Abstract
Treatment with individual anti‐androgens is associated with the development of hot‐spot mutations in the androgen receptor (AR). Here, we found that anti‐androgens‐mt‐ARs have similar binary structure to the 5α‐dihydrotestosterone‐wt‐AR. Phage display revealed that these ARs bound to similar peptides, including BUD31, containing an Fxx(F/H/L/W/Y)Y motif cluster with Tyr in the +5 position. Structural analyses of the AR‐LBD‐BUD31 complex revealed formation of an extra hydrogen bond between the Tyr+5 residue of the peptide and the AR. Functional studies showed that BUD31‐related peptides suppressed AR transactivation, interrupted AR N‐C interaction, and suppressed AR‐mediated cell growth. Combination of peptide screening and X‐ray structure analysis may serve as a new strategy for developing anti‐ARs that simultaneously suppress both wt and mutated AR function.
Keywords: FxxLF, BUD31, Crystallography, Anti-androgen withdrawal syndrome, Androgen receptor
Highlights
Anti‐androgens‐mutant‐ARs had similar binary structure to the DHT‐wild type‐AR.
These ARs bound to similar peptides with Fxx(F/H/L/W/Y)Y motifs.
BUD31 with FxxFY motif was a new AR coregulator.
BUD31‐related peptides could suppress AR related function.
Combined peptide screening and structure analysis for anti‐ARs was proposed.
1. Introduction
Anti‐androgen withdrawal syndrome (AWS), initially described in the context of hydroxyflutamide (HF) treatment of prostate cancer as the response to withdrawal of therapy (Kelly and Scher, 1993), has since been documented for other anti‐androgens, including bicalutamide (CDX), nilutamide, and chlormadinone (Akakura et al., 1998; Gomella et al., 1997; Small and Carroll, 1994). This syndrome has also been extended to steroid hormone withdrawal syndromes that occur with cessation of other steroid hormone treatments, including megestrol acetate (Dawson and McLeod, 1995), diethylstilbestrol (Bissada and Kaczmarek, 1995), and estramustine (Shibata et al., 1999). Approximately 30% of patients may experience this phenomenon, and withdrawing the anti‐androgen treatment may have short‐term benefits. The underlying mechanism is not clear, but mutation of the androgen receptor (AR) has frequently been reported. Some hot‐spot mutations have been linked to treatment with individual anti‐androgens, such as T877A, with hydroxyflutamide (HF) treatment, and W741(C/L), with bicalutamide (CDX) treatment (Hara et al., 2003). The ligand specificity of T877A AR is lost; thus, this receptor can be activated by both androgens and anti‐androgens (Miyamoto et al., 2004).
The AR is a ligand‐activated transcription factor that belongs to the nuclear receptor superfamily (Chang et al., 1988; Lubahn et al., 1988). The transcriptional activity of the AR is modulated by co‐regulators such as ARA70, also known as NCOA4 (nuclear receptor coactivator 4) and SHP, also known as NR0B2 (nuclear receptor subfamily 0, group B, member 2) (Heinlein and Chang, 2002). The AR and other nuclear receptors can bind to the canonical LxxLL motif in these co‐regulators (Darimont et al., 1998; Jouravel et al., 2007), with the AR preferring aromatic‐rich FxxLF‐like motifs. Studies of the first (+1) and the fifth (+5) amino acid positions of the FxxLF motif indicate that these residues are involved in mediating high affinity and specific interactions with the AR ligand‐binding domain (LBD) (Dubbink et al., 2006). Furthermore, interchanging F and Y at the +5 position, as in FxxLF, FxxLY and FxxFF, modulates coregulator binding activity (He et al., 2000; Hsu et al., 2003; van de Wijngaart et al., 2006). In fact, the FxxLF‐like motif exists in many AR coregulators and plays an important role in modulating coregulator‐mediated AR functions (Heinlein and Chang, 2002; Hsu et al., 2003; Yeh and Chang, 1996). This FxxLF‐like motif is also found in the AR N‐terminus and is pivotal in mediating the interaction of AR N‐ and C‐termini, which is required for full AR function (He et al., 2000, 2002).
Upon androgen binding to the AR, helix 12 undergoes a conformational change that allows opening of the AF2 (activation function 2) hydrophobic cleft formed by helices 3, 4, 5, and 12 of the LBD to accommodate the bulky peptide side chains. Specifically, the FxxLF peptide forms a short α‐helix with Phe+1, Leu+4, and Phe+5 that binds hydrophobic face down in the L‐shaped AF2 cleft of the AR. Electrostatic interactions involving Lys720 and Glu897 at either end of the AF2 cleft interact with the backbone carbonyl group of Phe+5 and the amine group of residue +1 or −1, respectively (Dubbink et al., 2004; Hur et al., 2004). The side chains of two key amino acid residues, Met734 and Met894, also help to form the hydrophobic binding pocket for the +1 and +5 position of the motif. In contrast, Leu+4 binds to a shallow, hydrophobic patch in the sidewall of the L‐shaped groove and is largely solvent exposed. The other intra‐motif and flanking amino acids support the interaction of the peptide with the AR (Hur et al., 2004).
Using 5α‐dihydrotestosterone (DHT) wild‐type AR‐DBD‐LBD (wt‐AR‐DBD‐LBD) and HF mutant T877A AR‐DBD‐LBD (HF‐T877A‐AR‐DBD‐LBD) as baits, we found that screened peptides were enriched for Tyr+5. Further characterization of the roles of the peptide motifs using both functional and structural approaches led us to conclude that the newly identified BUD31 peptide motif functions to suppress both DHT‐mediated AR and HF‐mediated T877A‐AR transactivation, thus potentially offering a better therapeutic approach for suppressing prostate cancer in patients with AWS.
2. Materials and methods
2.1. Materials and plasmids
DHT and CDX were obtained from Sigma Chemical Co. (St. Louis, MO) and HF was obtained from Schering‐Plough (Kenilworth, NH). The Ph.D.‐12 peptide library was purchased from New England Biolabs (Beverly, MA). The human prostate cancer cell lines, PC‐3 and LNCaP, were purchased from the American Type Culture Collection (Manassas, VA). The AR expression plasmids PCMV‐Flag‐AR and pCMX‐VP16‐AR were constructed as described previously (Nishimura et al., 2003; Yeh and Chang, 1996). BUD31 cDNA, prepared from human LNCaP cells, was cloned into the p3xFLAG‐CMV vector (Sigma Chemical Co.). Motif‐mutants of the BUD31 plasmid, p3X‐FLAG‐mt‐AxxAA, were generated by site‐directed mutagenesis using a kit obtained from Stratagene (La Jolla, CA). Peptides with the following sequences were synthesized by Genesis Biotech Inc. (Hsintien, Taiwan): Biotin‐BUD31p, Biotin‐KTRYI‐FDLFY‐KRKAY; Biotin‐3–18, Biotin‐NTNA‐FSRLF‐YPS; Biotin‐C320, Biotin‐SDSA‐FSRLY‐TRS; Transactivator of Transcription (TAT), fluorescein‐GGGYGRKKRRQRRRG; TAT‐BUD31p, fluorescein‐TAT‐KTRYI‐FDLFY‐KRKAY; TAT‐3–18, fluorescein‐TAT‐NTNA‐FSRLF‐YPS; TAT‐C320, TAT‐SDSA‐FSRLY‐TRS; and TAT‐3–14, fluorescein‐TAT‐DHSK‐LYSLL‐QSS. All peptides were dissolved in water. Anti‐AR (C19) and anti‐BUD31 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Sigma–Aldrich, respectively.
2.2. Co‐immunoprecipitation
The co‐immunoprecipitation (Co‐IP) procedure has been previously described in detail (Shyr et al., 2010). Here, LNCaP cells were transfected with 5 mg of p3xFLAG‐BUD31 and harvested after 48 h, and nuclear extracts were prepared for Co‐IP assays. A 400‐μl aliquot of each extract was first incubated with anti‐Flag or anti‐AR (C19) antibody (Santa Cruz Biotechnology) overnight at 4 °C, and then with Protein G Sepharose for an additional 16 h at 4 °C. Protein G Sepharose containing the immune complex was then washed three times with washing buffer (50 mM Tris–HCl pH 7.4, 100 mM NaCl, 5 mM CaCl2, 5 mM MgCl2, 0.1% Nonidet P‐40) and resuspended in sodium dodecyl sulfate (SDS)‐containing sample buffer. Flag‐ and AR‐associated complexes were immunoprecipitated with anti‐Flag and anti‐AR antibodies, respectively. The isolated protein complexes were fractionated, transferred to membranes, and immunoblotted with anti‐AR or anti‐BUD31 antibodies.
2.3. Patient enrollment
Primary prostate tumors were collected from patients undergoing a prostate tumor biopsy from 2007 to 2009. Written informed consent was obtained from all patients prior to surgery. The studies were approved by the Institutional Review Board of Chang Gung Memorial Hospital, Taoyuan, Taiwan.
2.4. Cloning, expression, and purification of human AR proteins and phage‐display procedure
AR‐DBD‐LBD cDNA (amino acids 548–919) and AR‐LBD cDNA (amino acids 663–919) were amplified from the AR expression vector pSG5‐AR by polymerase chain reaction (PCR) and inserted into the pET28c vector (Novagen, San Diego, CA). The pET28c‐T877A‐AR‐DBD‐LBD vector was generated using the Stratagene site‐directed mutagenesis kit. Protein expression and purification, and phage‐display procedures were described previously (Chang et al., 2005; Hsu et al., 2003; Hur et al., 2004).
2.5. Crystallization, data collection, and structural determination
AR‐LBD proteins were crystallized with the hanging‐drop method using a commercial kit. After extensive trials, individual crystals of the AR‐LBD protein were obtained after 2 d in a solution of 1.6 M magnesium sulfate heptahydrate in 0.1 mM MES buffer (pH 6.5), reaching a maximal size within 2 wk. Rhombus‐shaped AR‐LBD protein crystals (0.6 × 0.6 × 2.5 mm) were transferred to solutions containing different 3–6‐fold molar excess peptides and incubated for 1 h at room temperature. Finally, crystals were soaked in a protectant containing mother liquid and an additional 20% (v/v) glycerol for 20 s and placed in the cold stream at 100 K.
Data were collected using an ADSC Quantum 315r CCD detector at the BL13B at the National Synchrotron Radiation Research Center (NSRRC) in Hsinchu, Taiwan. The datasets were indexed and integrated using HKL2000 and SCALEPACK software (Otwinowski and Minor, 1997). The structures were determined by the molecular replacement method with the AMore program using the AR‐LBD ternary structure (PDB code: 1T73) as the search model (Navaza, 1994). Models were built with the Coot program (Emsley and Cowtan, 2004). Refinement was carried out with the REFMAC 5 program (Murshudov et al., 1997). Water molecules were added and checked using the ARP/WARP program (Perrakis et al., 1999). The refined model was checked using the PROCHECK program (Laskowski et al., 1993). Protein structural figures were prepared with the PyMOL program (Schrodinger, 2010). The coordinates have been deposited in the Protein Data Bank. Data collection and statistics are summarized in Supplementary Table 1.
2.6. Surface plasmon resonance
Synthetic biotinylated peptides were injected onto a streptavidin‐immobilized chip and allowed to bind; binding was detected in resonance units (400 resonance units for peptides only). Different concentrations (12–48 nM) of AR‐DBD‐LBD in an HBS‐P buffer (10 mM HEPES pH 7.4, 0.15 M NaCl, 0.05% surfactant P20) containing 10 nM DHT or 10 μM HF were injected at a flow rate of 30 μl/min, and binding to peptides, in resonance units, was measured. A blank flow cell served as the negative control, and background signals were subtracted from sample signals. Sensorgrams of association and dissociation were recorded and analyzed using BIAevaluation software 3.0 (Biacore AB, Uppsala, Sweden). A simple 1:1 Langmuir model was employed to fit the data. A two‐state conformational change model was used, as previously described (Warnmark et al., 2002).
2.7. Glutathione sepharose transferase (GST) pull‐down assay
Wild‐type (wt) and mutant (mt) GST‐BUD31 and GST control proteins were purified as described by the manufacturer of the GST pull‐down assay (Amersham Biosciences, Sunnyvale, CA). Five microliters of in vitro‐translated [35S] methionine‐labeled AR (full‐length, N‐terminus, or C‐terminus) was incubated with wt or mt GST‐BUD31 protein or GST control bound to glutathione‐Sepharose beads in the presence or absence of 10 nM DHT (Hsu et al., 2003). After extensive washing, the pull‐down complex was loaded onto an 8% or 10% polyacrylamide gel and visualized by autoradiography. The input represented 20% of the amount of labeled protein used in the pull‐down assay.
2.8. Mammalian two‐hybrid assay, transfection, and reporter gene assay
Procedures were performed as previously described in detail (Hsu et al., 2003). The PC‐3 cell line was used in mammalian two‐hybrid assays and reporter gene assays.
2.9. ChIP assay
LNCaP cells were grown in phenol‐red‐free RPMI‐1640 medium supplemented with 10% FBS. LNCaP cells were transfected with P3xFlag‐BUD31 plasmid using the Superfect reagent (Qiagen, Valencia, CA), as described by the manufacturer. After transfection, the cells were cultured overnight in phenol‐red‐free RPMI‐1640 supplemented with 8% charcoal‐dextran‐stripped FBS. Cells were treated with or without 10 nM DHT for 2 h before cross‐linking for 10 min with a final concentration of 1% formaldehyde in growth media. Cells were washed twice with ice‐cold PBS and pelleted. Chromatin immunoprecipitation (ChIP) assays were performed using a Magna ChIP kit (Millipore, Billerica, MA). Antibodies used in these experiments were anti‐AR (PG‐21; Millipore), anti‐BUD31 (H‐61; Santa Cruz Biotechnology), anti‐Flag M2 (Sigma), and anti‐RNA polymerase II (clone CTD4H8; Millipore). Two microliters of DNA extract (out of a total of 50 μl) was used in each PCR. The primer pairs used were as follows: ARE I/II (−459 to −121), 5′‐GCC AAG ACA TCT ATT TCA GGA GC‐3′ (forward) and 5′‐CCC ACA CCC AGA GCT GTG GAA GG‐3′ (reverse); and ARE III (−4288 to −3922), 5′‐GGG GTT TGT GCC ACT GGT GAG‐3′ (forward) and 5′‐GGG AGG CAA TTC TCC ATG GTT‐3′ (reverse) (Shatkina et al., 2003).
2.10. Cell growth assay
LNCaP cells were grown in RPMI medium containing 10% FBS. Cells were plated at 5 × 104 cells/well in 24‐well plates and incubated with or without 0.2 nM DHT and 10 μM peptide for 2–7 d. Cell growth was assessed using the MTT (3‐{4,5‐dimethylthiazol‐2‐yl}‐2,5‐diphenyltetrazolium bromide) assay (Hsu et al., 2005). At each time point, 50 μl of a 5‐mg/ml MTT solution was added to each well containing 500 μl of medium and incubated for 3 h; 500 μl of isopropyl alcohol was added to dissolve the reduced formazan product. The absorbance of each well was measured at a wavelength of 590 nm in a DU 640B spectrophotometer (Beckman, Fullerton, CA) according to the manufacturer's protocol. Values presented in figures are mean OD590 ± SD from at least three independent reaction wells.
2.11. Statistical analysis
Data are presented as means ± SEM. Differences between two groups were assessed using the unpaired two‐tailed Student's t test.
3. Results and discussion
3.1. Binding profiles of binary liganded–AR‐LBD complex structures
AWS, in which anti‐androgens become agonists, is found in anti‐androgen‐treated prostate cancer patients, who are frequently found to harbor the mutated T877A‐AR (with HF treatment) or mutated W741L‐AR (with CDX treatment) (Kelly et al., 1997; Miyamoto et al., 2004). These findings suggest that liganded wt and mutant ARs (DHT‐wt‐AR, HF‐T877A‐AR, and CDX‐W741L‐AR) might share similar structures that could recognize common AR‐associated peptide motifs. Therefore, structures of AR‐LBD co‐crystals with androgen or anti‐androgen might reveal structural variations among these mutants.
Three binary complex structures were obtained: DHT‐wt‐AR‐LBD, HF‐T877A‐AR‐LBD, and CDX‐W741L‐AR‐LBD. All the crystallographic parameters of these crystals belonged to the orthorhombic group with one molecule per asymmetric unit, results similar to those observed previously (Hur et al., 2004). Furthermore, all models were refined to R and free‐R values that conformed to a typical range. Crystallographic statistics are summarized in Supplemental Table 1.
To further verify the structural‐rearrangement effects trigged by androgen versus anti‐androgens, we overlapped these binary structures. The equivalent Cα atoms were identical (root mean square deviation [RMSD] < 0.5 Å) among these structures, indicating that these compounds propelled identical structural rearrangements of the AR architecture (Figure 1A). In addition to the overall structures, the binding sites of the androgens were also compared. DHT bound to four surrounding residues (Asn705, Gln711, Arg752, and Thr877) of the AR protein by forming hydrogen bonds. Binding contacts were similar for HF‐T877A and CDX‐W741L AR mutants, except that the contact involving Thr877 was lacking in the HF‐T877A mutant and the CDX‐W741L mutant formed an additional hydrogen bond with the O atom of Leu704 (Figure 1B).
Figure 1.
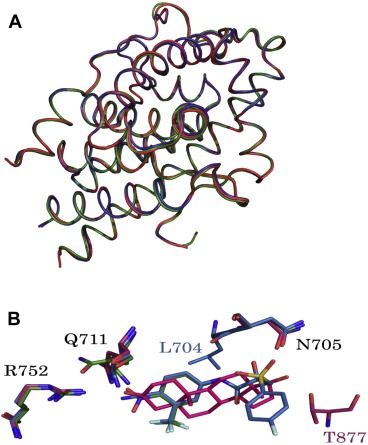
(A). Superposition of the overall structures of wt‐AR‐LBD (red), T877A‐AR‐LBD (green), and W741L (blue)‐AR‐LBD, drawn as ribbon models. (B). Overlapping crucial residues in the active site. Residues are shown as sticks. Nitrogen and oxygen atoms are colored blue and red, respectively. DHT, HF, and CDX are illustrated as red, green, and blue, respectively. Fluorine, nitrogen, oxygen, and phosphorous atoms are depicted as light green, blue, red, and orange, respectively.
3.2. Tyrosine in the +5 position of the peptide motif is enriched in peptides that associate with DHT‐wt‐AR‐DBD‐LBD and HF‐T877A‐AR‐DBD‐LBD
To test if these liganded‐AR complexes, which share a similar structure, recognize similar peptide motifs, we used bacterially expressed DHT‐wt‐AR‐DBD‐LBD and HF‐T877A‐AR‐DBD‐LBD proteins as baits to screen potential peptides using phage display. We found that peptides containing a tyrosine in the +5 position of the motif, including those with the sequence FxxLY, FxxFY, FxxHY, FxxWY and FxxYY, were frequently identified in the screened peptides (Table 1). We then used a mammalian two‐hybrid assay to confirm the interaction of these screened peptides with full‐length DHT‐wt‐AR or HF‐T877A‐AR and found that they were able to interact with the ARs. Some peptides screened using the HF‐T877A protein as bait, such as those with an FxxFY motif (A41, B37, and B45) or FxxHY motif (HF‐26), showed a tendency to interact more strongly with HF‐T877A‐AR than with DHT‐T877A‐AR in these assays (Table 1).
Table 1.
Results of selection for peptides that associate with HF‐T877A‐AR protein.
| −1 | +1 +4+5 | +6 | wt‐AR | T877A‐AR | ||||
|---|---|---|---|---|---|---|---|---|
| Ethanol | DHT | Ethanol | DHT | HF | ||||
| Gal‐4 | 1 | 1 | 1 | 1 | 1 | |||
| 1.B310 | SPTPH | FTRLF | HY | 8 | 17810 | 60 | 12100 | 6717 |
| 2.B426 | EASNSS | FARLF | H | 2 | 13500 | 10 | 9552 | 1513 |
| 3.C317 | TEPPAA | FRQLF | F | 5 | 6600 | 15 | 5227 | 4380 |
| 4.C314 | SS | FADLW | RNEAT | 7 | 1142 | 16 | 859 | 121 |
| 5.B49 | SK | FSQLY | SYEPP | 12 | 1788 | 24 | 1032 | 318 |
| 6.B423 | SPTRL | FDHLY | MT | 10 | 1216 | 20 | 401 | 168 |
| 7.C33 | ST | FESLY | NNMTA | 20 | 1328 | 15 | 1018 | 399 |
| 8.C320 | SDSA | FSRLY | TRS | 8 | 13365 | 20 | 12725 | 4650 |
| 9.A41 | SSGG | FERFY | IAQ | 13 | 4314 | 21 | 3065 | 3510 |
| 10.A48 | DPPTSK | FSTFY | Q | 8 | 32 | 13 | 12 | 18 |
| 11.A49 | VLPGST | FERFY | R | 8 | 25 | 11 | 26 | 19 |
| 12.B37 | HNTK | FSTFY | NMD | 12 | 9162 | 29 | 6245 | 8711 |
| 13.B45 | NTPL | FQTFY | NQR | 15 | 18010 | 90 | 12560 | 17240 |
| 14.C41 | S | FYDFY | NGASTP | 15 | 23 | 14 | 23 | 17 |
| 15.B32 | NTTDTL | FSQHY | R | 10 | 41 | 11 | 32 | 25 |
| 16.B34 | SP | FEEHY | LSTQS | 13 | 42 | 15 | 37 | 28 |
| 17.B411 | HPARTS | FETHY | M | 13 | 13 | 9 | 13 | 16 |
| 18.HF‐26 | HASP | FEMHY | LLG | 12 | 8905 | 27 | 8401 | 8904 |
| 19.B46 | ANSS | FRDWY | TSS | 10 | 792 | 14 | 376 | 1262 |
| 20.A47 | DSQYSS | FSRYY | K | 11 | 21 | 10 | 19 | 15 |
| 21.A310 | SHSS | FYQYY | LDN | 16 | 4771 | 28 | 3586 | 4461 |
| 22.B311 | HKASP | FNYYY | LN | 22 | 46 | 23 | 33 | 38 |
| 23.B414 | NPRTPTT | FERYY | 12 | 18 | 13 | 15 | 15 | |
| 24.A43 | PAPK | FKEFF | YAE | 4 | 8845 | 5 | 7352 | 8304 |
| 25.HF‐15 | HTSM | FESFF | RKE | 6 | 11500 | 20 | 13854 | 14523 |
| a CON‐1 | MYKP | HNHHQ | TSS | 1 | 2 | 2 | 4 | 5 |
| b #3‐18 | NTNA | FSRLF | YPS | 2 | 12530 | 9 | 10985 | 3890 |
| c ARN20 | RGA | FQNLF | QSV | 3 | 1310 | 4 | 1578 | 1423 |
| d D30 | HPTHSSR | LWELL | MEATPTM | 18 | 19860 | 415 | 18060 | 8803 |
| e BUD31 | KTRYI | FDLFY | KRKAY | 2 | 8945 | 4 | 11965 | 4870 |
CON‐1: negative control.
#3‐18.
ARN20.
D30: positive controls.
BUD31: fragment of novel AR coregulator candidate.
3.3. Peptide motif complex structures
Although the binary architectures of the AR variants were similar to those of the wt‐AR, the affinity between peptides and AR protein varied. To clarify how peptides preferentially interact with these proteins, we determined the ternary complex structures of the AR proteins in the presence of various peptides. As expected, an additional density map on the surface was found for each and was modeled to a short α‐helical peptide given by the indicated sequences. These peptides were coordinated by residues L720 and E894, suggesting that these peptides bound to the AF2 sites formed by helices 3 (H3), 5 (H5), and 12 (H12) (Hur et al., 2004).
Seventeen ternary complex structures were determined and divided into three groups: wt‐AR, T877A‐AR, and W741L‐AR. To gain further insight into the structural consequences of peptide association, we overlapped the binary (absence of peptides) and ternary (presence of peptides) structures. Superpositions of the equivalent Cα atoms were identical (RMSD < 0.5 Å) among ternary wt‐AR structures, indicating that these structures share identical folds owing to DHT binding. Moreover, association of the peptides did not influence the architecture of the wt‐AR. Similar results were obtained with respect to the ternary complex structures of AR variants. To further confirm these structural similarities, we assessed differences between wt‐AR and variants by overlapping the ternary wt‐AR‐FQNLF structure with that of T877A‐AR‐FQNLF and W741L‐AR‐FQNLF structures. RMSD values of 1.0 and 0.3 Å in the Cα backbone were observed for T877A‐AR‐FQNLF and W741L‐AR‐FQNLF structures, respectively, with respect to that of wt‐AR‐FQNLF, indicating that the folding of the overall structures is identical. These findings indicate that the structural rearrangements induced in mutant ARs by HF and CDX treatment are similar. Collectively, these results indicate that the similarity of the rearrangements that occur with AWS carries significant implications.
Although the peptides were unable to disturb the scaffolds of the ARs, the resolved structures revealed slight differences in the interactions between peptides and the AF2 site, such as accommodations in the +1 and +5 positions of the peptide. In the front‐end position (+1) of the AF2 site, the mobile Oδ2 atoms of residue E897 make variable hydrogen bond contacts with the N atoms of peptides at −1 or −2 positions. These contacts not only provide the major stabilizing influence, they also contribute to the variable affinities of peptides for protein (Figure 2A). Notably, residues M734 and M897 on either side of the +1 position provide the structural flexibility necessary to accommodate the +1 peptide position van der Waals interactions.
Figure 2.
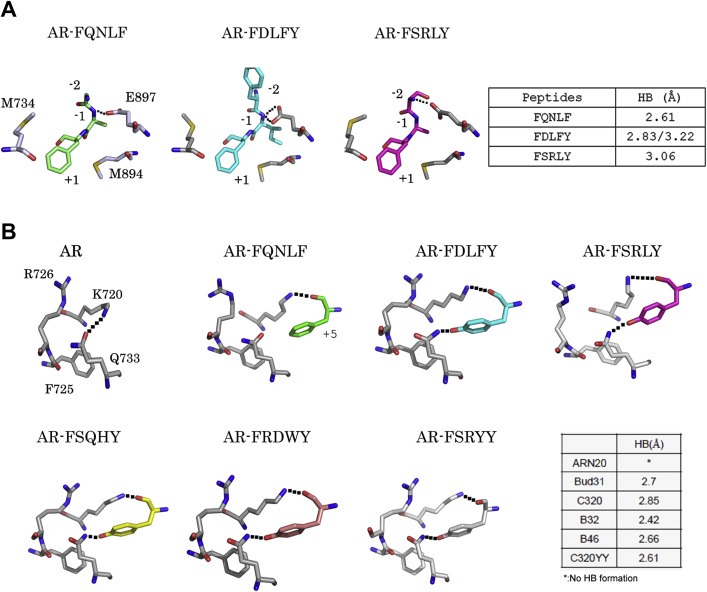
Interactions in the AF2 site. Interactions between the peptides and proteins in front‐end (A) and back‐end (B) areas. Peptides are color‐coded as follow: unbound, gray; FQNLF, green, FDLFY, cyan; FSRLY, magenta; FSQHY, yellow, and FSRYY, wheat. Residues are shown as sticks; carbon, nitrogen, and oxygen atoms are shown in gray, blue, and red, respectively. Hydrogen bond contacts are illustrated with black dashed lines. The lengths of the hydrogen bonds are summarized in the tables.
The back‐end interactions are remarkably distinct from those of the front‐end area. The hydrogen bond contacts in this area are redistributed after association of the peptides. In the absence of the peptide, the H3 and H5 helices are stabilized by a hydrogen bond with the Nε atom of Lys720 and the Oδ2 atom of Q733 of the protein (Figure 2B, AR). However, association of the peptides disrupts these stabilizing interactions in two ways. First, in the presence of the peptide, the N atoms at Lys720 turn toward to the O atoms of the +5 peptide residue, with which it forms a hydrogen bond, providing a new constraining bond between the peptide and the protein (Figure 2B, AR‐FQNLF). Second, with the change in the +5 peptide position to Tyr, the side chain of the Q733 residue flips the Nε atom over to face the hydroxyl group of the peptide (Figure 2B, AR‐FxxxY), enabling a hydrogen bond to form between the nitrogen atom and the hydroxyl group. This result indicates that Tyr in the +5 position of the peptide provides an extra constraining force through formation of a hydrogen bond interaction, which also enhances the association between peptide and protein.
Previous structural studies have demonstrated that ARs can accommodate short peptides through their conserved residues K720, M734, M894, and E897 (Hur et al., 2004). To examine this in the context of our resolved structures, we superposed the peptides and compared their arrangements within this binding pocket. These peptides bound to the wt‐AR protein overlapped well, exhibiting virtually identical positions of Cα atoms (Figure 3A); similar results were obtained for binding to T877A‐AR and W741L‐AR (Figure 3B and C). In addition to interactions at both ends of the peptides, the +4 position of the groove is suitable for accommodating bulky side chains of residues (Dubbink et al., 2004). Our resolved complex structures also demonstrated that bulky side chains of the +4 residue in the peptide adopt a trans conformation that extends into the groove. Moreover, peptides with aromatic bulky side chains in the +4 residue showed greater affinity for protein than peptides containing other hydrophobic residues (Table 2). The bulky side chain of +4 residues, including Trp and His, were exposed toward the broad groove. No hydrogen bond contacts were made between the side chain of the +4 peptide and the protein, whereas the orientation of this side chain was influenced by the surrounding hydrophobic residues, V713, V716 and K717. Therefore, the surrounding hydrophobic environment promotes a favorable orientation of the side chain in this groove. Remarkably, the side chains of residues in the +2 and +3 positions were superposed well and did not interact with the protein (Figure 3D).
Figure 3.
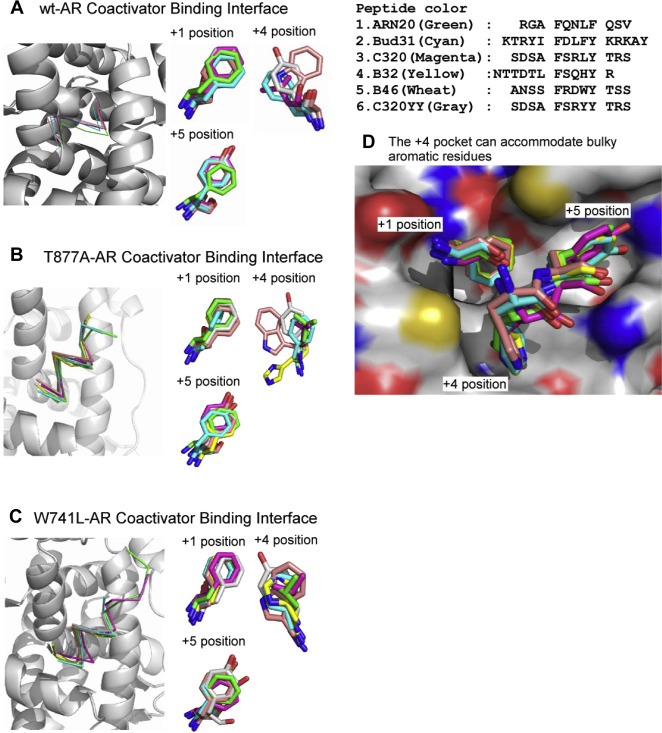
Structural profiles of AR peptide‐binding interfaces among variants. Cα atoms of the peptides, illustrated as thin stick structures, were superposed onto the AF2 site of the (A) wt‐AR, (B) T877A‐AR, and (C) W741L‐AR proteins, shown as ribbon diagrams. Hydrophobic side chains of the peptides were superposed (right). (D) Various peptides were superposed in the AF2 site. The protein and peptides are shown in surface representation and as thin sticks, respectively. Positively and negatively charged areas are blue and red, respectively. The atoms of nitrogen and oxygen are colored blue and red, respectively.
Table 2.
Comparisons of the kinetic constants for the interactions between the AR and peptides.
| Peptides | ka (1/Ms) | kd (1/s) | Ka (1/M) | Kd (M) |
|---|---|---|---|---|
| ARN20‐wt | 5.7E+4 | 3.0E‐3 | 1.9E+7 | 5.3E‐8 |
| ARN20‐LY | 2.8E+4 | 2.9E‐3 | 9.8E+6 | 1.0E‐7 |
| ARN20‐FY | 1.7E+5 | 6.5E‐3 | 2.6E+7 | 3.8E‐8 |
| ARN20‐HY | 3.7E+1 | 5.1E‐3 | 7.3E+3 | 1.4E‐4 |
| ARN20‐WY | 7.3E+3 | 3.6E‐3 | 2.0E+6 | 5.0E‐7 |
| ARN20‐YY | 9.3E+3 | 8.2E‐3 | 1.1E+6 | 8.8E‐7 |
| C320‐LY | 2.0E+5 | 5.9E‐3 | 3.4E+7 | 3.0E‐8 |
| C320‐LF | 5.9E+4 | 4.6E‐3 | 1.3E+7 | 7.8E‐8 |
| C320‐FY | 1.2E+5 | 6.0E‐3 | 1.9E+7 | 5.2E‐8 |
| C320‐HY | 1.4E+2 | 5.3E‐3 | 2.6E+4 | 3.8E‐5 |
| C320‐WY | 4.7E+4 | 6.5E‐3 | 7.2E+6 | 1.4E‐7 |
| C320‐YY | 9.7E+2 | 3.7E‐3 | 2.6E+5 | 3.8E‐6 |
Previous phage‐display screens indicated that the +4 position of peptides favored hydrophobic amino acids (Leu and Phe), whereas the +5 position favored aromatic amino acids (Phe and Tyr). Using surface plasmon resonance to determinate the kinetic constants for the association between the interacting molecules, we confirmed that the variant peptides are capable of associating with AR protein. However, the K d (M) values obtained from sensorgrams were remarkably different in –HY peptides (Table 2). A comparison of the results reported for variant peptides indicated that the –HY peptide associated faster with AR proteins than others peptides, indicating that the hydrophobicity of the side chain of the +4 residue influences binding affinity. These results, taken together with the major role of variations in electromagnetic forces in both ends in the overall attraction, suggest that the hydrophobic side chain modulates the affinity of peptides for proteins through contributions to intermolecular force and steric hindrance.
3.4. BUD31, containing an FxxFY motif, is a new AR co‐regulator
A computer search of existing protein databases found an 18‐kD protein, BUD31 (also known as EDG‐2/hG10) containing the FxxFY motif (61FDLFY65). Early studies detected BUD31 in human umbilical vein endothelial cells (Hla et al., 1995) and showed that it shared 93% homology with G10 from the African frog Xenopus. In yeast, BUD31 is involved in spliceosome assembly and promotes pre‐mRNA splicing (Saha et al., 2012). However, little is known about the role of BUD31 in humans. Therefore, we chose BUD31 and its FxxFY motif for further study.
In Co‐IP assays using anti‐AR or anti‐Flag antibodies to pull down AR‐BUD31 protein complexes in LNCaP cells expressing 3X‐Flag‐BUD31 (Figure 4A), we found that wt‐BUD31 with the FxxFY motif interacted with the GST‐AR, whereas mt BUD31 with an AxxAA motif did not (Figure 4B). These data suggest that BUD31 does indeed interact with the AR via its FxxFY motif. We also found that BUD31 interacted with the AR N‐terminus (Figure 4B). We then demonstrated that, in AR‐negative PC‐3 prostate cancer cells co‐transfected with AR and an AR‐responsive MMTV‐LUC reporter construct, treatment with 10 nM DHT resulted in AR transactivation (Figure 4C). Moreover, exogenous overexpression of BUD31‐wt‐FxxFY further enhanced AR transactivation in a dose‐dependent manner (Figure 4C). In contrast, expression of mutant BUD31 with an AxxAA motif had little effect on AR transactivation.
Figure 4.
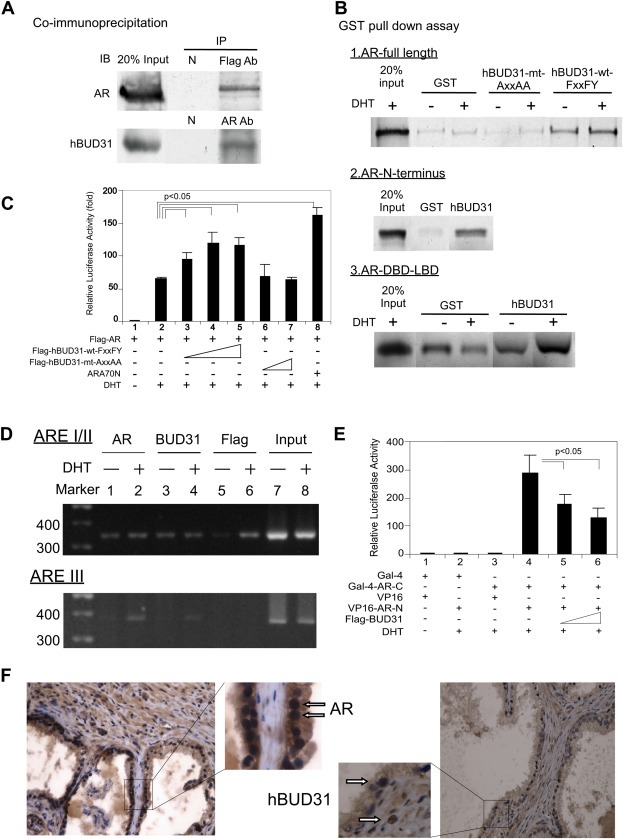
Interaction between AR and BUD31. (A) Co‐IP of Flag‐BUD31 with endogenous AR in the prostate cancer cell line LNCaP. Extracts of LNCaP cells overexpressing 3xFlag‐BUD31 were treated with 1 μM DHT. IP was performed using an anti‐AR (C19) or anti‐Flag antibody, or normal rabbit serum (negative control), followed by immunoblotting (IB) with antibodies to AR or BUD3. (B) BUD31 interacted with full‐length AR and N‐ and C‐terminal regions of the AR in GST pull‐down assays; mutation of the FxxFY motif to AxxAA in BUD31 reduced BUD31 interactions with the AR. BUD31 enhanced AR transactivation and blocked AR N–C interactions. (C) PC‐3 prostate cancer cells transfected with BUD31. PC‐3 cells in 24‐well plates were co‐transfected with 300 ng of MMTV‐LUC reporter plasmid and 0.5 ng of SV40‐Renilla luciferase plasmid together with 100 ng of pCMV‐Flag‐AR, 100–500 ng of p3xFLAG‐BUD31‐wt‐FxxFY or 100–500 ng p3xFLAG‐BUD31‐mt‐AxxAA; plasmid DNA was brought to a total of 1 μg with pCMV. After 16 h, ethanol or 10 nM DHT was added and cells were incubated for an additional 16 h. Relative LUC activity was determined using the dual luciferase system. ARA70 served as a positive control. (D) BUD31 interacts with AR in promoter regions of target genes. LNCaP cells were transfected with P3xflag‐BUD31 plasmid and cultured overnight. Soluble chromatin was prepared from LNCaP cells treated with or without DHT, and ChIP assays were performed using an antibody to AR (AR), BUD31 (BUD31), Flag (Flag), and RNA polymerase II. The final DNA extracts were amplified using the primers for ARE I/II and ARE III, described in Materials and methods. (E) BUD31 blocked AR N–C interactions. PC‐3 cells in 24‐well plates were transfected with 200 ng of pG5‐LUC reporter plasmid and 0.5 ng of SV40‐Renilla luciferase plasmid together with various combinations of 200 ng of Gal‐4‐AR‐C (amino acids 1–501), 200 ng of VP16‐AR‐N (amino acids 556–919), 200–400 ng of p3xFLAG‐BUD31, as indicated; pCMV was added as necessary to bring the total to 1 μg plasmid DNA. After 16 h, ethanol or 10 nM DHT was added and cells were incubated for an additional 16 h. Luciferase activity was assessed using a dual luciferase assay system. (F) BUD31 is expressed in benign prostatic hyperplasia tissue. Human prostate gland tissues were immunostained for AR and BUD31. The arrows indicate ARs (left panel) and BUD31 (right panel). The figures are representative of three benign prostatic gland hyperplasia tissues.
We further applied ChIP assays to determine whether BUD31 bound to endogenous ARs at promoter regions of target genes. ARs and BUD31 were constitutively associated with ARE I/II and were recruited to ARE III in a DHT‐dependent manner (Figure 4D). Importantly, we found that full‐length BUD31 suppressed interactions of AR N‐ and C‐termini (Figure 4E). Finally, immunohistochemical staining revealed that both the AR and BUD31 were present in epithelial and stromal cells of benign prostatic hyperplasia tissue (Figure 4F), suggesting the coexistence of the AR and the BUD31 peptide in the human prostate.
Taken together, the results shown in Figure 4A–F demonstrate that BUD31, containing an FxxFY motif, is a bona fide AR co‐activator that enhances AR transactivation in prostate cells.
3.5. The BUD31‐FxxFY peptide effectively suppresses DHT‐AR‐mediated cell growth
The above data suggest that the short peptide BUD31 could bind tightly to AR‐LBD and AR‐DBD‐LBD; therefore, we were interested in testing whether binding of this or other FxxFY motif‐like peptides to the AR modulated AR transactivation. As expected, BUD31 and other AR‐DBD‐LBD‐related peptides (see sequences in Figure 5A) suppressed AR transactivation in the presence of DHT and HF (Figure 5B).
Figure 5.
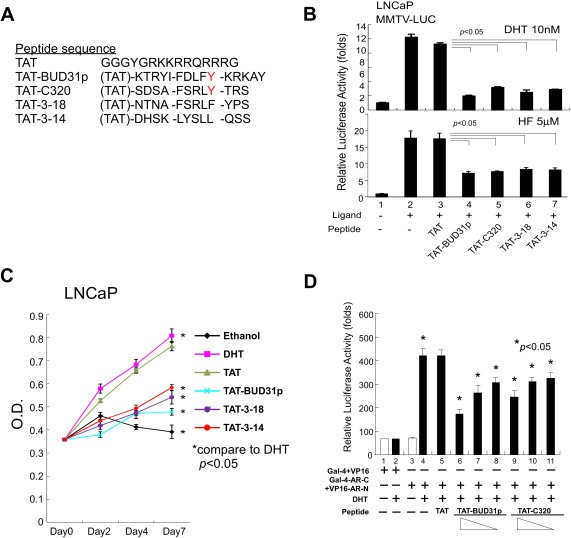
BUD31 peptide potently suppresses AR‐mediated function and growth of AR‐harboring prostate cancer cells, and blocks AR N–C interactions. Short peptides of wt‐BUD31, C320 (FxxLY motif), 3–18 (FxxLF motif) (Hsu et al., 2003), and 3–14 (LxxLL motif) peptides conjugated with TAT were synthesized and tested in LNCaP cells (B and C) and PC‐3 cells (D) in the presence and absence of 0.2 nM DHT. (A) Peptide sequences. (B–D) Test peptides suppress AR transactivation in the presence of DHT or HF (B), suppress LNCaP cell growth in vitro (C), and block AR N–C interactions (D).
We then examined the potential pathophysiological consequences of suppression of AR transactivation by BUD31‐related peptides by conjugating these peptides to the leading TAT sequence (which facilitates import of conjugated peptides into the cytosol) and transducing them into LNCaP cells. The results showed that BUD31 and related peptides suppressed LNCaP cell growth, with the BUD31 peptide exhibiting the greatest inhibitory effect (Figure 5C). Using an MMTV‐LUC reporter assay, we found that the BUD31 peptide suppressed AR N–C interactions in a dose‐dependent manner, suggesting a potential molecular mechanism by which BUD31 peptide‐AR interactions suppress AR transactivation (Figure 5D).
4. Conclusions
As a unique pathological feature of cancer, AWS could be a target for the development of new anti‐cancer treatment strategies. Here, we compared the binary structures of wt‐AR and its variants, and demonstrated that anti‐androgen‐mt‐AR complexes share a folding similar to that of DHT‐wt‐AR. This phenomenon explained the facile transition between androgen antagonist and agonist in this disease. Our ligand‐AR‐peptide ternary structure analysis revealed that the equivalent Cα atoms were almost identical, with RMSD values <0.5 Å within these structures, indicating that each altered AF2 site is very similar. The AF2 sites among these mt‐ARs were found to be fully functional, and therefore capable of recruiting identical peptide motifs as well as those in wt‐AR.
Among these AF2 sites, however, we observed different electrostatic interactions between the peptides and proteins. One charge clump residue, Glu897, was found to make HB contacts with the −1 or −2 positions of the peptides; this was due to the polar character of its side chain, and demonstrated an alternative means for connecting a peptide to the protein (Hur et al., 2004). In contrast, the 5th position of AR‐interacting peptides was generally tyrosine rich, and replacement of Phe with Tyr at this position obviously enhanced the peptide‐protein interaction, resulting in the formation of an additional HB between the hydroxyl of the 5th residue of the peptide and the Q733 residue of AR (Figure 2B). Both interactions strain the peptide and significantly enhanced the binding affinity.
Aside of the both ends of AF2 pocket, both the Val713 residue and its neighboring hydrophobic residues (e.g., Leu712 and Val716) may provide attractions to the side chain of the 4th residue of the AR‐interacting peptide. The side chains of these residues were found to have higher variety compared to the first and the 5th residue of the peptides as shown in 2, 3. Furthermore, the kinetic parameters of the peptide‐protein interaction may be influenced by the selectivity of this AF2 groove. For example, the presence of a histidine residue, with a characteristic hydrophilic motif in its side chain, in AR‐interacting peptides significantly decreased the ability of the peptide and protein to interact. This distinctive affinity suggested that the sequence preference for the associating peptide arose at least partially from the side chain of the 4th residue.
In addition to competition for ligand binding with the AR‐LBD, the AR‐targeting mechanism was found to involve the abrogation of AR N–C and AR‐coregulator interactions, which use the same AR cofactor‐binding groove (He et al., 2002; Trapman and Dubbink, 2007). The AR antagonist, EPI‐001 (Andersen et al., 2010), the protein, MAGE‐11 (melanoma antigen family A) (Bai et al., 2005), the AR N‐terminus itself (Quayle et al., 2007; Watson et al., 2010), the AR‐degrading chemical, ASC‐J9 (Yang et al., 2007), and AR‐LBD BF‐3‐blocking small molecules (Estebanez‐Perpina et al., 2007) have all been shown to disrupt AR‐related functions by interfering with AR N–C interactions and/or AR co‐regulator functions. However, some peptides and small molecules have been shown to suppress the AR‐mediated transcription and growth of prostate cells by directly targeting the AR cofactor‐binding groove (Axerio‐Cilies et al., 2011; Hsu et al., 2005; van de Wijngaart et al., 2011). Here, we showed that the FxxFY‐containing BUD31 peptide can block AR transcriptional activity, the AR N–C interaction, and AR‐mediated cancer cell growth.
Using a unique approach that combined phage‐display screening and X‐ray crystal structure analysis of the AR cofactor‐binding groove, we developed a new strategy for identifying anti‐AR drugs containing an FxxFY motif, and used this strategy to identify the AR co‐activator, BUD31, which was found to interact tightly with the AR. Recent studies have provided proof of principle for the notion that a peptidomimetic molecule can block AR‐related functions by disrupting peptide/cofactor‐AR interactions (Gunther et al., 2009; Hsu et al., 2014; Levine et al., 2012; Ravindranathan et al., 2013). Excellent studies have shown that the LxxLL‐mimetic small molecule, D2, can block AR nuclear translocation, the AR‐cofactor interaction and AR‐mediated cell growth, and may potentially influence the functions of other steroid hormone receptors (Ravindranathan et al., 2013). The above‐described peptidomimetic molecules sterically hinder the critical protein–protein interactions of AR, but do not suffer the same limitations as peptide therapeutics (e.g., facile degradation by peptidases, poor tissue penetration and lower target specificity). Our present data and the previous findings should pave the way for the future design of highly specific and efficient AR‐targeting small molecules.
Conflict of interest
The authors declare no conflicts of interest.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Acknowledgments
We thank Karen L. Wolf for proofreading. We also thank the staff at the National Synchrotron Radiation Research Center in Taiwan for access to the Beamline BL13B. Portions of this research were carried out at the National Synchrotron Radiation Research Center, a national user facility supported by the National Science Council of Taiwan, ROC. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Core Facility Program for Biotechnology.
Supplementary material 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.06.009.
Hsu Cheng-Lung, Liu Jai-Shin, Wu Po-Long, Guan Hong-Hsiang, Chen Yuh-Ling, Lin An-Chi, Ting Huei-Ju, Pang See-Tong, Yeh Shauh-Der, Ma Wen-Lung, Chen Chung-Jung, Wu Wen-Guey, Chang Chawnshang, (2014), Identification of a new androgen receptor (AR) co‐regulator BUD31 and related peptides to suppress wild‐type and mutated AR‐mediated prostate cancer growth via peptide screening and X‐ray structure analysis, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.06.009.
Grant support: This work was supported by grants from the NIH (CA127300), the George Whipple Professorship Endowment, Taiwan National Science Council (NSC98‐2314‐B‐182A‐053‐MY3), the Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (DOH102‐TD‐B‐111‐004), and the Chang Gung Memorial Hospital (CMRPG‐390061, CMRPG‐300171, CMRPG‐3B0421, CMRPG‐3B1261‐2, CMRPG‐3C0201).
Contributor Information
Wen-Guey Wu, Email: wgwu@life.nthu.edu.tw.
Chawnshang Chang, Email: chang@urmc.rochester.edu, Email: sexhormonerescter@gmail.com.
References
- Akakura, K. , Akimoto, S. , Furuya, Y. , Ito, H. , 1998. Incidence and characteristics of antiandrogen withdrawal syndrome in prostate cancer after treatment with chlormadinone acetate. Eur. Urol.. 33, 567–571. [DOI] [PubMed] [Google Scholar]
- Andersen, R.J. , Mawji, N.R. , Wang, J. , Wang, G. , Haile, S. , Myung, J.K. , Watt, K. , Tam, T. , Yang, Y.C. , Banuelos, C.A. , Williams, D.E. , McEwan, I.J. , Wang, Y. , Sadar, M.D. , 2010. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 17, 535–546. [DOI] [PubMed] [Google Scholar]
- Axerio-Cilies, P. , Lack, N.A. , Nayana, M.R. , Chan, K.H. , Yeung, A. , Leblanc, E. , Guns, E.S. , Rennie, P.S. , Cherkasov, A. , 2011. Inhibitors of androgen receptor activation function-2 (AF2) site identified through virtual screening. J. Med. Chem.. 54, 6197–6205. [DOI] [PubMed] [Google Scholar]
- Bai, S. , He, B. , Wilson, E.M. , 2005. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol. Cell. Biol.. 25, 1238–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissada, N.K. , Kaczmarek, A.T. , 1995. Complete remission of hormone refractory adenocarcinoma of the prostate in response to withdrawal of diethylstilbestrol. J. Urol.. 153, 1944–1945. [PubMed] [Google Scholar]
- Chang, C.S. , Kokontis, J. , Liao, S.T. , 1988. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science. 240, 324–326. [DOI] [PubMed] [Google Scholar]
- Chang, C.Y. , Abdo, J. , Hartney, T. , McDonnell, D.P. , 2005. Development of peptide antagonists for the androgen receptor using combinatorial peptide phage display. Mol. Endocrinol.. 19, 2478–2490. [DOI] [PubMed] [Google Scholar]
- Darimont, B.D. , Wagner, R.L. , Apriletti, J.W. , Stallcup, M.R. , Kushner, P.J. , Baxter, J.D. , Fletterick, R.J. , Yamamoto, K.R. , 1998. Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev.. 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, N.A. , McLeod, D.G. , 1995. Dramatic prostate specific antigen decrease in response to discontinuation of megestrol acetate in advanced prostate cancer: expansion of the antiandrogen withdrawal syndrome. J. Urol.. 153, 1946–1947. [PubMed] [Google Scholar]
- Dubbink, H.J. , Hersmus, R. , Verma, C.S. , van der Korput, H.A. , Berrevoets, C.A. , van Tol, J. , Ziel-van der Made, A.C. , Brinkmann, A.O. , Pike, A.C. , Trapman, J. , 2004. Distinct recognition modes of FXXLF and LXXLL motifs by the androgen receptor. Mol. Endocrinol.. 18, 2132–2150. [DOI] [PubMed] [Google Scholar]
- Dubbink, H.J. , Hersmus, R. , Pike, A.C. , Molier, M. , Brinkmann, A.O. , Jenster, G. , Trapman, J. , 2006. Androgen receptor ligand-binding domain interaction and nuclear receptor specificity of FXXLF and LXXLL motifs as determined by L/F swapping. Mol. Endocrinol.. 20, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Emsley, P. , Cowtan, K. , 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr.. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Estebanez-Perpina, E. , Arnold, L.A. , Nguyen, P. , Rodrigues, E.D. , Mar, E. , Bateman, R. , Pallai, P. , Shokat, K.M. , Baxter, J.D. , Guy, R.K. , Webb, P. , Fletterick, R.J. , 2007. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc. Natl. Acad. Sci. U. S. A.. 104, 16074–16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomella, L.G. , Ismail, M. , Nathan, F.E. , 1997. Antiandrogen withdrawal syndrome with nilutamide. J. Urol.. 157, 1366 [PubMed] [Google Scholar]
- Gunther, J.R. , Parent, A.A. , Katzenellenbogen, J.A. , 2009. Alternative inhibition of androgen receptor signaling: peptidomimetic pyrimidines as direct androgen receptor/coactivator disruptors. ACS Chem. Biol.. 4, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T. , Miyazaki, J. , Araki, H. , Yamaoka, M. , Kanzaki, N. , Kusaka, M. , Miyamoto, M. , 2003. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res.. 63, 149–153. [PubMed] [Google Scholar]
- He, B. , Kemppainen, J.A. , Wilson, E.M. , 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem.. 275, 22986–22994. [DOI] [PubMed] [Google Scholar]
- He, B. , Minges, J.T. , Lee, L.W. , Wilson, E.M. , 2002. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem.. 277, 10226–10235. [DOI] [PubMed] [Google Scholar]
- Heinlein, C.A. , Chang, C. , 2002. Androgen receptor (AR) coregulators: an overview. Endocr. Rev.. 23, 175–200. [DOI] [PubMed] [Google Scholar]
- Hla, T. , Jackson, A.Q. , Appleby, S.B. , Maciag, T. , 1995. Characterization of edg-2, a human homologue of the Xenopus maternal transcript G10 from endothelial cells. Biochim. Biophys. Acta. 1260, 227–229. [DOI] [PubMed] [Google Scholar]
- Hsu, C.L. , Chen, Y.L. , Yeh, S. , Ting, H.J. , Hu, Y.C. , Lin, H. , Wang, X. , Chang, C. , 2003. The use of phage display technique for the isolation of androgen receptor interacting peptides with (F/W)XXL(F/W) and FXXLY new signature motifs. J. Biol. Chem.. 278, 23691–23698. [DOI] [PubMed] [Google Scholar]
- Hsu, C.L. , Chen, Y.L. , Ting, H.J. , Lin, W.J. , Yang, Z. , Zhang, Y. , Wang, L. , Wu, C.T. , Chang, H.C. , Yeh, S. , Pimplikar, S.W. , Chang, C. , 2005. Androgen receptor (AR) NH2- and COOH-terminal interactions result in the differential influences on the AR-mediated transactivation and cell growth. Mol. Endocrinol.. 19, 350–361. [DOI] [PubMed] [Google Scholar]
- Hsu, C.L. , Liu, J.S. , Lin, A.C. , Yang, C.H. , Chung, W.H. , Wu, W.G. , 2014. Minoxidil may suppress androgen receptor-related functions. Oncotarget. 5, 2187–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur, E. , Pfaff, S.J. , Payne, E.S. , Gron, H. , Buehrer, B.M. , Fletterick, R.J. , 2004. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol.. 2, E274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouravel, N. , Sablin, E. , Arnold, L.A. , Guy, R.K. , Fletterick, R.J. , 2007. Interaction between the androgen receptor and a segment of its corepressor SHP. Acta Crystallogr. D Biol. Crystallogr.. 63, 1198–1200. [DOI] [PubMed] [Google Scholar]
- Kelly, W.K. , Scher, H.I. , 1993. Prostate specific antigen decline after antiandrogen withdrawal: the flutamide withdrawal syndrome. J. Urol.. 149, 607–609. [DOI] [PubMed] [Google Scholar]
- Kelly, W.K. , Slovin, S. , Scher, H.I. , 1997. Steroid hormone withdrawal syndromes. Pathophysiology and clinical significance. Urol. Clin. North Am.. 24, 421–431. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A. , Moss, D.S. , Thornton, J.M. , 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol.. 231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- Levine, P.M. , Imberg, K. , Garabedian, M.J. , Kirshenbaum, K. , 2012. Multivalent peptidomimetic conjugates: a versatile platform for modulating androgen receptor activity. J. Am. Chem. Soc.. 134, 6912–6915. [DOI] [PubMed] [Google Scholar]
- Lubahn, D.B. , Joseph, D.R. , Sullivan, P.M. , Willard, H.F. , French, F.S. , Wilson, E.M. , 1988. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 240, 327–330. [DOI] [PubMed] [Google Scholar]
- Miyamoto, H. , Rahman, M.M. , Chang, C. , 2004. Molecular basis for the antiandrogen withdrawal syndrome. J. Cell. Biochem.. 91, 3–12. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N. , Vagin, A.A. , Dodson, E.J. , 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr.. 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Navaza, J. , 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 50, 157–163. [Google Scholar]
- Nishimura, K. , Ting, H.J. , Harada, Y. , Tokizane, T. , Nonomura, N. , Kang, H.Y. , Chang, H.C. , Yeh, S. , Miyamoto, H. , Shin, M. , Aozasa, K. , Okuyama, A. , Chang, C. , 2003. Modulation of androgen receptor transactivation by gelsolin: a newly identified androgen receptor coregulator. Cancer Res.. 63, 4888–4894. [PubMed] [Google Scholar]
- Otwinowski, Z. , Minor, W. , 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. Part A. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis, A. , Morris, R. , Lamzin, V.S. , 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol.. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Quayle, S.N. , Mawji, N.R. , Wang, J. , Sadar, M.D. , 2007. Androgen receptor decoy molecules block the growth of prostate cancer. Proc. Natl. Acad. Sci. U. S. A.. 104, 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan, P. , Lee, T.K. , Yang, L. , Centenera, M.M. , Butler, L. , Tilley, W.D. , Hsieh, J.T. , Ahn, J.M. , Raj, G.V. , 2013. Peptidomimetic targeting of critical androgen receptor–coregulator interactions in prostate cancer. Nat. Commun.. 4, 1923 [DOI] [PubMed] [Google Scholar]
- Saha, D. , Banerjee, S. , Bashir, S. , Vijayraghavan, U. , 2012. Context dependent splicing functions of Bud31/Ycr063w define its role in budding and cell cycle progression. Biochem. Biophys. Res. Commun.. 424, 579–585. [DOI] [PubMed] [Google Scholar]
- Schrodinger, L.L.C. , 2010. The PyMOL Molecular Graphics System. Version 0.99 [Google Scholar]
- Shatkina, L. , Mink, S. , Rogatsch, H. , Klocker, H. , Langer, G. , Nestl, A. , Cato, A.C. , 2003. The cochaperone Bag-1L enhances androgen receptor action via interaction with the NH2–terminal region of the receptor. Mol. Cell. Biol.. 23, 7189–7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y. , Morita, T. , Kashiwagi, B. , Tomizawa, H. , Yamanaka, H. , 1999. Estramustine phosphate withdrawal syndrome with dramatic pain relief. J. Urol.. 162, 805 [DOI] [PubMed] [Google Scholar]
- Shyr, C.R. , Tsai, M.Y. , Yeh, S. , Kang, H.Y. , Chang, Y.C. , Wong, P.L. , Huang, C.C. , Huang, K.E. , Chang, C. , 2010. Tumor suppressor PAX6 functions as androgen receptor co-repressor to inhibit prostate cancer growth. Prostate. 70, 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, E.J. , Carroll, P.R. , 1994. Prostate-specific antigen decline after casodex withdrawal: evidence for an antiandrogen withdrawal syndrome. Urology. 43, 408–410. [DOI] [PubMed] [Google Scholar]
- Trapman, J. , Dubbink, H.J. , 2007. The role of cofactors in sex steroid action. Best Pract. Res. Clin. Endocrinol. Metab.. 21, 403–414. [DOI] [PubMed] [Google Scholar]
- Warnmark, A. , Treuter, E. , Gustafsson, J.A. , Hubbard, R.E. , Brzozowski, A.M. , Pike, A.C. , 2002. Interaction of transcriptional intermediary factor 2 nuclear receptor box peptides with the coactivator binding site of estrogen receptor alpha. J. Biol. Chem.. 277, 21862–21868. [DOI] [PubMed] [Google Scholar]
- Watson, P.A. , Chen, Y.F. , Balbas, M.D. , Wongvipat, J. , Socci, N.D. , Viale, A. , Kim, K. , Sawyers, C.L. , 2010. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc. Natl. Acad. Sci. U. S. A.. 107, 16759–16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wijngaart, D.J. , van Royen, M.E. , Hersmus, R. , Pike, A.C. , Houtsmuller, A.B. , Jenster, G. , Trapman, J. , Dubbink, H.J. , 2006. Novel FXXFF and FXXMF motifs in androgen receptor cofactors mediate high affinity and specific interactions with the ligand-binding domain. J. Biol. Chem.. 281, 19407–19416. [DOI] [PubMed] [Google Scholar]
- van de Wijngaart, D.J. , Dubbink, H.J. , Molier, M. , de Vos, C. , Jenster, G. , Trapman, J. , 2011. Inhibition of androgen receptor functions by gelsolin FxxFF peptide delivered by transfection, cell-penetrating peptides, and lentiviral infection. Prostate. 71, 241–253. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Chang, Y.J. , Yu, I.C. , Yeh, S. , Wu, C.C. , Miyamoto, H. , Merry, D.E. , Sobue, G. , Chen, L.M. , Chang, S.S. , Chang, C. , 2007. ASC-J9 ameliorates spinal and bulbar muscular atrophy phenotype via degradation of androgen receptor. Nat. Med.. 13, 348–353. [DOI] [PubMed] [Google Scholar]
- Yeh, S. , Chang, C. , 1996. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. U. S. A.. 93, 5517–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


