Abstract
The large T antigen (LT) protein of JCV and SV40 polyomaviruses is required to induce tumors in rodents and transform cells in culture. When both LTs are compared side-by-side in cell culture assays, SV40 shows a more robust transformation phenotype even though the LT sequences are highly conserved. A complete understanding of SV40’s enhanced transforming capabilities relative to JCV is lacking. When the least conserved region of the LT proteins, the variable linker and host range region (VHR), was removed, changes in T antigen expression and cellular p53 post-translational modifications occurred, but interaction with the pRB pathway was unaffected. Transformation assessed by growth in low serum was reduced after VHR truncation of the SV40, but not the JCV, T antigen. Conversely, anchorage independent transformation was enhanced only by truncation of the JCV VHR. This is the first report to link the SV40 or JCV VHR region to transformation potential.
Keywords: polyomavirus, SV40, JCV, large T antigen, transformation
Introduction
Polyomaviruses are a group of small, double-stranded DNA viruses with an icosahedral capsid that lacks an envelope. Over 40 polyomaviruses have been identified in multiple species, ranging from mammals to birds, and even amphibians and reptiles. The majority of human polyomaviruses (PyV) do not cause disease in healthy individuals, but some are the cause of disease when the immune system is compromised. One of the most studied disease-associated polyomaviruses is the human JC virus (JCV). A significant percent of the population is seropositive for JCV, however subsequent disease is overwhelmingly associated with immunocompromised individuals (1). In an immunocompromised setting, JCV can actively infect the central nervous system, causing progressive multifocal leukoencephalopathy as a result of the destruction of glial tissue in the brain. This disease is fatal, since few effective treatments are available. JCV is closely related to the primate polyomavirus SV40 (2), which is considered the model virus of this group. Since its discovery in 1960 (3), the study of SV40 has facilitated the understanding of many basic viral and cellular functions including DNA replication, cell cycling, and transcriptional control of gene expression (4).
All polyomaviruses share a similar genome structure, where a large non-coding region physically and temporally separates and controls the expression of the early and late transcribed regions of the genome. The early transcribed region (ER) is alternatively spliced to create at least two transcripts translated into a large T antigen and a small T antigen protein. The large T antigen (LT) is a multi-functional, multi-domain protein responsible for the replication of the viral genome as well as for the manipulation of the host cell environment to produce conditions conducive to replication (5, 6). The small T antigen (sT) and various other T antigen proteins also contribute indirectly to genome replication and directly to environmental manipulation (7–9). Later in infection, transcription of the right arm of the genome, or late region, is activated and viral capsid and exit facilitator proteins are expressed, which are necessary to generate a productive infection.
Some PyVs exhibit transformation potential only when expressed in a non-native host system (SV40, JCV, BKV, African green monkey PyV), while others induce tumor formation in the natural host (Merkel cell PyV (10, 11), murine PyV, and hamster PyV) (12). Transformation normally requires the expression of the LT, except for murine PyV in which the middle T antigen is essential (13). The current model of polyomavirus LT induced transformation of cells is based on studies with SV40 LT and has been reviewed extensively (5, 13, 14). Mutational analysis of SV40 LT has shown that the elements required for transformation include the J domain, LxCxE motif, and ATPase domain (see Fig 1A for domain structure of LT). The LxCxE motif and J domain are required, respectively, to bind cellular pRB proteins and release E2F transcription factors from an inhibitory complex with pRBs. Once E2Fs are free, they trigger expression of genes involved in cell cycling and proliferation. Both the ectopic proliferation and DNA damage induced directly by SV40 LT activate p53, a master regulator of cell-cycle checkpoints and inducer of cell death. Surface residues on the ATPase domain of SV40 LT bind and block the p53 DNA-binding domain, preventing transcriptional activation of p53-responsive genes and resulting in unchecked growth and transformation in SV40 LT-expressing cells.
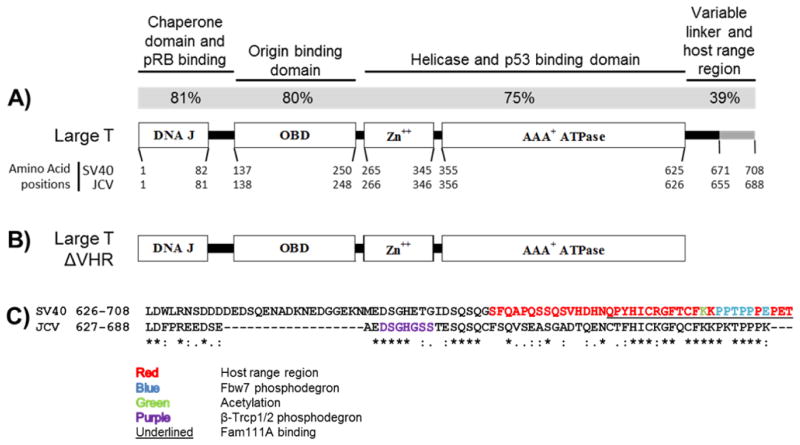
Figure 1. Structure and similarity of large T antigens.
A) Structure of the large T antigen protein. Functional domains (rectangles) within large T are depicted. The functions and percent amino acid identity (gray bar) of each domain within SV40 and JCV large T are indicated above the diagram. At the N-terminus are the DNA J domain and linker region, which are both required to fully inhibit pRB activity. The origin binding domain is required for the viral genome replication. The zinc binding domain and the ATPase domain together contribute to the helicase activity of large T. Cellular p53 binds large T through surface residues of the ATPase domain. The variable linker region and host range region are poorly characterized in comparison to the other regions of large T antigen. B) Structure of the large T antigen expressed from ERΔVHR constructs. The variable linker and host range region was removed from each TAg gene by PCR. These constructs express amino acids 1–625 of SV40 large T or amino acids 1–626 of JCV large T as well as other T antigen proteins (discussed in the Results). C) Protein sequence comparison of SV40 and JCV VHR regions. An alignment of amino acids was performed with LALIGN. The host range region of SV40 is highlighted in red letters, and was used to define the boundaries of the JCV host range region. Previously described motifs and post-translational modification sites are shown (described in the Discussion).
JCV and SV40 have been shown to cause tumors in rodent animal models and transformation of cells in culture, and both LTs are necessary and sometimes sufficient to induce tumorigenesis by targeting both pRB and p53 (14, 15). In spite of this similarity, SV40 T antigen is more efficient than JCV T antigen in side-by-side comparisons of transformation ability (16–20). The reason for this difference has yet to be fully defined. A comparison of the amino acid sequences reveals that JCV LT is about 70% identical to SV40 LT, and that they are up to 80% identical in specific shared functional domains (Figure 1A). Interestingly, the short C-terminal section immediately following the ATPase domain, known in SV40 LT as the variable linker and host range region (VHR), shows the least similarity between both LTs (Fig. 1A,C). We hypothesized that JCV LT VHR does not have the same biological functions as SV40 LT VHR due to structural disparity. We describe here the effects on cellular transformation produced by removing the VHR from JCV and SV40 T antigens.
Results
Establishment of a system to compare the early regions of SV40 and JCV
We cloned full length (SV40.ER and JCV.ER) and VHR-truncated (SV40.ERΔVHR or JCV.ERΔVHR) early regions for both SV40 and JCV into the same expression vector. All four constructs were introduced into murine embryonic fibroblasts (MEFs), and pools of cells that survived selection were collected. We compared the ability of VHR truncated early regions to transform versus the full length counterparts. Three individual pools of cells were generated and tested for each construct to ensure that the observed phenotypes within an individual pool were not random.
The early regions of both SV40 and JCV are capable of producing multiple protein products through alternative splicing mechanisms (Fig. 2A) (21, 22). Truncation of the JCV early region removes the 3′ splice acceptor sites for the T′165 and T′136 introns, ultimately preventing the corresponding proteins from being made. In addition, deletion of the VHR shortens the LT proteins to 625 and 626 aa in SV40 and JCV, respectively (Fig. 1B), causing a shift in apparent molecular size from ~94 to ~70 kD (Fig. 2B). Table 1 lists the expected protein products from each construct.
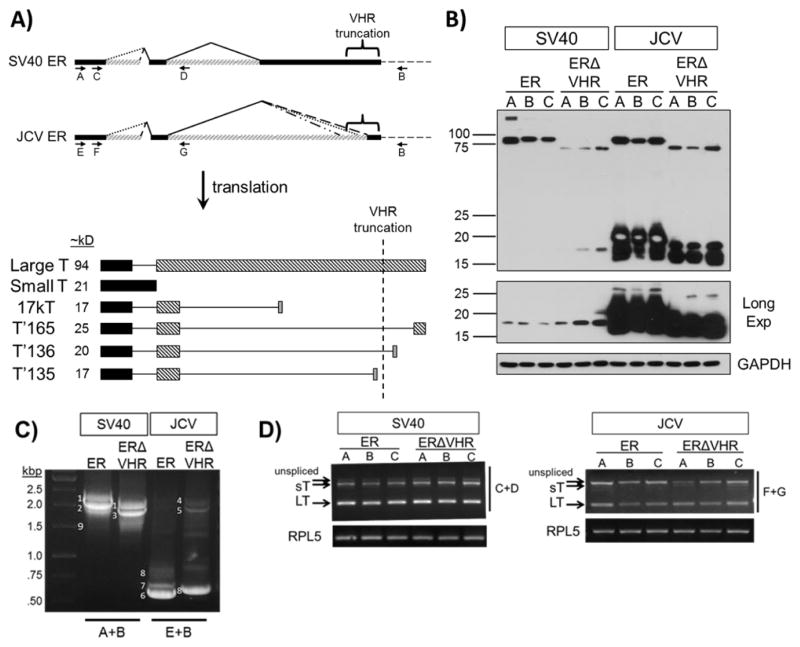
Figure 2. Expression of T antigens from full length or VHR-truncated SV40 and JCV early region in MEFs.
A) Transcripts and proteins encoded by the SV40 and JCV viral early region (ER). Both the SV40 and JCV ER produce multiple T antigen transcripts by alternative splicing (top panel). Translation of these transcripts results in a large T (LT), small T (sT), and one or more smaller T proteins (lower panel). Bars of the same color or pattern represent an expressed region of the protein within the same reading frame, while continuous lines represent regions that are removed due to alternative splicing of the early region pre-mRNA. Approximate sizes of the different T antigen proteins are listed. Removal of the VHR region in JCV prevents the expression of T′165 and T′136. Both SV40 and JCV VHR truncations produce a smaller LT protein. B) T antigen protein expression in MEFs from full length or VHR-truncated SV40 and JCV ER. Three individual pools are shown (labeled A, B, and C) for each construct. Equivalent amounts of whole cell lysates were separated by SDS-PAGE, transferred to PVDF, and blotted for T antigens with a mixture of SV40 (416, 419) and JCV specific antibodies (962, 2003). GAPDH, loading control. C) T antigen transcripts amplified by RT-PCR using primers that surround the entire open reading frame (A and B or E and B, indicated by arrows in panel A). Each band corresponds to a different T antigen transcript: 1) SV40 sT, 2) SV40 LT, 3) SV40 LT ΔVHR, 4) JCV sT, 5) JCV LT ΔVHR, 6) JCV T′165, 7) JCV T′136, 8) JCV T′135 9) SV40 17kT. D) RT-PCR specific for LT and sT transcripts (using primers C and D or F and G, indicated by the arrows in panel A). Rpl5 transcript levels were tested to verify that equal amounts of total cDNA were used for each sample.
Table 1. Protein products potentially expressed from early region constructs.
Possible protein products expressed from both SV40.ER and SV40.ERΔVHR constructs are equivalent. Truncation of the VHR in JCV.ER removes two 3′ splice sites, which prevents expression of the T′165 and T′136 proteins.
| Expected Protein Products | |
| SV40.ER | SV40.ERΔVHR |
| Large T | Large T ΔVHR |
| small T | small T |
| 17kT | 17kT |
| JCV.ER | JCV.ERΔVHR |
| Large T | Large T ΔVHR |
| small T | small T |
| T′165 T′136 T′135 |
T′135 |
Removal of the VHR alters the pattern of SV40 and JCV early region protein expression
We first determined the protein and messenger RNA expression of T antigen products in each pool of cells by western blot and RT-PCR, respectively. In SV40.ER expressing cells LT was the dominant product identified at both the protein and mRNA levels (Fig. 2B, lanes 1–3; Fig. 2C and D, SV40.ER lanes). Small T protein was only detected after a long exposure (Fig. 2B lanes 1–3), and the level of sT mRNA was lower than LT mRNA (Fig. 2D, SV40.ER lanes). LT protein expression was reduced when the VHR was removed from the SV40 early region, while sT protein expression increased (Fig. 2B lanes 4–6). The altered levels of LT and sT protein could not be attributed to changes in mRNA transcripts since there was no detectable difference in LT or sT transcripts between cell pools expressing SV40.ER and SV40.ERΔVHR (Fig. 2C and D, compare SV40.ER and SV40.ERΔVHR lanes). In preliminary experiments, we observed only a slight decrease in the half-life between full length and VHR truncated- LT protein (data not shown). This suggests that the stability of the protein was not significantly altered and should not account for the drastic decrease of SV40 LT protein in SV40.ERΔVHR cells.
We were unable to detect the SV40 17kT protein. This is not surprising, since the level of 17kT transcript was either extremely low or undetected in multiple experiments performed with all SV40-expressing pools (Fig. 2C and data not shown).
Unlike with SV40, LT was not the dominant protein seen in cells expressing the full length JCV early region. Rather, smaller T antigen proteins (sT, T′135, T′136, T′165) were expressed in the highest abundance (Fig. 2B, lanes 7–9), and this was reflected in the mRNA expression. The T′ transcripts were so abundant that we could not detect LT or sT transcripts using a set of primers that amplify all T antigen transcripts (Fig. 2A, primers E and B; Fig. 2C, JCV ER lane). The inability to detect JCV LT and sT transcript was mostly likely because the T′ transcripts absorbed most of the primers during PCR amplification. To address this issue, a set of primers designed to amplify only LT, sT, or unspliced transcripts was used for RT-PCR (Fig. 2A, primers F and G), and both LT and sT transcripts were detected in the JCV.ER samples (Fig. 2D, JCV ER lanes). Removal of the VHR from JCV early region resulted in a decrease in LT protein (Fig. 2B, lanes 10–12) but, there was no significant change in the level of LT mRNA (Fig. 2D, compare JCV.ER to JCV.ERΔVHR lanes). Similar to JC.ER pools, the total level of smaller T antigens was higher than LT in JCV.ERΔVHR-expressing cells for both protein and mRNA (Fig 2B, lanes 10–12; Fig 2C, JCV ERΔVHR lane).
The identity of smaller T antigen proteins expressed from JCV.ER and JCV.ERΔVHR was difficult to discern by migration in SDS-PAGE. Instead, cDNA size analysis and sequencing revealed that JCV.ER pools expressed sT and all three T′ transcripts, while JCV.ERΔVHR pools produced only sT and T′135 transcripts due to the deletion (Fig. 2C,D and data not shown).
Under our experimental conditions, the level of LT protein was similar between the SV40 and JCV ER-expressing cell pools (Fig. 2B, compare lanes 1–3 with lanes 7–9). However, expression of the VHR truncated JCV LT appeared to be higher than that observed for SV40 LTΔVHR in the corresponding pools (Fig. 2B, compare lanes 4–6 with lanes 10–12), and the expression of both truncated LT proteins was comparatively reduced to that of their full length counterparts (Fig. 2B).
Growth in low serum conferred by SV40 is hindered by removal of the VHR
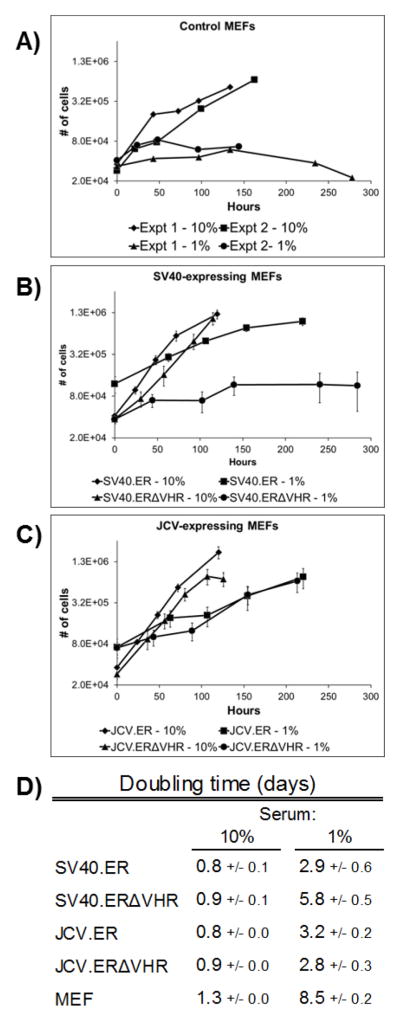
MEFs respond to a reduction in serum levels in culture medium by ceasing proliferation and eventually dying (Fig. 3A,D and data not shown), but unrestricted growth in normal (10%) or low (1%) serum conditions takes place in the presence of some oncogenes, including SV40 LT. All T antigen-expressing MEFs grew similarly in 10% serum (Fig. 3B,C,D), and the expression of either SV40.ER or JCV.ER allowed proliferation of MEFs at a similar rate in 1% serum (Fig. 3B,C,D). Truncation of the VHR from the JCV early region did not significantly alter the rate of cell growth in low serum (Fig. 3C,D). On the other hand, SV40.ERΔVHR was unable to allow cell growth in low serum. These cells doubled only once during the low serum treatment, similar to wild-type MEFs, whereas all other T antigen-expressing cells doubled at least twice while cultured in low serum for the same amount of time (Fig. 3B,C). Furthermore, the doubling time for the SV40.ERΔVHR MEFs was greater than that of all of the other T antigen-expressing cells (Fig 3D). We did not find any difference in the level of large T antigen expressed in cells cultured in low serum compared to those in normal serum for either SV40.FL or SV40.ERΔVHR cells (data not shown).
Figure 3. SV40.ERΔVHR-expressing MEFs show limited growth in reduced serum conditions.
Cells were grown in medium containing either 10% or 1% serum for up to 12 days. Duplicate wells were counted at regular intervals and averaged. Two experiments performed with wild-type MEFs are shown separately (A). For SV40 (B) and JCV samples (C), each point on the graph represents the average number of cells from 3 different cell pools expressing the same early region construct. Error bars denote the standard error of the mean. The doubling time was calculated from time points within the exponential phase of growth and the results are summarized in (D). Each value represents the average doubling time from 3 cell pools expressing the same early region construct. MEF values are the average of 2 independent experiments. The standard error of the mean is indicated.
Truncation of the JCV VHR region enhances anchorage independent transformation
Both SV40 and JCV early regions have independently been shown to induce anchorage independent growth. We analyzed the ability of full length and VHR-truncated viral early regions to induce this phenotype in MEFs. As expected, control MEF cells were unable to form a significant number of colonies when grown in agar suspension, but expression of all early region constructs resulted in formation of many colonies (Fig. 4A). In addition, the size of colonies formed in all early region expressing pools was significantly larger than colonies formed by control MEFs (Fig. 4B,C).
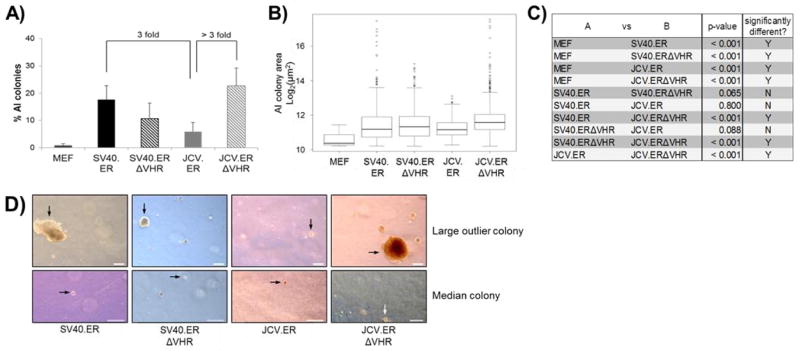
Figure 4. Removal of the VHR in JCV TAg increases anchorage independent colony formation.
Cells expressing different T antigens were suspended in agar medium, allowed to grow for 21 days, and all single cell and multi-cell events were counted and measured as described in methods. A) The number of colonies was divided by the total number of events to produce the percent of AI colonies per experiment. The average percent of AI colony formation for at least two pools expressing the same T antigen is shown (see text). Error bars represent 2 times the standard error of the mean. B) The sizes of AI colonies from all experiments of the indicated cell type were pooled and are shown in box-and-whisker format. The horizontal black line within the box represents the median colony size for each population. Extremely large individual outlier colonies are shown as open circles outside of the whiskers. C) All possible pairs of cell populations from (B) were tested to determine if the distribution of AI colony sizes between the two cell types was significantly different. The p-value from each test is displayed in the table. P-values less than 0.05 were considered significant. D) An image of the largest colony and a median-sized colony for each TAg-expressing population is presented. All images were taken at the same magnification. Images of median sized colonies were digitally magnified to better visualize the colony. Scale bar indicates 200 μm.
We found that SV40.ER pools generated three times more colonies than JCV.ER pools (Fig. 4A), and also showed a higher frequency of extremely large outlier colonies (Fig. 4B). However, the overall distribution of colony sizes was not significantly different between cells expressing these two constructs (Fig. 4C). This is consistent with previous reports using anchorage independence assays to compare SV40 and JCV T antigen transformation properties (16, 17, 19).
MEFs expressing the SV40 early region with or without the VHR showed similar numbers and sizes of anchorage independent colonies (Fig. 4A,B,C). On the other hand, removal of the VHR from JCV early region induced an increase in the number of colonies by more than 3 fold compared to cells expressing full length JCV early region (Fig 4A). Furthermore, the size for JCV.ERΔVHR colonies was significantly larger than that observed for JCV.ER colonies (Fig. 4B,C,D). This was reflected in both an overall increase in the number of large-sized colonies and in the high frequency of extremely large outlier colonies found in JCV.ERΔVHR cell pools (Fig. 4B).
The VHR region is not required to interfere with the pRB pathway
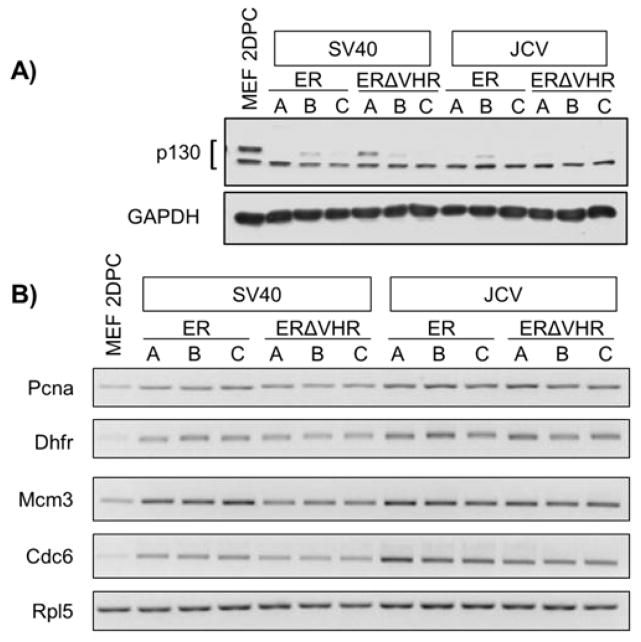
We assessed the ability of ERΔVHR products to perturb the pRB and p53 pathways, as both must be inhibited for full LT-mediated transformation to occur. Two markers were used to evaluate the integrity of the pRB pathway. First, western blot analysis was employed to assess the steady-state levels of the cellular protein p130, a member of the pRB family of proteins, which is reduced upon binding to SV40 LT (23–25). Levels of p130 in all ER- and ERΔVHR-expressing MEFs were decreased compared to control MEFs (Fig. 5A). Next, we examined the expression levels of E2F target genes, which are increased as a downstream effect of LT inhibition of pRB proteins (26). RT-PCR showed an increase in the level of transcript of a set of canonical E2F-dependent genes in all cells expressing ER or ERΔVHR constructs when compared with non-proliferating MEFs (Fig. 5B). These combined results suggest that the pRB pathway is effectively inhibited in all SV40 and JCV ER-expressing cells, irrespective of VHR truncation or level of LT produced.
Figure 5. Removal of the VHR does not alter the ability of T antigens to disrupt the pRB pathway.
A) Steady state levels of p130 decrease in MEFs upon expression of either ER or ERΔVHR constructs, as monitored by Western blot. GAPDH was used as a loading control. B) E2F-target gene expression is increased in all T antigen-expressing MEFs. The expression of four E2F-regulated genes was assessed by RT-PCR of RNA from 2 days post-confluent cells. Pcna: proliferating cell nuclear antigen; Dhfr: dihydrofolate reductase; Mcm3: minichromosome maintenance complex component 3; Cdc6: cell division cycle 6. Rpl5 is an endogenous control.
p53 is bound and stabilized in all T antigen expressing MEFs
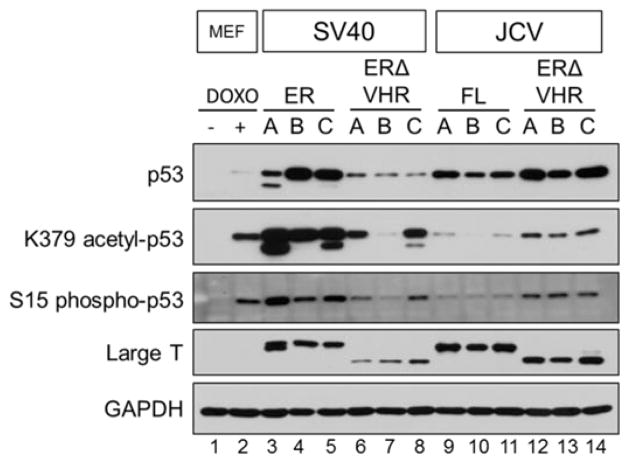
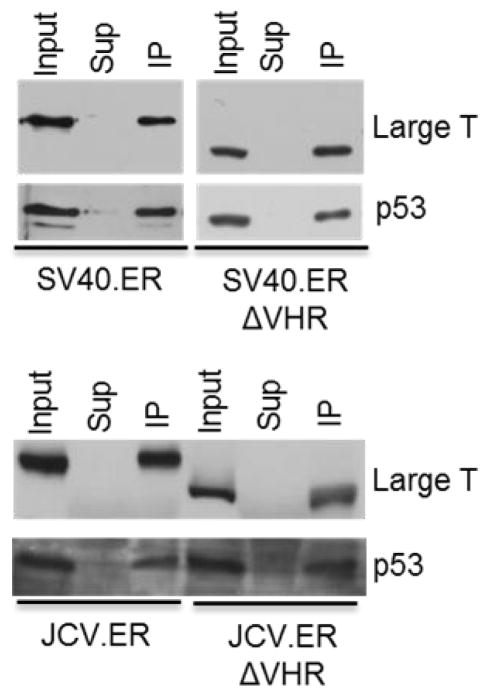
Inactivation of the p53 pathway by LT is critical to induce most transformation phenotypes. One of the consequences of LT expression is the accumulation of p53 protein to very high levels in the cell. However, this p53 product is not functional, as LT binds the DNA binding domain of p53 and so prevents it from activating genes whose expression would lead to cell death (27–29). All ER- and ERΔVHR-expressing MEFs showed increased levels of p53 when compared to wild-type MEFs and MEFs treated with doxorubicin, a DNA damaging agent and inducer of p53 (Fig. 6) (30, 31). Although lower levels of total p53 were observed in cells expressing SV40.ERΔVHR when compared with SV40.ER pools, this decrease is consistent with the corresponding decrease in the amount of LT (Fig. 6, compare lanes 3–5 with 6–8). In contrast, there was an increase in total p53 levels in JCV.ERΔVHR pools in comparison to JCV.ER pools, but in this case the LT expression levels remained similar or were slightly decreased when the VHR truncated LT was expressed (Fig. 6, compare lanes 9–11 to 12–14). Despite the observed differences regarding total p53 levels attained in these cells, the ability of large T antigen to bind p53 – with or without the VHR – was conserved in both SV40 and JCV as shown by immunoprecipitation (Fig. 7).
Figure 6. p53 is stabilized and shows different levels of activation in full length and VHR-truncated early region expressing MEFs.
Steady-state levels of total and post-translationally modified p53 protein were detected by western blot in T antigen expressing cells. Large T antigen levels were detected with a combination of SV40- and JCV-specific antibodies (see Methods) and are included for comparison. MEFs treated with doxorubicin were used as a positive control for p53 stabilization and activation. GAPDH, loading control.
Figure 7. VHR truncated large T antigens retain the ability to bind p53.
Protein extracts were immunoprecipitated with a p53-specific antibody (PAb 421, SV40.ER and SV40.ERΔVHR samples) or a T antigen specific antibody (PAb 416, JCV.ER and JCV.ERΔVHR samples). Co-precipitated proteins were resolved by SDS-PAGE. The presence of large T and p53 was monitored by western blot.
In cells transformed by SV40 LT, the p53 protein bound by the large T antigen shows post-translational markers of activation (32). We assessed the activation state of p53 in cells expressing the SV40 or JCV ER and ERΔVHR by examining K379-acetylation and S15-phosphorylation by western blot. Control MEFs were treated with doxorubicin as a positive control for p53 activation. Doxorubicin-treated cells showed a significant increase in both post-translational modifications of p53 when compared to untreated MEFs (Fig. 6, compare lanes 1 and 2). SV40.ER pools produced the highest levels of acetylated and phosphorylated p53 (Fig. 6, lanes 3–5). Similar to the total levels of p53, the amount of post-translationally modified p53 was decreased when the VHR was removed from SV40 (Fig 6, lanes 6–8). JCV.ER-expressing cells displayed a very low level of K379 acetylation and S15 phosphorylation of p53 (Fig. 6, lanes 9–11). Cells expressing the VHR truncated JCV early region showed increased levels of post-translationally modified p53 compared to full length JCV early region pools (Fig. 6, compare lanes 9–11 and 12–14), although these levels did not reach those observed in SV40.ER pools (Fig. 6, compare lanes 12–14 and 3–5). At present, it is unclear if variations in the p53 levels and modifications have a role in tumor induction by T antigens.
Discussion
Previous studies have indicated that the SV40 LT VHR contributes to viral productive infection by allowing viral growth in different host cells (33–35), and is required to permit growth of adenovirus in normally restrictive cells (36). While early studies suggested that the SV40 VHR is not required for immortalization or dense focus formation in rodent cells (37, 38), it remained to be seen whether this region contributes to other transformation phenotypes. SA12, BKV and JCV polyomavirus LTs also have a VHR, but their roles in viral infection or transformation have not been well studied. Though these 3 VHR sequences are highly conserved, there is little conservation with the SV40 VHR except for a stretch of 18 amino acids within the host range region. Within this homologous sequence there are conserved residues that have been shown in SV40 to be acetylated and that bind cellular proteins (Fig. 1C) (39–42). Little examination of the JCV LT VHR region has been performed. Only one report has identified a cellular protein, the SCF ubiquitin ligase component β-TrCP1/2 (43), which binds to the variable linker of JCV LT, and the binding motif involved is fully conserved in BKV and SA12. None of these interactions has been tested for the ability to affect transformation.
In order to explore contributions of the VHR to transformation, we examined the ability of SV40 and JCV wild-type LT and LT mutants lacking the VHR to alter the growth properties of primary MEFs. We took advantage of the co-linearity of SV40 and JCV LT amino acid sequences to design VHR truncations that would not interfere with the structure of other LT domains. The full length and truncated early regions were cloned into the same vector and expressed from the CMV promoter. T antigen-expressing MEFs were then characterized for the ability to grow in low serum and to form anchorage independent colonies.
Whereas normal MEFs proliferated slowly in 10% serum, were growth arrested in 1% serum, and could not form anchorage independent colonies, MEFs expressing the full length SV40 ER grew well at both serum concentrations and formed a significant number of AI colonies. Surprisingly, we found that cells expressing the SV40 VHR truncation failed to grow in 1% serum, but showed no change in the ability to induce anchorage independent transformation. Some reports suggest that the degree of transformation by LT is dependent on the level of LT expression (19, 44, 45), and we observed a clear decrease in LT expression from the VHR-truncated SV40 ER, suggesting that this may contribute to the observed decrease of growth in low serum. Alternatively, the VHR of LT could contribute directly to growth in low serum without affecting anchorage independence. Finally, the SV40 VHR mutant did not disrupt the interaction of LT with Rb proteins or binding to p53, which are required for LT to transform. However, it is possible that the VHR truncation alters the effect of LT on Rb or p53 in some subtle way that our tests did not detect.
In contrast to the SV40 VHR truncation, growth in low serum was not affected by the removal of the JCV VHR region. Both mutant and full length JCV early regions grew at similar rates in 10% and 1% serum. Unexpectedly, VHR truncation of the JCV ER greatly enhanced the number and size of anchorage independent colonies compared to the full length JCV ER. JCV LT binding to the pRB and p53 proteins was not affected by removal of the VHR, indicating that the VHR mutant did not change one of these fundamental pathways required for transformation. Like SV40, LT expression decreased when JCV VHR was removed, but, unlike SV40, there was an increase of transformation, which challenges previous reports linking higher LT levels to increased transformation. However, due to the multiple T antigen proteins produced by the JCV early region constructs, it remains to be determined if the altered pattern of expression of JCV T antigen proteins, the removal of the LT VHR region itself, or a combination of the two is responsible for the increased capacity to induce anchorage independent transformation. For example, we show that the JCV T′165 protein is expressed at high levels in JCV.ER cells and is absent in JCV.ERΔVHR cells. If T′165 is growth inhibitory, then this could explain the increase in anchorage independent growth seen in JCV.ERΔVHR cells. More experiments are needed to address this possibility as well as others.
Unfortunately, our attempts to express either the SV40 or JCV VHR region alone or in combination with full length or truncated early regions in MEFs were not successful. Also, efforts to express and test individual JCV T antigens were complicated by the limited life span of MEFs. As such, we were unable to pursue complementation studies or independent characterization of the JCV T antigen proteins or of the VHR alone.
Another possible factor contributing to the altered transformation induced by the SV40 and JCV truncated ERs is the level of p53 in the cells. For both truncated LTs, although the ability to bind p53 was not affected, the level of total, K379 acetylated, and S15 phosphorylated p53 changed depending on whether or not the VHR was present. The changes in total and activated p53 levels correlated with the observed changes in transformation; growth in low serum was prevented and p53 levels were decreased when the VHR was deleted from the SV40 early region, and removal of the VHR from JCV early region resulted in increased anchorage independent transformation and increased p53 levels. It is unclear how the level of p53 in the cells would contribute to the ability to transform, since the interaction of LT with p53 is known to confer a loss of p53 function. Our results suggest that p53 could be acquiring a gain of function activity when in a complex with LT, thus enhancing transformation, but further experiments are needed to test this hypothesis.
In summary, we have reported the first evidence linking the VHR region to transformation by any polyomavirus T antigen. Overall, our results indicate that the VHR from each virus early region confers different functional capabilities in terms of transformation, and that different transformation phenotypes are probably induced by distinct molecular mechanisms. Given the results in this report, it is possible that VHR or C-terminal tail regions found in polyomaviruses other than SV40 may contribute to transformation in different ways than SV40 LT.
Methods
Plasmids
The early regions from SV40 strain 776 (GenBank accession NC_001669) and JCV Mad-1 strain (GenBank accession NC_001699; received from Richard Frisque) were PCR amplified using specific primers (Table 2) and cloned into the pLenti6.3-TOPO vector (Invitrogen) following the manufacturer’s protocol. The full length early region includes nucleotides 2691–5163 of SV40 and 2603–5013 of JCV, while the VHR-truncated early regions contain nucleotides 2943–5163 of SV40 and 2792–5013 of JCV. An artificial Kozak sequence (ACC) was included before the start codon of all constructs to ensure efficient translation. The truncated early region constructs required the addition of a stop codon immediately after the early region sequence. Both insertions were added during PCR amplification of the viral early regions.
Table 2.
Primers used in the experiments described in this manuscript.
| Primers for Cloning | ||
| SV40.ER | F | ACCATGGATAAAGTTTTAAACAGAGA |
| R | TTATGTTTCAGGTTCAGGGG | |
| SV40.ERΔVHR | F | ACCATGGATAAAGTTTTAAACAGAGAGG |
| R | CTAAACTCCAATTCCCATAGC | |
| JCV.ER | F | ACCATGGACAAAGTGCTGAATAGGG |
| R | TTATTTTGGGGGAGGGGTCT | |
| JCV.ERΔVHR | F | ACCATGGACAAAGTGCTGAATAGGG |
| R | CTAAATGGGTCTCCCCATACCA | |
| Primers for RT-PCR | ||
| T Antigen | A | ACCATGGATAAAGTTTTAAACAGAGAGG |
| B | ACCGAGGAGAGGGTTAGGGAT | |
| C | CTGACTTTGGAGGCTTCTGG | |
| D | AAGGAAAGTCCTTGGGGTCT | |
| E | ACCATGGACAAAGTGCTGAATAGGG | |
| F | GCCTGATTTTGGTACATGGAA | |
| G | ACAGCTTGACTGAGGAATGC | |
| Pcna | F | AGGCTCTCAAAGACCTCATCAATG |
| R | CCTGTTCTGGGATTCCAAGTTG | |
| Dhfr | F | GTAGAGAACTCAAAGAACCACCACG |
| R | TTTTCCTCCTGGACCTCAGAGAG | |
| Mcm | F | CGCAGGAAGAATGAAAAGAGGG |
| R | CTGAGGAAGCAGGAAGTGAGAGTC | |
| Cdc6 | F | AGTTCTGTGCCCGCAAAGTG |
| R | AGCAGCAAAGAGCAAACCAGG | |
| Rpl5 | F | CCAAACGATTCCCTGGTTATGAC |
| R | GACGATTCCACCTCTTCTTCTTCAC | |
Cell culture and transfection
Harvest of murine embryonic fibroblasts from FVB mice was performed as previously reported (46). MEFs were maintained in DMEM medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. Where indicated, confluent MEFs were treated with a final concentration of 1 μg/ml of doxorubicin (Adriamycin, Sigma) in growth medium for 9 hours. The cells were then trypsinized and harvested for RNA or protein extraction as described below.
Plasmids were transfected into MEFs in 6-well cell culture plates using Fugene6 (Promega) following the manufacturer’s protocol with minimal modifications. A ratio of 1 μg plasmid to 3 μl Fugene was used. Forty-eight hours after transfection, cells were transferred to 10-cm tissue culture dishes and allowed to recover overnight before starting selection with 2 μg/ml blasticidin (Invitrogen). Cells were kept under selection until all mock-transfected MEFs died. All surviving cells from an individual transfection were pooled, and three independent pools were generated for each plasmid construct. All experiments were performed with cells between passages 3 and 10.
Western blot and immunoprecipitation
All cells were collected at 2 days post confluence and lysed with either RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Na Deoxycholate, 1% Triton X-100, 1 mM EDTA) for western blot analysis, or buffer containing 50 mM Hepes pH 7.9; 0.4M KCl; 0.5 mM EDTA; 0.1% NP40 and 10% glycerol for immunoprecipitation assays. Lysis and subsequent procedures were performed in the presence of protease and phosphatase inhibitors (1X Calbiochem protease inhibitor cocktail I, 1 mM pepstatin, 10 μM leupeptin, 1 mM Na3VO4, 1 mM NaF). Clarified lysates were collected and protein concentration was assessed by the Bio-Rad protein microassay.
For western blots, equal amounts of total protein were mixed with Laemmli buffer and heated at 95°C for 5 minutes to denature proteins. We routinely separated 15–30 μg of denatured protein by SDS-PAGE, then transferred proteins to PVDF, and performed western blot with indicated antibodies. To detect the different T antigen proteins a cocktail of 4 antibodies was used: anti-JCV T antigen specific monoclonals PAb 962 (47) and PAb 2003 (48) (both received from Richard Frisque) and anti-SV40 T antigen monoclonal antibodies PAb 416 and PAb 419 (49). The epitopes of JCV antibodies PAb 962 and 2003 are within amino acids 1–81, and they recognize all of the known JCV T antigen proteins. PAb 962 can recognize SV40 LT but not ST, while PAb 2003 does not recognize LT or ST of SV40. The anti-SV40 antibody PAb 416 can detect SV40 LT and 17kT, but not sT. PAb 419 recognizes LT, sT, and 17kT of SV40. This reactivity information is described in Table 1 of reference 48. The following antibodies were used to detect cellular proteins: p130 C-20 (SantaCruz sc-317x), p53 (PAb 421(49)), S15 phospho-p53 (Cell Signaling 9284S), K379 acetyl-p53 (Cell Signaling 2570S), GAPDH (US Biological G8140-11).
For immunoprecipitation assays, 25 μl of Protein G Dynabeads magnetic particles (Life Sciences) were first incubated for an hour at 4°C with 50 μl of anti-p53 PAb421 hybridoma supernatant, supplemented with PBS to achieve a final volume of 250 μl. Equivalent amounts of total protein extracts (50–100 μg) were then added to the different reactions and allowed to mix for 4 hours at 4°C. Pellets containing the immunoprecipitated complexes were retrieved with a magnetic stand and washed four times at 4°C with PBS plus protease inhibitors. Protein complexes were eluted from the magnetic beads by incubation with 20 μl 0.1 M glycine pH 2.8 for 10 min. at room temperature, and were subsequently neutralized with 2 μl 1M Tris-HCl pH 8.0 and prepared for SDS-PAGE electrophoresis.
Growth assays in variable serum conditions
3×104 cells were seeded onto 6-well tissue culture plates in DMEM + 10% FBS. Each experiment was performed in duplicate. For the low serum assay, the medium was replaced six hours later with DMEM + 1% FBS, after washing the cells once with PBS. Cells for both low and normal (10%) serum assays were incubated overnight before counting the first time point. This was considered time zero. Cells from two wells were counted with a hemacytometer and averaged every day (normal growth) or every other day (low serum growth) for up to 12 days. The average number of cells counted (y-axis) at each time point (x-axis) was plotted on a graph and the doubling time was calculated using two time points within the exponential growth phase according to the equation:
where T is the incubation time in hours, X2 is the number of cells from the later time point, and X1 is the number of cells from the earlier time point. Doubling time was divided by 24 to convert the value into doublings per day. Each pool expressing the same early region construct was considered a biological replicate. Wild-type MEFs were independently tested twice, constituting technical replicates. The doubling times from all replicates were averaged and the standard deviation was calculated using the Excel function “STDEV.S”. The standard error of the mean was derived by dividing the standard deviation by the square root of the number of replicates.
Anchorage independence assay
1.6×104 cells were suspended in 0.3% agarose (Invitrogen Ultrapure) in growth medium on top of a 0.5% agarose medium base layer in 35-mm tissue culture dishes, as previously reported (50). All experiments were performed in duplicate. Colony formation was observed after 21 days. The area of all single and multi- cell events was measured. The area cut-off value chosen (1165 μm2) to define a colony allowed for 1% or less anchorage independent (AI) colonies in the control MEF populations. The number of colonies from each plate was divided by the total number of cellular events to obtain the percentage of anchorage independent colonies formed. Each pool was tested twice and the percentages of AI colonies from both assays were averaged. Pools expressing the same early region construct were considered biological replicates and the percentages of AI colonies from all three pools were averaged. One of the JCV.ER-expressing pools (pool C) was deemed an outlier as it did not accurately represent the behavior of JCV early region as seen in pools A and B, or as previously reported (16, 17, 19). It was, therefore, excluded from the analysis and the percentages of AI colonies from the remaining two JCV.ER-expressing pools were averaged. The standard error of the mean was calculated as for the low serum growth assay.
A comparison of the size of colonies was performed by pooling the AI colonies from assays of the same cell type, then forming a box and whisker plot using the R statistical software package. The Mann-Whitney-Wilcoxon test for independent samples was applied to each possible pair of cell types also using the R software. The null hypothesis that the distribution of the two populations is the same was rejected if the p-value was less than 0.05.
Reverse transcription-PCR
Standard techniques were used to extract total RNA including the on-column DNA digestion, and to synthesize cDNA (Qiagen RNeasy kit, Qiagen RNAse-free DNase set (51)). Primers used for all RT-PCRs are listed in Table 2. Amplification of T antigen specific cDNAs using primers (A and B) and (E and B) from 1 μg of total cDNA was performed with the Roche Expand Long Template PCR system, using buffer 1 with the following cycling parameters: 94°C for 3 min, then 30 cycles of 94°C 30 sec, 54°C (A+B) or 55°C (E+B) for 30 sec, 68°C 3 min, a final extension was performed at 68°C for 10 min. Specific E2F-target gene cDNAs or T antigen specific cDNAs (primers C and D or F and G) were amplified from equal amounts of total cellular cDNA by PCR using GoTaq master mix (Promega) and the corresponding primers as previously reported (52). Annealing temperatures were as follows: Pcna 55°C, Dhfr 55°C, Mcm3 55°C, Cdc6 55°C, Rpl5 56°C, T antigen (C and D) and (F and G) 55°C. Primers used are listed in Table 2. PCR products were separated on a 2% agarose gel in 1x Tris-acetate-EDTA and stained with ethidium bromide. Images were captured with a Fujifilm LAS-3000 imager.
Highlights.
The variable linker and host range region was removed from SV40 and JCV T antigens.
Expression of T antigen proteins is altered in cells with truncated constructs.
Truncated SV40 prevents robust growth of cells in low serum.
Truncated JCV increases size and number of anchorage independent colonies.
This is the first report of the T antigen VHR region affecting transformation.
Acknowledgments
This work was supported by NIH grant CA98956 to J.M.P. NIH Institutional Training grant T32AI049820 supported N.T.M.S. We would like to thank Richard Frisque for invaluable JCV reagents, as well as thoughtful discussion and suggestions. We thank Paul Cantalupo, Han Na Choi, and Chikdu Shivalila for expert technical help.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hirsch HH, Kardas P, Kranz D, Leboeuf C. The human JC polyomavirus (JCPyV): virological background and clinical implications. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2013;121:685–727. doi: 10.1111/apm.12128. [DOI] [PubMed] [Google Scholar]
- 2.Pipas JM. Common and Unique Features of T Antigens Encoded by the Polyomavirus Group. Journal of virology. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eddy BE, Borman GS, Berkeley WH, Young RD. Tumors induced in hamsters by injection of rhesus monkey kidney cell extracts. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine; New York, NY. 1961. pp. 191–197. [DOI] [PubMed] [Google Scholar]
- 4.Howley PM, Livingston DM. Small DNA tumor viruses: large contributors to biomedical sciences. Virology. 2009;384:256–259. doi: 10.1016/j.virol.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An P, Saenz Robles MT, Pipas JM. Large T antigens of polyomaviruses: amazing molecular machines. Annual review of microbiology. 2012;66:213–236. doi: 10.1146/annurev-micro-092611-150154. [DOI] [PubMed] [Google Scholar]
- 6.Topalis D, Andrei G, Snoeck R. The large tumor antigen: a “Swiss Army knife” protein possessing the functions required for the polyomavirus life cycle. Antiviral research. 2013;97:122–136. doi: 10.1016/j.antiviral.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Khalili K, Sariyer IK, Safak M. Small tumor antigen of polyomaviruses: role in viral life cycle and cell transformation. Journal of cellular physiology. 2008;215:309–319. doi: 10.1002/jcp.21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollag B, Hofstetter CA, Reviriego-Mendoza MM, Frisque RJ. JC virus small T antigen binds phosphatase PP2A and Rb family proteins and is required for efficient viral DNA replication activity. PloS one. 2010;5:e10606. doi: 10.1371/journal.pone.0010606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prins C, Frisque RJ. JC Virus T′ proteins encoded by alternatively spliced early mRNAs enhance T antigen-mediated viral DNA replication in human cells. Journal of neurovirology. 2001;7:250–264. doi: 10.1080/13550280152403290. [DOI] [PubMed] [Google Scholar]
- 10.Arora R, Chang Y, Moore PS. MCV and Merkel cell carcinoma: a molecular success story. Current opinion in virology. 2012;2:489–498. doi: 10.1016/j.coviro.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spurgeon ME, Lambert PF. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. Virology. 2013;435:118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kean JM, Garcea RL. Polyomaviruses and Disease. In: Damania B, Pipas JM, editors. DNA Tumor Viruses. Springer Science+Business Media; New York, New York: 2009. pp. 53–74. [Google Scholar]
- 13.Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Cellular transformation by Simian Virus 40 and Murine Polyoma Virus T antigens. Seminars in cancer biology. 2009;19:218–228. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pipas JM. SV40: Cell transformation and tumorigenesis. Virology. 2009;384:294–303. doi: 10.1016/j.virol.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Frisque RJ, Hofstetter CA, Tyagarajan SK. Transforming Activities of JC Virus Early Proteins. Adv Exp Med Biol. 2006;577:288–309. doi: 10.1007/0-387-32957-9_21. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi H, Endo S, Suzuki S, Tanaka S, Sawa H, Ozaki Y, Sawamura Y, Nagashima K. JC virus large T protein transforms rodent cells but is not involved in human medulloblastoma. Neuropathology. 2001;21:129–137. doi: 10.1046/j.1440-1789.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 17.Bollag B, Chuke W, Frisque RJ. Hybrid Genomes of the Polyomaviruses JC Virus, BK Virus, and Simian Virus 40: Identification of Sequences Important for Efficient Transformation. Journal of virology. 1989;63:863–872. doi: 10.1128/jvi.63.2.863-872.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haggerty S, Walker DL, Frisque RJ. JC Virus-SImian Virus 40 Genomes Containing Heterologous Regulatory Signals and Chimeric Early Regions: Identification of Regions Restricting Transformation by JC Virus. Journal of virology. 1989;63:2180–2190. doi: 10.1128/jvi.63.5.2180-2190.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trowbridge PW, Frisque RJ. Analysis of G418-Selected Rat2 Cells Containing Prototype, Variant, Mutant, and Chimeric JC Virus and SV40. Genomes 1993. 1993;196:458–474. doi: 10.1006/viro.1993.1502. [DOI] [PubMed] [Google Scholar]
- 20.Tavis JE, Trowbridge PW, Frisque RJ. Converting the JCV T Antigen Rb Binding Domain to That of SV40 Does Not Alter JCV’s Limited Transforming Activity But Does Eliminate Viral Viability. Virology. 1994;199:384–392. doi: 10.1006/viro.1994.1136. [DOI] [PubMed] [Google Scholar]
- 21.Trowbridge PW, Frisque RJ. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. Journal of neurovirology. 1995;1:195–206. doi: 10.3109/13550289509113966. [DOI] [PubMed] [Google Scholar]
- 22.Zerrahn J, Knippschild U, Winkler T, Deppert W. Independent expression of the transforming amino-terminal domain of SV40 large T antigen from an alternatively spliced third SV40 early mRNA. EMBO J. 1993;12:4739–4746. doi: 10.1002/j.1460-2075.1993.tb06162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin JY, DeCaprio JA. SV40 large T antigen promotes dephosphorylation of p130. The Journal of biological chemistry. 2003;278:46482–46487. doi: 10.1074/jbc.M307044200. [DOI] [PubMed] [Google Scholar]
- 24.Stubdal H, Zalvide J, DeCaprio JA. Simian Virus 40 Large T Antigen Alters the Phosphorylation State of the RB-Related Proteins p130 and p107. Journal of virology. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stubdal H, Zalvide J, Campbell KS, Schweitzer C, Roberts TM, DeCaprio JA. Inactivation of pRB-Related Proteins p130 and p107 Mediated by the J Domain of Simian Virus 40 Large T Antigen. Molecular and Cellular Biology. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantalupo P, Saenz Robles MT, Rathi AV, Beerman RW, Patterson WH, Whitehead RH, Pipas JM. Cell-type specific regulation of gene expression by simian virus 40 T anitgens. Virology. 2009;386:183–191. doi: 10.1016/j.virol.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes & development. 2006;20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segawa K, Minowa A, Sugasawa K, Takano T, Hanaoka F. Abrogation of p53-mediated transactivation by SV40 large T antigen. Oncogene. 1993;8:543–548. [PubMed] [Google Scholar]
- 29.Jiang D, Srinivasan A, Lozano G, Robbins PD. SV40 T antigen abrogates p53-mediated transcriptional activity. Oncogene. 1993;8:2805–2812. [PubMed] [Google Scholar]
- 30.Lorenzo E, Ruiz-Ruiz C, Quesada AJ, Hernandez G, Rodriguez A, Lopez-Rivas A, Redondo JM. Doxorubicin induces apoptosis and CD95 gene expression in human primary endothelial cells through a p53-dependent mechanism. The Journal of biological chemistry. 2002;277:10883–10892. doi: 10.1074/jbc.M107442200. [DOI] [PubMed] [Google Scholar]
- 31.Attardi LD, de Vries A, Jacks T. Activation of the p53-dependent G1 checkpoint response in mouse embryo fibroblasts depends on the specific DNA damage inducer. Oncogene. 2004;23:973–980. doi: 10.1038/sj.onc.1207026. [DOI] [PubMed] [Google Scholar]
- 32.Borger DR, DeCaprio JA. Targeting of p300/CREB Binding Protein Coactivators by Simian Virus 40 Is Mediated through p53. Journal of virology. 2006;80:4292–4303. doi: 10.1128/JVI.80.9.4292-4303.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pipas JM. Mutations Near the Carboxyl Terminus of the Simian Virus 40 Large Tumor Antigen Alter Viral Host Range. Journal of virology. 1985;54:569–575. doi: 10.1128/jvi.54.2.569-575.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tornow J, Cole CN. Nonviable Mutants of Simian Virus 40 with Deletions Near the 3′ end of Gene A Define a Function for Large T Antigen Required After Onset of Viral DNA Replication. Journal of virology. 1983;47:487–494. doi: 10.1128/jvi.47.3.487-494.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole CN, Stacy TP. Biological Properties of Simian Virus 40 Host Range Mutants Lacking the COOH-Terminus of Large T Antigen. Virology. 1987;161:170–180. doi: 10.1016/0042-6822(87)90183-8. [DOI] [PubMed] [Google Scholar]
- 36.Cole CN, Crawford LV, Berg P. Simian Virus 40 Mutants with Deletions at the 3′ End of the Early Region Are Defective in Adenovirus Helper Function. Journal of virology. 1979;30:683–691. doi: 10.1128/jvi.30.3.683-691.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tevethia MJ, Pipas JM, Kierstead T, Cole C. Requirements for Immortalization of Primary Mouse Embryo Fibroblasts Probed with Mutants Bearing Deletions in the 3′ End of SV40 Gene A. Virology. 1988;162:76–89. doi: 10.1016/0042-6822(88)90396-0. [DOI] [PubMed] [Google Scholar]
- 38.Pipas JM, Peden KW, Nathans D. Mutational Analysis of Simian irus 40 T Antigen: Isolation and Characterization of Mutants with Deletions in the T-Antigen Gene. Molecular and Cellular Biology. 1983;3:203–213. doi: 10.1128/mcb.3.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulin DL, Kung AL, DeCaprio JA. p53 targets simian virus 40 large T antigen for acetylation by CBP. Journal of virology. 2004;78:8245–8253. doi: 10.1128/JVI.78.15.8245-8253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulin DL, DeCaprio JA. The carboxyl-terminal domain of large T antigen rescues SV40 host range activity in trans independent of acetylation. Virology. 2006;349:212–221. doi: 10.1016/j.virol.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 41.Welcker M, Clurman BE. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. The Journal of biological chemistry. 2005;280:7654–7658. doi: 10.1074/jbc.M413377200. [DOI] [PubMed] [Google Scholar]
- 42.Fine DA, Rozenblatt-Rosen O, Padi M, Korkhin A, James RL, Adelmant G, Yoon R, Guo L, Berrios C, Zhang Y, Calderwood MA, Velmurgan S, Cheng J, Marto JA, Hill DE, Cusick ME, Vidal M, Florens L, Washburn MP, Litovchick L, DeCaprio JA. Identification of FAM111A as an SV40 host range restriction and adenovirus helper factor. PLoS pathogens. 2012;8:e1002949. doi: 10.1371/journal.ppat.1002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reviriego-Mendoza MM, Frisque RJ. Interaction and co-localization of JC virus large T antigen and the F-box protein beta-transducin-repeat containing protein. Virology. 2011;410:119–128. doi: 10.1016/j.virol.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sompayrac L, Danna KJ. An amino-terminal fragment of SV40 T antigen transforms REF52 cells. Virology. 1992;191:439–442. doi: 10.1016/0042-6822(92)90206-5. [DOI] [PubMed] [Google Scholar]
- 45.Price TN, Moorwood K, James MR, Burke JF, Mayne LV. Cell cycle progression, morphology and contact inhibition are regulated by the amount of SV40 T antigen in immortal human cells. Oncogene. 1994;9:2897–2904. [PubMed] [Google Scholar]
- 46.Markovics JA, Carroll PA, Robles MT, Pope H, Coopersmith CM, Pipas JM. Intestinal dysplasia induced by simian virus 40 T antigen is independent of p53. Journal of virology. 2005;79:7492–7502. doi: 10.1128/JVI.79.12.7492-7502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tevethia SS, Epler M, Georgoff I, Teresky A, Marlow M, Levine AJ. Antibody response to human papovavirus JC (JCV) and simian virus 40 (SV40) T antigens in SV40 T antigen-transgenic mice. Virology. 1992;190:459–464. doi: 10.1016/0042-6822(92)91234-l. [DOI] [PubMed] [Google Scholar]
- 48.Bollag B, Prins C, Snyder EL, Frisque RJ. Purified JC virus T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology. 2000;274:165–178. doi: 10.1006/viro.2000.0451. [DOI] [PubMed] [Google Scholar]
- 49.Harlow E, Crawford L, Pim D, Williamson N. Monoclonal Antibodies Specific for Simian Virus 40 Tumor Antigens. Journal of virology. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun H, Taneja R. Analysis of transformation and tumorigenicity using mouse embryonic fibroblast cells. Methods Mol Biol. 2007;383:303–310. doi: 10.1007/978-1-59745-335-6_19. [DOI] [PubMed] [Google Scholar]
- 51.Saenz-Robles MT, Markovics JA, Chong JL, Opavsky R, Whitehead RH, Leone G, Pipas JM. Intestinal hyperplasia induced by simian virus 40 large tumor antigen requires E2F2. Journal of virology. 2007;81:13191–13199. doi: 10.1128/JVI.01658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rathi AV, Saenz Robles MT, Cantalupo PG, Whitehead RH, Pipas JM. Simian virus 40 T-antigen-mediated gene regulation in enterocytes is controlled primarily by the Rb-E2F pathway. Journal of virology. 2009;83:9521–9531. doi: 10.1128/JVI.00583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]