Abstract
To determine whether the EP4 receptor for prostaglandin E2 (PGE2) contributes to the tumor promoting activity of PGs in murine skin, EP4 over‐expressing mice (BK5.EP4) were generated and subjected carcinogenesis protocols. An initiation/promotion protocol resulted in 25‐fold more squamous cell carcinomas (SCCs) in the BK5.EP4 mice than wild type (WT) mice. An increase in SCCs also occurred following treatment with initiator alone or UV irradiation. The initiator dimethylbenz[a]anthracene caused cytotoxicity in BK5.EP4, but not WT mice, characterized by sloughing of the interfollicular epidermis, regeneration and subsequent SCC development. A comparison of transcriptomes between BK5.EP4 and WT mice treated with PGE2 showed a significant upregulation of a number of genes known to be associated with tumor development, supporting a pro‐tumorigenic role for the EP4 receptor.
Keywords: Prostaglandin E2, EP4 receptor, Tumor promotion, Skin tumors, Murine skin
Highlights
Transgenic mice overexpressing the EP4 receptor for prostaglandin E2 (PGE2) were generated.
The EP4 transgene conferred endogenous tumor promoting and progression activity in skin.
The transcriptomes of BK5.EP4 and wild type mice treated with PGE2 were compared.
Genes associated with tumor development were upregulated in BK5.EP4 mice.
Interleukin‐20 was among the most highly upregulated in BK5.EP4 mice.
1. Introduction
The development of non‐melanoma skin cancer (NMSC) is a complex process characterized by the acquisition of altered proliferation and invasiveness. These changes are classically defined as occurring in stages, i.e., initiation, promotion, and progression. In the two‐stage model of initiation and promotion in mouse skin, initiation is due to DNA mutations, conferred by low dose carcinogen, while promotion involves increased proliferation and inflammation. A high dose of carcinogen, including exposure to ultraviolet light (UV) alone can accomplish both initiation and promotion. One of the most common features of exposure to chemical carcinogens or UV is the induction of inflammation, which is characterized by the production and release of cytokines, growth factors and arachidonic acid metabolites, particularly the prostaglandins (PGs) (Rundhaug and Fischer, 2010).
The enzymes responsible for the production of PGs are referred to as the PGH synthases, of which there are two isoforms, cyclooxygenase‐1 (COX‐1) and cyclooxygenase‐2 (COX‐2). Although both forms exhibit similar COX activity with regard to arachidonic acid metabolism, they are differentially regulated such that COX‐1 is normally constitutively expressed in murine epidermis, while COX‐2, which is usually expressed at only very low levels, is highly inducible by a large number of irritating agents, including the tumor promoter 12‐tetradecanoylphorbol‐13‐acetate (TPA) (Fischer et al., 2007; Maldve and Fischer, 1996; Rundhaug and Fischer, 2010).
Both pharmacological and genetic approaches have shown that PGs are critical for NMSC development. Non‐steroidal anti‐inflammatory drugs that target both COX‐1 and COX‐2, as well as selective COX‐2 inhibitors, have strong chemopreventive activity against either chemical‐ or UV‐induced skin carcinogenesis (Fischer et al., 1999; Muller‐Decker, 2011; Tober et al., 2006). The role of PGs from COX‐2 in NMSC was further verified using mice in which one or more alleles of COX‐2 were knocked out (Rundhaug et al., 2007; Tiano et al., 2002).
The clear demonstration that the induction of COX‐2 and its primary product PGE2 are involved in NMSC raised questions concerning the mechanisms by which PGE2 promotes the growth of NMSC. PGE2 binds to and activates four G‐protein‐coupled E receptors, referred to as EP1, EP2, EP3 and EP4. Each EP receptor is linked to Gα subunits, such that activation of the different receptors leads to activation of different signal transduction pathways (Narumiya, 2009; Sugimoto and Narumiya, 2007). This led to investigations identifying which receptor is responsible for the tumor promoting action of PGE2.
Studies on the EP1 receptor, which activates phospholipase C and subsequently protein kinase C, the target of the tumor promoter TPA, showed that mice over‐expressing the EP1 receptor in the epidermis developed very aggressive squamous cell carcinomas (SCC) within a few weeks after treatment with 7,12‐dimethylbenz[a]anthracene (DMBA) (Surh et al., 2012). Over‐expression of the EP2 receptor, which is linked to the cAMP‐protein kinase A pathway (Sugimoto and Narumiya, 2007), significantly increased the number of both papillomas and SCCs after two‐stage DMBA‐TPA treatment (Sung et al., 2006). Conversely, knockout of the EP2 receptor reduced skin tumor numbers by 50% (Sung et al., 2005). Knockout of the EP3 receptor, of which there are multiple variants that inhibit cAMP production (Namba et al., 1993), had no effect on two‐stage DMBA‐TPA skin carcinogenesis (Sung et al., 2005). Like the EP2 receptor, EP4 activates adenylate cyclase, but also has been reported to activate the PI3K‐Akt pathway as well (Fujino et al., 2003). However, the contribution of the EP4 receptor to skin carcinogenesis has not been previously studied.
Here we describe the effect of over‐expressing the EP4 receptor, under control of a keratin 5 promoter, on both two‐stage DMBA‐TPA and single treatment DMBA‐ or UV‐induced skin tumor development. With all protocols, the number of SCCs was significantly higher in BK5.EP4 compared to wild type (WT) mice, which had very few tumors, suggesting that elevated EP4 levels confer endogenous tumor promoting, and especially progression, activity. A comparison of transcriptomes between WT and BK5.EP4 mice treated with PGE2 and BK5.EP4 mice treated with either EtOH or PGE2 was carried out. Ingenuity Pathway Analysis (IPA) revealed differentially expressed transcripts to be part of several networks that include keratinocyte differentiation‐ and inflammation‐associated genes, e.g., many keratins, Csf2rb, Coro2A, GPX4 and IL‐20, which further supports a pro‐tumorigenic role for the EP4 receptor.
2. Materials and methods
2.1. Generation of BK5.EP4 transgenic mice
Murine EP4 cDNA was excised from the pBluescript‐EP4 vector, which was kindly provided by Dr. Yukihiko Sugimoto (Kyoto University, Kyoto, Japan), by digestion with XhoI, followed by blunt end formation using Klenow (Roche, Indiannapolis, IN), then digested with NotI (Roche). The pGEM7Z‐BK5 vector generated in our lab containing the BK5 promoter in the pGEM7Z vector (Promega, Madison, WI) was digested with SnaBI and NotI (Roche). Both the EP4 cDNA fragment and the BK5 vector fragment were gel purified using Qiaex II (Qiagen; Valencia, CA) and ligation of the EP4 cDNA into the BK5 vector generated the BK5.EP4 vector. The EP4 transgene containing the BK5 promoter, a β‐globin intron, the EP4 cDNA, and an SV40 polyA tail was excised from the BK5.EP4 vector by digestion with SalI and ClaI (Roche). The transgene was microinjected into fertilized eggs of FVB mice and transferred to pseudopregnant FVB mice, which was carried out by the Transgenic Services Core at Science Park. An EP4 cDNA probe was generated by digestion of the pBluescript‐EP4 vector with XhoI and NotI (Roche), which then was α‐[32P]‐dCTP‐radiolabeled using random primed labeling (Decaprime II kit; Ambion, Austin, TX). The offspring were genotyped by Southern blot with this probe to identify 2 founders (lines 2 and 9) that were subsequently crossed to WT FVB mice. The heterozygous (±) BK5.EP4 mice, along with their WT littermates, were genotyped by polymerase chain reaction (PCR) using the oligomers 5'‐ACT ACA TCC TGG TCA TCA TCC TGC‐3' and 5'‐TCG TCT CTT TCT GCT CCT TGC G‐3'. Epidermal scrapes were collected from 3 mice of each genotype, total RNA extracted with Tri‐Reagent (Molecular Research Center, Inc., Cincinnati, OH), purified through RNeasy minicolumns (Qiagen) and the mRNA used for qPCR analysis of EP4 expression levels. All mice were housed in climate‐controlled quarters 22 ± 1 °C at 50% humidity with 12 h light/dark cycles; Science Park is AAALAC accredited.
2.2. Cyclic adenosine monophosphate assay (cAMP)
Five 7‐wk old WT and BK5.EP4 mice were topically treated once with 200 μl 95% ethanol (EtOH) or 30 μg PGE2 in 200 μl 95% EtOH and sacrificed 1 h after treatment. The epidermis was chipped and homogenized in 5% TCA, and the supernatant assayed as previously described(Sung et al., 2006) using a cAMP EIA kit from Cayman Chemical Co. (Ann Arbor, MI).
2.3. Tumor and short‐term experiments
Groups of shaved WT and BK5.EP4 mice 6–8 wks of age were subjected to one of 3 skin carcinogenesis protocols. For the two‐stage protocol, mice were initiated topically with 100 μg DMBA (Sigma, St. Louis, MO) in 200 μl acetone, and followed 1 wk later by twice weekly applications of 2.5 μg TPA (LKT Laboratories, St. Paul, MN) in 200 μl acetone. For the DMBA‐only protocol, mice were treated topically one time with 400 μg DMBA in 200 μl acetone (no TPA treatments). Exposure of WT and BK5.EP4 mice to UV (1080 or 1440 mJ/cm2) was carried out as previously described (Mikulec et al., 2013). Tumor incidence and tumor multiplicity were recorded weekly. Tumor multiplicity was calculated as the average number of skin tumors per mouse; tumor incidence was calculated as the percentage of mice with tumors. Tumor type was determined by histological analyses as papillomas or SCCs. Mice used in short‐term experiments were treated as descried above, sacrificed at designated times after treatment and their skins used for histological analysis or the generation of epidermal lysates for immunoblotting.
2.4. Histology and immunohistochemistry
Five 6–8 wk old WT and BK5.EP4 mice were topically treated once or 6 times with 200 μl acetone or 2.5 μg TPA in 200 μl acetone. At specified times after the last treatment, mice were sacrificed, dorsal skin removed and skin sections fixed in 10% formalin, embedded in paraffin and stained with either hematoxylin and eosin (H&E) or specific antibodies. Immunohistochemistry (IHC) was performed using anti‐CD31 (1:400; R&D Systems, Minneapolis, MN), anti‐macrophage antibody (Macrophage Marker Antibody) (1:50; Santa Cruz Biotechnology, Santa Cruz, CA), anti‐Ki67 (1:200; Dako, Carinteria, CA) and anti‐caspase 3 (1:500; R&D Systems), with either a 2‐ or 3‐step labeled polymer HRP detection system (GE Healthcare, Buckinghamshire, UK) on cross sections of dorsal skin. Angiogenesis was analyzed by counting the number of CD31‐positive vessels in 12 different fields using a 20× objective, while inflammation was analyzed by counting the number of macrophages in 15 different fields using a 40× objective. Proliferation was assessed by counting the number of Ki67‐positive basal cells in approximately 1000 total cells, while apoptosis was assessed by counting the number of caspase 3‐positive cells in approximately 1500 total cells as well as counting the number of hair follicles positive for caspase 3 in 100 total hair follicles. Stem cells were visualized by IHC using anti‐CD34 (1:50; eBioscience, San Diego, CA) and anti‐K15 (1:2000; Thermo Scientific, Waltham, MA) antibodies.
Tumors taken at the end of the DMBA‐only experiment, and acetone‐ or DMBA‐treated skin, were fixed in 10% formalin, embedded in paraffin, and stained with H&E. IHC was performed on acetone‐ and DMBA‐treated skin using anti‐Ki67, anti‐caspase 3, anti‐CD31, as described above, and anti‐MMP‐9 (1:500), anti MMP‐7 (1:50) and anti‐E‐cadherin (1:50) antibodies (Chemicon, Temecula, CA). Quantitation was performed as described above. The Histology and Tissue Processing Core at Science Park performed all histologies and IHCs.
2.5. Western blots
Three or more 7‐wk old WT and BK5.EP4 mice were topically treated with 200 μl acetone or 2.5 μg TPA in 200 μl acetone, and total protein was isolated from the epidermis at 3, 12, and 24 h after treatment as previously described (Sung et al., 2006). Fifty to 75 μg epidermal lysates were separated on either an 8% or 15% SDS‐polyacrylamide gel and transferred to polyvinylidene difluoride membranes (Thermo Scientific). Primary antibodies used were anti‐MMP‐9 (1:1000; Chemicon) for the 24 h samples, anti‐pErk1/2 (1:1000), anti‐Erk1/2 (1:1000), anti‐pStat3 (1:1000), and anti‐Stat3 (1:1000), all from Cell Signaling, Danvers, MA, for the 3 h samples, and anti‐actin‐HRP (1:1000; Santa Cruz Biotechnology) for all samples as loading controls. Antibodies against CYP1A1 and CYP1B1 (1:500, Santa Cruz Biotechnology) and aromatase (1:1000; Invitrogen, Carlsbad, CA) were also used. Secondary antibodies were anti‐mouse‐HRP (1:2500) and anti‐rabbit‐HRP (1:10000; GE Healthcare). Blots were blocked in 5% BSA in Tris‐buffered saline containing 0.1% Tween 20 (TBST), primary antibodies were incubated overnight in 5% BSA in TBST at 4 °C, and the secondary antibodies were incubated for an hour in 5% non‐fat milk in TBST at room temperature. The membranes were washed, and SuperSignal West Femto Maximum Sensitivity Substrate (Pierce Biotechnology, Inc., IL) was used to detect antibody binding.
2.6. Label‐retaining cells
Nine 3‐day old WT and 4 3‐day old BK5.EP4 pups were weighed and injected with 50 μg 5‐bromo‐2‐deoxyuridine (BrdU)/g body weight twice a day for 3 days. At 7 wks of age, skin was harvested, fixed in 10% formalin, and embedded in paraffin. IHC was performed using an anti‐BrdU antibody (1:500; Accurate Chemical and Scientific Corp., Westbury, NY) to observe the label‐retaining cells.
2.7. Stem cell isolation from the bulge region
Bulge region keratinocytes were isolated from WT and BK5.EP4 mice 7 wks of age as previously described (Trempus et al., 2007). Hair follicle cells were labeled with a CD34‐biotin antibody (eBioscience), a stem cell marker, followed with streptavidin coupled allophycocyanin (Invitrogen) as well as labeled with an anti‐α6 integrin‐PE antibody (R&D Systems) and analyzed using a Becton Dickson FACS Aria SORP (San Jose, CA) flow cytometer by the Cell and Tissue Analysis Facility Core at Science Park. Three separate experiments were performed.
2.8. Abrasion wound healing
Groups of 3 mice were shaved on the dorsal side and hair depilated with Nair (Church&Dwight Co., Princeton, NJ). The epidermis was then abraded using a sterile felt wheel as previously described (Morris et al., 2000). Wound diameters were measured every other day until the wounds completely healed.
2.9. RNA extraction and sequencing using RNA‐seq technology
Six 7‐wk old WT FVB and BK5.EP4 mice were topically treated once with 200 μl 95% EtOH or 30 μg PGE2 in 200 μl 95% EtOH. Total RNA was isolated 6 h after treatment using Tri‐Reagent, and RNAseq was performed by the Next Generation Sequencing Core at Science Park. Briefly, RNA quality was checked by on‐chip gel electrophoresis using the Agilent 2100 Bioanalyzer (Palo Alto, CA), and 100 ng of each RNA sample were transcribed into cDNA using Ovation RNA‐seq System V2 (NuGen, San Carlos, CA) and sheared using the Covaris S220 (Covaris Inc., Woburn, MA). One ?g of ∼180 base pair cDNA was used to construct a library by loading onto the SPRI‐TE Fragment Library System I (Becton Dickinson, Franklin Lakes, NJ), and libraries were enriched using 8 cycles of PCR then sequenced on an Illumina HiSeq 2000 (San Diego, CA). The reads were mapped to mouse genome (mm9) by TopHat (V2.0.6) (Kim et al., 2013). The number of fragments in each known gene from RefSeq database (Pruitt et al., 2012), downloaded from UCSC Genome Browser on March 09, 2012, was enumerated using htseq‐count from HTSeq package (V 0.5.3p9). The differential expression between BK5.EP4 and WT mice with or without PGE2 treatment was statistically accessed by R/Bioconductor package DESeq (V1.10.1)(Anders and Huber, 2010) and genes with FDR ≤0.05 and fold change ≥2 were deemed as differentially expressed. Additionally, differential expression was also assessed by R/Bioconductor edgeR (V3.0.4)(Robinson et al., 2010), and genes with FDR ≤0.05 and fold change ≥2.0 were deemed as differentially expressed.
2.10. Pathway analysis
The transcriptomes were further analyzed by IPA (Ingenuity Systems, Mountain View, CA). The expression values were uploaded onto IPA and theoretical networks were then constructed. A score for each possible network (the negative base‐10 logarithm of the p value) was calculated. Scores of 2 or higher indicated at least a 99% confidence of not being constructed by random chance.
2.11. Interleukin‐20 ELISA
Five 7‐wk old WT and BK5.EP4 mice were topically treated once with 200 μl 95% EtOH or 30 μg PGE2 in 200 μl 95% EtOH and sacrificed 8 h later. Protein was isolated from the epidermis as previously described (Sung et al., 2006), and the lysate assayed using an interleukin (IL)‐20 ELISA kit from USCN Life Science Inc. (Houston, TX) according to the manufacturer's instructions. Total protein was measured using the BCA protein assay (ThermoScientific) and the data expressed as pg IL‐20/mg protein.
2.12. Statistics
Unpaired t tests were done using GraphPad (La Jolla, CA) InStat, version 3 or Prism 6 software to determine the statistical significance for EP4 mRNA, cAMP and IL‐20 levels, as well as the number of caspase‐3 and Ki67‐positive cells and number of macrophages between WT and BK5.EP4 mice or between treatments. Welch's correction for the unpaired t test was used if standard deviations between the two groups were significantly different. p < 0.05 was considered significant.
3. Results
3.1. Characterization of BK5.EP4 transgenic mice
In order to determine the role of the EP4 receptor in mouse skin carcinogenesis, transgenic mice over‐expressing the EP4 receptor were generated (Figure 1A). Mouse EP4 receptor antibodies are unable to detect a band at the correct size, therefore Southern blot analysis was performed to establish which mice contained the EP4 transgene. BK5.EP4 mice were distinguished from WT mice by the appearance of prominent bands at approximately 5 Kb, 3 Kb, and 750 base pairs (Figure 1B). RNA analysis on untreated WT and BK5.EP4 mice showed a statistically significant (p < 0.01) 15‐fold increase in EP4 mRNA in the transgenic mice (Figure 1C). To determine the functionality of the EP4 transgene, a cAMP assay was utilized since activation of the EP4 receptor by PGE2 leads to an increase in cAMP levels (Figure 1D)(Fujino et al., 2003). No difference in cAMP levels was observed between WT and BK5.EP4 mice treated with ethanol, but after treatment with PGE2, the BK5.EP4 mice also showed a statistically significant increase (p < 0.005) in the cAMP levels compared to WT mice, indicating that the EP4 transgene is functional in the transgenic mice. BK5.EP4 pups were smaller than their WT littermates, although not statistically different, but this difference was gone by 10 weeks of age (data not shown). Histological analysis of untreated skin from both WT and BK5.EP4 mice also indicated normal morphology of the skin and hair follicles (Figure 1E), although a slight alteration of the hair cycle was observed in BK5.EP4 mice (data not shown).
Figure 1.
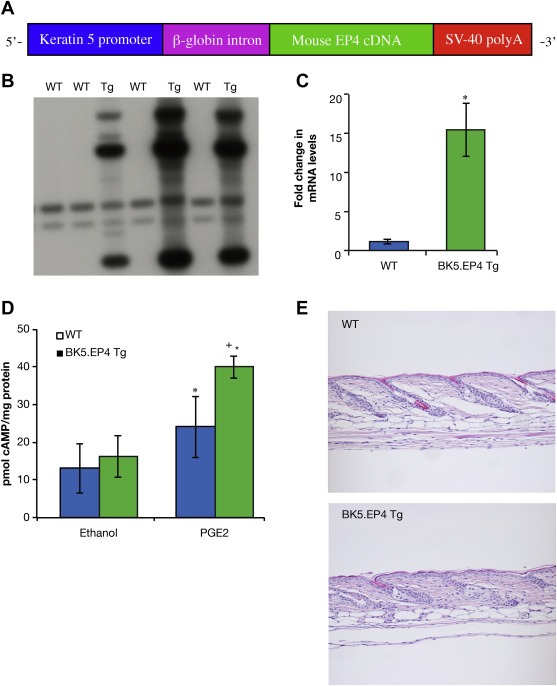
Expression of the EP4 transgene. A. Diagram of the bovine keratin 5 EP4 (BK5.EP4) construct. B. Southern blot showing integration of the BK5.EP4 construct. C. Increased EP4 mRNA levels in line 9 BK5.EP4 transgenic mice (n = 3; bars ± SD) (p < 0.01). D. Increased cAMP levels after vehicle or 30 μg PGE2 treatment (1 h) in vivo (n = 5 per group ± SD (WT PGE2 v. WT EtOH, p < 0.05; BK5.EP4 PGE2 v. WT PGE2, p < 0.005; BK5.EP4 PGE2 v. BK5.EP4 EtOH, p < 0.0001), assayed as described in Methods and Materials. E. Histology of representative H&E stained sections of skins from WT (top) and line 9 BK5.EP4 mice (bottom).
3.2. Effect of EP4 overexpression on two‐stage skin tumor development
In a short pilot experiment to test the sensitivity of the two transgenic lines, a relatively small number of WT and BK5.EP4 mice were subjected to a standard DMBA/TPA two‐stage carcinogenesis protocol. Both line 2 and line 9 BK5.EP4 mice showed a rapid increase in tumor formation compared to WT mice (Figure 2A), indicating that the difference from WT in tumor development is due to transgene and not to an insertional effect. A larger and longer experiment using line 9 transgenics showed that there were significant differences between WT and BK5.EP4 mice with regard to SCC development. While most of the tumors in the WT mice were predominantly papillomas (100% incidence at 14 weeks; data not shown), by the end of the experiment the transgenic mice had developed a large number of SCCs (Figure 2B), with a papilloma incidence similar to WT mice (data not shown). Histological analysis of SCC from WT and both lines of BK5.EP4 mice showed that there were no differences between genotypes (Figure 2C). These results suggest that the EP4 receptor plays a pro‐tumorigenic role in tumor development in mouse skin. Line 9 BK5.EP4 mice were used in all subsequent experiments.
Figure 2.
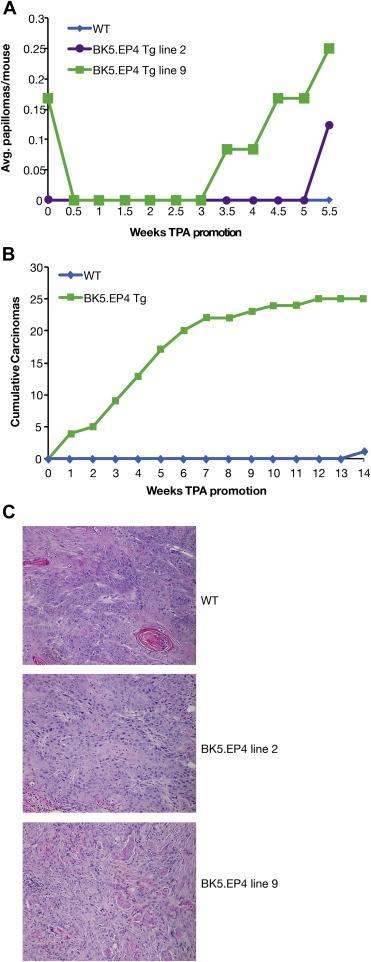
Enhanced tumor response in BK5.EP4 mice. A two‐stage DMBA/TPA protocol was carried out with WT and lines 2 and 9 of the BK5.EP4 mice. A. Papilloma multiplicity (average number of tumors per mouse; n = 13 WT, n = 8 line 2, n = 12 line 9 BK5.EP4 mice). B. Cumulative squamous cell carcinomas (SCCs) in line 9 BK5.EP4 (n = 28) and WT (n = 30) mice. C. Histology of H&E stained sections from SCCs from WT, line 2 and line 9 BK5.EP4 mice.
3.3. Overexpression of EP4 receptor increases proliferation in mouse skin
To determine the possible mechanisms by which the EP4 receptor increases skin tumorigenesis, WT and BK5.EP4 mice were treated once or six times with TPA, and their skin analyzed for Ki67 or caspase 3 expression to assess changes in proliferation and apoptosis, respectively (Suppl. Table 1). A statistically significant increase in Ki67‐positive basal cells was observed in BK5.EP4 mice with both acetone (25.8% vs 19.3%; p < 0.05) and TPA treatment (35.5% vs 26.4%; p < 0.05), compared to WT mice. No significant difference in caspase 3 expression was detected either in the epidermis or hair follicles of WT and BK5.EP4 mice after TPA treatment (Suppl. Table 1). Surprisingly, a decrease in infiltrating macrophages was observed in BK5.EP4 mice with both acetone (3.53 per field vs 6.43; ns) and 6 TPA treatments (21.12 per field vs 26.78; p < 0.05), compared to WT mice. These data suggest that the increased tumorigenesis seen in BK5.EP4 mice is due in large part to an increase in proliferation.
3.4. Effect of EP4 overexpression on DMBA‐only skin tumor development
Because one of the BK5.EP4 mice in the two‐stage tumor experiment developed a papilloma before TPA promotion was started (Figure 2A), a high‐dose DMBA‐only protocol was utilized. Visual inspection showed that the dorsal skin of the BK5.EP4 mice responded to DMBA with raised red areas; WT mice also developed some small red areas along the periphery of the treated area, but these subsided within weeks (Figure 3A). Both WT and BK5.EP4 mice responded to DMBA with marked hyperplasia by 48 h (Figure 3B and C). The major difference, however, was that the epidermis of the BK5.EP4 mice began detaching in places (Figure 3C, right panel), which persisted to day 5 (data not shown); no epidermal sloughing occurred in the WT mice. By day 15 there were palpable lesions on the transgenic mice that did not regress and subsequently developed into SCC's within 3–4 weeks. At 33 weeks all transgenic mice had growing SCC's, which necessitated their euthanasia, while only one WT mouse had a tumor (Figure 3D).
Figure 3.
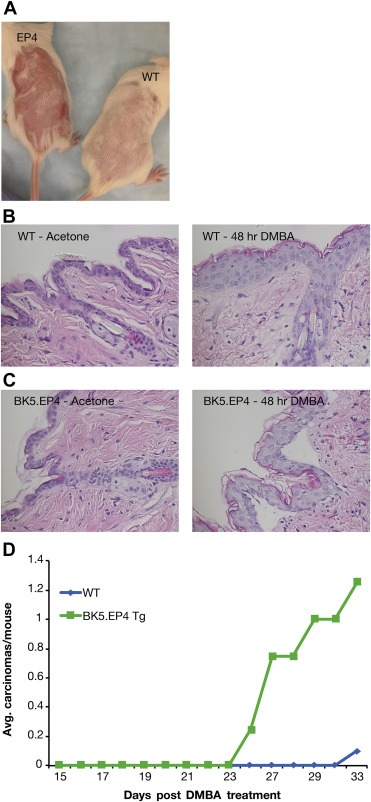
Response of WT and BK5.EP4 mice to a single treatment with 400 μg DMBA. A. Macroscopic appearance of dorsal area of treated WT and BK5.EP4 mice. B. Representative histology of H&E stained skin sections of WT (n = 3) mice 48 h after treatment with acetone or DMBA. C. Representative histology of H&E stained skin sections of BK5.EP4 (n = 3) mice 48 h after treatment with acetone or DMBA. D. Carcinoma multiplicity of WT (n = 11) and BK5.EP4 (n = 13) mice after a single treatment with DMBA.
3.5. DMBA‐induced proliferation, apoptosis and proteases
Potential differences in apoptosis and proliferation in response to DMBA were assessed. DMBA treatment caused an increase in apoptosis of interfollicular epidermal cells that was 3 times greater in the transgenic mice at 24 h (Figure 4A). This was likely responsible for the visible sloughing of areas of the epidermis at later time points (Figure 3C). Apoptosis was also greater in the hair follicles of transgenic mice, particularly at 48 h (Figure 4B). Apoptosis was followed by an increase in keratinocyte proliferation of both WT and transgenic mice that persisted for a week, which is characteristic of a regenerative response. By day 11, however, proliferation had substantially decreased in WT mice but not in transgenic mice (Figure 4C). This differential response was maintained for another week, after which proliferation was similar in both genotypes.
Figure 4.
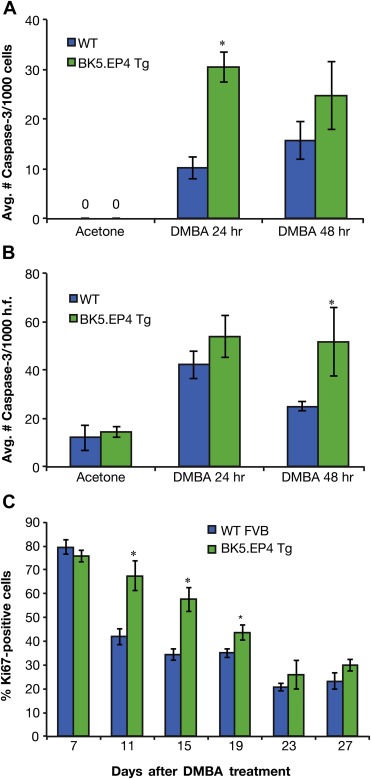
Proliferative, apoptotic and angiogenic response to DMBA treatment. A. Average number of caspase 3 positive cells/1000 cells in the interfollicular epidermis of WT and BK5.EP4 mice following acetone or DMBA (400 μg) treatment (n = 3 ± SD; DMBA 24 h BK5.EP4 v. WT, p < 0.01). B. Average number of caspase 3 positive cells/100 cells in hair follicles of acetone or DMBA treated WT and BK5.EP4 mice (n = 3 ± SD; DMBA 48 h BK5.EP4 v. WT, p < 0.02). C. Percentage of basal keratinocytes staining positive for Ki67, a marker of proliferation, in WT and BK5.EP4 mice following DMBA treatment (n = 3 ± SD; BK5.EP4 v. WT day 11, p < 0.005; BK5.EP4 v. WT day 15, p < 0.005; BK5.EP4 v. WT day 19, p < 0.02).
One of the most significant differences between genotypes was the large increase in angiogenesis, with regard to both vessel number and vessel diameters, in the transgenic mice around day 11, which was visually noticeable on the reverse side of the skin (Figure 5). A number of other possible changes or mechanisms could account for the dramatic difference between WT and BK5.EP4 mice in the toxic response to DMBA and in the tumor response, including alterations in DMBA metabolism, differences in wound healing and/or differences in stem cell populations. Immunoblotting showed (Suppl. Figure 1) no differences in the expression of aromatase, CYP1A1 or CYP1B1 between WT and transgenic mice, suggesting that the metabolism of DMBA was similar between genotypes. Similarly, a wounding experiment (Suppl. Figure 2) showed the same rate of healing in WT and transgenic mice. An analysis of keratinocyte stem cells showed (Suppl. Figure 3) that there were no significant differences in the number of CD34 or K15 positive cells in the hair follicles of transgenic and WT mice.
Figure 5.
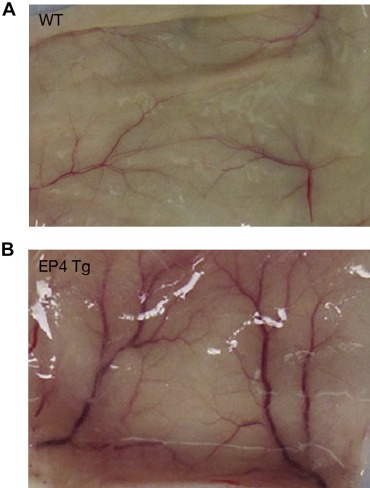
DMBA‐induced angiogenesis in BK5.EP4 mice. Macroscopic appearance of the backside of the skin of WT and BK5.EP4 mice 10 days after topical treatment with 400 μg DMBA. Photograph is representative of groups of 3 mice/genotype.
Potential differences in expression of several other proteins associated with tumorigenesis were investigated. Although DMBA induced MMP‐9 only slightly in WT mice, it robustly induced MMP‐9 in the transgenics in a sustained manner and prior to epidermal sloughing it was localized primarily to basal cells (Figure 6A). MMP‐7 was also upregulated early (by 24–48 h) in the transgenic mice (Figure 6B); by day 5 it was upregulated in the WT mice as well (data not shown). There was also greater E‐cadherin staining in the transgenic mice at 24 h, although the differences were less noticeable at later time points (Figure 6C).
Figure 6.
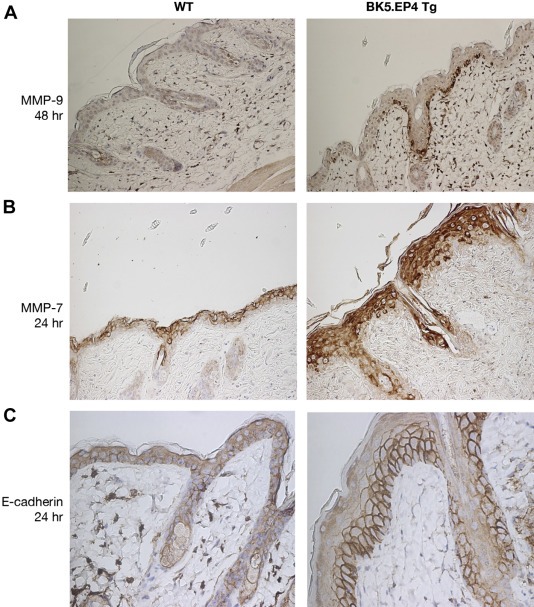
DMBA‐induced alterations in MMP‐9, MMP‐7 and E‐cadherin. WT and BK5.EP4 transgenic mice (n = 3 each) were treated with 400 μg DMBA, sacrificed after 24 and 48 h and after 5 days and processed for IHC (200× magnification). A. Representative skin sections stained with an antibody against MMP‐9 48 h after treatment. B. Representative skin sections stained with an antibody against MMP‐7 24 h after treatment. C. Representative skin section stained with an antibody against E‐cadherin 24 h after treatment.
3.6. Effect of EP4 overexpression on UV‐induced skin tumor development
To further confirm that the tumor promoting activity of the EP4 receptor was independent of DMBA per se, experiments were performed in which WT and BK5.EP4 mice were exposed to a single high dose of UV (1080 mJ/cm2). Although both genotypes responded to UV with erythema, by 7 wks the WT mice appeared normal while the transgenic mice had red raised areas similar to that seen with DMBA, but usually in the middle of the back (data not shown). By 12 wks 3 of the 4 transgenic mice had histologically verified SCCs (data not shown). No lesions or tumors developed on the WT mice. A second experiment using 1440 mJ/cm2 UV also produced tumors only on the transgenic mice (data not shown).
3.7. Analysis of the transcriptomes of WT and BK5.EP4 epidermis
To obtain information about the possible differential expression of genes that may contribute to the tumor promoting activity of the EP4 receptor, high‐throughput sequencing was carried out to compare BK5.EP4 mice treated with PGE2 or EtOH and BK5.EP4 mice treated with PGE2 with WT mice treated with PGE2. The up‐ or down‐regulated genes shown in Table 1 were generated using DESeq software (Anders and Huber, 2010). The expression levels of these genes were also found to be altered using EdgeR software (Robinson et al., 2010), which is less restrictive and thus identified a large number of other changes as well (data not shown). To further explore the observed differences in transcript levels between BK5.EP4 mice treated with PGE2 or EtOH and BK5.EP4 mice treated with PGE2 with WT mice treated with PGE2, IPA software was used to map the differentially expressed transcripts into biological networks. For transcripts from BK5.EP4 mice treated with either PGE2 or EtOH, the IPA software created 11 networks, with the first 7 having highly significant scores of 49, 39, 35, 30, 23, 17 and 12, respectively (Suppl. Table 2A; gene abbreviations defined in Suppl. Table 2B). Mapped within each of networks 1‐7 were 23, 16, 18, 16, 13, 11 and 8 differentially expressed transcripts (focus molecules). The biological functions associated with these networks include cell movement, cell‐to‐cell interaction, lipid metabolism, embryonic and organ development, cell cycle, and cell death and survival.
Table 1.
Top upregulated and downregulated transcripts in WT and BK5.EP4 mice.
| Gene | Gene name | log2 ratio | p‐value | Fold change |
|---|---|---|---|---|
| A. Top upregulated and downregulated transcripts for PGE 2 treated EP4 relative to WT mice | ||||
| Upregulated | ||||
| A130040M12Rik | Unclassified gene; VL30 family | 1.60859 | 5.20E‐06 | 3.05 |
| IL‐20 | Interleukin‐20 | No transcript | 2.33E‐07 | |
| Kcnk2 | Potassium channel, subfamily K, member 2; Trek1 | 2.32131 | 1.90E‐05 | 4.99 |
| Krtap16‐10b | Keratin associated protein 16‐10b | No transcript | 4.09E‐05 | |
| Ptger4 | Prostaglandin E receptor 4 (EP4) | 1.34283 | 7.99E‐06 | 2.54 |
| Serpinb6d | Serine (or cysteine) peptidase inhibitor, clade B member 6d | 2.11551 | 7.33E‐07 | 4.33 |
| Sprr4 | Small proline‐rich protein 4 | 1.40363 | 9.13E‐08 | 2.65 |
| Downregulated | ||||
| Akr1c13 | Aldo‐keto reductase family 1, member C13 | −2.42942 | 2.32E‐05 | 0.19 |
| Akr1c19 | Aldo‐keto reductase family 1, member C19 | −3.51993 | 3.74E‐05 | 0.09 |
| Cartpt | Cocaine‐and amphetamine‐regulated prepropeptide | −6.54051 | 1.76E‐05 | 0.01 |
| Hamp | Hepcidin antimicrobial peptide | −9.58007 | 1.50E‐05 | 0.04 |
| S100a8 | S100 calcium binding protein A8 | −2.73713 | 3.10E‐05 | 0.21 |
| Scd3 | Stearoyl‐coenzyme A desaturase 3 | −2.00069 | 4.03E‐05 | 0.25 |
| Steap | Six‐transmembrane epithelial antigen of prostate 4 | −2.25736 | 7.12E‐17 | 0.21 |
| B. Top upregulated and downregulated transcripts: EP4 PGE 2 relative to EP4 EtOH | ||||
| Upregulated | ||||
| Csf2rb | Colony stimulating factor 2 receptor beta | 1.33980634 | 3.94E‐06 | 2.53 |
| Mt4 | Metallothionein 4 | 1.21674309 | 9.05E‐07 | 2.32 |
| Pmepa1 | Prostate transmembrane protein, androgen induced 1 | 1.66276986 | 2.04E‐05 | 3.17 |
| Downregulated | ||||
| C230091D08Rik | Unclassified non‐coding RNA gene | −1.2859205 | 9.22E‐06 | 0.41 |
| Coro2a | Coronin, actin binding 2A | −1.9046218 | 1.27E‐05 | 0.27 |
| Gm9705 | Cytochrome P450 family; CYP4f37 | −2.3798049 | 1.32E‐07 | 0.19 |
| Mill1 | MHC I like leukocyte 1 | −1.340297 | 4.78E‐07 | 0.39 |
| Mmd | Monocyte to macrophage differentiation‐associated | −1.368169 | 1.31E‐05 | 0.39 |
| Slc39a6 | Solute carrier family 39 (metal ion transporter), member 6 | −1.226609 | 3.38E‐05 | 0.43 |
| Smoc2 | SPARC related modular calcium binding 2 | −1.632054 | 8.69E‐08 | 0.32 |
| Steap4 | Six‐transmembrane epithelial antigen of prostate, member 4 | −1.403364 | 8.15E‐08 | 0.38 |
| C. IL‐20 protein concentrations 8 h after treatment of WT and BK5.EP4 mice with EtOH or PGE2, as measured by ELISA | ||||
| Genotype | Treatment | x ± sd | ||
| WT | EtOH | 290.8 ± 1.12 | ||
| WT | PGE2 | 438.6 ± 30.28 | ||
| BK5.EP4 | EtOH | 378.3 ± 36.83 | ||
| BK5.EP4 | PGE2 | 1297.8 ± 135.36 | ||
Values represent the mean (n = 3–5 mice) ± std dev (pg IL‐20/?g protein). WT‐PGE2 differs significantly from WT‐EtOH (p < 0.001); BK5.EP4‐PGE2 differs significantly from BK5.EtOH and both WT groups (p < 0.01).
An IPA analysis was also carried out on transcripts from BK5.EP4 and WT mice treated with PGE2. The IPA software created 8 networks, with highly significant scores of 52, 51, 36, 31, 27, 26, 19 and 13 (Suppl. Table 3A; gene abbreviations defined in Suppl. Table 3B). Mapped within each network were 25, 25, 16, 17, 16, 16, 12 and 9 differentially expressed transcripts, respectively. The biological functions of these networks include dermatological diseases, developmental disorders, embryonic and organ development, lipid metabolism, cancer, cell death and survival, and cellular movement.
Many of the differentially expressed transcripts were different between the two comparison groups, but this was expected because PGE2 also activates the EP1, EP2 and EP3 receptors. There were, however, quite a number of differentially expressed transcripts that were the same between the comparison groups, including many keratin and keratin‐associated proteins, DKK3, STEAP4, DIO2, KCNK2, TCHH1, MAPK12, PMEPA1, FKBP, PAD13, SMOC2, GREB1, several SLC members, PLEKHH2, and SNAP25. The specific biological function and role of these genes remains to be determined.
3.8. Validation of IL‐20 upregulation in BK5.EP4 mice
To determine whether the upregulation of IL‐20 mRNA in PGE2‐treated BK5.EP4 epidermis resulted in upregulated IL‐20 protein, an ELISA was carried out. As shown in Table 1C, IL‐20 protein is induced a significant 1.5‐fold by PGE2 in WT mice and over 3‐fold in BK5.EP4 mice treated with PGE2, compared to EtOH treatment. Additionally, IL‐20 protein levels were higher in BK5.EP4 mice compared to WT with EtOH treatment.
4. Discussion
The EP4 receptor for PGE2 is similar to the EP1, EP2 and EP3 receptors in that they all have seven transmembrane segments and are coupled to Gα subunits of heterotrimeric G proteins, with a different Gα subunit for each receptor (Sugimoto and Narumiya, 2007). The EP4 receptor, however, has several unique structural features that suggest that it may have distinct biological functions. EP4 is also unique among the EP receptors in that it is rapidly internalized in response to PGE2 (Regan, 2003).
The EP1 and EP2 receptors have been the most extensively studied with regard to cancer development and progression in several organ systems, including colon and breast cancer development (Kawamori et al., 2005, 2001, 1999). With regard to skin cancer, the EP1 receptor confers tumor promoting and even stronger tumor progression activity in carcinogen‐initiated mice (Surh et al., 2011, 2012), and in UV‐induced carcinogenesis (Tober et al., 2006). EP1 overexpression in transgenic mice resulted in protein kinase C activation and upregulation of COX‐2, suggesting that its tumor promoting activity is via the same mechanism used by TPA (Surh et al., 2011, 2012).
The EP2 receptor has also been linked to the growth and progression of a number of cancers (Baba et al., 2010; Castellone et al., 2005; Kuo et al., 2009; Sonoshita et al., 2001), including skin (Sung et al., 2005). EP2 null mice develop 50% fewer papillomas than WT mice (and no SCCs) in a DMBA/TPA protocol (Sung et al., 2005). Conversely, overexpression of EP2 resulted in significantly more skin tumors, including more SCCs, than WT mice (Sung et al., 2006). In a UV protocol EP2 null mice also developed 50% fewer tumors, but these tumors were larger and more aggressive than those in WT mice (Brouxhon et al., 2007), suggesting that the role of EP2 may be carcinogen‐insult dependent.
The limited number of studies on the EP3 receptor indicated that it can have both pro‐ and anti‐inflammatory activity (Goulet et al., 2004; Honda et al., 2009). However, the EP3 receptor was not found to play a role in skin tumorigenesis, where inflammation is usually a driving factor (Shoji et al., 2005; Sung et al., 2005).
Like the other EP receptors, EP4 was reported to be upregulated in skin tumors from both UV and DMBA/TPA protocols (Lee et al., 2005; Neumann et al., 2007; Tober et al., 2007). Because the EP4 receptor was also shown to contribute to breast, kidney, and head and neck cancers (Abrahao et al., 2010; Ma et al., 2013; Wu et al., 2011; Xin et al., 2012), we hypothesized that EP4 would also contribute to skin tumor development. As shown in this report, the skins of the BK5.EP4 mice are essentially identical to WT mice at the microscopic level, albeit their hair cycles are slightly different. When subjected to the two‐stage DMBA/TPA protocol, the transgenic mice developed considerably more tumors than WT mice. More significantly, most of the tumors in the transgenic mice were SCCs, unlike in the wild type mice. Despite the fact that TPA treatment increased keratinocyte proliferation to a greater extent in the transgenic mice, the mechanism responsible for increased tumorigenesis is not clear. An unusual finding in the DMBA/TPA experiment was the development of a tumor in a transgenic mouse after DMBA but before TPA treatment was begun. This suggested that, like the EP1 receptor (Surh et al., 2012), EP4 could have endogenous tumor promoting activity. To test this, the response to DMBA alone was investigated. Using a single application of DMBA, it was visually apparent that the transgenic mice had a much more cytotoxic response than WT mice. Several avenues were investigated to attempt to elucidate the basis for this response. However, studies on the expression of enzymes that metabolize DMBA (CYP1A1, CYP1B1 and aromatase) as well as on keratinocyte stem cell (CD34 and ?6 integrin positive cells) numbers indicated no difference between transgenic and WT mice.
Exposure to UV instead of DMBA also produced a sloughing and angiogenic response in transgenic, but not WT, mice, indicating that the epidermis of the BK5.EP4 mice has a much lower apoptosis threshold than WT mice, as suggested by others (Chun and Langenbach, 2007). Overall, these studies suggest that a very strong regenerative response is responsible for the subsequent development of tumors.
To assess whether the EP4 receptor was activating the signaling pathways reported by others (Yoshida et al., 2013), we examined the levels of cAMP, PI3K/Akt and ERK1/2 activation and the upregulation of COX‐2. Although we found increased levels of cAMP in the epidermis of transgenic mice, we did not find activation of PI3K/Akt, ERK1/2 or Stat3, or the upregulation of COX‐2 (Suppl. Figure 1 and data not shown).
To elucidate possible mechanisms through which EP4 promotes the development and progression of skin tumors, the RNAseq approach was used to assess differences in gene expression in BK5.EP4 and WT mice, with and without PGE2 treatment. This is an unbiased approach, however, we expected to see changes in the expression of genes associated with inflammation because EP4 was reported to be involved in the activation of NFkB in keratinocytes (Soontrapa et al., 2011) and in the regulation of the proliferative and pro‐inflammatory genes TNFα, PGE2 synthase and cyclin D1 (Fujino et al., 2003). Even though these specific genes were not found to be significantly upregulated, several gene changes associated with inflammation and tumor development were noted.
Among the genes that were the most highly upregulated in EP4 transgenic mice treated with PGE2, compared to PGE2‐treated WT mice (Table 1A), were A130040M12Rik and interleukin‐20 (IL‐20). A130040M12Rik belongs to the VL30 family of murine retroelement noncoding RNAs and has been shown to regulate proto‐oncogene transcription and tumorigenesis (Wang et al., 2009, 2009). Interestingly, IL‐20 transcripts were below the limits of detection in WT mice but were highly upregulated in the BK5.EP4 mice. IL‐20 has been shown to play a role in skin inflammation, along with several related cytokines (Uto‐Konomi et al., 2012). The regenerative hyperplasia seen in psoriasis also has been linked to IL‐20 (Wang et al., 2012; Wegenka, 2010); this may be relevant to the robust regenerative response of BK5.EP4 mice to DMBA‐induced cell death, which in turn resembles the regenerative cell cycle progression in keratinocytes following TPA treatment (Kirkhus et al., 1992).
Some of the other upregulated genes, including Kcnk2 (TREK‐1), have been linked to ovarian and prostate cancer (Innamaa et al., 2013; Voloshyna et al., 2008). Serbinb6 is an inhibitor of kallikrein‐8 that modulates proliferation and differentiation of keratinocytes and has been implicated in the pathogenesis of psoriasis (Kishibe et al., 2007). Small proline‐rich protein (Sprr) family members are antioxidants and are expressed by migrating keratinocytes in healing wounds (Vermeij and Backendorf, 2010) and thus might be expected to be elevated in tissues that are susceptible to regeneration and tumor development.
The down‐regulation of S100a8 in the keratinocytes of PGE2‐treated BK5.EP4 mice was surprising because it is a pro‐inflammatory mediator and is elevated in a variety of cancers (Gebhardt et al., 2006). Because the biological function for S100a8 (and its partner S100a9) is unknown, it is difficult to determine the relationship between its down‐regulation and increased sensitivity to tumor development in BK5.EP4 mice.
A comparison of differentially expressed transcripts between BK5.EP4 mice treated either with PGE2 or with EtOH also showed that several of the genes that were upregulated after PGE2 treatment are related to cancer development. Csf2rb is the common beta chain of the high affinity receptor for IL‐3, IL‐5 and GM‐CSF (Woodcock et al., 1994), which is known to promote the progression of tumors in a number of organs including murine skin (Aziz et al., 2006; Gutschalk et al., 2013). Metallothionein‐4 (MT‐4) is involved in differentiation of stratified squamous epithelium (Vasak and Meloni, 2011) suggesting that EP4 is involved in keratinocyte differentiation. Pmepa1 expression has not been previously reported in keratinocytes, however, it is over‐expressed in colorectal, breast and ovarian cancer (Giannini et al., 2003; Shi et al., 2012) and promotes proliferation via suppression of Smad3/4 signaling (Liu et al., 2011), suggesting a possible mechanism/pathway through which EP4 promotes tumor development.
Among the down‐regulated genes were several of interest. Coronin2a (Coro2a), a component of the nuclear receptor co‐repressor complexes, contributes to de‐repression of inflammatory response genes (Huang et al., 2011). Mill1, an MHC family member expressed in hair follicles, represses wound healing (Rabinovich et al., 2008); the reduction in Mill1 expression may thus contribute to the robust wound healing response to the toxicity of DMBA in the transgenic mice. SMOC2 is known to stimulate the attachment of keratinocytes to matrices (Maier et al., 2008); reduction in SMOC2 would likely contribute to cell migration and a dysplastic/hyperplastic epidermis. STEAP4 is an oxidoreductase that mediates apoptosis and glucose metabolism (Gomes et al., 2012), although its function in skin has not been clearly elucidated.
IPA analysis of the differentially expressed transcripts identified molecular networks with biological functions that are consistent with the tumor promoting phenotype of the BK5.EP4 mice. Further studies will be needed to determine which networks and their components are critical to the skin tumor promoting activity of the EP4 receptor.
Overall, the genes upregulated by over‐expression and activation of the EP4 receptor are primarily those that are associated with increased proliferation and inflammation and thus likely contribute to the robust regenerative response of the epidermis of DMBA‐treated BK5.EP4 mice. This response is reminiscent of the protein kinase C epsilon transgenic mice in which complete epidermal necrosis occurs after TPA treatment, followed by regeneration beginning from the hair follicles. It was proposed that prolonged hyperplasia of the hair follicle results in an expansion of initiated cells leading to subsequent development of SCCs (Li et al., 2005). Similarly, apoptotic cell death has been shown to promote wound healing and tissue regeneration via a pathway referred to as the “phoenix rising” pathway (Li et al., 2010). In the case of the BK5.EP4 mice, this pathway is facilitated by the upregulation of pro‐inflammatory and proliferation‐related genes.
In summary, the EP4 receptor for PGE2 confers endogenous tumor promoting and even greater tumor progression activity in murine skin. This receptor causes the upregulation of a number of genes involved in the proliferative and inflammatory aspects of tumor development and thus is a reasonable target for the prevention and therapeutic treatment of skin cancer.
Conflict of interests
None declared.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
SFig. 1Over‐expression of the EP4 receptor does not alter expression of the polycyclic aromatic hydrocarbon enzymes aromatase, CYP1A1 or CYP1B1 in skin.
SFig. 2Healing of abrasional wounds is not altered in BK5.EP4 transgenic mice.
SFig. 3Keratinocyte stem cell numbers, as denoted by CD34 and K15 positive cells in the hair follicle, are not altered by over‐expression of the EP4 receptor.
Acknowledgments
We are grateful to Shawna Johnson and Laura Denton for help in formatting this manuscript. We acknowledge the Histology and Tissue Processing Facility Core, the Molecular Biology Facility Core, and the Next Generation Sequencing Core at Science Park in Smithville, Texas. This study also made use of the Research Animal Support Facilities, and the Genetically Engineered Mouse Facility, supported by the National Institutes of Health grants CA100140, P30 CA16672‐30 DHHS ⁄ NCI Cancer Center Support Grant (CCSG) and CPRIT Core Facility Support Grant RP120348.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.06.013.
Simper Melissa S., Rundhaug Joyce E., Mikulec Carol, Bowen Rebecca, Shen Jianjun, Lu Yue, Lin Kevin, Surh Inok, Fischer Susan M., (2014), The tumor promoting activity of the EP4 receptor for prostaglandin E2 in murine skin, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.06.013.
References
- Abrahao, A.C. , Castilho, R.M. , Squarize, C.H. , Molinolo, A.A. , dos Santos-Pinto, D. , Gutkind, J.S. , 2010. A role for COX2-derived PGE2 and PGE2-receptor subtypes in head and neck squamous carcinoma cell proliferation. Oral Oncol. 46, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Huber, W. , 2010. Differential expression analysis for sequence count data. Genome Biol. 11, R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, M.H. , Wheeler, D.L. , Bhamb, B. , Verma, A.K. , 2006. Protein kinase C delta overexpressing transgenic mice are resistant to chemically but not to UV radiation-induced development of squamous cell carcinomas: a possible link to specific cytokines and cyclooxygenase-2. Cancer Res. 66, 713–722. [DOI] [PubMed] [Google Scholar]
- Baba, Y. , Nosho, K. , Shima, K. , Goessling, W. , Chan, A.T. , Ng, K. , Chan, J.A. , Giovannucci, E.L. , Fuchs, C.S. , Ogino, S. , 2010. PTGER2 overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Cancer Epidemiol. Biomarkers Prev. 19, 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouxhon, S. , Konger, R.L. , VanBuskirk, J. , Sheu, T.J. , Ryan, J. , Erdle, B. , Almudevar, A. , Breyer, R.M. , Scott, G. , Pentland, A.P. , 2007. Deletion of prostaglandin E2 EP2 receptor protects against ultraviolet-induced carcinogenesis, but increases tumor aggressiveness. J. Invest. Dermatol. 127, 439–446. [DOI] [PubMed] [Google Scholar]
- Castellone, M.D. , Teramoto, H. , Williams, B.O. , Druey, K.M. , Gutkind, J.S. , 2005. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science 310, 1504–1510. [DOI] [PubMed] [Google Scholar]
- Chun, K.S. , Langenbach, R. , 2007. A proposed COX-2 and PGE(2) receptor interaction in UV-exposed mouse skin. Mol. Carcinog. 46, 699–704. [DOI] [PubMed] [Google Scholar]
- Fischer, S.M. , Lo, H.H. , Gordon, G.B. , Seibert, K. , Kelloff, G. , Lubet, R.A. , Conti, C.J. , 1999. Chemopreventive activity of celecoxib, a specific cyclooxygenase-2 inhibitor, and indomethacin against ultraviolet light-induced skin carcinogenesis. Mol. Carcinog 25, 231–240. [PubMed] [Google Scholar]
- Fischer, S.M. , Pavone, A. , Mikulec, C. , Langenbach, R. , Rundhaug, J.E. , 2007. Cyclooxygenase-2 expression is critical for chronic UV-induced murine skin carcinogenesis. Mol. Carcinog 46, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino, H. , Xu, W. , Regan, J.W. , 2003. Prostaglandin E2 induced functional expression of early growth response factor-1 by EP4, but not EP2, prostanoid receptors via the phosphatidylinositol 3-kinase and extracellular signal-regulated kinases. J. Biol. Chem. 278, 12151–12156. [DOI] [PubMed] [Google Scholar]
- Gebhardt, C. , Nemeth, J. , Angel, P. , Hess, J. , 2006. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 72, 1622–1631. [DOI] [PubMed] [Google Scholar]
- Giannini, G. , Ambrosini, M.I. , Di Marcotullio, L. , Cerignoli, F. , Zani, M. , MacKay, A.R. , Screpanti, I. , Frati, L. , Gulino, A. , 2003. EGF- and cell-cycle-regulated STAG1/PMEPA1/ERG1.2 belongs to a conserved gene family and is overexpressed and amplified in breast and ovarian cancer. Mol. Carcinog 38, 188–200. [DOI] [PubMed] [Google Scholar]
- Gomes, I.M. , Maia, C.J. , Santos, C.R. , 2012. STEAP proteins: from structure to applications in cancer therapy. Mol. Cancer Res. 10, 573–587. [DOI] [PubMed] [Google Scholar]
- Goulet, J.L. , Pace, A.J. , Key, M.L. , Byrum, R.S. , Nguyen, M. , Tilley, S.L. , Morham, S.G. , Langenbach, R. , Stock, J.L. , McNeish, J.D. , Smithies, O. , Coffman, T.M. , Koller, B.H. , 2004. E-prostanoid-3 receptors mediate the proinflammatory actions of prostaglandin E2 in acute cutaneous inflammation. J. Immunol. 173, 1321–1326. [DOI] [PubMed] [Google Scholar]
- Gutschalk, C.M. , Yanamandra, A.K. , Linde, N. , Meides, A. , Depner, S. , Mueller, M.M. , 2013. GM-CSF enhances tumor invasion by elevated MMP-2, -9, and -26 expression. Cancer Med. 2, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, T. , Matsuoka, T. , Ueta, M. , Kabashima, K. , Miyachi, Y. , Narumiya, S. , 2009. Prostaglandin E(2)-EP(3) signaling suppresses skin inflammation in murine contact hypersensitivity. J. Allergy Clin. Immunol. 124, 809–812. e802 [DOI] [PubMed] [Google Scholar]
- Huang, W. , Ghisletti, S. , Saijo, K. , Gandhi, M. , Aouadi, M. , Tesz, G.J. , Zhang, D.X. , Yao, J. , Czech, M.P. , Goode, B.L. , Rosenfeld, M.G. , Glass, C.K. , 2011. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature 470, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innamaa, A. , Jackson, L. , Asher, V. , van Schalkwyk, G. , Warren, A. , Keightley, A. , Hay, D. , Bali, A. , Sowter, H. , Khan, R. , 2013. Expression and effects of modulation of the K2P potassium channels TREK-1 (KCNK2) and TREK-2 (KCNK10) in the normal human ovary and epithelial ovarian cancer. Clin. Transl. Oncol. 15, 910–918. [DOI] [PubMed] [Google Scholar]
- Kawamori, T. , Kitamura, T. , Watanabe, K. , Uchiya, N. , Maruyama, T. , Narumiya, S. , Sugimura, T. , Wakabayashi, K. , 2005. Prostaglandin E receptor subtype EP(1) deficiency inhibits colon cancer development. Carcinogenesis 26, 353–357. [DOI] [PubMed] [Google Scholar]
- Kawamori, T. , Uchiya, N. , Nakatsugi, S. , Watanabe, K. , Ohuchida, S. , Yamamoto, H. , Maruyama, T. , Kondo, K. , Sugimura, T. , Wakabayashi, K. , 2001. Chemopreventive effects of ONO-8711, a selective prostaglandin E receptor EP(1) antagonist, on breast cancer development. Carcinogenesis 22, 2001–2004. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. , Salzberg, S.L. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkhus, B. , Glaso, M. , Clausen, O.P. , 1992. Multivariate flow cytometry of epidermal regeneration provoked by a skin irritant and a tumor promoter. Cytometry 13, 267–274. [DOI] [PubMed] [Google Scholar]
- Kishibe, M. , Bando, Y. , Terayama, R. , Namikawa, K. , Takahashi, H. , Hashimoto, Y. , Ishida-Yamamoto, A. , Jiang, Y.P. , Mitrovic, B. , Perez, D. , Iizuka, H. , Yoshida, S. , 2007. Kallikrein 8 is involved in skin desquamation in cooperation with other kallikreins. J. Biol. Chem. 282, 5834–5841. [DOI] [PubMed] [Google Scholar]
- Kuo, K.T. , Wang, H.W. , Chou, T.Y. , Hsu, W.H. , Hsu, H.S. , Lin, C.H. , Wang, L.S. , 2009. Prognostic role of PGE2 receptor EP2 in esophageal squamous cell carcinoma. Ann. Surg. Oncol. 16, 352–360. [DOI] [PubMed] [Google Scholar]
- Lee, J.L. , Kim, A. , Kopelovich, L. , Bickers, D.R. , Athar, M. , 2005. Differential expression of E prostanoid receptors in murine and human non-melanoma skin cancer. J. Invest. Dermatol. 125, 818–825. [DOI] [PubMed] [Google Scholar]
- Li, F. , Huang, Q. , Chen, J. , Peng, Y. , Roop, D.R. , Bedford, J.S. , Li, C.Y. , 2010. Apoptotic cells activate the “phoenix rising” pathway to promote wound healing and tissue regeneration. Sci. Signal. 3, ra13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wheeler, D.L. , Alters, W. , Chaiswing, L. , Verma, A.K. , Oberley, T.D. , 2005. Early epidermal destruction with subsequent epidermal hyperplasia is a unique feature of the papilloma-independent squamous cell carcinoma phenotype in PKCepsilon overexpressing transgenic mice. Toxicol. Pathol. 33, 684–694. [DOI] [PubMed] [Google Scholar]
- Liu, R. , Zhou, Z. , Huang, J. , Chen, C. , 2011. PMEPA1 promotes androgen receptor-negative prostate cell proliferation through suppressing the Smad3/4-c-Myc-p21 Cip1 signaling pathway. J. Pathol. 223, 683–694. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Holt, D. , Kundu, N. , Reader, J. , Goloubeva, O. , Take, Y. , Fulton, A.M. , 2013. A prostaglandin E (PGE) receptor EP4 antagonist protects natural killer cells from PGE-mediated immunosuppression and inhibits breast cancer metastasis. Oncoimmunol 2, e22647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, S. , Paulsson, M. , Hartmann, U. , 2008. The widely expressed extracellular matrix protein SMOC-2 promotes keratinocyte attachment and migration. Exp. Cell Res. 314, 2477–2487. [DOI] [PubMed] [Google Scholar]
- Maldve, R. , Fischer, S.M. , 1996. Multifactor regulation of prostaglandin H synthase-2 in murine keratinocytes. Mol. Carcinog 17, 207–216. [DOI] [PubMed] [Google Scholar]
- Mikulec, C.D. , Rundhaug, J.E. , Simper, M.S. , Lubet, R.A. , Fischer, S.M. , 2013. The chemopreventive efficacies of nonsteroidal anti-inflammatory drugs: the relationship of short-term biomarkers to long-term skin tumor outcome. Cancer Prev. Res. 6, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R.J. , Tryson, K.A. , Wu, K.Q. , 2000. Evidence that the epidermal targets of carcinogen action are found in the interfollicular epidermis of infundibulum as well as in the hair follicles. Cancer Res. 60, 226–229. [PubMed] [Google Scholar]
- Muller-Decker, K. , 2011. Cyclooxygenase-dependent signaling is causally linked to non-melanoma skin carcinogenesis: pharmacological, genetic, and clinical evidence. Cancer Met. Rev. 30, 343–361. [DOI] [PubMed] [Google Scholar]
- Namba, T. , Sugimoto, Y. , Negishi, M. , Irie, A. , Ushikubi, F. , Kakizuka, A. , Ito, S. , Ichikawa, A. , Narumiya, S. , 1993. Alternative splicing of C-terminal tail of prostaglandin E receptor subtype EP3 determines G-protein specificity. Nature 365, 166–170. [DOI] [PubMed] [Google Scholar]
- Narumiya, S. , 2009. Prostanoids and inflammation: a new concept arising from receptor knockout mice. J. Mol. Med. 87, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Neumann, M. , Dulsner, E. , Furstenberger, G. , Muller-Decker, K. , 2007. The expression pattern of prostaglandin E synthase and EP receptor isoforms in normal mouse skin and preinvasive skin neoplasms. Exp. Dermatol. 16, 445–453. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. , Tatusova, T. , Brown, G.R. , Maglott, D.R. , 2012. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich, B.A. , Ketchem, R.R. , Wolfson, M. , Goldstein, L. , Skelly, M. , Cosman, D. , 2008. A role for the MHC class I-like Mill molecules in nutrient metabolism and wound healing. Immunol. Cell Biol. 86, 489–496. [DOI] [PubMed] [Google Scholar]
- Regan, J.W. , 2003. EP2 and EP4 prostanoid receptor signaling. Life Sci. 74, 143–153. [DOI] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. , Smyth, G.K. , 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundhaug, J.E. , Fischer, S.M. , 2010. Molecular mechanisms of mouse skin tumor promotion. Cancers 2, 436–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundhaug, J.E. , Mikulec, C. , Pavone, A. , Fischer, S.M. , 2007. A role for cyclooxygenase-2 in ultraviolet light-induced skin carcinogenesis. Mol. Carcinog 46, 692–698. [DOI] [PubMed] [Google Scholar]
- Shi, Z.Z. , Zhang, Y.M. , Shang, L. , Hao, J.J. , Zhang, T.T. , Wang, B.S. , Liang, J.W. , Chen, X. , Zhang, Y. , Wang, G.Q. , Wang, M.R. , 2012. Genomic profiling of rectal adenoma and carcinoma by array-based comparative genomic hybridization. BMC Med. Genomics 5, 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, Y. , Takahashi, M. , Takasuka, N. , Niho, N. , Kitamura, T. , Sato, H. , Maruyama, T. , Sugimoto, Y. , Narumiya, S. , Sugimura, T. , Wakabayashi, K. , 2005. Prostaglandin E receptor EP3 deficiency modifies tumor outcome in mouse two-stage skin carcinogenesis. Carcinogenesis 26, 2116–2122. [DOI] [PubMed] [Google Scholar]
- Sonoshita, M. , Takaku, K. , Sasaki, N. , Sugimoto, Y. , Ushikubi, F. , Narumiya, S. , Oshima, M. , Taketo, M.M. , 2001. Acceleration of intestinal polyposis through prostaglandin receptor EP2 in Apc(Delta 716) knockout mice. Nat. Med. 7, 1048–1051. [DOI] [PubMed] [Google Scholar]
- Soontrapa, K. , Honda, T. , Sakata, D. , Yao, C. , Hirata, T. , Hori, S. , Matsuoka, T. , Kita, Y. , Shimizu, T. , Kabashima, K. , Narumiya, S. , 2011. Prostaglandin E2-prostaglandin E receptor subtype 4 (EP4) signaling mediates UV irradiation-induced systemic immunosuppression. Proc. Nat. Acad. Sci. U S A 108, 6668–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, Y. , Narumiya, S. , 2007. Prostaglandin E receptors. J. Biol. Chem. 282, 11613–11617. [DOI] [PubMed] [Google Scholar]
- Sung, Y.M. , He, G. , Fischer, S.M. , 2005. Lack of expression of the EP2 but not EP3 receptor for prostaglandin E2 results in suppression of skin tumor development. Cancer Res. 65, 9304–9311. [DOI] [PubMed] [Google Scholar]
- Sung, Y.M. , He, G. , Hwang, D.H. , Fischer, S.M. , 2006. Overexpression of the prostaglandin E2 receptor EP2 results in enhanced skin tumor development. Oncogene 25, 5507–5516. [DOI] [PubMed] [Google Scholar]
- Surh, I. , Rundhaug, J. , Pavone, A. , Mikulec, C. , Abel, E. , Fischer, S.M. , 2011. Upregulation of the EP1 receptor for prostaglandin E2 promotes skin tumor progression. Mol. Carcinog 50, 458–468. [DOI] [PubMed] [Google Scholar]
- Surh, I. , Rundhaug, J.E. , Pavone, A. , Mikulec, C. , Abel, E. , Simper, M. , Fischer, S.M. , 2012. The EP1 receptor for prostaglandin E2 promotes the development and progression of malignant murine skin tumors. Mol. Carcinog 51, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiano, H.F. , Loftin, C.D. , Akunda, J. , Lee, C.A. , Spalding, J. , Sessoms, A. , Dunson, D.B. , Rogan, E.G. , Morham, S.G. , Smart, R.C. , Langenbach, R. , 2002. Deficiency of either cyclooxygenase (COX)-1 or COX-2 alters epidermal differentiation and reduces mouse skin tumorigenesis. Cancer Res. 62, 3395–3401. [PubMed] [Google Scholar]
- Tober, K.L. , Thomas-Ahner, J.M. , Kusewitt, D.F. , Oberyszyn, T.M. , 2007. Effects of UVB on E prostanoid receptor expression in murine skin. J. Invest. Dermatol. 127, 214–221. [DOI] [PubMed] [Google Scholar]
- Tober, K.L. , Wilgus, T.A. , Kusewitt, D.F. , Thomas-Ahner, J.M. , Maruyama, T. , Oberyszyn, T.M. , 2006. Importance of the EP(1) receptor in cutaneous UVB-induced inflammation and tumor development. J. Invest. Dermatol. 126, 205–211. [DOI] [PubMed] [Google Scholar]
- Trempus, C.S. , Morris, R.J. , Ehinger, M. , Elmore, A. , Bortner, C.D. , Ito, M. , Cotsarelis, G. , Nijhof, J.G. , Peckham, J. , Flagler, N. , Kissling, G. , Humble, M.M. , King, L.C. , Adams, L.D. , Desai, D. , Amin, S. , Tennant, R.W. , 2007. CD34 expression by hair follicle stem cells is required for skin tumor development in mice. Cancer Res. 67, 4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto-Konomi, A. , Miyauchi, K. , Ozaki, N. , Motomura, Y. , Suzuki, Y. , Yoshimura, A. , Suzuki, S. , Cua, D. , Kubo, M. , 2012. Dysregulation of suppressor of cytokine signaling 3 in keratinocytes causes skin inflammation mediated by interleukin-20 receptor-related cytokines. PloS One 7, e40343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasak, M. , Meloni, G. , 2011. Chemistry and biology of mammalian metallothioneins. J. Biol. Inorg. Chem. 16, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Vermeij, W.P. , Backendorf, C. , 2010. Skin cornification proteins provide global link between ROS detoxification and cell migration during wound healing. PloS One 5, e11957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshyna, I. , Besana, A. , Castillo, M. , Matos, T. , Weinstein, I.B. , Mansukhani, M. , Robinson, R.B. , Cordon-Cardo, C. , Feinmark, S.J. , 2008. TREK-1 is a novel molecular target in prostate cancer. Cancer Res. 68, 1197–1203. [DOI] [PubMed] [Google Scholar]
- Wang, F. , Smith, N. , Maier, L. , Xia, W. , Hammerberg, C. , Chubb, H. , Chen, C. , Riblett, M. , Johnston, A. , Gudjonsson, J.E. , Helfrich, Y. , Kang, S. , Fisher, G.J. , Voorhees, J.J. , 2012. Etanercept suppresses regenerative hyperplasia in psoriasis by acutely downregulating epidermal expression of interleukin (IL)-19, IL-20 and IL-24. Br. J. Dermatol. 167, 92–102. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Cui, Y. , Zhang, G. , Garen, A. , Song, X. , 2009. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc. Nat. Acad. Sci. USA 106, 16794–16798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Colby, J.K. , Rengel, R.C. , Fischer, S.M. , Clinton, S.K. , Klein, R.D. , 2009. Overexpression of cyclooxygenase-2 (COX-2) in the mouse urinary bladder induces the expression of immune- and cell proliferation-related genes. Mol. Carcinog 48, 1–13. [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Kawamori, T. , Nakatsugi, S. , Ohta, T. , Ohuchida, S. , Yamamoto, H. , Maruyama, T. , Kondo, K. , Ushikubi, F. , Narumiya, S. , Sugimura, T. , Wakabayashi, K. , 1999. Role of the prostaglandin E receptor subtype EP1 in colon carcinogenesis. Cancer Res. 59, 5093–5096. [PubMed] [Google Scholar]
- Wegenka, U.M. , 2010. IL-20: biological functions mediated through two types of receptor complexes. Cytokine Growth Factor Rev 21, 353–363. [DOI] [PubMed] [Google Scholar]
- Woodcock, J.M. , Zacharakis, B. , Plaetinck, G. , Bagley, C.J. , Qiyu, S. , Hercus, T.R. , Tavernier, J. , Lopez, A.F. , 1994. Three residues in the common beta chain of the human GM-CSF, IL-3 and IL-5 receptors are essential for GM-CSF and IL-5 but not IL-3 high affinity binding and interact with Glu21 of GM-CSF. EMBO J. 13, 5176–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Zhang, Y. , Frilot, N. , Kim, J.I. , Kim, W.J. , Daaka, Y. , 2011. Prostaglandin E2 regulates renal cell carcinoma invasion through the EP4 receptor-Rap GTPase signal transduction pathway. J. Biol. Chem. 286, 33954–33962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, X. , Majumder, M. , Girish, G.V. , Mohindra, V. , Maruyama, T. , Lala, P.K. , 2012. Targeting COX-2 and EP4 to control tumor growth, angiogenesis, lymphangiogenesis and metastasis to the lungs and lymph nodes in a breast cancer model. Lab Invest. 92, 1115–1128. [DOI] [PubMed] [Google Scholar]
- Yoshida, K. , Fujino, H. , Otake, S. , Seira, N. , Regan, J.W. , Murayama, T. , 2013. Induction of cyclooxygenase-2 expression by prostaglandin E stimulation of the prostanoid EP4 receptor via coupling to G and transactivation of the epidermal growth factor receptor in HCA-7 human colon cancer cells. Eur. J. Pharmacol. 18, 408–417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
SFig. 1Over‐expression of the EP4 receptor does not alter expression of the polycyclic aromatic hydrocarbon enzymes aromatase, CYP1A1 or CYP1B1 in skin.
SFig. 2Healing of abrasional wounds is not altered in BK5.EP4 transgenic mice.
SFig. 3Keratinocyte stem cell numbers, as denoted by CD34 and K15 positive cells in the hair follicle, are not altered by over‐expression of the EP4 receptor.


