Abstract
Compost-assisted phytostabilization has recently emerged as a robust alternative for reclamation of metalliferous mine tailings. Previous studies suggest that root-associated microbes may be important for facilitating plant establishment on the tailings, yet little is known about the long-term dynamics of microbial communities during reclamation. A mechanistic understanding of microbial community dynamics in tailings ecosystems undergoing remediation is critical because these dynamics profoundly influence both the biogeochemical weathering of tailings and the sustainability of a plant cover. Here we monitor the dynamics of soil microbial communities (i.e. bacteria, fungi, archaea) during a 12-month mesocosm study that included 4 treatments: 2 unplanted controls (unamended and compost-amended tailings) and 2 compost-amended seeded tailings treatments. Bacterial, fungal and archaeal communities responded distinctively to the revegetation process and concurrent changes in environmental conditions and pore water chemistry. Compost addition significantly increased microbial diversity and had an immediate and relatively long-lasting buffering-effect on pH, allowing plants to germinate and thrive during the early stages of the experiment. However, the compost buffering capacity diminished after six months and acidification took over as the major factor affecting plant survival and microbial community structure. Immediate changes in bacterial communities were observed following plant establishment, whereas fungal communities showed a delayed response that apparently correlated with the pH decline. Fluctuations in cobalt pore water concentrations, in particular, had a significant effect on the structure of all three microbial groups, which may be linked to the role of cobalt in metal detoxification pathways. The present study represents, to our knowledge, the first documentation of the dynamics of the three major microbial groups during revegetation of compost-amended, metalliferous mine tailings.
Keywords: Mine tailings, phytostabilization, microbial community structure, metal(loid)s, acidification, mesocosms
1. Introduction
Mine tailings are a primary component of mine wastes produced during ore processing for metal extraction. Legacy mine tailings are a major source of environmental contamination due to historical inefficiencies of mining technologies, which left relatively high concentrations of toxic metal(loid)s in the tailings (Dybowska et al., 2006). Due to small particle size, limited quantities of essential nutrients and organic matter, high metal(loid) content, acidic pH, and the lack of a normal soil structure, these mine tailings generally do not support plant growth or a normal soil microbial community (Alvarenga et al., 2008; Mendez and Maier, 2008b). The inability to support plants is exacerbated in arid environments due to climatic conditions and high levels of salinity (Mendez and Maier, 2008a). Thus, mine tailings in arid environments are especially susceptible to wind dispersion and water erosion (Kabas et al., 2011; Meza-Figueroa and Maier, 2009).
Several methods for the assisted growth of plants on mine tailing piles and other metal-contaminated sites in arid environments have been proposed to minimize contaminant dispersion through revegetation (Alvarenga et al, 2008; Mendez et al., 2007; Mendez and Maier, 2008ab, Tordoff et al., 2000; Wong, 2003). Phytostabilization is one such method that employs plants to control dust emissions, stabilize tailings materials against water erosion, and reduce water percolation through contaminated materials by enhanced evapotranspiration. The use of plants to control dust emissions and minimize wind and water erosion of bare soils, especially in arid environments, is well documented (Gyssels et al., 2005; Grantz et al., 1998; Kort et al., 1998). In the case of phytostabilization, the plants used are specifically selected for their capacity to immobilize metal contaminants in the root zone rather than accumulating the metals in the shoot tissues (Mendez and Maier, 2008ab; Solís-Domínguez et al., 2012). Solís-Domínguez et al. (2012) demonstrated decreases in tailings aqueous extractable metals following 60 days of plant growth in compost-amended tailings.
Several studies have suggested that root-associated microbes may be important for phytostabilization as microbes can facilitate plant establishment on tailings, enhance plant biomass production and potentially serving as bioindicators of revegetation status (Grandlic et al., 2008; Ma et al., 2011; Mendez et al., 2008; Solís-Domínguez et al., 2011). Still, little is known about the long-term dynamics of microbial communities during the assisted revegetation of mine tailings and, specifically, how the microbial communities may: 1) respond to the remediation treatment, 2) influence the stabilization of the metal contaminants, and 3) influence the success of plant establishment.
Previous work has shown that tailings amendment and plant growth during the revegetation of mine wastes have a significant impact on the composition, abundance and stability of portions of the soil microbial communities (Pérez-de-Mora et al., 2006; Li et al., 2013; Mummey et al., 2002; Rosario et al., 2007). Pérez-de-Mora et al. (2006) and Li et al. (2013) independently showed that plant selection for the revegetation of metal-contaminated land had a stronger effect on microbial community composition and diversity than the type of soil amendment used. Additional studies have shown that plant growth and addition of soil amendments can significantly increase the abundance of heterotrophs and decrease the abundance of chemolithotrophs in mine tailings, thus improving soil health. (Mendez et al., 2007; Solís-Domínguez et al., 2012).
An important limitation of previous revegetation-research efforts is the focus on a single microbial group, usually bacteria, rather than simultaneously analyzing multiple microbial groups (e.g. fungi, archaea) (Gremion et al., 2004; Pérez-de-Mora et al., 2005; Solís-Domínguez et al., 2011). Numerous studies have demonstrated that the dominant soil microbial groups, bacteria, fungi, and archaea, behave differently in their response to environmental stressors, and in their influence on the functioning of an ecosystem (Hayden et al., 2012; Navarrete et al., 2010; Pereira-e-Silva et al., 2012; Reed and Martiny, 2007). Therefore, it is crucial that a comprehensive study of the dynamics of microbial communities in mine tailings during revegetation monitors multiple microbial groups simultaneously, as each group may respond differently to various aspects of the remediation treatment.
The objective of this study was to evaluate the relationship between microbial community structure and phytostabilization outcomes in a well-instrumented mesocosm experiment using metalliferous acid mine tailings. We hypothesize that changes in key environmental parameters resulting from compost amendment and plant establishment will drive group-specific changes in the structure of root-associated bacterial, fungal and archaeal communities during revegetation of acidic mine tailings. To test this hypothesis we employed community DNA fingerprinting analysis of small subunit bacterial, archaeal and fungal RNA genes in combination with multivariate analysis of the influence of environmental parameters on community structure in a one year greenhouse experiment.
2. Materials and methods
2.1. Iron King mine tailings
Tailings used for this study were collected and homogenized from both the oxidized surface layer and the underlying reduced zone at the Iron King Mine and Humboldt Smelter superfund site (IKMHSS), in Dewey-Humboldt, Arizona (34°30’02.11”N, 112°15’08.75”W). The IKMHSS was added to the National Priorities List in 2008 due to the exposure risks associated with high levels of arsenic (2590 mg kg−1) and lead (2200 mg kg−1) in the surface layer of the tailings pile (EA-EST, 2010). The oxidized surface layer (0 to 25 cm) of the tailings is highly acidic (pH 2.3 to 3.7) while the reduced zone (> 25 cm) has a higher pH (5.5 to 6.3). The IKMHSS tailings also have high levels of salinity, low organic carbon (0.014%) and nitrogen (0.043%) content, and a stressed microbial community dominated by chemolithoautotrophic microbial populations (Solís-Domínguez et al., 2012). These microbial populations are primarily iron- and sulfur-oxidizers, which diverge substantially from the heterotrophic populations commonly found in healthy soil ecosystems (Mendez et al., 2008; Tan et al., 2008). Chemolithoautotrophic microbial activity is supported by the pyritic composition of the tailings and is the driving force behind the low pH of the surface tailings layer, a condition that influences metal(loid)s solubility and limits plant growth on the tailings (Mendez et al., 2007; Schippers et al., 2010; Solís-Domínguez et al., 2012). The net acid producing potential (NAPP) was determined for the tailings used in this mesocosm study as described by Solís-Domínguez et al. (2011). NAPP=acid potential (AP)-acid neutralizing capacity (ANC).
2.2. Iron King mine tailings and setup of greenhouse experiment
A 12-month greenhouse mesocosm study was conducted to evaluate changes in physical, chemical and biological parameters during plant establishment in compost-amended IKMHSS mine tailings. The experiment consisted of 4 treatments that were monitored for 1 year under controlled conditions at the Controlled Environment Agricultural Center (CEAC) at The University of Arizona (Tucson, AZ). Twelve large polypropylene containers (ProPlastics, Chandler, AZ, USA), measuring 1 m in diameter and 0.5 m in depth, were custom-built to serve as mesocosms (Figure 1). Each mesocosm was equipped with pore water samplers that operated under constant tension (5–15 kPa), with a pore size of 2 µm and a sampling area of 33 cm2. Samplers were placed at 10 cm depth intervals from 5 cm to 35 cm. The following four treatments were examined in triplicate and arranged in a spatially randomized design (Figure 1): (i) Tailings only (TO); (ii) Tailings mixed with 15% dry-weight/dry-weight (w/w) compost (TC); (iii) Tailings mixed with 15% (w/w) compost and seeded with Buchloe dactyloides (buffalo grass) (BG); and (iv) Tailings mixed with 15% (w/w) compost and seeded with Atriplex lentiformis (quail bush) (QB). These native plant species were selected based on their salinity tolerance and ability to grow in IKMHSS tailings amended with 15% (w/w) compost without accumulating elevated levels of metals in their shoots (Solís-Domínguez et al., 2012).
Figure 1.
Greenhouse mesocosm experiment. (A) Schematic diagram showing the spatially randomized arrangement of the mesocosm treatments at the greenhouse (TO, tailings only; TC, tailings + 15% w/w compost; BG, tailings + 15% w/w compost + buffalo grass; QB, tailings + 15% w/w compost + quail bush). (B) Picture of mesocosm containers prior to the start of the experiment. (C) Buffalo grass plants at t3. (D) Quail bush plants at t3.
Surface and subsurface tailings were homogenized using a rotating cement mixer in a 3:1 ratio of oxidized surface tailings (collected from 0–20 cm depth) and reduced subsurface tailings (collected from > 35 cm depth) in order to create a substrate representative of the variable top 40 cm of the IKMHSS tailings pile. Amended treatments contained the tailings mixture homogenized with 15% (w/w) compost made from a mixture of composted cattle manure and green waste (Arizona Dairy Compost LLC, Anthem, AZ) and composted steer manure (El Toro De-Odorized Steer Manure, Tempe, AZ). All materials were sieved to 0.5 cm. The unamended tailings were packed to a depth of 40 cm in the TO mesocosms and to 20 cm in the TC, BG and QB mesocosms. The TC, QG and QB mesocosms were then topped with 20 cm of the compost-amended tailings mixture. The BG treatment was seeded at 8.8 g m−2 (7 g mesocosms−1) and the QB treatment at 5.5 g m−2 (4.3 g mesocosms−1) with approximately 123 and 63 seeds g−1, respectively. The mesocosms were irrigated immediately following seeding at a rate of 5–10 L per week and the first irrigation event represented time 0 for the study.
2.3. Mesocosm sampling and processing
Core samples from the soil profiles were collected from the mesocosms at 3 (t3), 6 (t6), 9 (t9), and 12 months (t12) by manually inserting a polycarbonate core sampler liner (3 cm in diameter) into the soil. Time 0 (t0) analyses were performed on triplicate samples of TO and TC materials collected immediately following homogenization. Data from the TC-t0 samples were used to represent all the amended treatments at time 0 of the study. Cores were collected from random locations in the TO and TC mesocosms at each time point beginning at t3. For planted treatments an average plant was harvested from each mesocosm by cutting the shoot at the surface of the soil. The core sampler was then centered over the stem to maximize the retrieval of roots and rhizosphere-influenced soil. All cores were stored on ice after collection and transported to the laboratory for further processing. The top 15 cm of the core samples was aseptically removed, the roots separated from the soil and the soil homogenized in sterile bags before being stored at −20°C for microbial analysis. Subsamples of the homogenized core samples were used to estimate the soil water content (moisture).
Pore water collected from the 5 cm and 15 cm pore water samplers was filtered (0.45 µm syringe filtered with Acrodisc GHP membrane) before chemical and physical analysis. Filtered samples were used to measure pH (USEPA method 150.2) and electrical conductivity (EC) (EPA method 120.1). Major anions were quantified with ion chromatography (IC, Dionex, DX-500, following USEPA method 300.0). Total organic carbon and total nitrogen were analyzed by automated high temperature wet combustion TC/TN analyzer (Shimadzu TOC-VCSH high sensitivity carbon analyzer, USEPA method 415.3). The filtered samples were then acidified (pH<2 with UHP HNO3) for analysis of total dissolved metal(loid)s by inductively coupled plasma mass spectrometer (ICP-MS, Perkin Elmer Elan DRC-II, following USEPA method 200.8). The values of the parameters measured from the 5 cm and 15 cm pore water samples were averaged prior to further statistical analysis to correspond to the 15 cm deep homogenized soil core used for microbial analysis.
2.4. Analysis of microbial communities
2.4.1. DNA extraction and SSU RNA gene PCR
Community DNA was extracted from 0.5 g core subsamples using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, Solon OH, USA) with modifications to the manufacturer’s protocol to enhance DNA yield. Both vortexing and centrifugation of the Lysing Matrix tube was increased to 15 min, the spin filters containing the binding matrix were air-dried under a laminar flow hood for 1 h prior to DNA elution with preheated (60°C) ultrapure water. The DNA extracts were quantified using a TBS-380 Fluorometer (Turner BioSystems, Sunnyvale, CA, USA) with PicoGreen dye (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s directions.
SSU rRNA genes from the DNA extracts were amplified to analyze the root-associated bacterial, fungal and archaeal community structures using Denaturing Gradient Gel Electrophoresis (DGGE). A 40-bp GC clamp was added to the reverse primer for each of the following reactions (Ferris et al., 1996; May et al., 2001). For bacteria, the V7/V8 variable region of the 16S rRNA gene was amplified following a modified protocol described by Colores et al. (2000) using primers 1070(f) and 1406(r) (Ferris et al., 1996). The amplification protocol was 95 °C for 5 min followed by 30 cycles of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 60 s followed by a 72 °C extension for 7 min. For fungi, approximately 350 bp of the 18S rRNA gene was amplified using primers GC-Fung-38(f) and NS1–368(r) (May et al., 2001). The amplification protocol was 95 °C for 5 min followed by 35 cycles of 94 °C for 45 s, 50 °C for 45 s, and 72 °C for 90 s, followed by a 72 °C extension for 7 min. For archaea, approximately 571 bp of the 16S rRNA gene including the hypervariable regions V3, V4 and V5 (Casamayor et al. 2002) was amplified with the primers GC-A344(f) and A915(r) (Casamayor et al., 2000). The amplification protocol was 95 °C for 5 min followed by 40 cycles of 94 °C for 60 s, 55 °C for 60 s, and 72 °C for 90 s, followed by a 72 °C extension for 7 min. The master mix for each 25 µL PCR reaction contained 2.5 µL of 10X Dream Taq™ buffer containing 20 mM MgCl2 (Fermentas, Lithuania), 0.2 mM dNTP, 0.4 µM of each primer, 0.4 µg µL−1 unacetylated albumin bovine serum (Sigma, St. Louis,MO), 0.625 U Dream Taq™ DNA polymerase (Fermentas, Lithuania) and 200 pg of template DNA for bacteria and fungi and 300 pg of DNA for archaea.
2.4.2. Community fingerprinting analysis
DGGE analysis was performed using the Dcode Universal Mutation Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Acrylamide gels (7%) were prepared with 45 to 65% (for bacteria), 20 to 45% (for fungi) and 40 to 60% (for archaea) urea-formamide denaturing gradients according to the manufacturer’s protocol. Gel lanes were loaded with 5–15 µl PCR product depending on the agarose electrophoresis band intensity. The parameters of the DGGE run and the analysis of the gel images are described elsewhere (Solís-Domínguez et al., 2011). The Standard-Based Polynomial Interpolation (SBPIn) method (Valentín-Vargas et al., 2013) was used to align DGGE profiles from multiples gels for reliable across-gel comparisons.
2.5. Statistical Analysis
2.5.1. Non-Metric Multidimensional Scaling (NMDS)
Non-Metric Multidimensional Scaling (NMDS) was used to analyze changes in microbial community structure over time as previously described (Valentín-Vargas et al., 2012). A non-parametric multivariate analysis of variance (NPMANOVA) was conducted to test the significance of treatment differences in overall microbial community structure and to confirm the results observed in the NMDS plot. Both the NMDS and NPMANOVA were based on similarity matrices calculated with the Jacquard similarity index as implemented in PAST 2.16 (Hammer et al., 2001).
2.5.2. Principal Component Analysis (PCA)
Elemental and organic carbon pore water profiles from the four mesocosm treatments were compared using Principal Component Analysis (PCA). The data was square-root transformed before performing the PCA to minimize the effect of the different scales and to dampen the influence of outliers. The scaling for this indirect gradient analysis was focused on inter-sample distances and the ordination was centered by species (Lepš and Šmilauer, 2003). Analyses were conducted using CANOCO 4.5 (ter Braak and Šmilauer, 2002).
2.5.3. Canonical Correspondence Analysis (CCA)
Canonical Correspondence Analysis (CCA) was used to evaluate the effect of environmental parameters measured during the mesocosm experiment on the microbial community structure. The CCA scaling focused on inter-sample distances to optimize the position of the samples in the ordination diagram, and was performed using binary microbial community data as biological variables against square-root transformed environmental parameters as explanatory variables. The presence of both plants and compost in the system were also included as nominal variables. CCA ordination diagrams were interpreted as described by ter Braak and Šmilauer (2002). The statistical significance of the CCA analyses was tested by a Monte Carlo permutation test (1000 unrestricted random permutations; p < 0.05) of residuals from a reduced model against the null hypothesis that microbial community composition was unrelated to the measured environmental parameters (ter Braak and Wiertz, 1994). To facilitate the interpretation of the CCA ordinations, the environmental data was divided into two groups: (i) physicochemical and nominal variables (pH, TOC, TDS, EC, moisture, plants, and compost), and (ii) elemental parameters (K, Na, Mg, Ca, Mn, Se, Cd, Co, Cu, Ni, Al, Zn, Pb, As, Fe, N [as TN], and S [as SO4]). CCA was carried out using CANOCO 4.5 (ter Braak and Šmilauer, 2002).
3. Results
3.1. Plant growth in the mesocosms
QB and BG germination was observed within the first week of the experiment and the plants in both treatments grew slowly but well for the first 3 months. However, the BG plants began to show signs of stress by t3, with severe die-back observed by t12. The QB plants survived the duration of the experiment, however they began to show signs of stress as early as t6.
3.2. Treatment effect on the temporal dynamics of microbial community structure
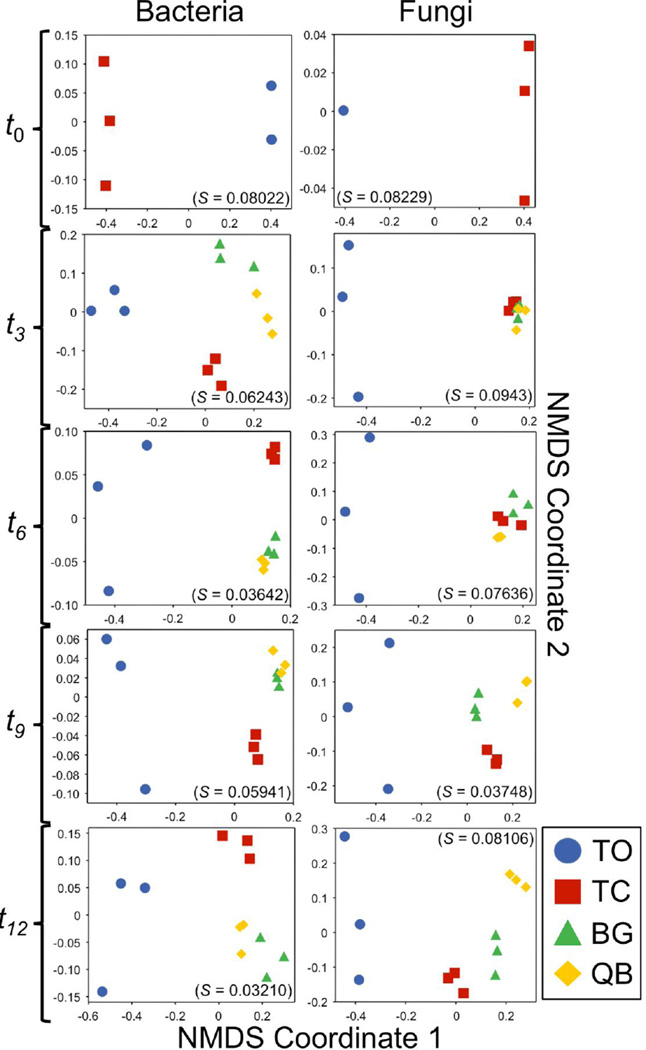
The NMDS analysis for bacterial and fungal communities (Figure 2) showed a clear separation in the ordination space between the TC and TO treatments at t0, which was statistically significant (NPMANOVA, p < 0.05). This confirms that the addition of compost to the tailings had an immediate and significant effect on the community composition of these two microbial domains. The separation of the TO treatment from compost-amended treatments remained throughout the 12-month duration of the study. Compost also affected within-treatment variability of replicates; replicates in the compost-amended treatments diverged less from each other over time than replicates in the TO treatment. Compost-amendment also lead to an immediate and persistent doubling of band number in the bacterial and fungal DGGE profiles relative to the TO profiles (Table 1).
Figure 2.
Non-Metric Multidimensional Scaling (NMDS) ordination diagrams of temporal variations in bacterial (left column) and fungal (right column) community structure. The ordinations are based on Jaccard similarity matrices calculated from the presence/absence data obtained from the DNA fingerprinting analysis. Each scatter point in a plot represents the structure of a microbial community in a particular sample (i.e. mesocosm replicate). The spatial separation between points approximates the similarity between their communities in terms of phylotype composition. All NMDS ordinations showed stress values below the preferred threshold of 0.1.
Table 1.
Mean number of bands detected in DGGE profiles per mesocosm treatment
| Microbial Group |
Treatment | Band number a | ||||
|---|---|---|---|---|---|---|
| t0 | t3 | t6 | t9 | t12 | ||
| Bacteria | TO | 21 ± 0 | 23 ± 2 | 18 ± 5 | 22 ± 2 | 18 ± 1 |
| TC | 45 ± 3 | 43 ± 3 | 46 ± 1 | 45 ± 2 | 50 ± 1 | |
| BG | – | 43 ± 1 | 58 ± 1 | 45 ± 2 | 43 ± 2 | |
| QB | – | 40 ± 3 | 55 ± 2 | 47 ± 1 | 46 ± 1 | |
| Fungi | TO | 19 ± 0 | 18 ± 1 | 14 ± 1 | 20 ± 4 | 24 ± 5 |
| TC | 37 ± 1 | 38 ± 1 | 34 ± 5 | 32 ± 2 | 36 ± 2 | |
| BG | – | 41 ± 3 | 40 ± 3 | 32 ± 2 | 47 ± 2 | |
| QB | – | 37 ± 2 | 37 ± 2 | 32 ± 1 | 38 ± 2 | |
| Archaea | TO | 0 | 0 | 0 | 0 | 1 ± 1 |
| TC | 15 ± 0 | 15 ± 0 | 15 ± 0 | 23 ± 1 | 14 ± 10 | |
| BG | – | 15 ± 0 | 15 ± 0 | 21 ± 1 | 24 ± 9 | |
| QB | – | 15 ± 0 | 15 ± 0 | 23 ± 2 | 30 ± 2 | |
Values are means ± SD (n=3)
The bacterial and fungal communities responded differently to plant establishment in the compost-amended treatments. For bacteria, a separation in NMDS community plots between the planted treatments (BG, QB) and the TC treatment was evident as early as t3, suggesting a relatively fast response of the bacterial communities to plant growth (Figure 2). This separation continued for the duration of the experiment (p < 0.05). In contrast, the fungal community NMDS revealed that the BG and QB treatments did not separate from the TC treatment until t9. Further analysis of the NMDS plots showed no evidence of a bacterial community response to the die-off of the buffalo grass plants that began at t3 and continued throughout the experiment. In contrast, the BG fungal community structure began to separate from that of the QB treatment at t9 and by t12 it clustered more closely with the TC communities than the QB treatment. Interestingly, the final fungal band number for BG was significantly higher than for QB and TC despite the fact that the community structure of TC and BG appeared more similar. Explanations for this complex dynamic are addressed in the discussion.
Archaeal community DGGE profiles showed a third pattern quite distinct from the bacteria and fungi. Archaea were not detected in the TO treatment throughout the experiment with the exception of a single mesocosm at t12 with just one band (Table 1). In contrast, fifteen bands were observed in all TC profiles, indicating that compost provided a significant archaeal inoculum that persisted during the experiment. As with fungi, the data indicate no archaeal community response to plant establishment. The archaeal profiles for TC, BG, and QB treatments were identical from t0 to t6. However, the DGGE profiles of these communities diverged and exhibited a significant increase in band number at t9 (Supplementary Figure 1, Table 1). The community response continued to diverge at t12 as indicated by the DGGE profiles. Band number decreased to t6 levels for the TC community, remained the same for the BG and increased in the QB treatment (Table 1).
3.3. Pore water sample analysis
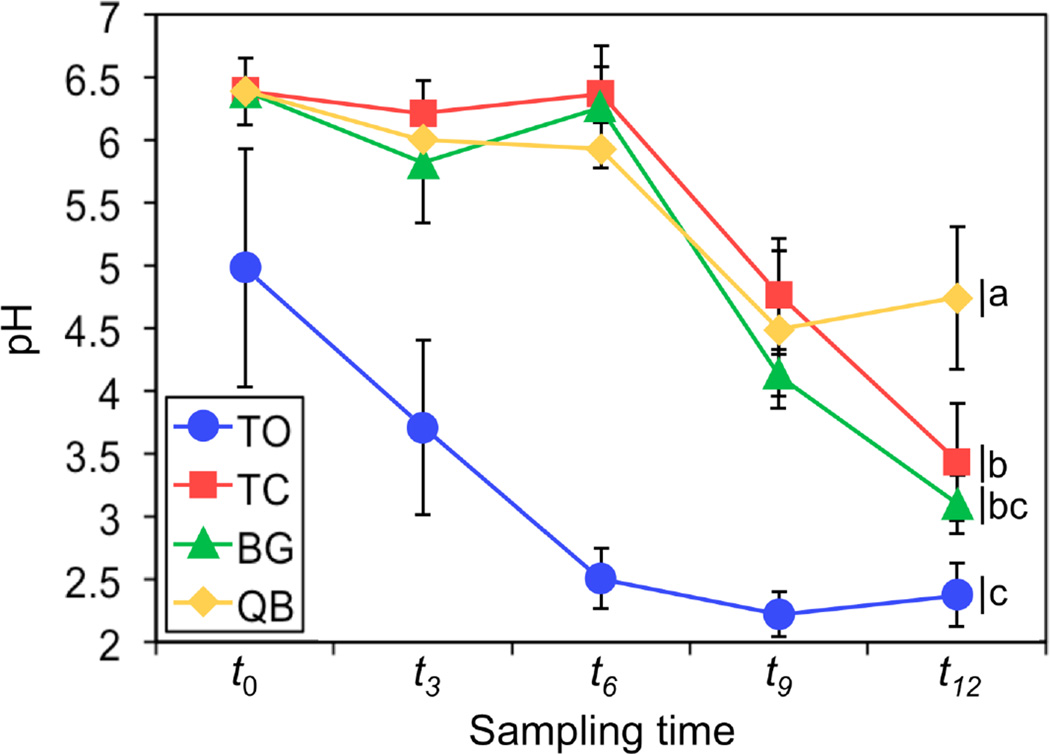
The results (mean values and standard deviations) of the pore water analysis are summarized in Supplementary Table 1. The pore water pH of the unamended TO treatment decreased sharply from 5.0 to 2.5 during the first six months of the study and then stabilized between 2 and 2.5 for the remaining six months. TC treatment data revealed that compost addition caused an immediate increase in pH from 5.0 to 6.4 (Figure 3). The average pH of all compost-amended treatments remained relatively stable until t6, but as the experiment progressed further, the positive effect of compost on pH yielded to a rapid acidification of the tailings in the TC and BG treatments. The decline in pH corresponded to the increase in observed stress and eventual die-off of the buffalo grass plants. The QB treatment also showed a decline in pH between t6 and t9, but the pH then stabilized between t9 and t12 at a pH that was significantly higher than any of the other treatments (Figure 3).
Figure 3.
Temporal variations in pH of pore water samples. Values represent samples collected at 5 cm and 15 cm deep along the mesocosms profile and averaged prior to further statistical analysis to correspond to the 15 cm deep homogenized soil core used for microbial analysis. Each symbol represents the mean for a particular time point and treatment combination. Error bars show standard deviation (n = 6). A one-way ANOVA was performed for the t12 results (F = 18.020; p = 0.0006). Means with different letters are significantly different at p < 0.05 (Tukey’s HSD test; n = 6).
The NAPP analysis of tailings materials revealed that the 3:1 mixture of surface and subsurface tailings had a NAPP of 4.67 moles acid kg−1 tailings, and an ANC:AP ratio of 0.014. NAPP values < −20 are considered non-acid generating and values >20 are considered acid producing. The ratio of ANC and AP is used when NAPP values are < 20 but positive, where it is expected that an ANP/AGP ratio greater than 3:1 will not generate acid and a ratio less than 1:1 is expected to generate acid (Hutchinson et al., 1992; Brodie et al., 1991). Based on the 15% compost amendment rate, the tailings + compost had a NAPP of 3.8 with an ANC:AP = 0.056 (ANC of compost was 1.5). The NAPP of the surface tailings alone was 1.6 with no measurable ANC. This analysis confirms that both the TO and compost amended tailings were acid generating.
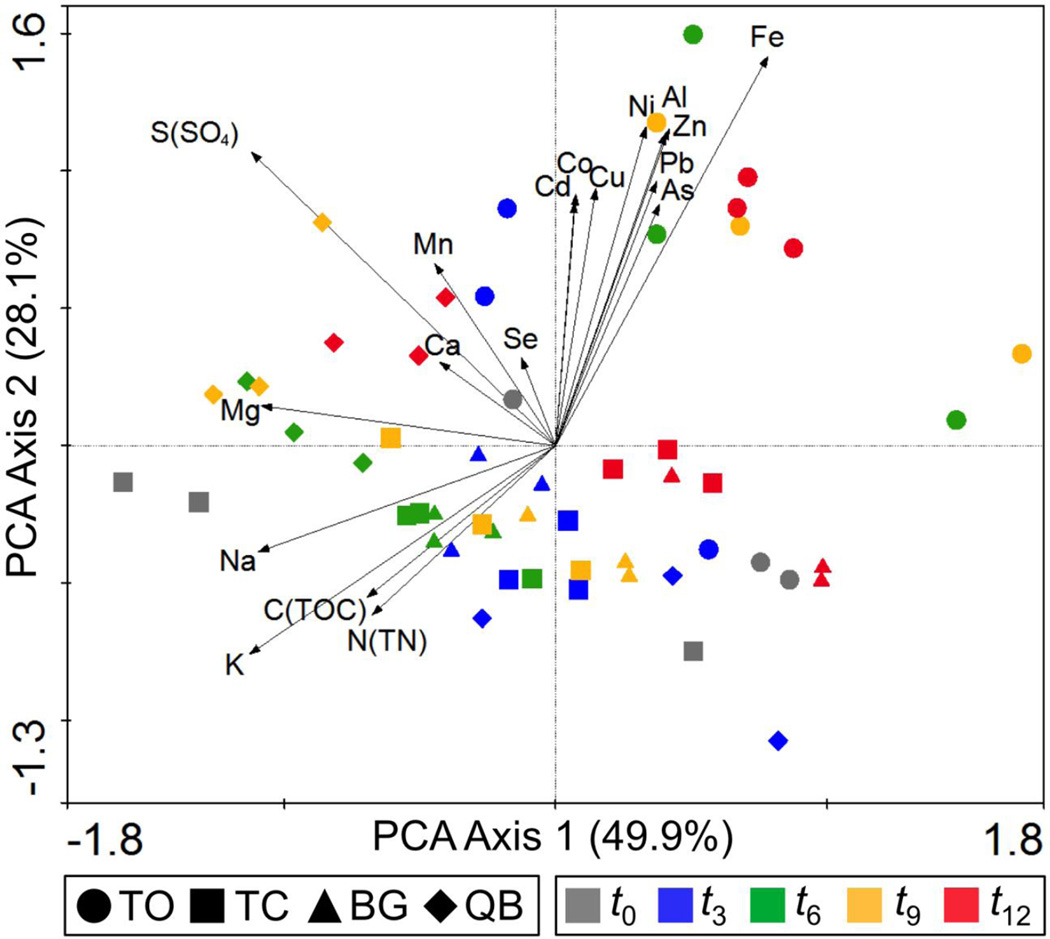
The effect of treatment on pore water chemistry was examined using PCA (Figure 4). Results show a separation between TO and compost-amended samples that began at t3 and became more pronounced as the experiment progressed. The results also show a strong positive correlation between TO samples at later times (i.e. t6, t9, t12) and higher pore water concentration of most metal(loid)s. This correlation corresponds to the acidification pattern of the respective mesocosms. The compost-amended treatments were negatively correlated with pore water metal(loid) concentration for the majority of the study. The PCA also revealed that most TO samples had a negative correlation with TOC and TN, while most compost-amended samples were positively correlated with the same parameters, suggesting that compost was the main source of these nutrients in the mesocosms.
Figure 4.
Principal Component Analysis (PCA) biplot showing the temporal variations in pore water chemical composition. Chemical parameters: K, Na, Mg, Ca, Mn, Se, Cd, Co, Cu, Ni, Al, Zn, Pb, As, Fe, C (as TOC), N (as TN), and S (as SO4). Arrows represent the relationship (direction and strength) of the chemical parameters with the samples. The direction of an arrow in PCA indicates an increase in that variable. The position of a sample along the length of an arrow indicates the concentration of the parameter represented by the arrow in that sample relative to the other samples. The value given on each axis label represents the percentage of the total variance explained by that axis.
3.4. Relationship between pore water chemistry and microbial community structure
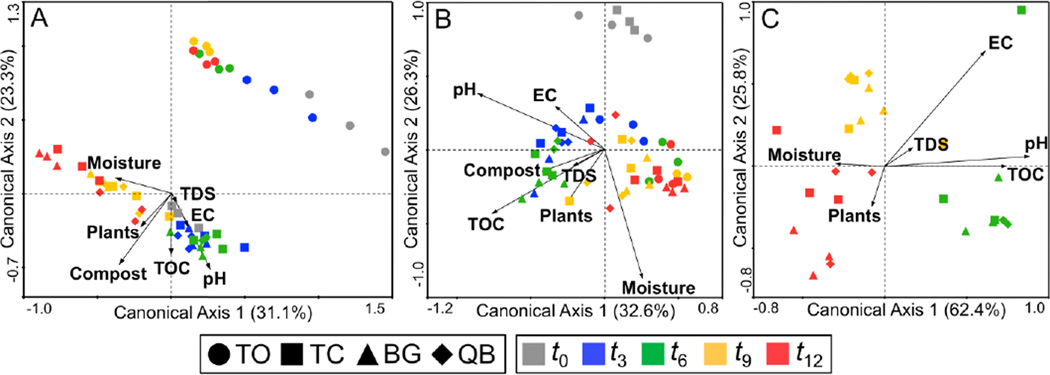
A series of CCA ordinations was performed to quantify the relative impact of selected environmental variables on the observed changes in microbial community structure. The first set of CCA ordinations evaluating physicochemical and nominal variables (pH, TOC, TDS, EC, moisture, plants and compost) revealed that for bacterial community structure the two most important parameters were compost and pH, while for fungal and archaeal communities, the top two parameters were pH and moisture, and EC and pH, respectively (Figure 5). pH, in particular, stands out as an important explanatory parameter influencing community structure for all three microbial groups. All three groups were positively correlated with pH through t6, but negatively correlated with pH for t9 and t12, which corresponds with the time points associated with the acidification of all compost-amended mesocosms (Figure 3). While not the most dominant factor, pore water TOC concentrations also figured prominently in the CCA analysis of all three domains and should be considered as an important nutrient source for heterotrophic populations. TOC levels peaked at t3 for BG and t6 for TC and QB (Supplementary Table 1). The CCA of compost-amended treatments showed positive correlations between TOC and community structure at 3 and 6 months for bacteria and fungi and at 6 months for archaea, but not at 9 and 12 months when TOC concentrations had decreased by over 50% (Figure 5).
Figure 5.
Canonical Correspondence Analysis (CCA) biplots of correlation between temporal changes in microbial community structure (symbols) and major environmental and nominal variables (arrows); (A) bacteria; (B) fungi; (C) archaea. Environmental variables were pH, moisture, electrical conductivity (EC), total organic carbon (TOC), and total dissolved solids (TDS) and nominal variables were plants, and compost. TO samples and the compost nominal variable were omitted from the archaeal CCA ordination due to the absence of archaeal populations in the TO unamended treatment. The value given on each axis label represents the percentage of the total variance explained by that axis. Unrestricted Monte-Carlo permutation tests were performed (1000 permutations) to determine the statistical significance of the relationship between the environmental variables and the canonical axes (all ordinations were statistically significant, p < 0.05).
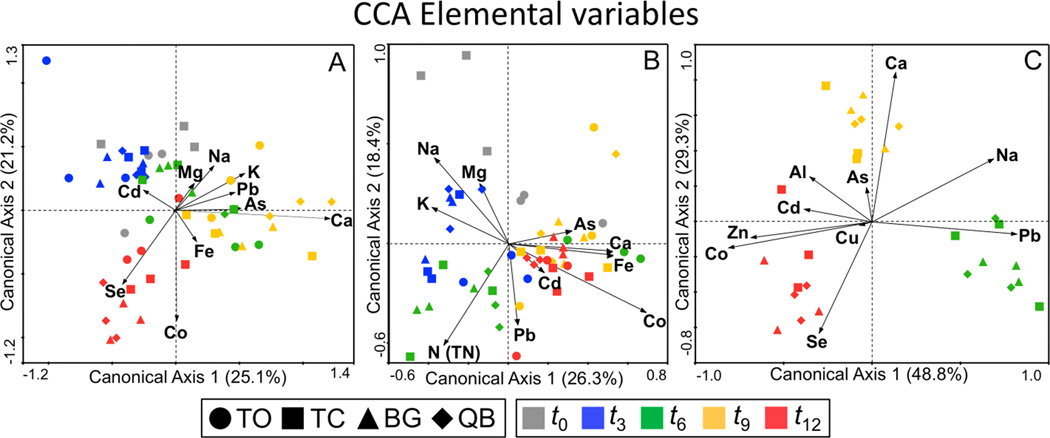
The second set of CCA ordinations evaluated the influence of pore water chemistry on microbial community dynamics. The chemical parameters included in these CCA ordinations (i.e. Na, Mg, Al, K, Ca, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Cd, Pb, S [SO4], N [TN]) were selected based on 2 main criteria: biological importance and potential toxicity. An automatic forward selection procedure (ter Braak and Šmilauer, 2002) was used to select and plot the 10 most relevant variables for each microbial group from the original set of 17 elements. Six variables were shared in common for all three microbial groups (i.e,. Na, Ca, Co, As, Cd, Pb,) and a total of 14 parameters were included in the three CCA ordinations combined (Figure 6). The most important explanatory elements as identified by the relative magnitude of the vectors were Ca and Co for bacteria, Co and N(TN) for fungi, and Ca, Co and Pb for archaea.
Figure 6.
Canonical Correspondence Analysis (CCA) biplots of correlation between temporal changes in microbial community structure (symbols) and pore water elemental parameters (arrows); (A) bacteria; (B) fungi; (C) archaea. Elemental variables were K, Na, Mg, Ca, Mn, Se, Cd, Co, Cu, Ni, Al, Zn, Pb, As, Fe, N (as TN), and S (as SO4). An automatic forward selection procedure was used to select and plot only the 10 most significant variables for each ordination. TO samples were omitted from the archaeal ordination because archaea were not detected. The value given on each axis label represents the percentage of the total variance explained by that axis. Unrestricted Monte-Carlo permutation tests were performed (1000 permutations) to determine the statistical significance of the relationship between the elemental variables and the canonical axes (all ordinations were statistically significant, p < 0.05).
4. Discussion
This study provides an extended temporal analysis of the response of bacterial, archaeal and fungal communities to compost amendment, plant establishment, and the geochemical dynamics of a year-long controlled assisted phytostabilization in IKMHSS mine tailings. Seeds were planted directly into compost-amended, acidic, high-metal content tailings. Results revealed that compost addition had an immediate and relatively long-lasting buffering-effect on pH, allowing plants to germinate and thrive for the first three months of the experiment. After six months the buffering capacity of the compost appeared to be exhausted and increasing acidification took over as the major driver affecting plant growth and survival. The pH of the QB treatment at t12 was significantly higher than all other treatments, demonstrating the potential control on tailings acidification imposed by successful plant establishment. This plant effect supports a pattern previously observed in both laboratory and short-term greenhouse studies (Solís-Domínguez et al., 2012).
4.1 Microbial community response to compost amendment and plant establishment
Compost addition produced an immediate and significant increase in bacterial and fungal community diversity, and established an archaeal community profile (not visible in TO samples), resulting in a long-lasting change in the structure of all three microbial communities. The results confirmed that compost amendment introduces entirely new bacterial, fungal and archaeal populations to the tailings ecosystem and suggested that this inoculum promotes a more diverse and stable microbial community.
The effect of plant establishment on the three microbial domains was less consistent. The results showed that the bacterial community responded quickly and maintained a differentiation between planted and unplanted treatments throughout the study. In contrast, a plant effect was not observed for the fungal community until midway through the experiment and archaea showed no clear plant effect within the study’s timeframe. Bacteria and fungi are known to respond differently to plants in natural ecosystems (Garbeva et al., 2004; Buée et al., 2009). Simple root exudates are almost exclusively degraded by fast-growing heterotrophic bacteria that colonize the rhizosphere in high numbers (Buée et al., 2009; Buyer et al., 2002; de Boer et al., 2005). In contrast, fungal populations gain a competitive advantage under acidic conditions and their contribution to root exudate degradation increases with acidification (de Boer et al., 2005). Recall that at t6 the pH declined sharply in planted treatments.
We hypothesize that in the present study, it is likely that between t0 and t6 fast-growing bacterial populations monopolized the degradation of root exudates, while saprotrophic fungal populations dominated the degradation of complex organic compounds derived from the compost (Buée et al., 2009). As the pH declined, the relative contribution of fungal populations to the degradation of root exudates increased, which was reflected in the separation of BG and QB fungal communities from the TC communities at t9 and t12 (Figure 2). Further, we contend that the closer association observed between the BG and TC fungal communities at t12 relative to QB is due to the fact that QB plant health and presumably root-exudate production was significantly greater at this time than for BG. As noted previously, fungal band number at this timepoint was similar for TC and QB and significantly higher for BG despite the fact that NMDS indicated that the TC and BG community structures were more similar. Previous research has demonstrated that increasing concentrations of lingo-cellulosic substrates in soil from decaying vegetative matter result in an increase in the richness and biodiversity of saprophytic fungal populations (Lonsdale et al., 2008; Schutter and Dick, 2001), thus the BG-specific band increase may reflect a response of substrate-specific fungal populations associated with the BG die-off. Taken together, the two analyses suggest that QB plant health had the most significant effect on fungal community structure at t12 and that fungal communities may reflect the above ground condition of the plants, especially since pore water TOC concentrations had decreased substantially by this time. However future research must confirm this hypothesis as the observed differences between TC, BG, and QB at t12 were not significant in this study.
4.2 Influence of geochemical parameters on microbial community dynamics
The CCA identified pH as a dominant chemical variable influencing the microbial community structure of bacteria, fungi and archaea (Figure 5). The results indicate that the compost inoculum provided the most significant and immediate influence on microbial community structure, but once the buffering capacity of the compost was exhausted acidification became the major explanatory variable governing community composition. Recall that the 3:1 tailings mix included reduced subsurface tailings known to be rich in sulfide minerals (e.g. pyrite, sphalerite, arsenopyrite) (Root et al., 2010). Relatively slow abiotic sulfide oxidation of the reduced tailings component would be expected to occur at circumneutral pH, but as the pH decreases to less than 5, rapid microbially-mediated processes typically dominate (Akcil and Koldas, 2006). The observed temporal shift in bacterial community structure with respect to pH beginning at t9 (Figure 5A) could partially reflect an increase in sulfur- and iron-oxidizing populations in all compost-amended mesocosms. In addition, previous studies have suggested that pH is one of the strongest determinants of soil microbial community composition (Fierer and Jackson, 2006; Högberg et al., 2007; Lauber et al., 2009) and that different microbial groups respond differently to changes in pH in the ecosystem (Högberg et al., 2007; Nicol et al., 2008; Rousk et al., 2009; 2010). Studies have shown that bacterial communities are usually more stable and diverse at circumneutral pH while fungal communities usually tolerate a wider range of pH values and even thrive in more acidic environments (Högberg et al., 2007; Rousk et al., 2010). The acidification process can have both direct (e.g. inhibition of enzymes) and indirect (e.g. increased metal toxicity) effects on neutrophilic microbial populations (Fierer and Jackson, 2006). The observed dramatic shift in bacterial community composition that accompanied the sudden decrease in pore water pH suggests that pore water pH could be used as a future temporal indicator of changes in community dynamics for studies focused on biotic influences on tailings acidification.
Archaeal community dynamics were less clearly defined, as demonstrated by the patterns of DGGE bands and the fluctuations in community structure, especially between t9 and t12 (Supplementary Figure 1, Table 1). The significant changes in community structure associated with pH and EC were detected at the point when both the pH and EC showed a significant decline. Pore water EC was lowest in all compost-amended treatments at t12 (Supplementary Table 1) when the greatest negative correlation was observed between archaeal community structure and EC (Figure 5C). We hypothesize that the archaeal populations dominant in the compost-amended treatments at the beginning of the experiment may have been species of haloarchaea (Soppa, 2006) that are favored in alkaline environments with elevated EC. We contend that these populations lost their competitive edge with the decrease in EC and pH (Walsh et al., 2005) and were replaced by a shift in community structure to increasing numbers of non-halophylic, acid tolerant archaeal species. Previous studies suggest that change in pH is a major factor influencing archaeal community dynamics in soil ecosystems (Görres et al., 2013; Pereira-e-Silva et al., 2012). Moreover, other studies suggest that members of the Thaumarchaeota (e.g. ammonia oxidizers), Euryarchaeota (e.g. iron oxidizers) and Crenarchaeota (e.g. sulfur oxidizers) are actually favored in acidic soils (Erguder et al., 2009; Kondrat‧eva et al., 2012; Lehtovirta et al., 2009; Nicol et al., 2008). Thus, the increase in archaeal band number in the planted treatments as the pH declined (Table 1) may correspond to increases in some of these acidophilic archaeal populations (Kondrat’eva et al., 2012).
The data do not indicate whether the dramatic change in bacterial, archaeal and fungal community structure between t6 and t9 precipitated the drop in pH or revealed a community response to acidification. It is conceivable that prokaryotic populations may be contributing to iron- and sulfur-oxidation reactions responsible for driving the observed acidification and fungi are simply exploiting the heterotrophic competitive advantage gained by the change in pH. This hypothesis is supported by the observed decrease in pore water TOC at this time point which would favor the autotrophic iron- and sulfur-oxidating bacterial and archaeal communities. Regardless of the specific community dynamics, plant health was directly affected by the acidification so the inter-relationship dynamics associated with changes in microbial structure, substrate pH and plant health must be carefully evaluated in future studies with pH serving as an indicator of ecosystem stability.
4.3 Influence of pore water chemistry on microbial community structure
The PCA suggests that the incorporation of compost influenced pore water metal(loid) concentrations, as composted treatments were generally negatively correlated with pore water metal(loid)s, whereas a positive correlation was observed between TO samples at t6, t9 and t12 and pore water metal(loid)s concentrations (Figures 3 and Figure 5). The positive correlation was strongest for the more acidic pore water samples. The general effect of pH on metal(loid) solubility is well known; in soil environments the solubility of toxic metal(loid)s (e.g. Cu, Zn, As, Cd, Pb) increases as the pH of the soil decreases (Chuan et al., 1996; Rieuwerts et al., 1998). Taken together these results suggest that the effect of compost on metal(loid) solubility may involve: (i) changing the pH which in turn controls the solubility of metal(loid)s (Madejón et al., 2006; Solís-Domínguez et al., 2012; Walker et al., 2004); (ii) limiting acid generation by decreasing the rate of microbial-mediated oxidation of ferrous and sulfide minerals (Johnson and Hallberg, 2005; Kim et al., 1999); (iii) and promoting metal(loid) sequestration processes, such as direct precipitation or co-precipitation of the metal(loid)s with various organic compounds and minerals (e.g. carbonates, oxides, hydroxides) or metal(loid) adsorption onto biotic and abiotic surfaces (Bolan and Duraisamy, 2003; Guo et al., 2006; Kumpiene et al., 2008; Park et al., 2011). Interestingly, a subtle trend towards increased pore water metal(loid)s was observed for TC, BG, and QB by t12 as demonstrated by a migration of data points towards the metal(loid) vectors, a pattern associated with both acidification and decreased pore water TOC. Thus, the acidification in all treatments appeared to be associated with increased leaching of numerous metal(loid) species.
In response to these observations, a second CCA was used to evaluate the influence of pore water chemistry on the community structure of the respective microbial communities. Co, Cd, Pb, and As were four of the six elements identified as significant influences on the community structure of all three domains (Figure 6). These results suggest that metal toxicity played an important role in shaping microbial community structure especially as an indirect consequence of mesocosm acidification. Interestingly, Ca and Co were among the most significant parameters analyzed for both bacteria and archaea while Co and TN were most significant for fungi (Figure 6). The bioavailability of metal(loid)s and other trace elements in soils is known to have a significant effect on the composition of soil microbial communities, usually because of either their biological importance or their toxicity (Gao et al., 2010; Khan et al., 2010; Liao and Xie, 2007).
The significance of N to fungi alone (Figure 6B) may reflect the fact that saprotrophic fungal populations are more sensitive to C:N ratios and usually require higher C:N ratios than prokaryotic populations (Fierer et al., 2004; Lauber et al., 2008). Previous studies suggest that N concentrations in soils and the amendment of soils with an N source (e.g. compost) usually have a stronger effect on the structure of fungal communities than that of prokaryotic communities (Bardgett et al., 1999; Buée et al., 2009; Frey et al., 2004). The analysis indicated that the influence of N was strongest at time points when pore water TN concentrations were highest (Figure 6B, Supplemental Table 1), potentially reducing the competitiveness of fungal populations which require relatively low concentrations of N in order to degrade complex organic compounds, such as lignin (Bittman et al., 2005; Tuomela et al., 2000).
Cobalt was the one element highly significant to bacteria, archaea and fungi with pore water concentrations ranging from 1.52 ± 3.91 mg L−1 at t0 to 7.30 ± 3.10 mg L−1 at t12. Cobalt has been reported to be toxic to microbial communities (Silver and Phung-le, 2005). Further it has been reported that Co potentially inhibits microbially-mediated sulfate reduction in metal-contaminated environments by outcompeting iron during the synthesis of metalloproteins (Ranquet et al., 2007; Ram et al., 2000). This is important when considering that sulfate reduction processes could mitigate the acidification of mine tailing systems and help prevent the generation of acid mine drainage (Johnson and Hallberg, 2005). Potentially more relevant in metal-contaminated environments, however, is the function of Co as an essential enzyme cofactor for microbial metabolism; even though its rarity in nature and the competition with other elements (e.g. Fe, Mg) makes it one of the less common transition metals in cells (Kobayashi and Shimizu, 1999; Okamoto and Eltis, 2011). Nonetheless, Co is the key transition metal involved in the synthesis of the important coenzyme cobalamin (better known as vitamin B12) in prokaryotic cells (Martens et al., 2002; Taylor and Sullivan, 2008). Cobalamin serves as an essential cofactor for the catalysis of transmethylation reactions that mediate important biosynthesis processes inside microbial cells, such as the synthesis of the amino acid methionine (Taylor and Sullivan, 2008). The capacity of cobalamin to facilitate transmethylation reactions, could be crucial for the survival of prokaryotes in metalliferous mine tailing environments given that it can mediate the methylation and volatilization of many metal(loid)s as a cell detoxification pathway (Ekstrom and Morel, 2008; Mason, 2012; Meyer et al., 2008; Nies, 1999). Some metal(loid)s that can be methylated by means of cobalamin for detoxification are: As (Gebel, 2002), Se (Gadd, 2004), Mg (Meyer et al., 2008), Pb (Ehrlich, 1997), and Cd (Mason, 2012). Further research is needed to determine whether the significance of Co to mine tailings ecosystems is related to its biological importance as an integral component of detoxification mechanisms or to its toxicity.
Finally, calcium was the most significant pore water element driving changes in the structure of both bacterial and archaeal communities. Ca in its mineral form is known to interact and adsorb toxic ions, including Cd, As, Pb and Se, in contaminated environments (Cravotta-Iii and Trahan, 1999; Bradl, 2004; Gadd, 2010; García-Sánchez et al., 1999; Sheoran and Sheoran, 2006). It is also important to note that in natural soil ecosystems the pH is tightly correlated to the concentration of Ca minerals (Warton and Matthiessen, 2005). Thus, the importance of Ca to microbial community structure in the mesocosms may be related to its capacity to complex toxic ions in the soil and mitigate the increase in toxicity promoted by the observed decline in pH.
5. Conclusions
The present study represents, to our knowledge, the first documentation of the structural dynamics of the three major microbial groups (i.e. bacteria, fungi, archaea) during revegetation of compost-amended metalliferous mine tailings. The differential response of bacterial, archaeal and fungal communities to plant establishment and mineral weathering in these mesocosm experiments highlights the importance of analyzing the dynamics of all three groups when determining the influence of microbial communities on the progress of plant establishment and biogeochemical weathering of tailings materials during phytoremediation. The data suggest that both bacteria and archaea may influence acidification processes and that pore water pH can be used as a temporal indicator of changes in the community dynamics of bacteria, fungi, and archaea. Relatively few studies have examined the influence of multiple environmental drivers on all three microbial groups during the reclamation of mine tailings, but we have shown here the significance of this multifaceted analysis. Further, our results reveal some intriguing patterns such as the responses of all three microbial groups to elevated pore water concentrations of Co and/or Ca. These relationships must be further investigated to understand their potential relevance to acidification and plant health. Future research will focus on the specific taxonomic and functional profiles associated with the structural changes documented in this study.
Supplementary Material
Highlights.
Bacterial, fungal and archaeal communities respond differently to revegetation
The three microbial groups responded differently to environmental fluctuations
Revegetation and compost mitigated tailings acidification and metal solubilization
Cobalt and pH were major drivers of change in microbial community structure
Acknowledgments
This research was supported by Grants P42 ES04940 and R01 ES017079 from the National Institute of Environmental Health Sciences Superfund Research Program, NIH (USA). Additional financial support for AVV was provided by an American Society for Microbiology-Robert D. Watkins fellowship and an Alfred P. Sloan Foundation scholarship. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of this manuscript. We wish to thank Steven Schuchardt for providing access to the IKMHSS, Scott White for coordinating the collection of the tailing samples, and Karis Nelson, Corin Hammond, and all who were involved in the collection of the tailings and setting-up of the mesocosm experiment.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akcil A, Koldas S. Acid Mine Drainage (AMD): causes, treatment and case studies. J Clean Prod. 2006;14:1139–1145. [Google Scholar]
- Alvarenga P, Gonçalves AP, Fernandes RM, de Varennes A, Vallini G, Duarte E, et al. Evaluation of composts and liming materials in the phytostabilization of a mine soil using perennial ryegrass. Sci Total Environ. 2008;406:43–56. doi: 10.1016/j.scitotenv.2008.07.061. [DOI] [PubMed] [Google Scholar]
- Bardgett RD, Mawdsley JL, Edwards S, Hobbs PJ, Rodwell JS, Davies WJ. Plant species and nitrogen effects on soil biological properties of temperate upland grasslands. Funct Ecol. 1999;13:650–660. [Google Scholar]
- Bittman S, Forge TA, Kowalenko CG. Responses of the bacterial and fungal biomass in a grassland soil to multiyear applications of dairy manure slurry and fertilizer. Soil Biol Biochem. 2005;37:613–623. [Google Scholar]
- Bolan NS, Duraisamy VP. Role of inorganic and organic soil amendments on immobilization and phytoavailability of heavy metals: a review involving specific case studies. J Soil Res. 2003;41:533–555. [Google Scholar]
- Bradl HB. Adsorption of heavy metal ions on soils and soils constituents. J Colloid Interface Sci. 2004;277:1–18. doi: 10.1016/j.jcis.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Brodie MJ, Broughton LM, Robertson AM. MEND Program, editors. Conference proceedings- Second International Conference on the Abatement of Acidic Drainage (Montreal, Quebec, Canada) Ottawa: Quebec MiningAssociation; 1991. A conceptual rock classification system for waste management and a laboratory method for ARD prediction from rock piles; pp. 119–135. [Google Scholar]
- Buée M, De Boer W, Martin F, Van Overbeek L. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil. 2009;321:189–112. [Google Scholar]
- Buyer JS, Roberts DP, Russek-Cohen E. Soil and plant effects on microbial community structure. Can J Microbiol. 2002;48:955–964. doi: 10.1139/w02-095. [DOI] [PubMed] [Google Scholar]
- Casamayor EO, Massana R, Benlloch S, Ovreas L, Diez B, Goddard VJ, et al. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol. 2002;4:338–348. doi: 10.1046/j.1462-2920.2002.00297.x. [DOI] [PubMed] [Google Scholar]
- Casamayor EO, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–408. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuan MC, Shu GY, Liu JC. Solubility of heavy metals in a contaminated soil: effects of redox potential and pH. Water Air Soil Poll. 1996;90:543–556. [Google Scholar]
- Colores GM, Macur RE, Ward DM, Inskeep WP. Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl Environ Microbiol. 2000;66:2959–64. doi: 10.1128/aem.66.7.2959-2964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravotta-Iii CA, Trahan MK. Limestone drains to increase pH and remove dissolved metals from acidic mine drainage. Appl Geochem. 1999;14:581–506. [Google Scholar]
- de Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Dybowska A, Farago M, Valsami-Jones E, Thornton I. Remediation strategies for historical mining and smelting sites. Sci Prog. 2006;89:71–138. doi: 10.3184/003685006783238344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EA-EST (EA Engineering, Science and Technology Inc.) [Accessed on April 4, 2013];Remedial Investigation Report: Iron King Mine- Humboldt Smelter Superfund Site, Dewey-Humboldt, Yavapai County, Arizona. 2010 Available from: http://yosemite.epa.gov/r9/sfund/r9sfdocw.nsf/3dc283e6c5d6056f88257426007417a2/9ff58681f889089c882576fd0075ea2f!OpenDocument.
- Ehrlich HL. Microbes and metals. Appl Microbiol Biot. 1997;48:687–692. [Google Scholar]
- Ekstrom EB, Morel FM. Cobalt limitation of growth and mercury methylation in sulfate-reducing bacteria. Environ Sci Technol. 2008;42:93–99. doi: 10.1021/es0705644. [DOI] [PubMed] [Google Scholar]
- Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Muyzer G, Ward DM. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. Global patterns in belowground communities. Ecol Lett. 2009;12:1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- Frey SD, Knorr M, Parrent JL, Simpson RT. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. Forest Ecol Manag. 2004;196:159–171. [Google Scholar]
- Gadd GM. Microbial influence on metal mobility and application for bioremediation. Geoderma. 2004;122:109–119. [Google Scholar]
- Gadd GM. Metals, minerals and microbes: geomicrobiology and bioremediation. Annu Rev Microbiol. 2010;156:609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhou P, Mao L, Zhi Y, Zhang C, Shi W. Effects of plant species coexistence on soil enzyme activities and soil microbial community structure under Cd and Pb combined pollution. J Environ Sci (China) 2010;22:1040–1048. doi: 10.1016/s1001-0742(09)60215-1. [DOI] [PubMed] [Google Scholar]
- Garbeva P, van Veen JA, van Elsas JD. Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- García-Sánchez A, Alastuey A, Querol X. Heavy metal adsorption by different minerals: application to the remediation of polluted soils. Sci Total Environ. 1999;242:179–188. [Google Scholar]
- Gebel TW. Arsenic methylation is a process of detoxification through accelerated excretion. Int J Hyg Environ Health. 2002;205:505–508. doi: 10.1078/1438-4639-00177. [DOI] [PubMed] [Google Scholar]
- Görres CM, Conrad R, Petersen SO. Effect of soil properties and hydrology on Archaeal community composition in three temperate grasslands on peat. FEMS Microbiol Ecol. 2013;85:227–240. doi: 10.1111/1574-6941.12115. [DOI] [PubMed] [Google Scholar]
- Grandlic CJ, Mendez MO, Chorover J, Machado B, Maier RM. Plant growth-promoting bacteria for phytostabilization of mine tailings. Environ Sci Technol. 2008;42:2079–2084. doi: 10.1021/es072013j. [DOI] [PubMed] [Google Scholar]
- Grantz DA, Vaughn DL, Farber RJ, Kim B, Ashbaugh L, VanCuren T, Campbell R. Wind barriers suppress fugitive dust and soil-derived airborne particles in arid regions. J Environ Qual. 1998;27:946–952. [Google Scholar]
- Gremion F, Chatzinotas A, Kaufmann K, Sigler Von W, Harms H. Impacts of heavy metal contamination and phytoremediation on a microbial community during a twelve-month microcosm experiment. FEMS Microbiol Ecol. 2004;48:273–283. doi: 10.1016/j.femsec.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Guo G, Zhou Q, Ma LQ. Availability and assessment of fixing additives for the in situ remediation of heavy metal contaminated soils: a review. Environ Monit Assess. 2006;116:513–528. doi: 10.1007/s10661-006-7668-4. [DOI] [PubMed] [Google Scholar]
- Gyssels G, Poesen J, Bochet E, Li Y. Impact of plant roots on the resistance of soils to erosion by water: a review. Prog Phys Geog. 2005;29:189–217. [Google Scholar]
- Hammer Ø, Harper D, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9. [Google Scholar]
- Hayden HL, Mele PM, Bougoure DS, Allan CY, Norng S, Piceno YM, et al. Changes in the microbial community structure of bacteria, archaea and fungi in response to elevated CO(2) and warming in an Australian native grassland soil. Environ Microbiol. 2012;14:3081–3096. doi: 10.1111/j.1462-2920.2012.02855.x. [DOI] [PubMed] [Google Scholar]
- Högberg MN, Högberg P, Myrold DD. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia. 2007;150:590–5901. doi: 10.1007/s00442-006-0562-5. [DOI] [PubMed] [Google Scholar]
- Hutchinson IPG, Ellison RD. Mine Waste Management. Michigan: Lewis Publishers; 1992. [Google Scholar]
- Johnson DB, Hallberg KB. Acid mine drainage remediation options: a review. Sci Total Environ. 2005;338:3–14. doi: 10.1016/j.scitotenv.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Kabas S, Acosta JA, Zornoza R, Faz-Cano A, Carmona DM, Martinez-Martinez S. Integration of landscape reclamation and design in a mine tailing in Cartagena-La Unión, SE Spain. Int J Energ Environ. 2011;2:301–308. [Google Scholar]
- Khan S, Hesham AE-L, Qiao M, Rehman S, He ZJ. Effects of Cd and Pb on soil microbial community structure and activities. Environ Sci Pollut Res Int. 2010;17:288–296. doi: 10.1007/s11356-009-0134-4. [DOI] [PubMed] [Google Scholar]
- Kim SD, Kilbane JJ, Cha DK. Prevention of acid mine drainage by sulfate reducing bacteria: organic substrate addition to mine waste piles. Environ Eng Sci. 1999;16:139–145. [Google Scholar]
- Kobayashi M, Shimizu S. Cobalt proteins. Eur J Biochem. 1999;261:1–9. doi: 10.1046/j.1432-1327.1999.00186.x. [DOI] [PubMed] [Google Scholar]
- Kondrat’eva TF, Pivovarova TA, Tsaplina IA, Fomchenko NV, Zhuravleva AE, Murav’ev MI, et al. Diversity of the communities of acidophilic chemolithotrophic microorganisms in natural and technogenic ecosystems. Microbiol. 2012;81:1–24. [PubMed] [Google Scholar]
- Kort J, Collins M, Ditsch D. A review of soil erosion potential associated with biomass crops. Biomass Bioenerg. 1998;14:351–359. [Google Scholar]
- Kumpiene J, Lagerkvist A, Maurice C. Stabilization of As, Cr, Cu, Pb and Zn in soil using amendments-- a review. Waste Manag. 2008;28:215–225. doi: 10.1016/j.wasman.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Strickland MS, Bradford MA, Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem. 2008;40:2407–2415. [Google Scholar]
- Lehtovirta LE, Prosser JI, Nicol GW. Soil pH regulates the abundance and diversity of Group 1.1c Crenarchaeota. FEMS Microbiol Ecol. 2009;70:367–376. doi: 10.1111/j.1574-6941.2009.00748.x. [DOI] [PubMed] [Google Scholar]
- Lepš J, Šmilauer P. Multivariate Analysis of Ecological Data Using CANOCO. 1st ed. New York: Cambridge University Press; 2003. [Google Scholar]
- Li J, Zheng Y, Yan J, Li H, Wang X, He J, et al. Effects of different regeneration scenarios and fertilizer treatments on soil microbial ecology in reclaimed opencast mining areas on the loess plateau, China. PLoS ONE. 2013;8:e63275. doi: 10.1371/journal.pone.0063275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Xie XM. Effect of heavy metals on substrate utilization pattern, biomass, and activity of microbial communities in a reclaimed mining wasteland of red soil area. Ecotoxicol Environ Saf. 2007;66:217–223. doi: 10.1016/j.ecoenv.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Lonsdale D, Pautasso M, Holdenrieder O. Wood-decaying fungi in the forest: conservation needs and management options. Eur J For Res. 2008;127:1–22. [Google Scholar]
- Ma Y, Prasad MN, Rajkumar M, Freitas H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol Adv. 2011;29:24–58. doi: 10.1016/j.biotechadv.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Madejón E, Pérez-de-Mora A, Felipe E, Burgos P, Cabrera F. Soil amendments reduce trace element solubility in a contaminated soil and allow regrowth of natural vegetation. Environ Pollut. 2006;139:40–52. doi: 10.1016/j.envpol.2005.04.034. [DOI] [PubMed] [Google Scholar]
- Martens JH, Barg H, Warren M, Jahn D. Microbial production of vitamin B12. Applied Microbiol Biotechnol. 2002;58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- Mason RP. The methylation of metals and metalloids in aquatic systems. In: Dricu A, editor. Methylation - From DNA, RNA and Histones to Diseases and Treatment. Croatia: InTech; 2012. pp. 271–302. [Google Scholar]
- May LA, Smiley B, Schmidt MG. Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can J Microbiol. 2001;47:829–841. doi: 10.1139/w01-086. [DOI] [PubMed] [Google Scholar]
- Mendez MO, Glenn EP, Maier RM. Phytostabilization potential of quailbush for mine tailings: growth, metal accumulation and microbial community changes. J Environ Qual. 2007;36:245–253. doi: 10.2134/jeq2006.0197. [DOI] [PubMed] [Google Scholar]
- Mendez MO, Maier RM. Phytostabilization of mine tailings in arid and semiarid environments- an emerging remediation technology. Environ Health Persp. 2008a;116:278–283. doi: 10.1289/ehp.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez MO, Maier RM. Phytoremediation of mine tailings in temperate and arid environments. Rev Environ Sci Bio/Technol. 2008b;7:47–59. [Google Scholar]
- Mendez MO, Neilson JW, Maier RM. Characterization of a bacterial community in an abandoned semiarid lead-zinc mine tailing site. Appl Environ Microbiol. 2008;74:3899–3907. doi: 10.1128/AEM.02883-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J, Michalke K, Kouril T, Hensel R. Volatilisation of metals and metalloids: An inherent feature of methanoarchaea? Syst Appl Microbiol. 2008;31:81–87. doi: 10.1016/j.syapm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Meza-Figueroa D, Maier RM, de la O-Villanueva M, Gómez-Alvarez A, Moreno-Zazueta A, Rivera J, et al. The impact of unconfined mine tailings in residential areas from a mining town in a semi-arid environment: Nacozari, Sonora, Mexico. Chemosphere. 2009;77:140–147. doi: 10.1016/j.chemosphere.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummey DL, Stahl PD, Buyer JS. Microbial biomarkers as an indicator of ecosystem recovery following surface mine reclamation. Appl Soil Ecol. 2002;21:251–259. [Google Scholar]
- Navarrete AA, Cannavan FS, Taketani RG, Tsai SM. A molecular survey of the diversity of microbial communities in different Amazonian agricultural model systems. Diversity. 2010;2:787–709. [Google Scholar]
- Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Nies DH. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Eltis LD. The biological occurrence and trafficking of cobalt. Metallomics. 2011;3:963–970. doi: 10.1039/c1mt00056j. [DOI] [PubMed] [Google Scholar]
- Park JH, Lamb D, Paneerselvam P, Choppala G, Bolan N, Chung JW. Role of organic amendments on enhanced bioremediation of heavy metal(loid) contaminated soils. J Hazard Mater. 2011;185:549–574. doi: 10.1016/j.jhazmat.2010.09.082. [DOI] [PubMed] [Google Scholar]
- Pereira-e-Silva MC, Dias AC, van Elsas JD, Salles JF. Spatial and temporal variation of archaeal, bacterial and fungal communities in agricultural soils. PLoS ONE. 2012;7:e51554. doi: 10.1371/journal.pone.0051554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-de-Mora A, Burgos P, Madejón E, Cabrera F, Jaeckel P, Schloter M. Microbial community structure and function in a soil contaminated by heavy metals: effects of plant growth and different amendments. Soil Biol Biochem. 2006;38:327–341. [Google Scholar]
- Pérez-de-Mora A, Ortega-Calvo JJ, Cabrera F, Madejón E. Changes in enzyme activities and microbial biomass after “in situ” remediation of a heavy metal-contaminated soil. Appl Soil Ecol. 2005;28:125–137. [Google Scholar]
- Ram MS, Singh L, Suryanarayana M, Alam SI. Effect of iron, nickel and cobalt on bacterial activity and dynamics during anaerobic oxidation of organic matter. Water Air Soil Pollut. 2000;117:305–312. [Google Scholar]
- Ranquet C, Ollagnier-de-Choudens S, Loiseau L, Barras F, Fontecave M. Cobalt stress in Escherichia coli: The effect on the iron-sulfur proteins. J Biol Chem. 2007;282:30442–30451. doi: 10.1074/jbc.M702519200. [DOI] [PubMed] [Google Scholar]
- Reed HE, Martiny JB. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol. 2007;62:161–170. doi: 10.1111/j.1574-6941.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- Rieuwerts JS, Thornton I, Farago ME, Ashmore FR. Factors influencing metal bioavailability in soils: preliminary investigations for the development of a critical loads approach for metals. Chem Spec Bioavailab. 1998;10:61–75. [Google Scholar]
- Root RA, Hayes SM, Schowalter C, Chorovoer J. Coupled arsenic and sulfur speciation in semi-arid tailings. Geochim Cosmochim Acta. 2010;74(Supplement):A881. [Google Scholar]
- Rosario K, Iverson SL, Henderson DA, Chartrand S, McKeon C, Glenn EP, et al. Bacterial community changes during plant establishment at the San Pedro River mine tailings site. J Environ Qual. 2007;36:1249–1259. doi: 10.2134/jeq2006.0315. [DOI] [PubMed] [Google Scholar]
- Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- Rousk JJ, Brookes PCP, Bååth EE. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol. 2009;75:1589–1596. doi: 10.1128/AEM.02775-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheoran AS, Sheoran V. Heavy metal removal mechanism of acid mine drainage in wetlands: A critical review. Hydrometallurgy. 2006;19:105–116. [Google Scholar]
- Schippers A, Breuker A, Blazejak A, Bosecker K, Kock D, Wright TL. The biogeochemistry and microbiology of sulfidic mine waste and bioleaching dumps and heaps, and novel Fe(II)-oxidizing bacteria. Hydrometallurgy. 2010;104:342–350. [Google Scholar]
- Schutter M, Dick R. Shifts in substrate utilization potential and structure of soil microbial communities in response to carbon substrates. Soil Biol Biochem. 2001;33:1481–1491. [Google Scholar]
- Silver S, Phung-le T. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J Ind Microbiol Biotechnol. 2005;32:587–605. doi: 10.1007/s10295-005-0019-6. [DOI] [PubMed] [Google Scholar]
- Solís-Domínguez FA, Valentín-Vargas A, Chorover J, Maier RM. Effect of arbuscular mycorrhizal fungi on plant biomass and the rhizosphere microbial community structure of mesquite grown in acidic lead/zinc mine tailings. Sci Total Environ. 2011;409:1009–1016. doi: 10.1016/j.scitotenv.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís-Domínguez FA, White SA, Hutter TB, Amistadi MK, Root RA, Chorover J, et al. Response of key soil parameters during compost-assisted phytostabilization in extremely acidic tailings: effect of plant species. Environ Sci Technol. 2012;46:1019–1127. doi: 10.1021/es202846n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J. From genomes to function: haloarchaea as model organisms. Annu Rev Microbiol. 2006;152:585–590. doi: 10.1099/mic.0.28504-0. [DOI] [PubMed] [Google Scholar]
- Tan GL, Shu WS, Hallberg KB, Li F, Lan CY, Zhou WH, et al. Culturable and molecular phylogenetic diversity of microorganisms in an open-dumped, extremely acidic Pb/Zn mine tailings. Extremophiles. 2008;12:657–664. doi: 10.1007/s00792-008-0171-9. [DOI] [PubMed] [Google Scholar]
- Taylor GT, Sullivan CW. Vitamin B12 and cobalt cycling among diatoms and bacteria in Antarctic sea ice microbial communities. Limnol Oceanorg. 2008;53:5862–5877. [Google Scholar]
- ter Braak CJF, Wiertz J. On the statistical analysis of vegetation change: a wetland affected by water extraction and soil acidification. J. Veg Sci. 1994;5:361–372. [Google Scholar]
- ter Braak CJF, Šmilauer P. CANOCO reference manual and CanoDraw for Windows user’s guide: Software for canonical community ordination. V 4.5. New York: Microcomputer Power; 2002. [Google Scholar]
- Tordoff GM, Baker AJ, Willis AJ. Current approaches to the revegetation and reclamation of metalliferous mine wastes. Chemosphere. 2000;41:219–228. doi: 10.1016/s0045-6535(99)00414-2. [DOI] [PubMed] [Google Scholar]
- Tuomela M, Vikman M, Hatakka A, Itavaara M. Biodegradation of lignin in a compost environment: a review. Bioresour Technol. 2000;72:169–183. [Google Scholar]
- Valentín-Vargas A, Chorover J, Maier RM. A New Standard-Based Polynomial Interpolation (SBPIn) method to address gel-to-gel variability for the comparison of multiple denaturing gradient gel electrophoresis profile matrices. J Microbiol Methods. 2013;92:173–177. doi: 10.1016/j.mimet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentín-Vargas A, Toro-Labrador G, Massol-Deyá AA. Bacterial community dynamics in full-scale activated sludge bioreactors: operational and ecological factors driving community assembly and performance. PLoS ONE. 2012;7:e42524. doi: 10.1371/journal.pone.0042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DJ, Clemente R, Bernal MP. Contrasting effects of manure and compost on soil pH, heavy metal availability and growth of Chenopodium album L. in a soil contaminated by pyritic mine waste. Chemosphere. 2004;57:215–224. doi: 10.1016/j.chemosphere.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Walsh DA, Papke RT, Doolittle WF. Archaeal diversity along a soil salinity gradient prone to disturbance. Environ Microbiol. 2005;7:1655–1666. doi: 10.1111/j.1462-2920.2005.00864.x. [DOI] [PubMed] [Google Scholar]
- Warton B, Matthiessen JN. The crucial role of calcium interacting with soil pH in enhanced biodegradation of metam-sodium. Pest Manag Sci. 2005;61:856–862. doi: 10.1002/ps.1095. [DOI] [PubMed] [Google Scholar]
- Wong MH. Ecological restoration of mine degraded soils, with emphasis on metal contaminated soils. Chemosphere. 2003;50:775–780. doi: 10.1016/s0045-6535(02)00232-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.