Abstract
Background
Pre-harvest sprouting (PHS) of wheat grain leads to a reduction in grain yield and quality. The availability of markers for marker-assisted selection (MAS) of PHS resistance will serve to enhance breeding selection and advancement of lines for cultivar development. The aim of this study was to identify candidate regions and develop molecular markers for PHS resistance in wheat. This was achieved via high density mapping of single nucleotide polymorphism (SNP) markers from an Illumina 90 K Infinium Custom Beadchip in a doubled haploid (DH) population derived from a RL4452/‘AC Domain’ cross and subsequent detection of quantitative trait loci (QTL) for PHS related traits (falling number [FN], germination index [GI] and sprouting index [SI]). SNP marker sequences flanking QTL were used to locate colinear regions in Brachypodium and rice, and identify genic markers associated with PHS resistance that can be utilized for MAS in wheat.
Results
A linkage map spanning 2569.4 cM was constructed with a total of 12,201 SNP, simple sequence repeat (SSR), diversity arrays technology (DArT) and expressed sequence tag (EST) markers. QTL analyses using Multiple Interval Mapping (MIM) identified four QTL for PHS resistance traits on chromosomes 3B, 4A, 7B and 7D. Sequences of SNPs flanking these QTL were subject to a BLASTN search on the International Wheat Genome Sequencing Consortium (IWGSC) database (http://wheat-urgi.versailles.inra.fr/Seq-Repository). Best survey sequence hits were subject to a BLASTN search on Gramene (www.gramene.org) against both Brachypodium and rice databases, and candidate genes and regions for PHS resistance were identified. A total of 18 SNP flanking sequences on chromosomes 3B, 4A, 7B and 7D were converted to KASP markers and validated with matching genotype calls of Infinium SNP data.
Conclusions
Our study identified candidate genes involved in abscissic acid (ABA) and gibberellin (GA) metabolism, and flowering time in four genomic regions of Brachypodium and rice respectively, in addition to 18 KASP markers for PHS resistance in wheat. These markers can be deployed in future genetic studies of PHS resistance and might also be useful in the evaluation of PHS in germplasm and breeding material.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-014-0340-1) contains supplementary material, which is available to authorized users.
Keywords: Wheat, Pre-harvest sprouting, Quantitative trait loci, Candidate genes
Background
Preharvest sprouting is observed across all major wheat growing regions in the world. In western Canada, the average annual losses due to PHS are approximately $100 million [1]. Insufficient seed dormancy is one major factor contributing to pre-harvest sprouting losses, particularly under humid, wet weather conditions at harvest. PHS resistant/tolerant wheat cultivars and land races have been identified globally, with origins mainly in Canada, USA, Australia, China, Japan, South Africa, Kenya and New Zealand [2]. Canadian red-seeded spring wheat cultivars (AC Domain, AC Majestic, Columbus, Pasqua, Waskada, Harvest) and white spring wheat genotypes (AC Vista, Snowbird, Snowstar, Kanata, HY361) are known to carry resistance to PHS, all having derived their resistance alleles from a red-seeded breeding line RL4137 [1,3].
Of the three PHS traits, FN [4,5] is most commonly used to quantify PHS [6] and indirectly measures the activity of the enzyme α-amylase that breaks down starch in germinating grains. Degradation of grain-starch as the result of greater α-amylase activitys result in lower FN values and are an indirect indication of low levels of PHS resistance or dormancy. Two other important traits for the characterization of PHS are GI [7,8] and SI [9]. While GI values deduced from seed-germination tests in petri dishes are a direct measure of seed dormancy, SI values obtained via artificial wetting of intact wheat spikes, detect dormancy and properties of the inflorescence that affect PHS [5].
Quantitative trait loci (QTL) linked to PHS traits have been reported on all 21 hexaploid wheat chromosomes [10-13], mainly on 3A [14-17], 3B and 3D [17-19], 4A [2,20-24], 5A [25,26], 6B and 7D [27]. Of these, the PHS QTL on 4A has consistently been identified in several different mapping populations. The RL4452/‘AC Domain’ DH population has been extensively characterized for QTL detection of PHS [28], agronomic [29] and quality traits [30], in several past studies that involved a small number of molecular markers. These studies relied mainly on SSR marker data for the preparation of genetic maps and locating QTL on chromosomes. With low costs and rapid advancements in sequencing technology, thousands of molecular markers, mainly SNPs have become available in wheat. Additionally, access to genome sequence information for rice [31] and Brachypodium [32] will now facilitate comparative mapping for the identification of genes underlying various important quantitative traits in wheat.
Interaction among PHS QTL (QxQ, QTL epistasis), and the environment (QxE, QxQxE) have been reported from various studies [18,33-35] aimed at understanding the complex genetic structure of QTL. As chromosomal locations of PHS QTL are not uniform across populations, obtaining a consensus on the precise genomic location of important trait QTL is required for fine mapping and cloning studies. Meta-QTL or Meta-analysis [36] integrates several QTL studies of a common trait to provide a meaningful estimate of the exact location and number of QTL for that given trait. Eight PHS QTL on chromosomes 3A, 3B, 3D and 4A were identified in a Meta-QTL study [37] involving 15 different populations (five DH; nine recombinant inbred line [RIL]; one backcross [BC]).
A high level of genome-synteny exists among wheat, Brachypodium and rice, with wheat being more closely related to Brachypodium than to rice [38,39]. Conservation or collinearity of genetic markers [40,41] and greater structural similarities in the coding regions of orthologous genes [39] of wheat and Brachypodium have been reported. However, given differences in gene content in orthologous regions of wheat, Brachypodium [41] and rice [42], it might be beneficial to use both genomic sequences of Brachypodium and rice in comparative mapping studies for map based cloning and gene discovery in wheat.
Our study deployed SNP markers from a 90K Infinium iSelect Custom Beadchip [43], in addition to available SSR, DArT and ESTs, to generate high density genetic maps for the identification of PHS resistance QTL. Sequences corresponding to polymorphic SNPs flanking PHS QTL were analyzed against genomic sequences of Brachypodium and rice. The objectives of our research were a) to identify candidate genes and regions in Brachypodium and rice that are orthologous to PHS resistance QTL intervals in wheat, and b) to utilize sequences of SNPs flanking PHS QTL to develop KASP markers for MAS of PHS resistance.
Results
Linkage mapping
A total of 12,201 SNP, SSR, DArT and EST markers were mapped to all 21 wheat chromosomes. The resulting linkage map spanning 2569.4 cM is reported in Additional file 1. Of the 12,201 markers, 11,282 or 92.5% were SNPs, while the remaining 919 or 7.5% comprised SSR, DArT and EST markers. The largest number of SNP markers (6,291) were distributed across the B genome, followed by 4,125 SNPs mapped to the A genome, and 1,785 SNP markers on the D genome (Table 1).
Table 1Table 1.
Cumulative map-lengths of A, B and D genome chromosomes alongside corresponding genome-wise distribution of SNP markers mapped in the hexaploid DH population of RL4452/‘AC Domain’
| Genome | Map length (cM) | Mapped markers | SNPs | SSRs, DArTs & ESTs | % SNPs |
|---|---|---|---|---|---|
| A | 888.4 | 4125 | 3816 | 309 | 92.5 |
| B | 940.6 | 6291 | 5871 | 420 | 93.3 |
| D | 740.4 | 1785 | 1595 | 190 | 89.4 |
| (A + B + D) | 2569.4 | 12201 | 11282 | 919 | 92.5 |
QTL analysis
PHS datasets were analyzed with both MIM and simple interval mapping (SIM; data not shown) methods. As results of both methods were very similar, only those of MIM were reported in this study. The MIM [44] analysis identified four QTL with significant effects, located across chromosomes 3B, 4A, 7B and 7D. Each of these four QTL appeared in two or more environments and had peak LOD scores greater than the critical threshold LOD at 5% significance levels (α0.05) [45]. Coincident QTL for GI, SI and FN were located on chromosome 4A. Across trials, RL4452 alleles on 3B and 7B provided PHS resistance as they reduced SI. However, ‘AC Domain’ alleles also provided PHS resistance as they increased FN on 4A and 7D (with the exception of the Glenlea 2005 trial in which they reduced FN on 7D) and reduced SI and GI on 4A (Table 2).
Table 2.
Results of Multiple Interval Mapping (MIM): four QTL for PHS traits ( GI, SI, FN ) identified on chromosomes 3B, 4A, 7B.1 and 7D.2 in a DH population of RL4452/‘AC Domain’ replicated in multi-year environments (Glenlea and Winnipeg in Manitoba; Swift Current in Saskatchewan)
| QTL | Trial dataset | Chromosome (Linkage gp.) | QTL peak location (cM) | Additive a | % PV ( R 2 ) | LOD | α0.05 |
|---|---|---|---|---|---|---|---|
| Germination Index (GI) | |||||||
| QGi.crc-4A | Glenlea2005 | 4A | 59.3 | −0.04 | 27.6 | 12.83 | 3.86 |
| QGi.crc-4A | Winnipeg2004 | 4A | 59.5 | −0.05 | 58.1 | 34.56 | 3.93 |
| QGi.crc-4A | Winnipeg2005 | 4A | 59.4 | −0.02 | 29.6 | 13.93 | 3.86 |
| Sprouting Index (SI) | |||||||
| QSi.crc-3B | Glenlea2005 | 3B | 63.6 | 0.43 | 12.7 | 5.39 | 3.96 |
| QSi.crc-3B | Winnipeg2004 | 3B | 70.2 | 0.53 | 16.1 | 6.97 | 3.95 |
| QSi.crc-4A | Glenlea2005 | 4A | 59.3 | −0.57 | 20.5 | 9.12 | 3.96 |
| QSi.crc-4A | Winnipeg2004 | 4A | 56.8 | −0.85 | 32.1 | 15.38 | 3.95 |
| QSi.crc-4A | Winnipeg2005 | 4A | 58.0 | −0.44 | 12.7 | 5.41 | 3.90 |
| QSi.crc-4A | Swift Current2003 | 4A | 58.0 | −0.49 | 10.5 | 4.41 | 3.94 |
| QSi.crc-7B | Swift Current2003 | 7B.1 | 55.6 | 0.78 | 20.5 | 9.12 | 3.94 |
| QSi.crc-7B | Swift Current2004 | 7B.1 | 56.4 | 0.59 | 11.8 | 4.99 | 3.92 |
| Falling Number (FN) | |||||||
| QFn.crc-4A | Glenlea2005 | 4A | 64.2 | 22.49 | 11.2 | 4.71 | 3.83 |
| QFn.crc-4A | Winnipeg2004 | 4A | 56.2 | 45.45 | 25.8 | 11.85 | 3.95 |
| QFn.crc-7D | Glenlea2003 | 7D.2 | 18.9 | 33.40 | 13.2 | 5.64 | 3.99 |
| QFn.crc-7D | Glenlea2005 | 7D.2 | 20.2 | −33.49 | 20.6 | 9.19 | 4.13 |
aPositive or negative additive values relate to allele effects of the AC Domain parent.
Candidate regions and genes for PHS resistance
Sequences of SNPs flanking QTL for PHS resistance on chromosomes 3B, 4A, 7B and 7D were subjected to BLASTN searches on the IWGSC and Gramene databases and returned hits to candidate regions in Brachypodium and rice (Table 3). Genetic and physical maps displaying orthologous regions for PHS resistance in wheat, Brachypodium and rice are given in Figures 1a and b. A 7.8 cM QTL interval on chromosome 3B was orthologous to a ~7.0 Mb region (46,936,013 – 53,904,697 bp) on chromosome 2 of Brachypodium (Bradi2) and to a ~8.7 Mb (27,906,608 – 36,656,340 bp) region on chromosome 1 of rice (Os01). The 4A QTL interval was 12.2 cM and was orthologous to a ~0.52 Mb region (481,247 – 1,030,837 bp) on chromosome 1 of Brachypodium (Bradi1) and to a ~6.9 Mb (29,401,950 – 36,320,679 bp) region on chromosome 3 of rice (Os03). On chromosome 7B.1, the QTL interval spanned 1.7 cM and was orthologous to a ~1.8 Mb region (42,620,688 – 44,420,413 bp) on chromosome 1 of Brachypodium (Bradi1) and to a ~1 Mb (5,588,196 – 6,603,975 bp) region on chromosome 6 of rice (Os06). The QTL interval on 7D.2 was 7.7 cM and was orthologous to a ~2.0 Mb region (47,249,027 – 49,335,697 bp) on chromosome 1 of Brachypodium (Bradi1), and a ~0.5 Mb region (2,558,015 – 3,079,059 bp) on chromosome 6 of rice (Os06).
Table 3.
Genetic map locations of SNP markers flanking PHS QTL on chromosomes 3B, 4A, 7B.1 and 7D.2 in a wheat DH population of a RL4452/‘AC Domain’ cross and their corresponding physical locations/candidate regions in Brachypodium distachyon and rice
| SNP marker | Map | Survey sequence | BLASTN hits to Brachypodium genes | BLASTN hits to Rice genes |
|---|---|---|---|---|
| (cM) | Contig no. | (genomic regions in parenthesis) | (genomic regions in parenthesis) | |
| Chromosome 3B | ||||
| wsnp_Ku_c6825_11858665 | 63.0 | 10469056 | Bradi2g46510 (46,936,013-46,952,333) | LOC_Os01g48680 (27,906,608-27,920,980) |
| wsnp_Ex_c4769_8510104 | 63.0 | 10613849 | Bradi2g46590 (47,003,547-47,009,237) | LOC_Os01g48790 (27,983,688-27,990,383) |
| RAC875_rep_c113906_294 | 64.0 | 10557485 | Bradi2g51030 (50,699,047-50,702,962) | LOC_Os01g56200 (32,367,683-32,371,816) |
| BobWhite_c46650_260 | 64.0 | 10441023 | Bradi2g51017 (50,685,769-50,695,622) | LOC_Os01g56190 (32,350,513-32,360,765) |
| Kukri_c4310_489 | 64.6 | 10759762 | Bradi2g51040 (50,703,620-50,708,573) | LOC_Os02g13910 (7,558,777-7,568,835) |
| TA002966-0294 | 65.1 | 10635317 | Bradi2g46710 (47,135,003-47,136,451) | LOC_Os01g56580 (32,615,694-32,622,894) |
| 10712014 | Bradi2g49590 (49,632,703-49,638,684) | LOC_Os01g54100 (31,111,291-31,116,151) | ||
| BS00078127_51 | 65.7 | 10754454 | Bradi2g51530 (51,119,031-51,123,083) | LOC_Os01g56810 (32,788,487-32,792,751) |
| Kukri_c21818_519 | 66.2 | 10455881 | Bradi2g51620 (51,191,828-51,198,372) | LOC_Os01g56910 (32,869,293-32,878,216) |
| wsnp_Ra_rep_c74606_72470419 | 66.8 | 10523702 | Bradi2g51710 (51,287,497-51,313,181 | LOC_Os01g57082 (32,984,982-32,994,519) |
| IACX3871 | 66.8 | 10521243 | Bradi2g51890 (51,441,692-51,446,044) | LOC_Os01g57450 (33,200,667-33,201,485) |
| Excalibur_c73633_120 | 67.3 | 10673653 | Bradi2g48430 (48,731,037-48,732,308) | LOC_Os01g52260 (30,042,527-30,043,938) |
| wsnp_Ex_c5547_9774195 | 68.4 | 10770075 | Bradi2g53020 (52,250,581-52,257,598) | LOC_Os01g59670 (34,514,117-34,520,887) |
| wsnp_Ex_rep_c69664_68618163 | 68.4 | 10477393 | Bradi2g52540 (51,883,735-51,889,623) | LOC_Os01g58680 (33,919,393-33,924,664) |
| wsnp_Ku_rep_c72700_72370664 | 69.0 | 10484009 | Bradi2g53340 (52,475,967-52,481,992) | LOC_Os01g60180 (34,803,492-34,804,046) |
| RAC875_rep_c116515_181 | 69.0 | 1068363 | Bradi2g53130 (52,329,608-52,334,764) | LOC_Os01g59880 (34,629,359-34,635,205) |
| BobWhite_rep_c64944_264 | 69.6 | 1040995 | Bradi2g53970 (52,969,054-52,973,550) | LOC_Os01g61400 (35,505,448-35,508,543) |
| Tdurum_contig38427_237 | 70.2 | 10658322 | Bradi2g55100 (53,817,575-53,821,406) | LOC_Os01g63250 (36,656,340-36,660,768) |
| Tdurum_contig27495_111 | 70.2 | 10538814 | Bradi2g53450 (52,567,117-52,569,109) | LOC_Os01g60430 (34,946,618-34,949,027) |
| Kasp3B(survey)_17 | 70.8 | 10495803 | Bradi2g55230 (53,904,697-53,906,640) | LOC_Os03g60200 (34,238,474-34,241,647) |
| Chromosome 4A | ||||
| BS00068243_51 | 53.8 | 7023446 | Bradi2g12660 (11,006,410-11,009,518) | LOC_Os01g28244 (15,823,709-15,829,849) |
| CD920298 | 58.6 | 7174272 | Bradi1g00600 (481,247-482,062) | LOC_Os03g64290 (36,320,679-36,333,253) |
| Kukri_c12563_52 | 59.3 | 7128338 | Bradi1g51817 (50,293,482-50,308,189) | LOC_Os05g37500 (21,943,044-21,959,786) |
| Bradi1g00760 (565,638-570,467)! | LOC_Os03g63370 (35,809,964-35,814,672)! | |||
| BS00072025_51 | 59.3 | 7168762 | Bradi1g00730 (555,714-559,377) | LOC_Os03g64210 (36,281,400-36,283,271) |
| RAC875_c21369_425 | 59.8 | 7070429 | Bradi1g00820 (594,037-597,877) | LOC_Os03g64190 (36,265,672-36,271,489) |
| IAAV3132 | 59.8 | 7114346 | Bradi1g01007 (695,876-702,209) | LOC_Os03g63920 (36,110,059-36,119,639) |
| wsnp_Ex_c5470_9657856 | 60.4 | 7174581 | Bradi1g01070 (731,493-733,959) | LOC_Os03g51390 (29,401,950-29,403,115) |
| RAC875_c25124_182 | 61.6 | 7061368 | Bradi1g01227 (825,624-828,017) | LOC_Os03g63680 (35,968,492-35,970,517) |
| wsnp_Ku_c4924_8816643 | 62.7 | 501046 | Bradi1g52230 (50,605,616-50,611,584) | LOC_Os02g29140 (17,257,940-17,266,066) |
| 3540051 | Bradi1G00720 (552,185-555,346)! | - | ||
| 864232 | - | LOC_Os03g60710 (34,502,945-34,508,158)! | ||
| Excalibur_c24511_1196 | 63.2 | 7119833 | Bradi1g49910 (48,564,700-48,565,690) | LOC_Os06g16640 (9,564,124-9,566,967) |
| 7139864 | Bradi1g00820 (594,037-597,877)! | - | ||
| 5949088 | - | LOC_Os03g53500 (30,679,685-30,689,230)! | ||
| Tdurum_contig13489_292 | 63.8 | 7124315 | Bradi1g01500 (976,919-979,161) | LOC_Os03g63470 (35,855,445-35,860,549) |
| wsnp_JD_c38619_27992279 | 66.0 | 7098863 | Bradi1g01580 (1,030,837-1,034,525) | LOC_Os03g63410 (35,826,263-35,830,205) |
| Chromosome 7B.1 | ||||
| CAP7_c10566_170 | 55.3 | 3116911 | Bradi1G46150 (44,420,413-44,423,001)! | LOC_Os06g10710 (5,588,196-5,594,757) |
| BobWhite_rep_c64768_264 | 55.3 | 3032904 | Bradi1G46137 (44,416,953-44,419,121) | LOC_Os06g10760 (5,619,105-5,621,750) |
| Tdurum_contig84962_256 | 55.3 | 3032904 | Bradi1G46137 (44,416,953-44,419,121) | LOC_Os06g10760 (5,619,105-5,621,750) |
| BS00022498_51 | 55.3 | 3115694 | Bradi1G46060 (44,341,065-44,348,362) | LOC_Os06g10880 (5,677,080-5,682,126) |
| wsnp_Ex_c908_1754208 | 56.4 | 3153345 | Bradi1g45210 (43,434,039-43,436,397) | LOC_Os06g12270 (6,603,975-6,604,635) |
| Tdurum_contig68347_605 | 56.4 | 3153345 | Bradi1G45210 (43,434,039-43,436,397) | LOC_Os06g12270 (6,603,975-6,604,635) |
| LOC_Os06g12280 (6,605,479-6,608,454) | ||||
| RFL_Contig124_558 | 57 | 3126436 | Bradi1g44967 (43,073,188-43,080,744)! | - |
| BobWhite_c46772_564 | 57 | 3109791 | Bradi1G44860 (42,951,596-42,953,323) | LOC_Os06g12990 (7,118,829-7,120,448) |
| Bradi1G44850 (42,949,245-42,951,551) | - | |||
| GENE-4333_211 | 57 | 3153554 | Bradi1G44790 (42,899,346-42,900,477) | - |
| Tdurum_contig51087_573 | 57 | 3165147 | Bradi1G44440 (42,620,688-42,629,717) | LOC_Os06g13820 (7,661,691-7,670,035) |
| Chromosome 7D.2 | ||||
| RAC875_c1829_321 | 14.3 | 3849095 | Bradi1g48660 (47,326,685-47,327,292) | LOC_Os06g06460 (3,040,092-3,041,121) |
| Kukri_c32845_116 | 14.3 | 3964075 | Bradi1g50860 (49,335,697-49,339,907) | LOC_Os06g05660 (2,558,015-2,562,242) |
| TA002473-0717 | 14.3 | 3929478 | Bradi1g49140 (47,871,489-47,874,424) | LOC_Os06g05700 (2,579,088-2,581,726) |
| wsnp_CAP8_rep_c9647_4198594 | 22.0 | 3945994 | Bradi1g48610 (47,249,027-47,255,499)! | LOC_Os06g06560 (3,079,059-3,086,808) |
!Weak hit to genomic regions in Brachpodium or rice that is orthologous to the QTL interval for PHS resistance in wheat.
Best hits that do not correspond to the candidate region in Brachpodium or rice are in italics.
Figure 1.
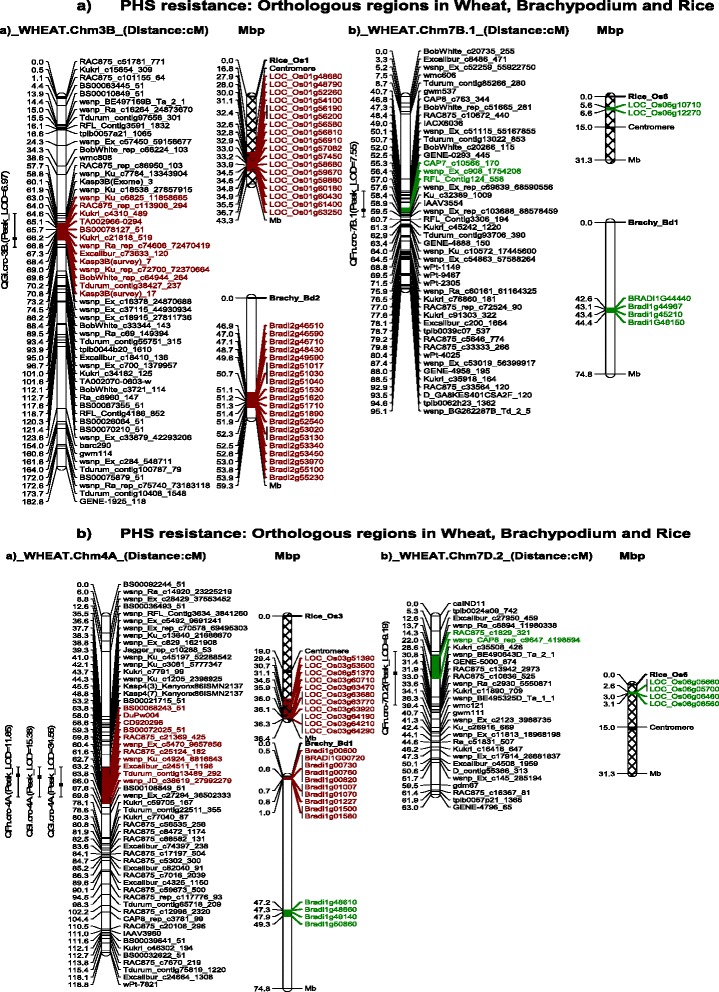
Location of QTL and syntenic regions in Brachpodium and rice. a. Location of QTL and flanking markers for PHS resistance on a) wheat chromosome 3B and its candidate regions on Brachypodium Bd2 and rice Os1, and b) chromosome 7B.1 and its candidate regions on Brachypodium Bd1 and rice Os6. b. Location of QTL and flanking markers for PHS resistance on a) wheat chromosome 4A and its candidate regions on Brachypodium Bd1 and rice Os3, and b) chromosome 7D.2 and its candidate regions on Brachypodium Bd1 and rice Os6.
Brachypodium/rice candidates for PHS resistance orthologous to consensus regions on wheat chromosomes 3B, 4A, 7B and 7D (Additional file 2) were identified. In the 3B region there are 895 genes in the Brachypodium orthologous region and 1375 in the rice region. The 4A region had 98 genes in the Brachypodium region and 1159 in rice, while the 7B region had 148 in Brachypodium and 155 in rice and the 7D region had 235 in Brachypodium and 88 in rice. Genes involved in ABA and GA metabolism as well as those affecting flowering time were present in the QTL regions. Among these were Bradi2g49795/Os01g54490 (FT PEBP [phosphatidylethanolamine - binding protein] homologous to Flowering Locus T gene), Os01g61100, Os01g63030 (Far-red impaired responsive [FAR1] family protein) orthologous to chromosome 3B, Bradi1g00950/Os03g63970 (gibberellin 20 [GA20] oxidase putative expressed protein), Os03g56630, Os03g62660 (Far-red impaired responsive [FAR1] family protein) orthologous to 4A, Bradi1g46060/Os06g10880 (ABF3/ABF2 - abscissic acid responsive elements) orthologous to 7B, Bradi1g48640, Bradi1g48650, Bradi1g48822, Bradi1g48816 (Far-red impaired responsive [FAR1] family protein), Bradi1g48690, Bradi1g50240 (VRN1-AP2/B3 - like transcriptional factor family protein) and Bradi1g48830/Os06g06320 (Vrn3/FT PEBP [phosphatidylethanolamine - binding protein] homologous to Flowering Locus T gene) orthologous to chromosome 7D.
Development and validation of KASP primers
A total of 18 KASP markers, five each for chromosomes 3B and 7B.1, and four each for chromosomes 4A and 7D.2 (Table 4) were developed from sequences of SNPs flanking QTL for PHS resistance. Genetic map locations of individual KASP markers were identical to the respective SNP from which they were derived. Primer sets of all 18 KASP markers are listed in Additional file 3. Further, we validated the conversion of these 18 KASPs from matching genotype calls of Infinium SNP data on 183 DH progeny genotypes. Four DH progeny genotypes of the RL4452/‘AC Domain’ cross were identified to carry PHS resistance on chromosomes 3B, 4A, 7B and 7D (Additional file 4).
Table 4.
A list of 18 Competitive Allele-Specific PCR (KASP) markers developed for MAS of PHS from SNPs flanking PHS QTL on chromosomes 3B, 4A, 7B and 7D in a DH population of a RL4452/‘AC Domain’ cross
| Sl. | KASP marker | Source SNP | Chr | PHS trait |
|---|---|---|---|---|
| 1. | Kasp3B_wsnp_Ku_rep_c72700_72370664 | wsnp_Ku_rep_c72700_72370664 | 3B | SI |
| 2. | Kasp3B_RAC875_rep_c116515_181, | RAC875_rep_c116515_181, | 3B | SI |
| 3. | Kasp3B_BobWhite_rep_c64944_264 | BobWhite_rep_c64944_264 | 3B | SI |
| 4. | Kasp3B_wsnp_Ex_c16378_24870688 | wsnp_Ex_c16378_24870688 | 3B | SI |
| 5. | Kasp3B_RAC875_c530_354 | RAC875_c530_354 | 3B | SI |
| 6. | Kasp4A_BS00072025_51 | BS00072025_51 | 4A | GI, SI, FN |
| 7. | Kasp4A_Kukri_c12563_52 | Kukri_c12563_52 | 4A | GI, SI, FN |
| 8. | Kasp4A_RAC875_c21369_425 | RAC875_c21369_425 | 4A | GI, SI, FN |
| 9. | Kasp4A_wsnp_Ex_c16175_24619793 | wsnp_Ex_c16175_24619793 | 4A | GI, SI, FN |
| 10 | Kasp7B_wsnp_Ex_c908_1754208 | wsnp_Ex_c908_1754208 | 7B.1 | SI |
| 11 | Kasp7B_RFL_Contig124_558 | RFL_Contig124_558 | 7B.1 | SI |
| 12 | Kasp7B_RAC875_c1638_165 | RAC875_c1638_165 | 7B.1 | SI |
| 13 | Kasp7B_wsnp_Ex_rep_c69639_68590556 | wsnp_Ex_rep_c69639_68590556 | 7B.1 | SI |
| 14 | Kasp7B_Ku_c32389_1009 | Ku_c32389_1009 | 7B.1 | SI |
| 15 | Kasp7D_Excalibur_c22419_460 | Excalibur_c22419_460 | 7D.2 | FN |
| 16 | Kasp7D_RAC875_c1829_321 | RAC875_c1829_321 | 7D.2 | FN |
| 17 | Kasp7D_Kukri_c32845_116 | Kukri_c32845_116 | 7D.2 | FN |
| 18 | Kasp7D_wsnp_CAP8_rep_c9647_4198594 | wsnp_CAP8_rep_c9647_4198594 | 7D.2 | FN |
Discussion
The objectives of our research were to identify candidate regions for PHS resistance QTL of wheat and develop KASP markers (for MAS) from sequences of SNPs flanking such QTL. This is an important step in the process of map-based cloning of genes that underlie important quantitative traits like PHS resistance. Our objectives were achieved using 11,282 SNPs from the 90 k Infinium Custom Beadchip to develop a high density linkage map in the RL4452/‘AC Domain’ mapping population and subsequently detect QTL for PHS resistance on chromosomes 3B, 4A, 7B and 7D. Comparative mapping utilizing sequences of SNPs flanking PHS resistance QTL enabled identification of candidate genes and regions in Brachypodium and rice. The resulting 18 KASP markers can be deployed in future genetic studies of PHS, and in evaluation of PHS in germplasm and breeding material.
Of the 12,201 mapped markers, 11,282 or 92.5% were SNP markers, while the remaining 919 or 7.5% were SSR, DArT and EST markers. The B genome chromosomes accounted for the largest number of 6291 SNP markers, followed by the A genome with 4125 SNPs, and the D genome with 1785 SNP markers. A likely explanation for larger numbers of B genome SNP markers could be the greater genetic diversity of B genome species when compared to the A and D genome species [46,47]. A faster rate of evolution of the B genome due to greater polymorphism and duplication events, in addition to greater genetic diversity brought about by cross pollination were cited [48-50] as possible explanations for findings of a greater number of ESTs associated with more unique loci on the B genome when compared to the A and D genomes.
PHS datasets were analyzed with both MIM and SIM (data not shown) methods. Because results of both methods were very similar, only those of the MIM analyses were reported. As QTL identified using MIM were robust and supported by SIM results, it is unlikely that additional large effect QTL involved in epistatic interactions might have been detected using other QTL mapping methods that detect both main effect (M-QTL) and epistatic QTL (E-QTL). Further, a Meta-QTL study [37] reporting PHS QTL on 4A and group 3 chromosomes support significant PHS QTL identified on chromosome 3B and 4A of our study.
The most consistent of the four PHS QTL identified on chromosomes 3B, 4A, 7B and 7D were located on chromosome 4A; GI, SI and FN trait QTL each accounting for 58.1%, 32.1% and 25.8% of the phenotypic variation in their respective traits. The QTL for these PHS traits were coincident and maybe associated with the same gene(s). These findings might suggest that chromosome 4A is involved in regulation of PHS trait QTL in our test population. Previous reports of the association of PHS traits with chromosome 4A [2,20-24], support the importance of this QTL for PHS
In addition to a major SI QTL on 4A, two other QTL for SI were identified on chromosomes 3B and 7B.1. Both SI QTL on 3B and 7B.1 were detected in two of six environments. QTL that provide tolerance to late maturity α- amylase (LMA) have been mapped on 3BS and 7BL in an Australian wheat cross Cranbrook/Halberd [51]. In both studies, the SSR markers Xwmc623, Xwmc808, Xgwm72, Xwmc612, Xgwm285, Xwmc693, Xwmc1 (3B LMA QTL interval) and Xgwm577, Xwmc273, Xwmc276 (7B LMA QTL interval) also flanked corresponding PHS QTL intervals on chromosomes 3B.1 and 7B.1 respectively (data not shown). Further, alleles of a regulator gene Vp-1B on 3B have been reported to influence grain dormancy in Chinese wheat varieties [19]. In a follow up study [52], the VP-1B locus was validated in a white-grained Chinese landrace Wanxianbaimaizi (high seed dormancy and PHS tolerance) using SSR markers and a gene-specific primer Vp1. A CIM analysis identified a seed dormancy QTL QSd.ahau-3B on 3B flanked by Vp1 which is linked to an SSR marker Xwmc446 that also happens to flank the PHS QTL interval on chromosome 3B of our study. The above findings suggest that PHS and LMA QTL on chromosomes 3B and 7B are likely the same.
‘AC Domain’ alleles contributed to increasing the FN on 7D (linkage group 7D.2), with the exception of the Glenlea 2005 trial, wherein a negative additive score was observed for the FN. While the FN QTL on chromosome 7D is unique to our study, a significant time to maturity (Mat) QTL (PV = 26%) also on 7D, and a positive contribution of the RL4452 allele, has been reported previously by [29] in the same RL4452/‘AC Domain’ population. The authors reported an SSR marker Xgwm130 tightly linked to this QTL, which is distally located on 7DL, and is 1.1 cM from the QTL peak of our study. In the Glenlea 2005 trial (with a negative additive score for FN), the average FN (LS Mean) score of 183 DH progeny was the lowest of the four trials (data not shown). The low FN score at this location might suggest greater levels of PHS of ‘AC Domain’ genotypes, probably brought on by wet weather conditions at the maturity stages or during the three weeks preceding harvest [53]. As QTL locations of both these Mat and FN traits nearly coincide and are influenced by negative and positive additive effects (with the exception of the FN QTL of the Glenlea 2005 trial) of ‘AC Domain’ alleles respectively, the action of a pleiotrophic locus regulating both FN and Mat could be assumed. At Glenlea in 2005 it is possible that the lower FN for the Domain allele is due to adverse weather conditions at maturity or that the 7D QTL identified here might not actually be a PHS QTL, but rather a pleiotrophic effect of the Mat QTL on PHS.
Flanking marker intervals of a given PHS trait (GI, SI or FN) QTL were not always the same across trials/datasets. It is quite likely that the respective underlying genes influencing each of these traits are the same; difference in QTL interval location being mainly due to environment or experimental error from differences in class means of individual trial data sets [54]. Alternatively, the possibility of two closely linked loci controlling the same trait cannot be ruled out.
BLASTN searches with sequences of SNP markers flanking PHS QTL on chromosomes 3B, 4A, 7B and 7D revealed candidate regions in Brachypodium and rice genomes. The QTL interval on chromosome 3B was orthologous to regions on Bradi2 and the long arm of Os01, while QTL intervals on chromosomes 4A were orthologous to regions on Bradi1 and the short arm of Os03. QTL intervals on chromosome 7B.1 and 7D.2 were orthologous to regions on Bradi1 and the short arm of Os06 of rice. The above findings of orthology between wheat/rice chromosomes: 3B/Os01, 4A/Os03 and 7B&7D/Os06 concur with previous reports [42,55-57] of wheat/rice chromosomal region similarities revealed via comparative mapping with DNA probes and ESTs. Further, orthologies between PHS QTL intervals of 4A, 7B, 7D and genomic regions of Bradi1, and 3B/Bradi2 in our study will be refined to tease out individual genes responsible for variation in PHS resistance. The availability of information on whole-genome 454 assembled gene sequences of Chinese spring [58] and gene-orthologies among the said wheat and Brachypodium chromosomes established using 5003 ESTs mapped to wheat deletion bins [32] will serve as useful references to complement our efforts.
Eighteen KASP markers were developed from SNP sequences flanking QTL for PHS resistance. Identical genotype calls of Infinium SNP data enabled validation of the 18 KASP markers and identified four (of 183) progeny genotypes of the RL4452/‘AC Domain’ population possessing PHS resistance on all four QTL on 3B, 4A, 7B and 7D (Additional file 4). Criteria for selection of these genotypes was based on findings of our study: ‘AC Domain’ (allele 'A') reduced GI and SI on 4A, increased FN on 4A and 7D, while RL4452 (allele 'B') reduced SI on chromosomes 3B and 7B. Further, these 18 KASP markers can be deployed in future genetic studies of PHS, and in evaluation of PHS in germplasm and breeding material.
Genes present in Brachypodium and rice in orthologous regions corresponding to the QTL were identified (Additional file 2). The 3B region is large and contains over 800 genes in Brachypodium and over 1300 in rice. More markers are needed to reduce the size of the region and the emerging reference sequence of chromosome 3B (http://wheat-urgi.versailles.inra.fr/Seq-Repository/Reference-sequence) will be a valuable resource. There are a number of ABA-inducible genes (2 Brachypodium and 3 rice) which could be a starting point to search for additional markers.
The 4A and 7B regions contain many fewer genes in Brachypodium and rice than the 3B region. Gibberellin 20 oxidase (GA20 – oxidase) [59] on Bradi1/Os03 orthologous to 4A and abscissic acid responsive elements (ABF2, ABF3) [60-62] on Bradi1/Os06 orthologous to chromosome 7B are candidates worth further study. GA20 - oxidase has previously been considered as a candidate gene underlying PHS QTL on 4A [63].
On chromosome 7D the QTL was coincident with a previously identified maturity QTL in the same population (29). Genes affecting flowering time are present in the orthologous regions in Brachypodium and rice. These include the Far-red impaired responsive (FAR1) related proteins [64] on chromosome Bradi1, as well as VRN1-AP2/B3-like transcription factors [65,66] on Bradi1 and phosphatidylethanolamine - binding protein (PEBP) homologous to the Flowering Locus T gene [67,68] on Bradi1/Os06, orthologous to chromosome 7D.
Because our study utilized a large number of sequence-based SNPs not available for previous mapping studies, the resulting genetic maps and QTL flanking SNP markers are a novel and current resource for identification of underlying genes based on synteny and collinearity to model species Brachypodium and rice. Further, the identification of candidate genes and regions for PHS in Brachypodium and rice will enable a targeted focus for selection of candidate genes whose physiological/biological functions are linked to or influence variation in PHS traits under study. Such candidate gene-specific PCR markers will be developed and validated via mapping to the QTL intervals for PHS resistance in wheat.
Conclusions
In our study we utilized SNPs from a wheat 90 K Infinium iSelect Custom Beadchip that permitted detection and assignment of significant PHS resistance QTL to specific chromosomal locations on genetic maps. Sequences of SNPs flanking PHS resistance QTL enabled identification of candidate genes and regions for PHS in Brachypodium and rice via comparative mapping. The 18 KASP markers resulting from this study can be suitably deployed in future genetic studies of PHS and might also be useful in the evaluation of PHS in germplasm and breeding material.
Methods
Plant material, experimental layout and trait phenotyping
A total of 193 DH progeny genotypes derived from a cross RL4452/‘AC Domain’ were used to develop the genetic linkage map. Of these, trait data was available on 183 DH lines for detection of QTL across the genome. Data on three PHS traits (GI, SI and FN) was collected from six trials: Glenlea (2003; 2005), Winnipeg (2004; 2005) and Swift Current (2003; 2004), in Manitoba and Saskatchewan Canada. The phenotyping methods, experimental design and layout for each of these traits are described in [6,28].
Molecular markers and genotyping
Infinium SNPs and PCR based markers
The 90 K Infinium iSelect Custom Wheat Beadchip identified 12,351 polymorphic markers that were added to existing SSR, DArT and EST markers for the RL4452/‘AC Domain’ cross. Of these, a total of 12,201 markers (11282 SNPs; 919 SSRs, DArTs and ESTs) were used in the construction of genetic maps. Further, co-segregating markers were removed from the set of 12,201 markers and QTL analysis was carried out (one marker per bin) with 1054 markers.
Linkage mapping
Genotypic data of 193 DH progeny, screened with 12,201 markers (SSR, SNP, DArT and ESTs), were used to construct genetic maps for all 21 chromosomes. Bins of co-segregating markers were identified with MSTMap [69], and the most informative marker per bin was retained for mapping with MapDisto® [70]. Linkage groups were created using a minimum LOD score of 4 and maximum recombination fraction (RF) of 0.25. Recombination fractions were converted into centiMorgan (cM) map distances using the Kosambi mapping function.
QTL analysis
Multi-year trial data collected at six environments on three PHS traits (GI, SI, FN) were used for QTL mapping with QGene version 3.0 software [71]. Trait data and molecular phenotypes of 183 DH progeny assessed with 1054 markers were subject to MIM and SIM (data not shown) analyses. QTL with LOD scores exceeding critical threshold values at 5% (α0.05), at two or more environments were deemed significant. Threshold values for trait QTL were obtained through permutation analyses involving 1000 iterations. Further, marker–trait regression (r2) values were interpreted as the percent phenotypic variation (% PV) explained due to respective QTL.
Identification of candidate genes and regions in Brachypodium and rice
Sequences of SNPs flanking QTL for PHS resistance traits (GI, SI, FN) on chromosomes 3B, 4A, 7B and 7D were subject to a BLASTN (Basic search) on the IWGSC database (http://wheat-urgi.versailles.inra.fr/Seq-Repository). Further, best survey sequence hits were subject to a BLASTN search (Maximum E-value 10) on Gramene (www.gramene.org) against both Brachypodium and rice databases to obtain candidate regions for PHS resistance. QTL intervals were deduced from centiMorgan map distances between SNP markers flanking QTL peaks of a given PHS resistance trait (GI, SI or FN). Consensus candidate regions for PHS resistance were arrived at from best hits (of PHS QTL flanking SNP sequences) to genes and genomic regions in Brachypodium and rice. A few of the SNP markers returned hits to non-candidate regions/chromosomes prompting the selection of weaker hits to the consensus candidate regions. MapChart 2.2 [72] was used to construct genetic and physical maps of orthologous regions in wheat, Brachypodium and rice. Candidate genes in Brachypodium and rice corresponding to QTL intervals for PHS resistance on chromosomes 3B, 4A, 7B and 7D of wheat were obtained from the online PlantGDB database (http://www.plantgdb.org/).
KASP markers
Sequences of SNP markers flanking QTL for PHS resistance on chromosomes 3B, 4A, 7B and 7D were converted to KASP markers. PrimerPicker Lite for KASP version 0.25 (KBioscience®) was used to generate KASP primer sets from QTL flanking SNP sequences. Protocols for the preparation and running of KASP reactions, and PCR conditions are given in the KASP manual (http://www.kbioscience.co.uk/). A FLUOstar Omega plate reader (BMG LABTECH® Offenburg Germany) with KlusterCaller™ software was used to visualize KASP marker polymorphisms.
Availability of supporting data
All the supporting data are available as additional files.
Acknowledgements
This study was conducted as part of the Canadian Triticum Advancement through Genomics (CTAG) project. The authors thank Genome Canada, Genome Prairie, the Western Grains Research Foundation, the Province of Saskatchewan, and Alberta Innovates for funding this project.
Additional files
A linkage map constructed using 193 DH progeny genotypes of a RL4452/‘AC Domain’ cross evaluated with 12,201 polymorphic markers (11282 SNPs and 919 PCR markers).
Brachypodium and rice candidates corresponding to QTL intervals for PHS resistance on chromosomes 3B, 4A, 7B and 7D in a DH population of a RL4452/‘AC Domain’ cross.
List of 18 Competitive Allele-Specific PCR (KASP) primers derived from sequences of SNPs flanking QTL for PHS resistance on chromosomes 3B, 4A, 7B and 7D.
Validation of 18 Competitive Allele-Specific PCR (KASP) markers designed from Illumina iSelect markers flanking QTL in a RL4452/‘AC Domain’ population.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MCJ, CAM, ALC (designed and edited the manuscript); ALC, MCJ (conducted the experiment and drafted the manuscript); GH (provided the RL4452/‘AC Domain’ DH population and edited the manuscript); CJP (conducted the 90 K genotyping, edited the manuscript and is the lead of the CTAG project that funded part of this work); FMY, RM (carried out bioinformatics and data sorting work); CAM (performed MST mapping and SNP clustering). All authors read and approved the final manuscript.
Contributor Information
Adrian L Cabral, Email: adrian.cabral@nrc-cnrc.gc.ca.
Mark C Jordan, Email: mark.jordan@agr.gc.ca.
Curt A McCartney, Email: curt.mccartney@agr.gc.ca.
Frank M You, Email: frank.you@agr.gc.ca.
D Gavin Humphreys, Email: gavin.humphries@agr.gc.ca.
Ron MacLachlan, Email: prm626@mail.usask.ca.
Curtis J Pozniak, Email: curtis.pozniak@usask.ca.
References
- 1.DePauw RM, Knox RE, Singh AK, Fox SL, Humphreys DG, Hucl P. Developing standardized methods for breeding preharvest sprouting resistant wheat, challenges and successes in Canadian wheat. Euphytica. 2012;188:7–14. doi: 10.1007/s10681-011-0611-y. [DOI] [Google Scholar]
- 2.Ogbonnaya FC, Imtiaz M, DePauw RM. Haplotype diversity of preharvest sprouting QTLs in wheat. Genome. 2007;50:107–118. doi: 10.1139/g06-142. [DOI] [PubMed] [Google Scholar]
- 3.DePauw RM, Clarke FR, Fofana B, Knox R, Humphreys G, Cloutier S. RL4137 contributes preharvest sprouting resistance to Canadian wheats. Euphytica. 2009;168(3):347–361. doi: 10.1007/s10681-009-9933-4. [DOI] [Google Scholar]
- 4.Hagberg S. Note on a simplified rapid method for determining alpha-amylase activity. Cereal Chem. 1961;38:202–203. [Google Scholar]
- 5.Paterson AH, Sorrells ME, Obendorf RL. Methods of evaluation for preharvest sprouting resistance in wheat breeding programs. Can J Plant Sci. 1989;69:681–689. doi: 10.4141/cjps89-084. [DOI] [Google Scholar]
- 6.Humphreys DG, Noll J. Methods for characterization of preharvest sprouting resistance in a wheat breeding program. Euphytica. 2002;126(1):61–65. doi: 10.1023/A:1019671622356. [DOI] [Google Scholar]
- 7.Hagemann MG, Ciha AJ. Evaluation of methods used in testing winter wheat susceptibility to preharvest sprouting. Crop Sci. 1983;24:249–253. doi: 10.2135/cropsci1984.0011183X002400020010x. [DOI] [Google Scholar]
- 8.Reddy LV, Metzger RJ, Ching TM. Effect of temperature on seed dormancy of wheat. Crop Sci. 1985;25:455–458. doi: 10.2135/cropsci1985.0011183X002500030007x. [DOI] [Google Scholar]
- 9.McMaster GJ, Derera NF. Methodology and sample preparation when screening for sprouting damage in cereals. Cereal Res Commun. 1976;4:251–254. [Google Scholar]
- 10.Mohan A, Kulwal P, Singh R, Kumar V, Mir RR, Kumar J, Prasad M, Balyan HS, Gupta PK. Genome-wide QTL analysis for pre-harvest sprouting tolerance in bread wheat. Euphytica. 2009;168(3):319–329. doi: 10.1007/s10681-009-9935-2. [DOI] [Google Scholar]
- 11.Munkvold JD, Tanaka J, Benscher D, Sorrells ME. Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor Appl Genet. 2009;119(7):1223–1235. doi: 10.1007/s00122-009-1123-1. [DOI] [PubMed] [Google Scholar]
- 12.Kulwal PL, Mir RR, Kumar S, Gupta PK. QTL analysis and molecular breeding for seed dormancy and pre-harvest sprouting tolerance in bread wheat. J Plant Biol. 2010;37:59–74. [Google Scholar]
- 13.Jaiswal V, Mir RR, Mohan A, Balyan HS, Gupta PK. Association mapping for pre-harvest sprouting tolerance in common wheat (Triticum aestivum L.) Euphytica. 2012;188(1):89–102. doi: 10.1007/s10681-012-0713-1. [DOI] [Google Scholar]
- 14.Osa M, Kato K, Mori M, Shindo C, Torada A, Miura H. Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor Appl Genet. 2003;106(8):1491–1496. doi: 10.1007/s00122-003-1208-1. [DOI] [PubMed] [Google Scholar]
- 15.Kulwal PL, Kumar N, Gaur A, Khurana P, Khurana JP, Tyagi AK, Balyan HS, Gupta PK. Mapping of a major QTL for pre-harvest sprouting tolerance on chromosome 3A in bread wheat. Theor Appl Genet. 2005;111(6):1052–1059. doi: 10.1007/s00122-005-0021-4. [DOI] [PubMed] [Google Scholar]
- 16.Mori M, Uchino N, Chono M, Kato K, Miura H. Mapping QTLs for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor Appl Genet. 2005;110(7):1315–1323. doi: 10.1007/s00122-005-1972-1. [DOI] [PubMed] [Google Scholar]
- 17.Fofana B, Humphreys DG, Rasul G, Cloutier S, Brûlé-Babel A, Woods S, Lukow OM, Somers DJ. Mapping quantitative trait loci controlling pre-harvest sprouting resistance in a red × white seeded spring wheat cross. Euphytica. 2009;165(3):509–521. doi: 10.1007/s10681-008-9766-6. [DOI] [Google Scholar]
- 18.Kulwal PL, Singh R, Balyan HS, Gupta PK. Genetic basis of pre-harvest sprouting tolerance using single-locus and two-locus QTL analyses in bread wheat. Funct Integr Genomic. 2004;4(2):94–101. doi: 10.1007/s10142-004-0105-2. [DOI] [PubMed] [Google Scholar]
- 19.Chang C, Zhang HP, Feng JM, Yin B, Si HQ, Ma CX. Identifying alleles of Viviparous-1B associated with pre-harvest sprouting in micro-core collections of Chinese wheat germplasm. Mol Breeding. 2010;25(3):481–490. doi: 10.1007/s11032-009-9346-z. [DOI] [Google Scholar]
- 20.Flintham J, Adlam R, Bassoi M, Holdsworth M, Gale M. Mapping genes for resistance to sprouting damage in wheat. Euphytica. 2002;126(1):39–45. doi: 10.1023/A:1019632008244. [DOI] [Google Scholar]
- 21.Mares D, Mrva K, Cheong J, Williams K, Watson B, Storlie E, Sutherland M, Zou Y. A QTL located on chromosome 4A associated with dormancy in white-and red-grained wheats of diverse origin. Theor Appl Genet. 2005;111(7):1357–1364. doi: 10.1007/s00122-005-0065-5. [DOI] [PubMed] [Google Scholar]
- 22.Torada A, Ikeguchi S, Koike M. Mapping and validation of PCR-based markers associated with a major QTL for seed dormancy in wheat. Euphytica. 2005;143(3):251–255. doi: 10.1007/s10681-005-7872-2. [DOI] [Google Scholar]
- 23.Ogbonnaya FC, Imtiaz M, Ye G, Hearnden PR, Hernandez E, Eastwood RF, Ginkel MV, Shorter SC, Winchester JM. Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor Appl Genet. 2008;116(7):891–902. doi: 10.1007/s00122-008-0712-8. [DOI] [PubMed] [Google Scholar]
- 24.Chen CX, Cai SB, Bai GH. A major QTL controlling seed dormancy and pre-harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Mol Breeding. 2008;21(3):351–358. doi: 10.1007/s11032-007-9135-5. [DOI] [Google Scholar]
- 25.Groos C, Gay G, Perretant MR, Gervais L, Bernard M, Dedryver F, Charmet G. Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white × red grain bread-wheat cross. Theor Appl Genet. 2002;104(1):39–47. doi: 10.1007/s001220200004. [DOI] [PubMed] [Google Scholar]
- 26.Singh R, Matus-Ca´diz M, Baga M, Hucl P, Chibbar RN. Identification of genomic regions associated with seed dormancy in white-grained wheat. Euphytica. 2010;174:391–408. doi: 10.1007/s10681-010-0137-8. [DOI] [Google Scholar]
- 27.Roy JK, Prasad M, Varshney RK, Balyan HS, Blake TK, Dhaliwal HS, Edwards KJ, Gupta PK. Identification of a microsatellite on chromosomes 6B and a STS on 7D of bread wheat showing an association with preharvest sprouting tolerance. Theor Appl Genet. 1999;99(1):336–340. doi: 10.1007/s001220051241. [DOI] [Google Scholar]
- 28.Rasul G, Humphreys DG, Brûlé-Babel A, McCartney CA, Knox RE, DePauw RM, Somers DJ. Mapping QTLs for pre-harvest sprouting traits in the spring wheat cross RL4452/‘AC Domain’. Euphytica. 2009;168(3):363–378. doi: 10.1007/s10681-009-9934-3. [DOI] [Google Scholar]
- 29.McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N, Noll J, Cloutier S, McCallum BD. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × 'AC Domain'. Genome. 2005;48(5):870–883. doi: 10.1139/g05-055. [DOI] [PubMed] [Google Scholar]
- 30.McCartney CA, Somers DJ, Lukow O, Ames N, Noll J, Cloutier S, Humphreys DG, McCallum BD. QTL analysis of quality traits in the spring wheat cross RL4452 × ‘AC Domain’. Plant Breed. 2006;125(6):565–575. doi: 10.1111/j.1439-0523.2006.01256.x. [DOI] [Google Scholar]
- 31.International Rice Genome Sequencing Project The mapbased sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 32.The International Brachypodium Initiative Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463(7282):763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Bai G, Cai S, Chen C. Dissection of genetic components of preharvest sprouting resistance in white wheat. Mol Breeding. 2010;27(4):511–523. doi: 10.1007/s11032-010-9448-7. [DOI] [Google Scholar]
- 34.Kulwal P, Kumar N, Kumar A, Gupta RK, Balyan HS, Gupta PK. Gene networks in hexaploid wheat: interacting quantitative trait loci for grain protein content. Funct Integr Genomic. 2005;5(4):254–259. doi: 10.1007/s10142-005-0136-3. [DOI] [PubMed] [Google Scholar]
- 35.Imtiaz M, Ogbonnaya FC, Oman J, van Ginkel M. Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics. 2008;178(3):1725–1736. doi: 10.1534/genetics.107.084939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goffinet B, Gerber S. Quantitative trait loci: a meta-analysis. Genetics. 2000;155(1):463–473. doi: 10.1093/genetics/155.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyagi S, Gupta PK. Meta-analysis of QTLs involved in pre-harvest sprouting tolerance and dormancy in bread wheat. Triticeae Genomics Genetics. 2012;3(2):9–24. [Google Scholar]
- 38.Faris JD, Zhang Z, Fellers JP, Gill BS. Micro-colinearity between rice, Brachypodium, and Triticum monococcum at the wheat domestication locus Q. Funct Integr Genomic. 2008;8(2):149–164. doi: 10.1007/s10142-008-0073-z. [DOI] [PubMed] [Google Scholar]
- 39.Huo N, Vogel JP, Lazo GR, You FM, Ma Y, McMahon S, Dvorak J, Anderson OD, Luo M-C, Gu YQ. Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Mol Biol. 2009;70(1–2):47–61. doi: 10.1007/s11103-009-9456-3. [DOI] [PubMed] [Google Scholar]
- 40.Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- 41.Bossolini E, Wicker T, Knobel PA, Keller B. Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J. 2007;49:704–717. doi: 10.1111/j.1365-313X.2006.02991.x. [DOI] [PubMed] [Google Scholar]
- 42.Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin, Mahmoud A, Ma X, Gustafson PJ, Qi LL, Echalier B, Gill BS, Matthews DE, Lazo GR, Chao S, Anderson OD, Edwards H, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvorak J, Zhang D, Nguyen HT, Peng J, Lapitan NLV, Gonzalez-Hernandez JL, Anderson JA, Hossain K, Kalavacharla V, et al. Comparative DNA Sequence Analysis of Wheat and Rice Genomes. Genome Res. 2003;13:1818–1827. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, International Wheat Genome Sequencing Consortium, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo M-C, Dvorak J, et al: Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array.Plant Biotech J 2014, ᅟ:ᅟ. doi:10.1111/pbi.12183. [DOI] [PMC free article] [PubMed]
- 44.Kao CH, Zeng ZB, Teasdale RD. Multiple interval mapping for quantitative trait loci. Genetics. 1999;152(3):1203–1216. doi: 10.1093/genetics/152.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao S, Dubcovsky J, Dvorak J, Luo MC, Baenziger SP, Matnyazov R, Clark DR, Talbert LE, Anderson JA, Dreisigacker S, Glover K, Chen J, Campbell K, Bruckner PL, Rudd JC, Haley S, Carver BF, Perry S, Sorrells ME, Akhunov ED. Population-and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.) BMC Genomics. 2010;11(1):727. doi: 10.1186/1471-2164-11-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan MC, Somers DJ, Banks TW. Identifying regions of the wheat genome controlling seed development by mapping expression quantitative trait loci. Plant Biotech J. 2007;5(3):442–453. doi: 10.1111/j.1467-7652.2007.00253.x. [DOI] [PubMed] [Google Scholar]
- 48.Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci U S A. 2002;99:8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akhunov ED, Akhunova AR, Linkiewicz AM, Dubcovsky J, Hummel D, Lazo GR, Chao S, Anderson OD, David J, Qi L, Echalier B, Gill BS, Miftahudin, Gustafson JP, La Rota M, Sorrells ME, Zhang D, Nguyen HT, Kalavacharla V, Hossain K, Kianian SF, Peng J, Lapitan NLV, Wennerlind EJ, Nduati V, Anderson JA, Sidhu D, Gill KS, McGuire PE, Qualset CO, et al. Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc Natl Acad Sci U S A. 2003;100:10836–10841. doi: 10.1073/pnas.1934431100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvorak J, Linkiewicz AM, Ratnasiri A, Dubcovsky J, Bermudez-Kandianis CE, Greene RA, Kantety R, La Rota CM, Munkvold JD, Sorrells SF, Sorrells ME, Dilbirligi M, Sidhu D, Erayman M, Randhawa HS, Sandhu D, Bondareva SN, Gill KS, Mahmoud AA, Ma XF, Miftahudin, Gustafson JP, Conley EJ, et al. A Chromosome Bin Map of 16,000 Expressed Sequence Tag Loci and Distribution of Genes Among the Three Genomes of Polyploid Wheat. Genetics. 2004;168:701–712. doi: 10.1534/genetics.104.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNeil MD, Diepeveen D, Wilson R, Barclay I, McLean R, Chalhoub B, Appels R. Haplotype analyses in wheat for complex traits: tracking the chromosome 3B and 7B regions associated with late maturity alpha amylase (LMA) in breeding programs. Crop Pasture Sci. 2009;60(5):463–471. doi: 10.1071/CP08340. [DOI] [Google Scholar]
- 52.Chang C, Feng JM, Si HQ, Yin B, Zhang HP, Ma CX. Validating a novel allele of viviparous-1 (Vp-1Bf) associated with high seed dormancy of Chinese wheat landrace, Wanxianbaimaizi. Mol Breeding. 2010;25(3):517–525. doi: 10.1007/s11032-009-9350-3. [DOI] [Google Scholar]
- 53.Mares DJ. Pre-harvest sprouting in wheat. I. Influence of cultivar, rainfall and temperature during grain ripening. Crop Pasture Sci. 1993;44(6):1259–1272. doi: 10.1071/AR9931259. [DOI] [Google Scholar]
- 54.Szalma SJ, Snook ME, Bushman BS, Houchins KE, McMullen MD. Duplicate Loci as QTL. Crop Sci. 2002;42(5):1679–1687. doi: 10.2135/cropsci2002.1679. [DOI] [Google Scholar]
- 55.Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M. Conservation of genome structure between rice and wheat. Nat Biotechnol. 1994;12(3):276–278. doi: 10.1038/nbt0394-276. [DOI] [Google Scholar]
- 56.Van Deynze AE, Nelson JC, Yglesias ES, Harrington SE, Braga DP, McCouch SR, Sorrells ME. Comparative mapping in grasses. Wheat relationships. Mol Gen Genet. 1995;248(6):744–754. doi: 10.1007/BF02191715. [DOI] [PubMed] [Google Scholar]
- 57.Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci U S A. 1998;95(5):1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D’Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, Trick M, Bancroft I, Gu Y, Huo N, Luo M-C, Sehgal S, Gill B, Kianian S, Anderson O, Kersey P, Dvorak J, McCombie WR, Hall A, Mayer KFX, Edwards KJ, Bevan MW, Hall N. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 2012;491(7426):705–710. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lange T, Hedden P, Graebe JE. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc Natl Acad Sci U S A. 1994;91(18):8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi H, Hong J, Ha J, Kang J, Kim S. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 61.Kim S, Ma J, Perret P, Li Z, Thomas T. Arabidopsis ABI5 subfamily members have distinct DNA binding and transcriptional activities. Plant Physiol. 2002;130:688–697. doi: 10.1104/pp.003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S, Kang J-Y, Cho D-I, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 63.Tyagi S, Gupta PK. Meta-analysis of QTLs Involved in Pre-harvest Sprouting Tolerance and Dormancy in Bread Wheat. Triticeae Gen Genet. 2012;3:9–24. [Google Scholar]
- 64.Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Gene Dev. 1999;13(15):2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A. 2003;100(10):6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 67.Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43(10):1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 68.Ciaffi M, Paolacci AR, D'Aloisio E, Tanzarella OA, Porceddu E. Identification and characterization of gene sequences expressed in wheat spikelets at the heading stage. Gene. 2005;346:221–230. doi: 10.1016/j.gene.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 69.Wu Y, Bhat PR, Close TJ, Lonardi S. Efficient and Accurate Construction of Genetic Linkage Maps from the Minimum Spanning Tree of a Graph. PLoS Genet. 2008;4(10):e1000212. doi: 10.1371/journal.pgen.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lorieux M: MapDisto: fast and efficient computation of genetic linkage maps.Mol Breeding 2012, ᅟ:ᅟ. in press (doi:10.1007/s11032-012-9706-y).
- 71.Joehanes R, Nelson JC. QGene 4.0 an extensible Java QTL-analysis platform. Bioinformatics. 2008;24:2788–2789. doi: 10.1093/bioinformatics/btn523. [DOI] [PubMed] [Google Scholar]
- 72.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]


