat from that of Yokoyama et al., who found that an MLL-AFF4 oncogene that does not encode the MLLT3 binding domain does transform murine HPCs albeit less efficiently than wild-type MLL-AFF4 [5]. The reason for this discrepancy is not clear but it compelled us to address whether the AFF4(K717D) mutation had additional deleterious effects on the protein apart from hindering MLLT3 binding.
Reasoning that the effect of this mutation on transformation is due solely to impaired AFF4 – MLLT3 binding, we predicted that co-expression of a mutant MLLT3 D544R protein which restores the electrostatic interaction within the respective binding domains would also restore the full transforming activity of MLL-AFF4 (K717D). We transduced c-kit+ HPCs with retroviral vectors expressing wild-type and mutant MLL-AFF4 in combination with vectors expressing wild-type and mutant MLLT3. Again, MLL-AFF4(K717D), when co-expressed with wild-type MLLT3, only weakly supported the growth of colonies after the third week of replating. However, the number of colonies found at the same time point was increased approximately six-fold among HPCs transduced with both mutant genes, MLL-AFF4(K717D) and MLLT3(D544R) strongly supporting a model in which transformation of c-kit+ HPCs by MLL-AFF4 is critically dependant on the ability of MLL-AFF4 to recruit MLLT3 (Figure 3F). As predicted, Meis1 expression was up-regulated by co-expression of both mutant genes (Figure 3G). We were surprised by the behavior of Hoxa9 which was expressed at high levels when MLL-AFF4(K717D) was paired with wild-type MLLT3 (Figure 3H). The experiment was performed in quadruplicate with similar results and the combined data are presented. Thus, these findings suggest that forced expression of MLLT3, and to a lesser degree MLLT3(D544R) activate Hoxa9 gene expression in c-kit+ HPCs. The effect of Mllt3 on Hox gene expression has been previously noted in Mllt3 null mice which exhibit a homeotic phenotype [17]. Despite activation of Hoxa9 by MLLT3, MLL-AFF4(K717D) failed to efficiently transform cells, due at least in part by weak activation of Meis1. The results of the transformation assays have additional implications. AFF1 and AFF4 homo- and heterodimerize through a binding domain that is apart from the K717D mutation. By way of homo-/heterodimerization of AFF1/4, two molecules of MLLT3 (or MLLT1) are recruited to the protein complex. Our data indicate that only one molecule of MLLT1/3 is sufficient for transformation by binding to the endogenous wild-type AFF1/4 contained in the MLL-AFF4(K717D) complex.
We attempted to perform the reciprocal experiment, specifically to rescue the impaired transforming capacity of MLL-MLLT3(D544R) by co-expressing AFF1(K764E). However, we found that retroviral expression of either wild-type AFF1 or AFF1(K764E) was toxic to mouse HPCs precluding further analysis (data not shown).
4. Conclusion
In conclusion, our findings extend those of others [5,6] revealing the critical importance of the physical interactions of AFF1 and its homologs as well as MLLT3 and its family members in MLL leukemia. Pinpointing a critical salt bridge within the binding motifs of these proteins in addition to the available structural data of the AFF1 – MLLT3 proteins in complex may aid in the development of small molecule inhibitors that can block the protein interaction to therapeutic advantage.
Supplementary Material
Highlights.
A single salt-bridge is important for binding of AFF1/4 with MLLT3.
MLL-MLLT3- and MLL-AFF4-induced transformation depends on this salt-bridge. Restoration of electrostatic interactions in mutant proteins rescues transformation.
Acknowledgments
This work was supported by the National Institutes of Health grant CA098459 to C.S.H. and CA155238 to J.H.B., C.S.H., and N.J.Z-L.
The work described in the manuscript was supported by the National Institutes of Health grant CA 155328 to John H. Bushweller, Charles S. Hemenway, and Nancy J. Zeleznik-Le.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. doi:10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 2.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. doi:10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. doi:10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. doi:10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. doi:10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci U S A. 2011;108:15751–15756. doi: 10.1073/pnas.1111498108. doi:10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monroe SC, Jo SY, Sanders DS, Basrur V, Elenitoba-Johnson KS, Slany RK, Hess JL. MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia. Exp Hematol. 2011;39:77–86. doi: 10.1016/j.exphem.2010.09.003. e71-75. doi:10.1016/j.exphem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taki T, Kano H, Taniwaki M, Sako M, Yanagisawa M, Hayashi Y. AF5q31, a newly identified AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia with ins(5;11)(q31;q13q23) Proc Natl Acad Sci U S A. 1999;96:14535–14540. doi: 10.1073/pnas.96.25.14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavau C, Du C, Thirman M, Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. Embo j. 2000;19:4655–4664. doi: 10.1093/emboj/19.17.4655. doi:10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17:62–68. doi: 10.1038/nsmb.1714. doi:10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leach BI, Kuntimaddi A, Schmidt CR, Cierpicki T, Johnson SA, Bushweller JH. Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure. 2013;21:176–183. doi: 10.1016/j.str.2012.11.011. doi:10.1016/j.str.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan RS, Nesbit JB, Marrero L, Erfurth F, LaRussa VF, Hemenway CS. The synthetic peptide PFWT disrupts AF4-AF9 protein complexes and induces apoptosis in t(4;11) leukemia cells. Leukemia. 2004;18:1364–1372. doi: 10.1038/sj.leu.2403415. doi:10.1038/sj.leu.2403415. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. doi:10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar AR, Li Q, Hudson WA, Chen W, Sam T, Yao Q, Lund EA, Wu B, Kowal BJ, Kersey JH. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. doi:10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlovsky K, Kalinkovich A, Rozovskaia T, Shezen E, Itkin T, Alder H, Ozer HG, Carramusa L, Avigdor A, Volinia S, Buchberg A, Mazo A, Kollet O, Largman C, Croce CM, Nakamura T, Lapidot T, Canaani E. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc Natl Acad Sci U S A. 2011;108:7956–7961. doi: 10.1073/pnas.1103154108. doi:10.1073/pnas.1103154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. doi:10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins EC, Appert A, Ariza-McNaughton L, Pannell R, Yamada Y, Rabbitts TH. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol Cell Biol. 2002;22:7313–7324. doi: 10.1128/MCB.22.20.7313-7324.2002. doi: 10.1128/MCB.22.20.7313-7324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.