Abstract
In complex multicellular systems, such as the brain or the heart, the ability to selectively perturb and observe the response of individual components at the cellular level and with millisecond resolution in time, is essential for mechanistic understanding of function. Optogenetics uses genetic encoding of light sensitivity (by the expression of microbial opsins) to provide such capabilities for manipulation, recording, and control by light with cell specificity and high spatiotemporal resolution. As an optical approach, it is inherently scalable for remote and parallel interrogation of biological function at the tissue level; with implantable miniaturized devices, the technique is uniquely suitable for in vivo tracking of function, as illustrated by numerous applications in the brain. Its expansion into the cardiac area has been slow. Here, using examples from published research and original data, we focus on optogenetics applications to cardiac electrophysiology, specifically dealing with the ability to manipulate membrane voltage by light with implications for cardiac pacing, cardioversion, cell communication, and arrhythmia research, in general. We discuss gene and cell delivery methods of inscribing light sensitivity in cardiac tissue, functionality of the light-sensitive ion channels within different types of cardiac cells, utility in probing electrical coupling between different cell types, approaches and design solutions to all-optical electrophysiology by the combination of optogenetic sensors and actuators, and specific challenges in moving towards in vivo cardiac optogenetics.
Keywords: Optogenetics, channelrhodopsin, gene delivery, cell delivery, cardiomyocytes, fibroblasts
1. Introduction
Cardiac electrical activity is the product of a well-orchestrated system of functionally-distinct and spatially-distributed cells within the heart. Full understanding of the role of these distinct structures in normal and pathological conditions has been hindered due to lack of tools to selectively manipulate them in vivo, independently, and with high spatiotemporal resolution. Optogenetics (Boyden et al., 2005; Deisseroth et al., 2006; Nagel et al., 2005), the genetic modification of mammalian cells and tissues by microbial opsins (light-gated ion channels and pumps), offers potential solutions. This new method allows for cell-selective and spatiotemporally precise optical control of biological function, including manipulation of membrane voltage, intracellular concentrations, receptor control, gene expression etc. Over the last decade, optogenetics has been used widely in neuroscience to probe brain function in health and disease (Mattis et al., 2012; Yizhar et al., 2011). As a versatile optical interrogation tool, optogenetics is likely to have a similar impact on cardiac research, particularly cardiac electrophysiology, arrhythmias, cell signaling, and drug discovery (Ambrosi and Entcheva, 2014b; Boyle et al., 2014; Entcheva, 2013).
When expressed in mammalian cells, the microbial opsins at the core of optogenetics, used to modulate membrane voltage, can produce either depolarizing (excitatory) or hyperpolarizing (inhibitory) currents. Excitatory opsins, such as channelrhodopsin2 (ChR2)(Nagel et al., 2003), can provide currents sufficient for triggering action potentials, whereas inhibitory opsins, such as Archaerhodopsin (Arch) and Halorhodopsin (Halo), can suppress activity (Chow et al., 2010). Upon illumination with the appropriate wavelength, depending on the channel, molecules flow passively with (ChR2 – cations) or are actively pumped against (Arch – H+; Halo – Cl−) the electrochemical gradient to affect transmembrane potential within milliseconds. As optogenetics is a fluid field of research, the toolbox of available opsins is expanding rapidly (Entcheva, 2013; Yizhar et al., 2011). Efforts have been focused on optimizing light sensitivity, speed, and spectral response. Single amino acid substitutions have resulted in ChR2 mutants with, for instance, higher conductances (H134R(Nagel et al., 2005) and T159C(Berndt et al., 2011)), calcium permeability (CatCh (Kleinlogel et al., 2011)), and response time (ChETA(Gunaydin et al., 2010)). In addition, mutants with red-shifted excitation spectra are being explored (Lin et al., 2013) which allow for deeper light penetration in in vivo applications, especially important in dense cardiac tissues. In cardiac applications, the most commonly-used excitatory mutant is ChR2-(H134R) as it encodes a 2–3x increase in channel conductance over the wild-type channel with minimal sacrifice of kinetic performance (Lin, 2012).
Cardiac applications of optogenetics (Entcheva, 2013) in mammalian cells include work by Bruegmann et al., who have generated transgenic mice with ChR2 expression throughout the body, including the heart, and demonstrated the disruption of normal cardiac activity with short light pulses, as well as inducing focal arrhythmias with longer light pulses in open-chest hearts (Bruegmann et al., 2010). In vitro uses have thus far explored the utility of expressing ChR2 through viral means applied at the embryonic stage (Abilez et al., 2011; Bruegmann et al., 2010), viral transduction of adult cardiomyocytes (Ambrosi and Entcheva, 2014a; Williams et al., 2013), as well as through a cell-delivery approach where nonexcitable cells, expressing ChR2, are coupled to cardiomyocytes (CMs) thus inscribing light sensitivity to the cardiac syncytium (Jia et al., 2011). Recent computational modeling also provides insights about ChR2 function (i.e. voltage- and light-sensitivities) and its effects on conduction and excitability in a variety of cardiac cell types (i.e. atrial, ventricular, and Purkinje), (Williams et al., 2013) as well as in whole heart models (Abilez et al., 2011; Boyle et al., 2013; Wong et al., 2012).
This review paper focuses on several aspects of the application of optogenetic tools to cardiac research, exclusively dealing with manipulation of membrane voltage. More specifically, using examples from published work and original data from our laboratory, we discuss gene and cell delivery methods of inscribing light sensitivity in cardiac tissue, functionality of ChR2 within different types of cardiac cells, utility of probing coupling between different cell types, approaches and design solutions to all-optical electrophysiology by the combination of optogenetic sensors and actuators, and specific challenges in moving towards in vivo cardiac optogenetics.
2. Cell-Specific Optogenetic Targeting in the Heart and Its Utility
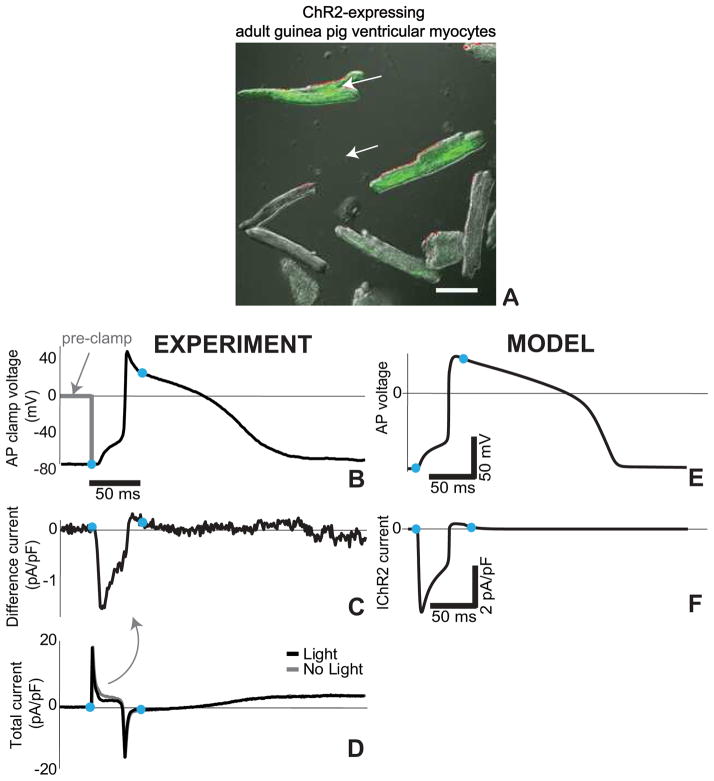
When expressed in cardiomyocytes, the excitatory opsin, ChR2, produces an inward current sufficient to elicit an action potential upon a light pulse. Recently, we demonstrated ChR2-(H134R)-eYFP viral transduction of adult guinea pig cardiomyocytes (Figure 1A) and recorded optically-triggered action potentials with 50 ms long light pulses (470 nm) (Figure 1B). We sought to specifically isolate the contribution of the ChR2 current during an action potential – data not available in the published neuroscience or cardiac optogenetics literature. Since ChR2 current (IChR2) is both light- and voltage-sensitive, its contribution during a dynamically changing membrane voltage is not trivial, and cardiomyocytes with different action potential morphology, e.g. atrial vs. ventricular, would have very different optogenetic response. As a variant of the classic “action potential (AP) clamp”(Doerr et al., 1990; Llinas et al., 1982), used to reveal the contribution of various voltage-dependent ion currents during an action potential, we developed an “optical AP clamp” (Williams et al., 2013). An optically-generated action potential (Figure 1B) is stored and later used to clamp voltage and record the total membrane current under dark and light conditions (Figure 1D), in synchronous application of a light pulse. IChR2 is the difference between the currents recorded under light and dark conditions (Figure 1C,D). This experimentally-obtained current is used to validate a computational model for ChR2 and its behavior in cardiomyocytes (Figure 1E,F) (Williams et al., 2013). The combination of computational optogenetics with experimentation, as illustrated in this example, will prove critical in optimizing opsin delivery to the myocardium in a cell-specific manner for in vivo use (Boyle et al., 2013).
Figure 1. ChR2 current during the cardiac action potential via AP clamp.
A. Adult guinea pig ventricular cardiomyocytes after 48h of viral infection with Ad-ChR2(H134R)-EYFP, green fluorescence indicates ChR2 expression; scale bar is 50 μm. Experimental (B–D) and modeling (E–F) traces for guinea pig ventricular cells. B. Optically-triggered action potential (50 ms pulse at 470nm, 1.5 mW/mm2) used for the AP clamp; dotted line indicates the voltage clamp conditions upon application of the waveform; blue dots indicate the beginning and end of the optical pulse. C. Extracted IChR2 as the difference current from the total current traces (panel D) recorded in dark conditions and with a light pulse. E. Analogous optically-triggered action potential in a model of a guinea pig ventricular cell. F. The underlying IChR2 according to the model. Reproduced with permission from (Williams et al., 2013).
2.1 Opsin expression in cardiac cells and tissues
Generation of light-sensitive cardiac tissue can be achieved in two ways: (1) through direct genetic modification of some (or all) cardiomyocytes or (2) through cell delivery, i.e. the introduction of opsin-expressing cells (i.e. stem cells or fibroblasts) which can couple with native cardiomyocytes through gap junctions. Neuroscience applications of optogenetics rely on the widespread availability of cell-specific promoters and have primarily focused on viral delivery of opsins into neural cells (Diester et al., 2011; Liewald et al., 2008; Zhang et al., 2007). For cardiac applications, it would not only be beneficial to selectively target cardiomyocytes (excluding transgene expression in other cardiac cell types such as fibroblasts and endothelial cells) (Addis et al., 2013; Pacak et al., 2008), but also to selectively target specific cardiac cell types, e.g. components of the conduction system, such as the sinoatrial node and Purkinje fibers, for optogenetic control. Although conduction system-specific gene expression has been identified (i.e. HCN4, Cx40, and the more recently characterized Contactin-2 (Pallante et al., 2010)), promoters have yet to be found to facilitate transgene expression confined to these structures.
Without specific promoters, transgene expression can also be localized by spatial targeting, i.e. opsin-expressing cells or viruses can be directly injected or perfused into the heart tissues. Direct injection may result in a cluster-like conglomeration of opsin-expressing cells (Prasad et al., 2011a; Rosen et al., 2007), whereas cardiac perfusion (or systemic delivery in the case of a virus with a cardiac-specific promoter) may yield a more diffuse pattern of transgene expression (Prasad et al., 2011b).
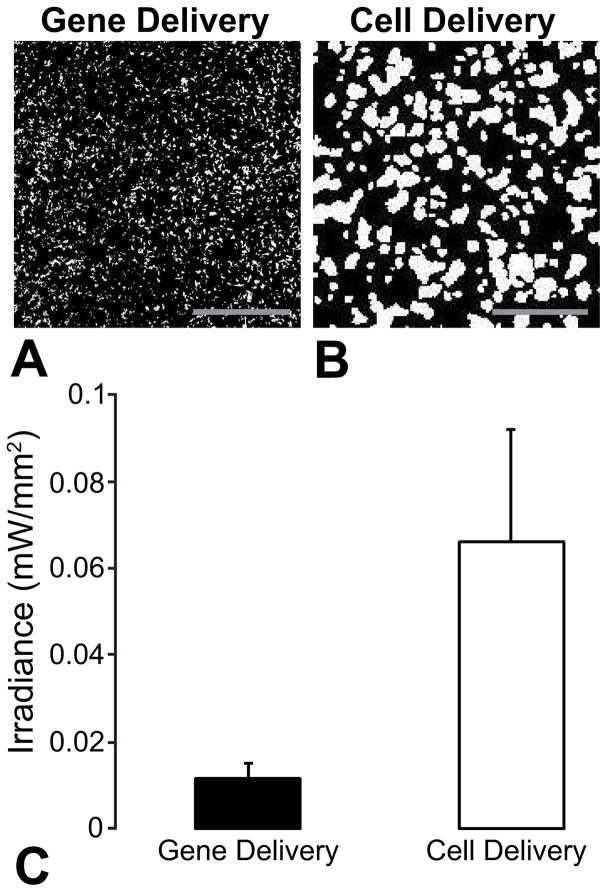
We have used both viruses and exogenous “donor” cells expressing ChR2 as means to create optically-sensitive cardiac cells and syncytia. A cell delivery approach to optogenetics, termed a tandem-cell-unit approach (TCU), can be realized, where non-excitable, “donor” cells expressing ChR2 electrically couple to cardiomyocytes through gap junctions to confer optical sensitivity to the syncytium (Jia et al., 2011). Quantitative comparison of the optical energy requirements for both approaches – direct viral ChR2 gene delivery into cardiomyocytes (using adenoviruses) vs. cell delivery (TCU) – shows lower energy requirements for excitation (lower irradiance) in the gene delivery case for comparable ratios of ChR2-expressing cells to unmodified cells (54%, gene delivery vs 59% for cell delivery) for spatially-disperse ChR2 expression (Figure 2). However, the performance of the two approaches varies with spatial localization of the opsin expression (Ambrosi et al., 2013). The ability to experimentally pattern gene expression (Ambrosi and Entcheva, 2014a) and to computationally capture the response of the tissue under viral and cell delivery can inform future efforts in cardiac optogenetics in vivo, as well as in gene therapy for cardiac applications, in general (Boyle et al., 2013; Entcheva, 2013).
Figure 2. Optical excitability of cardiac syncytium in vitro as a function of delivery mode.
A,B. Binarized images of transgene distributions resulting from adenoviral delivery of ChR2-eYFP (ChR2 density, 54%) and cell delivery via ChR2-eYFP-expressing HEK cells (ChR2 density, 59%). White pixels represent cells expressing ChR2-eYFP and black pixels represent unmodified cells. Scale bars are 1 mm. C. Optical excitation thresholds at 50 ms pulse width for both modes of delivery. Data presented as mean ± SEM.
2.2 Optical excitability of different cardiac cell types
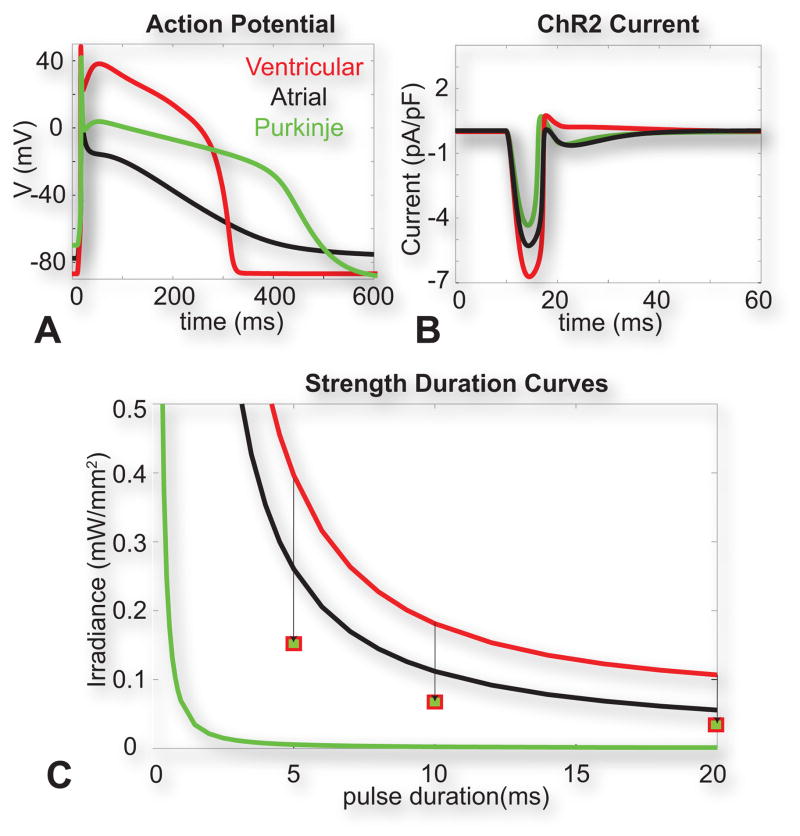
The differential response of different cardiomyocyte cell types to light can be used as a guide when considering targets for low-energy optical pacing of the heart. Using computational modeling to simulate the expression of ChR2 in human atrial, ventricular, and Purkinje cells (analogous to cell population-specific ChR2 expression with the use of relevant promoters), we show that a 10 ms, 0.5 mW/mm2 blue light pulse is sufficient to trigger physiologically-shaped action potentials in each cell type (Figure 3A) (Williams et al., 2013). Optical excitation threshold testing revealed that ventricular myocytes required the most energy for excitation at all pulse durations, whereas Purkinje cells required the least optical energy, likely due to differential, native expression of the potassium inward rectifier IK1, as a main opposing current upon excitation by electrical or optical stimuli (Figure 3B). These data, combined with whole-heart simulations (Boyle et al., 2013) suggest that Purkinje cells (and the conduction system) may be a suitable target for in vivo, low-energy optical pacing. Experimental data from open-chest in vivo experiments in mice (Bruegmann et al., 2010) corroborate differential response of atrial and ventricular cells to optical stimulation, importantly highlighting the effects of tissue structure and coupling in this response – better-coupled ventricular tissue required less optical energy than atrial tissue even though, at the single cell level, atrial cells were easier to excite.
Figure 3. Optical excitation in human cardiac cell types.
A. Optically-triggered action potentials (10 ms pulse at 470nm, 0.5 mW/mm2) in human ventricular, atrial and Purkinje cells. B. Underlying ChR2 current upon the action potential generation for the three cell types. C. Strength-duration curves for the three cell types. Squares show simulated values for optical excitation threshold in ventricular myocytes with a Purkinje formulation of IK1. Reproduced with permission from (Williams et al., 2013).
2.3 Opsin expression in cardiac fibroblasts
Cardiac fibroblasts are the most numerous cell type in the heart (60–70%), with multiple important roles, including contributions in extracellular matrix maintenance, structural and mechanical support, paracrine signaling, and controlled electrotonic influences on myocytes (Camelliti et al., 2005; Rohr, 2012; Vasquez and Morley, 2012). Because of their critical role in arrhythmogenesis during myocardial remodeling post-injury, there is great interest in tools that can selectively probe fibroblast-myocyte interactions.
Compared to cardiomyocytes (neonatal and adult) and to cell lines (HEK293T), primary cardiac fibroblasts were found to be much more resistant to ChR2 gene delivery using an adenoviral vector with a generic CAG promoter, and at the same time, more tolerant to high viral doses. For example, using a custom-developed Ad-CAG-ChR2(H134R)-eYFP vector (Ambrosi and Entcheva, 2014a), a dose of 25 MOI (multiplicity of infection) was sufficient to transduce >98% of the neonatal rat ventricular myocytes but showed practically zero expression in cardiac fibroblasts. De facto, this viral approach yielded myocyte-specific ChR2-expression, in our experience. We then pursued optimization of fibroblast targeting with ChR2. Robust expression (>50%) was obtained by infection in suspension, proper timing, high viral titers (1012 transducing units/ml), and high MOI (103 units/cell). ChR2 expression in primary cardiac fibroblasts was stable, persisting for at least 6 weeks, without detrimental effects to cell viability and proliferation (Yu et al., 2013). This is in contrast to severe negative effects and cell death reported in astrocytes when high viral titers > 1011 transducing units/ml of AAV–sGFAP–ChR2 were used (Figueiredo et al., 2011).
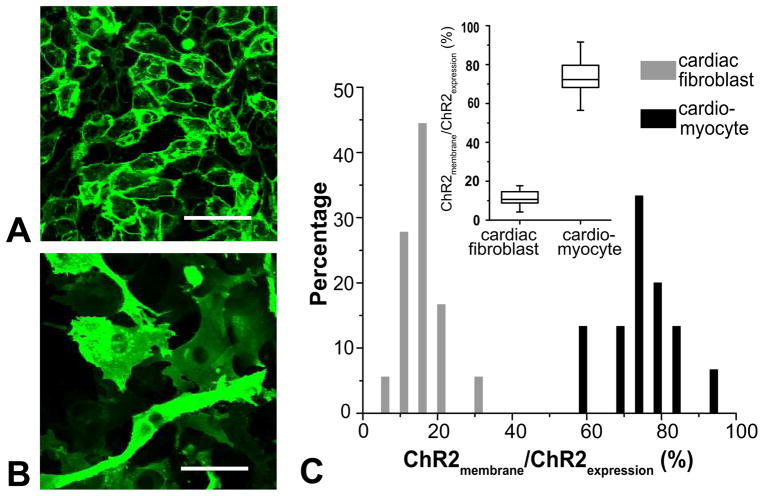
Furthermore, ChR2 internalization and trafficking within cardiomyocytes and cardiac fibroblasts was found to be different (Figure 4). While cardiomyocytes displayed preferential membrane localization, i.e. 73±9.38% of total ChR2 expression was localized to cardiomyocyte cell membranes (separately labeled by amphiphilic dye di-8-ANEPPS), in fibroblasts, membrane-localized ChR2 contributed only 12±5.6% of the total expression.
Figure 4. ChR2 expression in cardiomyocytes and cardiac fibroblasts.
A. ChR2-eYFP expression localizes to the cell membrane in cardiomyocytes. B. ChR2-eYFP expression is more diffuse cytoplasmic. C. Quantification of ChR2 expression patterns in cardiomyocytes and fibroblasts; inset is a summary box plot. Line inside box represents median; bars above and below box represents max and min; box represents the 25th and 75th percentile. Scale bars are 50 μm.
To take full advantage of the cell-specific targeting afforded by optogenetic approaches and to be able to study interactions between cardiomyocytes and light-sensitive fibroblasts in vivo, specific genetic markers are needed. Unlike available promoters to target glial cells in neural tissue (GFAP (Gourine et al., 2010)) and even glial subtypes (astrocytes by gfaABC1D (Shigetomi et al., 2013)), no specific promoter for cardiac fibroblasts has been identified yet. However, recent work has resulted in transgenic Cre mice with non-myocyte mesenchymal cell targeting in cardiac tissue (Acharya et al., 2011; Takeda et al., 2010), which may enable approaches to ChR2 expression in cardiac fibroblasts and the transition of this work in vivo.
2.4 Applications of optogenetics to study cell-cell interactions
Using the TCU strategy in conjunction with light-sensitization of cardiac fibroblasts, a quantitative assay of electrical coupling between fibroblasts and cardiomyocytes can be developed, using threshold excitation energy as a read-out. We have demonstrated that light-induced ChR2 currents in cardiac fibroblasts elicit depolarization in coupled cardiomyocytes, and that an inverse relationship exists between the optical excitation energy and the coupling strength, i.e. ChR2-expressing donor cells (HEK or fibroblasts) poorly coupled to cardiomyocytes require higher optical energies in order to elicit action potentials in the latter (Jia et al., 2011; Yu et al., 2013). Unlike dual cell patch clamp, which can only measure intercellular coupling currents in isolated cell pairs, an optogenetics-based coupling assay can be readily integrated to evaluate coupling within cardiac syncytia and whole tissues, and it can be applied in a cell-specific manner. An example application of this technique can be to track potential changes (increase) in electrical coupling between fibroblasts and myocytes following myocardial infarction when increased fibroblast proliferation from either activation or the scarring process can present excessive electronic loads on the myocytes and can increase susceptibility to arrhythmias (Rohr, 2009; Rohr, 2012; Vasquez and Morley, 2012; Zlochiver et al., 2008). Considering the controversy surrounding electrotonic coupling between cardiomyocytes and fibroblasts in vivo (Kohl and Gourdie, 2014) and the importance of such interactions in disease conditions, this new direct cell-specific optogenetic approach to quantify electrical coupling can provide answers. Similarly, the optogenetic TCU strategy can be useful in assessing cell delivery and tissue graft integration in the myocardium during cardiac tissue repair procedures. If the donor cells/tissue are selectively transduced to be light-sensitive, such testing can be done by optical actuation.
3. All-Optical Interrogation of Cardiac Electrophysiology
Optogenetics, offering both genetically-encoded optical sensors and actuators, enables all-optical interrogation of electrical activity in a completely contactless manner. Such approaches are inherently parallelizable and amenable to implementations of feedback control. Therefore, they can be particularly useful as a new generation of high-throughput drug screening methods, as cardiac liability testing (electrophysiological testing of all new drugs) requires physical contact with the cells and is therefore not easily scalable. Furthermore, the combined use of optogenetic sensors and actuators permits cell-specific in vivo studies, not possible previously.
Some key challenges must be considered to achieve all-optical cardiac electrophysiology. First, the spectral accommodation of at least three wavelengths needs to be resolved: for the actuator, for excitation of the sensor, and for the sensor emission. In addition, the system must have sufficient spatiotemporal resolution to invoke and track electrical events, including typically millisecond temporal resolution, which imposes high demands in terms of light-gathering ability. Unlike in structural imaging, signal to noise ratios (SNRs) cannot be improved by merely increasing integration time. In neuroscience, various implantable systems have been implemented in live animals over the last couple of years (Voigts et al., 2013), using the skull as a mounting platform. To extend such tools to cardiac use, special solutions must be found to address minimally-invasive access, motion artifacts, and photon scattering and absorption while remaining small enough to navigate through the heart.
3.1 Spectral challenges: Available sensors and actuators
For ex vivo cardiac optical mapping, two key classes of synthetic fast fluorescent dyes are used: voltage (potentiometric) and calcium. Commonly used voltage sensors include the styryl dyes RH-237, Di-4-ANEPPS, and Di-8-ANEPPS. These dyes are not particularly efficient; for example, Di-4 has a limited quantum efficiency and has at best a 10% relative change in fluorescence per 100 mV (Herron et al., 2012). In addition, the blue-green wavelengths used for excitation of these dyes undergo large amounts of scattering, particularly in tissue, and overlap spectrally with the wavelengths used for the optogenetic actuators, e.g. ChR2. More recently, new near-infrared (650 nm) dyes, such as di-4-ANBDQPQ, have been used in blood perfused systems, as their absorption peak is >70nm higher than that of hemoglobin (Herron et al., 2012). In addition to potentiometric dyes, calcium dyes are of particular interest for cardiac imaging due to the link between electrical activity and intracellular calcium (Entcheva and Bien, 2006; Herron et al., 2012). Unlike potentiometric dyes, commonly used calcium dyes such as Fluo-4, Rhod-4, and Fura-2 have much stronger optical signals (better SNRs). However, the use of Ca2+ recordings as surrogates for electrical activity (action potentials) is not always applicable, especially when the action potential upstrokes are of interest. Furthermore, the affinity of such dyes for Ca2+ is of crucial importance in their accurate representation of the morphology of the Ca2+ transients. For example, higher affinity dyes can unintentionally prolong the recorded transient and consequently confound interpretation by obliterating the true kinetics (Efimov et al., 2004; Entcheva and Bien, 2006; Herron et al., 2012).
Future in vivo applications will require the use of optogenetic sensors, i.e. genetically-encoded voltage and calcium indicators. Commonly used genetically-encoded calcium indicators include GCaMP variants, which use GFP as the fluorescent reporter (Mutoh and Knöpfel, 2013). The creation of genetically-encoded, fast voltage sensors has been challenging, yet some successful probes have recently been generated. Commonly-used classes of voltage-sensitive fluorescent proteins (VSFP) include VSFP3 (based on a phosphatase voltage-sensing domain) with mKate as the reporter. (Mutoh and Knöpfel, 2013) Recently, genetically-encoded opsins, such as Arch, have been adapted as voltage sensors (Maclaurin et al., 2013).
For actuation, light-sensitive ion channels, such as the blue-wavelength excited (470 nm) depolarizing (excitatory) channel ChR2 and the yellow-wavelength (570 nm) hyperpolarizing (inhibitory) channels Halo and ArchT are used (Packer et al., 2013; Smedemark-Margulies and Trapani, 2013). When combined, depolarizing and hyperpolarizing channels and pumps allow for precise bidirectional cellular manipulation – either silencing or activation by light (Chow et al., 2010; Zhang et al., 2007). Like fluorescence imaging, optical stimulation deep within tissue is primarily limited by photon scattering and absorption, particularly for shorter-wavelength absorbing light sensitive channels, such as ChR2. Figure 5A shows the activation spectrum for ChR2, as well as the excitation and emission spectra for two compatible optical sensors: the calcium-sensitive probe Rhod-4 and the red-shifted voltage sensor Di-4-ANBDQPQ. Many of these probes and actuators have broad absorption and emission spectra which can overlap causing issues with excitaiton and stimulation. For example, ChR2 has an intermediate deactivating green state at 520 nm after activation with blue light, meaning excitation light for dyes such as Rhod-4 could potentially affect ChR2 kinetics (Bamann et al., 2008), though in our experience such interference is negligible. An interesting activation of multiple probes within the same field of view can be achieved by altering the temporal and spectral properties of the actuators, as demonstrated with the new opsin variants Chrimson and Chronos, a red-shifted ChR2 variant and a more light-sensitive, faster ChR2 variant, respectively (Klapoetke et al., 2014).
Figure 5.
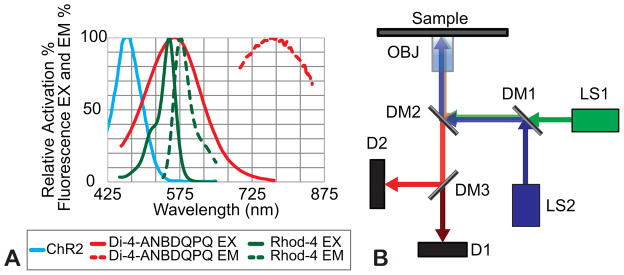
A. Activation spectra for ChR2, and excitation (EX) and emission (EM) spectra for the voltage sensitive dye Di-4-ANBDQPQ and the calcium sensitive dye Rhod-4. B. Conceptual schematic of the optical path for all-optical interrogation.
3.2 Engineering Solutions: Optical Setup
The “spectral congestion” issue arising from an all-optical approach is challenging, and must be addressed with either using laser lights sources or narrow-band optical filters and dichroic mirrors. Figure 5B shows a conceptual layout of an all-optical system, used in our laboratory, built around a standard microscope. The excitation source, LS1, is a green, 530 nm, LED (M530L3; Thorlabs) collimated using an aspheric condenser lens and filtered by the bandpass filter (ET535/50x; Chroma). The stimulation source, LS2, is a 470 nm high-powered mounted LED (M470L3; Thorlabs) collimated using an aspheric lens. Both light sources are combined using the dichroic mirror DM1 (T495lpxr; Chroma) and expanded using a telescopic lens pair to fill the back aperture of the objective (40x Nikon CFI Plan Fluor/20x Nikon CFI Super Plan Fluor). The combined beams are reflected up to the sample plane through the objective using the dichroic mirror DM2 (T565dcxr; Chroma), which allows for the separation of the stimulation/excitation sources and the emitted fluorescence signal. Collected light from the sample is sent to both detectors D1 and D2 simultaneously using the dichroic mirror DM3 (T600lpxr; Chroma). In one implementation, detector D2 is used for video tracking of contractions (MyoCam; Ionoptix) in conjunction with transillumination. Fluorescence measurements (of calcium or voltage) are obtained using D1, which in this implementation is a PMT (9136B; ET-Enterprises) after an emission filter F2 (ET605/70m; Chroma). Example traces comparing electrical and optical pacing in neonatal rat cardiomyocytes, with both optical readout of Rhod-4 and simultaneous optical tracking of mechanical contractions, can be seen in Figure 6. We have been the first to combine optogenetic stimulation and ultra-high resolution fast optical mapping to track excitation waves in cardiac tissue and quantitatively compare the response of cardiac tissue for both electrical and optical actuation (Jia et al., 2011).
Figure 6.
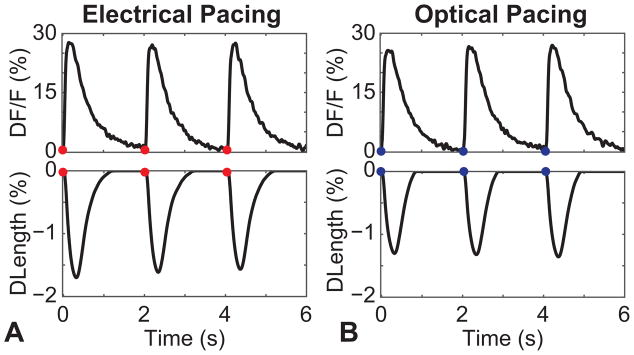
All-optical measurements in cardiomyocytes using ChR2 as actuator and Rhod-4 as a sensor – example traces, comparing: A. electrical pacing and B. optical pacing of ChR2 expressing neonatal cardiomyocytes. Top: Rhod-4 signal (Ca2+); Bottom: Contractility measurements obtained through videotracking simultaneously with the Ca2+ data.
4. Towards Cardiac Optogenetics In Vivo
Optogenetic studies in the brain are able to take advantage of fast optical systems, chronically fixed to the animal’s skull for in vivo interrogation. Long-term studies of optical actuation and electrial readout of triggered neuronal signals have been performed in freely moving, active rodents using head-mountable devices consisting of single electrodes or electrode arrays for readout along with optical fibers to deliver light for stimulation (Stark et al., 2012; Voigts et al., 2013; Wang et al., 2012). Studies in awake rhesus monkeys have also been performed using fiber optic systems (Ozden et al., 2013), as well as implanting optically-clear windows for easy light delivery and penetration by fine micropipettes for electrical recording during optogenetic studies (Ruiz et al., 2013). In contrast, the only cardiac application in vivo thus far (Bruegmann et al., 2010) has been in anesthetized open-chest animals, with epicardial stimulation. The neuroscience-used methods mentioned above are not easily extendable to cardiac studies, where there are no solid reference structures to mount the device; and and open-chest or open-heart studies are not applicable for use in intact animals. In addition, access to structures of interest, e.g. the conduction system, is not easily achievable from the epicardial surface, as scattering and absorption limit the penetration depth. Motion artifacts pose a great challenge compared to brain studies. However, both neuroscience and cardiac applications of optogenetics share the need for miniaturized optics.
4.1 Optical properties of cardiac tissue (absorption and scattering)
In addition to spectral overlaps of sensors and actuators, the optical properties of the heart and blood must be considered when designing an optical system for in vivo use. Recently, absorption and scattering data were obtained for the rat heart (Mesradi et al., 2013). As expected, absorption data mimics that of hemoglobin, with an absorption peak at 450 nm and 550 nm for the visible spectrum, while scattering follows standard Rayleigh scattering, decreasing monotonically with increases in wavelength (44217*λ−1.613). Because fluorescence measurements are depth-integrated, scattering and absorption can distort the measured optical signals. This can be partially rectified by using photon transport models, however they commonly assume homogeneous media (Mitrea et al., 2011). Thus, to most effectively perform optical actuation and sensing on hard-to-access cardiac structures, e.g. the conduction system, one has to find access routes beyond the epicardial surface.
4.2 Endoscopic routes to specific cardiac structures
In the clinical setting, relevant structures of the conduction system, such as sinoatrial (SA) node, the atrioventricular (AV) node, and bundle of His, can be accessed using catheterization procedures for both diagnostic and interventional electrophysiology. Catheters are typically chosen based on the size of the blood vessels along the way and the tortuosity required to navigate the route to the site of interest. Angioscopy, optical coherence tomography (OCT), and spectroscopic imaging are commonly used endoscopically for diagnostic purposes. For imaging with optical wavelengths, a blood-free environment is necessary, typically achieved using distal balloons to occlude blood flow or flushing clear fluid in front of the imager; near-IR light can also be used to image through blood based on lower scattering (Honda and Fitzgerald, 2008).
The conduction system is typically accessed through the right side of the heart because of lower risk due to lower pressure. For example, in humans, access to the SA node, AV node, and Bundle of His can be achieved through the right side of the heart using a 6–7 French (2–2.3 mm) catheter, which can be inserted through the femoral vein via the inferior vena cava into the right atrium; access through the superior vena cava is also possible by the brachial cut-down approach. In adult rats, access to the right side of the heart can be performed in a similar manner, where a 1.4 French (0.47 mm) catheter can be inserted through the right jugular vein into the RV (Deten et al., 2003). Such procedures have been used to measure pressure in the pulmonary arteries of rats (Deten et al., 2003) and directly measure LV performance (Pacher et al., 2008) using commercially available sensors and catheter systems, specifically designed for rats from Millar instruments.
4.3 Miniaturization and optical solutions: grin lenses, fiber optics
For designing all-optical endoscopic systems for in vivo use, the upper limit on the diameter of the system will need to be approximately 2 mm in humans and 0.5 mm in rodents. Optogenetic studies in the brain commonly involve inserting optical fibers directly into the tissue for optical stimulation, but optical readout is not performed. Injectable optrode systems have been used in freely-moving mice, which included a micro-inorganic LED (μ-ILED; 6.45 μm thick, 50×50 μm2) and a micro-inorganic photodiode (μ-IPD; 200 × 200 μm2), however the μ-IPD was only used to measure the irradiance of stimulation light (Kim et al., 2013). Insertion of such a probe directly into the heart wall is challenging, and catheterization procedures would still need to be performed for probe placement. In addition, wires for powering the device and collecting data would still be required. Although completely wireless systems have been used for implantable pacemakers in rodents (Laughner et al., 2013), they require operation within range of transmitter coils, and may lack enough power to operate the whole system.
Using optical fibers to remotely deliver and collect light removes size and power constraints from light sources and detectors, and helps improve SNRs. Flexible, stretchable μLED could be used for light delivery (McCall et al., 2013; Xu et al., 2014), however fibers would still be needed for optical readout. In addition to high-sensitivity cameras, SNRs can be improved by maximizing light collection efficiency by using high numerical aperture optics. For microendoscopic imaging in rodent brains, special objectives using gradient index (GRIN) micro-lenses have been developed, as they can be fabricated in diameters between 350–1,000 μm while having NAs between 0.4 – 0.6 and lengths on the order of millimeters (Flusberg et al., 2005). These low-cost lenses lack spherical aberrations and focus light due to having a continuously varying index of refraction in the radial direction of the rod, causing the light to propagate in a sinusoidal manner with respect to the rod’s axis (Barretto et al., 2009). These systems show promise for use for imaging in the heart, as they have been already used for imaging microcirculation and Ca2+ dynamics in freely moving mice (Flusberg et al., 2008). By adapting such a system to accommodate a third wavelength for optical actuation and optimizing the system to fit the size constraints of the heart, when combined with high-sensitivity detectors, an entirely fiber-based system for long-term optical actuation and sensing in vivo can be realized.
4.4 Inscribing light-sensitivity in the heart in vivo
The availability of transgenic mice with cardiac cell population-specific opsin expression is an attractive experimental model for exploring optogenetics as a new technical tool for addressing questions regarding pacing, arrhythmias, and cardioversion in a cell-specific manner. In neuroscience, a plethora of neural cell population-specific promoters have been identified and have resulted in the development of lines of transgenic mice (Asrican et al., 2013), many of which are commercially-available through the Jackson Laboratory. In contrast, in cardiac research, there is lack of available promoters for strictly specific gene expression in structures in the conduction system, for example the SAN or the Purkinje fibers only. But some inducible Cre mouse models are available for SA node and AV node localization by HCN4 (Hoesl et al., 2008), or for Purkinje fibers (also transducing atria and coronary vessels) by Cx40 (Beyer et al., 2011) and using Cre-loxP technology, opsins can be selectively expressed in those structures. Optogenetically targeting cell-specific locations in the heart, such as the His-bundle and the Purkinje system, may be a new route to optimized low-energy therapy, as suggested by our computational work (Boyle et al., 2013).
As an alternative to the transgenic animal approach, cardiac optogenetics in vivo can be realized using current gene therapy approaches. Of most interest clinically is the use of adeno-associated viruses (AAV), which have several advantages over other viral vectors, such as adenovirus and lentivirus, for long-term transgene expression with minimal side effects. In particular, AAVs have low immunogenicity, can transduce non-dividing cells, and are unable to replicate without the presence of a helper virus (Williams et al., 2010). There are also currently twelve characterized AAV serotypes each with differential tissue tropism, which can add a layer of specificity to transgene expression in addition to the contribution of a gene promoter. For instance, AAV9 has been reported to be most effective at transducing the heart (Bish et al., 2008), whereas AAV2 has been most widely-applied in the brain (Markakis et al., 2010). Although the success of transgene expression is intimately linked with viral dosing, (Prasad et al., 2011b) AAV serotype efficiency is not only directed by tissue tropism, but may also be species-dependent. (Yue et al., 2008)
We have recently begun serotype testing AAVs in the rat heart under both in vitro and in vivo conditions. As shown in Figure 7, AAV serotype 1 can transduce both neonatal rat cardiomyocytes in vitro (Figure 7A), as well as ventricular cardiomyocytes in the in vivo adult heart (Figure 7B). In humans, there is currently an ongoing cardiac-related clinical trial, testing the efficacy of delivering SERCA2a using AAV1 as a means to reduce and/or delay heart failure-related hospitalizations (CUPID trial).(Jessup et al., 2011; Zsebo et al., 2014) So far, this study has reported no adverse effects due to viral delivery, providing evidence that AAV-mediated transgene delivery may be a viable vehicle for long-term opsin expression in the heart.
Figure 7. In vitro and in vivo cardiac expression of AAV1.
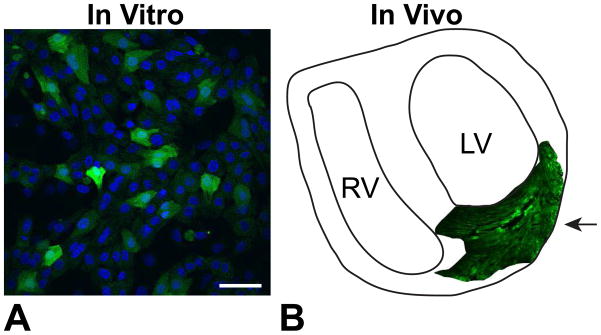
A. Neonatal rat ventricular cardiomyocytes infected with AAV1 in vitro at MOI 1000. Green fluorescence indicates eGFP reporter and blue indicates DAPI nuclear staining. Scale bar is 50 μm. B. Schematic of a slice of an adult rat heart infected with 1×1011 AAV1 particles, delivered via intramyocardial injection 4 weeks earlier, as indicated by the arrow.
Analogous to our in vitro cell delivery (TCU approach) (Jia et al., 2011), an in vivo translational strategy would ideally involve the use of autologous sources of somatic cells, such as fibroblasts or stem cells, thus mitigating immune responses. There are several ongoing clinical trials using a similar approach, including SCIPIO (Bolli et al., 2011; Chugh et al., 2012) and CADUCEUS (Makkar et al., 2012; Malliaras et al., 2014). In both of these trials the effects of the injected cells persisted up to at least one year demonstrating the potential for long-term opsin expression.
In addition to the vehicle types by which opsins can be delivered to the heart (i.e. viruses or genetically-modified cells), the routes of administering them must also be optimized. Both viruses or cells can be administered by direct myocardial injection (a delivery which typically results in a cluster-like conglomeration of opsin-expressing cells (Prasad et al., 2011a; Rosen et al., 2007)) or by perfusion directly into the heart or systemically (which may yield a more diffuse pattern of transgene expression (Prasad et al., 2011b)). Alternatively, viral delivery may be achieved through a relatively new technique termed gene painting (Kikuchi et al., 2005), where a poloxamer-F127 gel containing the virus of interest and a low concentration of trypsin is physically painted across the tissue, e.g. the epicardial surface of the heart. Although the penetration depth of expression must be further explored in order to ensure sufficient transgene expression for optical excitability, this technique has been successfully applied in large animals (swine) with implications for an efficient means of transgene delivery.
5. Conclusions and Future Directions
The ability for cell type-specific optogenetic actuation and sensing, the high spatiotemporal resolution and the contactless nature of optogenetics affords unique possibilities for cardiac electrophysiology in vivo, including but not limited to:
Low-energy pacing strategies can be explored by optogenetic investigations. Specifically, optical stimulation can be targeted to strategic structures of the conduction system, e.g. His bundle, for efficient, low-energy pacing, as suggested computationally (Boyle et al., 2013). The pacing rates of innate pacemakers (of the SAN or AVN cells) can also be selectively manipulated by light.
Understanding the role of specific cell types and structures in arrhythmia induction, maintenance and termination will be possible by selectively activating or inhibiting their electric contribution optically (not achievable by other means). Purkinje fibers have been implicated in pro-arrhythmic events either during the VF induction or at later stages of VF maintenance; selective dynamic suppression or activation of the Purkinje system in vivo can provide new insights and offer unique cardioversion ideas.
Neural-cardiac interactions can be probed functionally by selectively manipulating sympathetic or parasympathetic inputs by light during normal pacing and during arrhythmia events. Such manipulations are not possible by electrical stimulation, which is non-specific or by chemical stimulation, which lacks temporal and spatial resolution. Therefore, the optogenetic interrogation provides a new tool for mechanistic dissection of the neural-cardiac interface.
The TCU approach to optogenetics can provide new ways to test for electrical communication between non-excitable cells, e.g. fibroblasts, and cardiomyocytes in vivo in normal and pathological conditions. The optical energy needed to trigger full excitation through light-sensitive cardiac fibroblasts can be used to quantify their coupling to the cardiomyocytes, as discussed above. Similarly, electrical integration of exogenous cells in vivo can be tested for assessing cell therapy efficiency in cardiac repair or other cardiac treatments.
Consequently, optogenetics offers new cell-specific tools to dissect cardiac function, with high spatiotemporal resolution for both actuation and sensing using genetically-encoded light-sensitive proteins that are well-tolerated by cardiac tissue and easy to express. Such all-optical interrogation is scalable and lends itself to implementation of feedback control. We listed potential applications and questions that are not possible to address using existing electrical or chemical methods of perturbation. In addition to the manipulation of membrane voltage by excitatory or inhibitory microbial opsins, discussed here, optogenetic tools have expanded to provide cell-specific, precise dynamic manipulation of cell signaling (Airan et al., 2009; Ye et al., 2011) or feedback control of gene expression by light-induced transcriptional effectors (Konermann et al., 2013). For example, recently, Beiert et al. (Beiert et al., 2014) presented a technique to optically perturb Gq signaling in cardiomyocytes, by expression and optogenetic actuation of melanopsin, a light-sensitive G-protein coupled receptor protein to allows dynamic modulation of intracellular calcium via the IP3 pathway.
Most interesting and rewarding applications of optogenetics will come from in vivo use, as demonstrated in neuroscience. Overall, the combined development of cell-specific gene delivery (promoters and vectors) and minimally invasive access for all-optical actuation and readout are needed to move cardiac optogenetics in vivo, and to fully realize its potential.
6. Methods
Gene and Cell Delivery
Primary cardiac cell culture was done as described earlier (Ambrosi and Entcheva, 2014a; Jia et al., 2011). Prior to monolayer plating, neonatal rat ventricular myocytes (NRVM) were either infected with a predetermined dose (MOI 25) of in-house developed adenovirus (Ad-ChR2-eYFP, based on the expression cassette of plasmid #20940; Addgene, Cambridge, MA) for the gene delivery model or mixed with ChR2-expressing HEK cells for the cell delivery model. Distributions were created by global adenoviral infection in the gene delivery model and by mixing ratios (100:1) of light-sensitive HEK cells to light-insensitive NRVMs in the cell delivery model. Light-sensitive cardiac fibroblasts were created as follows: in-suspension cells were incubated for 2.5 hours in 2% FBS M199 culture medium containing Ad-CAG-ChR2-eYFP. After centrifuging, the virus was removed and the cell pellet was re-suspended in fresh culture medium (FBS 10%) and plated for future use.
In Vitro and In Vivo AAV Infection
After isolation, NRVMs were infected with predetermined doses (MOI 2000) of adeno-associated viral serotype 1 (AAV1) acquired from the Vector Core at University of Pennsylvania (Philadelphia, PA). The expression cassette of each viral serotype contained the transgene for eGFP driven by a ubiquitous CAG promoter. Five days post-infection NRVMs were imaged using an Olympus Fluoview FV1000 confocal imaging system to assess transgene (eGFP) expression.
For in vivo AAV delivery, animals were anesthetized with 4% isofluorane, intubated, and placed on a ventilator with 3.5% isofluorane supplemented with oxygen. A left thoracotomy was performed to expose the heart and two independent injections of 1×1011 AAV1 particles (10 μL total volume) were made using a 30-gauge needle (Hamilton Company, Reno, NV) in the LV free wall. After 4 weeks, the animals were euthanized by deep isofluorane anesthesia and the heart was removed for analysis. Hearts were embedded and frozen in Tissue-Tek OCT compound (Electron Microscopy Sciences, Hatfield, PA) and stored at −80°C. The tissues were cryosectioned at 16 μm and mounted on Superfrost Plus glass slides (Fisher Scientific). Confocal imaging was then performed to assess the extent of eGFP expression.
Optical Measurements
For confocal imaging, cardiomyocytes or cardiac fibroblasts infected with Ad-ChR2 were plated on glass bottom dishes and imaged using Olympus FluoView™FV1000 confocal system. The samples were stained using 14 μM di-8ANEPPS to outline the cell membrane. Quantification of ChR2 expression pattern was done using the image analysis toolbox in Matlab. After background subtraction, co-localization between the ChR2 and the di-8 signal was quantified.
Functional optical measurements were performed microscopically on monolayers stained with Quest Rhod4 AM (10 μM; AAT Bioquest, Sunnyvale, CA) on an inverted fluorescence microscope (Nikon TE-2000U) with a custom-built imaging system, as discussed. Macroscopic optical mapping was done to determine optical excitation thresholds using a system as described previously (Jia et al., 2011). Optical stimulation was delivered to the monolayer using a fiberoptics-coupled diode-pumped solid-state (DPSS) laser (470nm; Shanghai Laser, Shanghai, China) gated by a Myopacer Cell Stimulator (IonOptix, Milton, MA). Energies for optical stimulation thresholds are reported as irradiance (mW/mm2).
Acknowledgments
This work was supported by NIH-NHLBI grant R01-HL-111649 (E.E.), an Institutional NRSA T32-DK07521 (C.M.A.), and partially by a NYSTEM grant C026716 to the Stony Brook Stem Cell Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abilez OJ, Wong J, Prakash R, Deisseroth K, Zarins CK, Kuhl E. Multiscale computational models for optogenetic control of cardiac function. Biophys J. 2011;101:1326–34. doi: 10.1016/j.bpj.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis. 2011;49:870–7. doi: 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–9. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- Ambrosi CM, Chen K, Kalra J, Chen X, Zhu W, Yu J, Jiang YP, Lin RZ, Cohen IS, Entcheva E. Book Distribution of donor cells alters energy requirements for optogenetic control of the heart. City: 2013. Distribution of donor cells alters energy requirements for optogenetic control of the heart. [Google Scholar]
- Ambrosi CM, Entcheva E. Optogenetic control of cardiomyocytes by viral delivery. Methods in Molecular Biology. 2014a doi: 10.1007/978-1-4939-1047-2_19. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosi CM, Entcheva E. Optogenetics’ promise: pacing and cardioversion by light? Future Cardiol. 2014b;10:1–4. doi: 10.2217/fca.13.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrican B, Augustine GJ, Berglund K, Chen S, Chow N, Deisseroth K, Feng G, Gloss B, Hira R, Hoffmann C, Kasai H, Katarya M, Kim J, Kudolo J, Lee LM, Lo SQ, Mancuso J, Matsuzaki M, Nakajima R, Qiu L, Tan G, Tang Y, Ting JT, Tsuda S, Wen L, Zhang X, Zhao S. Next-generation transgenic mice for optogenetic analysis of neural circuits. Front Neural Circuits. 2013;7:160. doi: 10.3389/fncir.2013.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral Characteristics of the Photocycle of Channelrhodopsin-2 and Its Implication for Channel Function. Journal of Molecular Biology. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- Barretto RP, Messerschmidt B, Schnitzer MJ. In vivo fluorescence imaging with high-resolution microlenses. Nat Methods. 2009;6:511–2. doi: 10.1038/nmeth.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiert T, Bruegmann T, Sasse P. Optogenetic activation of Gq signalling modulates pacemaker activity of cardiomyocytes. Cardiovasc Res. 2014;102:507–16. doi: 10.1093/cvr/cvu046. [DOI] [PubMed] [Google Scholar]
- Berndt A, Schoenenberger P, Mattis J, Tye KM, Deisseroth K, Hegemann P, Oertner TG. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci U S A. 2011;108:7595–600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer S, Kelly RG, Miquerol L. Inducible Cx40-Cre expression in the cardiac conduction system and arterial endothelial cells. Genesis. 2011;49:83–91. doi: 10.1002/dvg.20687. [DOI] [PubMed] [Google Scholar]
- Bish LT, Morine K, Sleeper MM, Sanmiguel J, Wu D, Gao G, Wilson JM, Sweeney HL. Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther. 2008;19:1359–68. doi: 10.1089/hum.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–57. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Boyle PM, Entcheva E, Trayanova NA. See the light: can optogenetics restore healthy heartbeats? And, if it can, is it really worth the effort? Expert Rev Cardiovasc Ther. 2014;12:17–20. doi: 10.1586/14779072.2014.864951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle PM, Williams JC, Ambrosi CM, Entcheva E, Trayanova NA. A comprehensive multiscale framework for simulating optogenetics in the heart. Nat Commun. 2013;4:2370. doi: 10.1038/ncomms3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruegmann T, Malan D, Hesse M, Beiert T, Fuegemann CJ, Fleischmann BK, Sasse P. Optogenetic control of heart muscle in vitro and in vivo. Nat Methods. 2010;7:897–900. doi: 10.1038/nmeth.1512. [DOI] [PubMed] [Google Scholar]
- Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res. 2005;65:40–51. doi: 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh AR, Beache GM, Loughran JH, Mewton N, Elmore JB, Kajstura J, Pappas P, Tatooles A, Stoddard MF, Lima JA, Slaughter MS, Anversa P, Bolli R. Administration of cardiac stem cells in patients with ischemic cardiomyopathy: the SCIPIO trial: surgical aspects and interim analysis of myocardial function and viability by magnetic resonance. Circulation. 2012;126:S54–64. doi: 10.1161/CIRCULATIONAHA.112.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–6. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deten A, Millar H, Zimmer H-G. Catheterization of pulmonary artery in rats with an ultraminiature catheter pressure transducer. American Journal of Physiology - Heart and Circulatory Physiology. 2003;285:H2212–H2217. doi: 10.1152/ajpheart.00315.2003. [DOI] [PubMed] [Google Scholar]
- Diester I, Kaufman MT, Mogri M, Pashaie R, Goo W, Yizhar O, Ramakrishnan C, Deisseroth K, Shenoy KV. An optogenetic toolbox designed for primates. Nat Neurosci. 2011;14:387–97. doi: 10.1038/nn.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr T, Denger R, Doerr A, Trautwein W. Ionic currents contributing to the action potential in single ventricular myocytes of the guinea pig studied with action potential clamp. Pflugers Arch. 1990;416:230–7. doi: 10.1007/BF00392058. [DOI] [PubMed] [Google Scholar]
- Efimov IR, Nikolski VP, Salama G. Optical imaging of the heart. Circ Res. 2004;95:21–33. doi: 10.1161/01.RES.0000130529.18016.35. [DOI] [PubMed] [Google Scholar]
- Entcheva E. Cardiac optogenetics. Am J Physiol Heart Circ Physiol. 2013;304:H1179–91. doi: 10.1152/ajpheart.00432.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entcheva E, Bien H. Macroscopic optical mapping of excitation in cardiac cell networks with ultra-high spatiotemporal resolution. Progress in biophysics and molecular biology. 2006;92:232–257. doi: 10.1016/j.pbiomolbio.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Figueiredo M, Lane S, Tang F, Liu BH, Hewinson J, Marina N, Kasymov V, Souslova EA, Chudakov DM, Gourine AV, Teschemacher AG, Kasparov S. Optogenetic experimentation on astrocytes. Experimental Physiology. 2011;96:40–50. doi: 10.1113/expphysiol.2010.052597. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Cocker ED, Piyawattanametha W, Jung JC, Cheung EL, Schnitzer MJ. Fiber-optic fluorescence imaging. Nat Methods. 2005;2:941–50. doi: 10.1038/nmeth820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Nimmerjahn A, Cocker ED, Mukamel EA, Barretto RP, Ko TH, Burns LD, Jung JC, Schnitzer MJ. High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat Methods. 2008;5:935–8. doi: 10.1038/nmeth.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourine AV, Kasymov V, Marina N, Tang F, Figueiredo MF, Lane S, Teschemacher AG, Spyer KM, Deisseroth K, Kasparov S. Astrocytes Control Breathing Through pH-Dependent Release of ATP. Science. 2010;329:571–575. doi: 10.1126/science.1190721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–92. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- Herron TJ, Lee P, Jalife J. Optical Imaging of Voltage and Calcium in Cardiac Cells & Tissues. Circ Res. 2012;110:609–623. doi: 10.1161/CIRCRESAHA.111.247494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesl E, Stieber J, Herrmann S, Feil S, Tybl E, Hofmann F, Feil R, Ludwig A. Tamoxifen-inducible gene deletion in the cardiac conduction system. J Mol Cell Cardiol. 2008;45:62–9. doi: 10.1016/j.yjmcc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Honda Y, Fitzgerald PJ. Frontiers in intravascular imaging technologies. Circulation. 2008;117:2024–2037. doi: 10.1161/CIRCULATIONAHA.105.551804. [DOI] [PubMed] [Google Scholar]
- Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, Yaroshinsky A, Zsebo KM, Dittrich H, Hajjar RJ Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease I. Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation. 2011;124:304–13. doi: 10.1161/CIRCULATIONAHA.111.022889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Valiunas V, Lu Z, Bien H, Liu H, Wang HZ, Rosati B, Brink PR, Cohen IS, Entcheva E. Stimulating cardiac muscle by light: cardiac optogenetics by cell delivery. Circ Arrhythm Electrophysiol. 2011;4:753–60. doi: 10.1161/CIRCEP.111.964247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, McDonald AD, Sasano T, Donahue JK. Targeted modification of atrial electrophysiology by homogeneous transmural atrial gene transfer. Circulation. 2005;111:264–70. doi: 10.1161/01.CIR.0000153338.47507.83. [DOI] [PubMed] [Google Scholar]
- Kim T-i, McCall JG, Jung YH, Huang X, Siuda ER, Li Y, Song J, Song YM, Pao HA, Kim R-H, Lu C, Lee SD, Song I-S, Shin G, Al-Hasani R, Kim S, Tan MP, Huang Y, Omenetto FG, Rogers JA, Bruchas MR. Injectable, Cellular-Scale Optoelectronics with Applications for Wireless Optogenetics. Science. 2013;340:211–216. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, Xie Y, Yan Z, Zhang Y, Chow BY, Surek B, Melkonian M, Jayaraman V, Constantine-Paton M, Wong GK-S, Boyden ES. Independent optical excitation of distinct neural populations. Nat Meth. 2014 doi: 10.1038/nmeth.2836. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinlogel S, Feldbauer K, Dempski RE, Fotis H, Wood PG, Bamann C, Bamberg E. Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nat Neurosci. 2011;14:513–8. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- Kohl P, Gourdie RG. Fibroblast-myocyte electrotonic coupling: Does it occur in native cardiac tissue? J Mol Cell Cardiol. 2014 doi: 10.1016/j.yjmcc.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–6. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughner JI, Marrus SB, Zellmer ER, Weinheimer CJ, MacEwan MR, Cui SX, Nerbonne JM, Efimov IR. A Fully Implantable Pacemaker for the Mouse: From Battery to Wireless Power. PLoS ONE. 2013;8:e76291. doi: 10.1371/journal.pone.0076291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liewald JF, Brauner M, Stephens GJ, Bouhours M, Schultheis C, Zhen M, Gottschalk A. Optogenetic analysis of synaptic function. Nat Methods. 2008;5:895–902. doi: 10.1038/nmeth.1252. [DOI] [PubMed] [Google Scholar]
- Lin JY. Optogenetic excitation of neurons with channelrhodopsins: light instrumentation, expression systems, and channelrhodopsin variants. Prog Brain Res. 2012;196:29–47. doi: 10.1016/B978-0-444-59426-6.00002-1. [DOI] [PubMed] [Google Scholar]
- Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013 doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas R, Sugimori M, Simon SM. Transmission by presynaptic spike-like depolarization in the squid giant synapse. Proc Natl Acad Sci U S A. 1982;79:2415–9. doi: 10.1073/pnas.79.7.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclaurin D, Venkatachalam V, Lee H, Cohen AE. Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin. Proceedings of the National Academy of Sciences. 2013;110:5939–5944. doi: 10.1073/pnas.1215595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marban L, Mendizabal A, Johnston PV, Russell SD, Schuleri KH, Lardo AC, Gerstenblith G, Marban E. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malliaras K, Makkar RR, Smith RR, Cheng K, Wu E, Bonow RO, Marban L, Mendizabal A, Cingolani E, Johnston PV, Gerstenblith G, Schuleri KH, Lardo AC, Marban E. Intracoronary cardiosphere-derived cells after myocardial infarction: evidence of therapeutic regeneration in the final 1-year results of the CADUCEUS trial (CArdiosphere-Derived aUtologous stem CElls to reverse ventricUlar dySfunction) J Am Coll Cardiol. 2014;63:110–22. doi: 10.1016/j.jacc.2013.08.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis EA, Vives KP, Bober J, Leichtle S, Leranth C, Beecham J, Elsworth JD, Roth RH, Samulski RJ, Redmond DE., Jr Comparative transduction efficiency of AAV vector serotypes 1–6 in the substantia nigra and striatum of the primate brain. Mol Ther. 2010;18:588–93. doi: 10.1038/mt.2009.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O’Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, Yizhar O, Deisseroth K. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9:159–72. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall JG, Kim T-i, Shin G, Huang X, Jung YH, Al-Hasani R, Omenetto FG, Bruchas MR, Rogers JA. Fabrication and application of flexible, multimodal light-emitting devices for wireless optogenetics. Nat Protocols. 2013;8:2413–2428. doi: 10.1038/nprot.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesradi M, Genoux A, Cuplov V, Abi Haidar D, Jan S, Buvat I, Pain F. Experimental and analytical comparative study of optical coefficient of fresh and frozen rat tissues. Journal of Biomedical Optics. 2013;18:117010–117010. doi: 10.1117/1.JBO.18.11.117010. [DOI] [PubMed] [Google Scholar]
- Mitrea BG, Caldwell BJ, Pertsov AM. Imaging electrical excitation inside the myocardial wall. Biomedical optics express. 2011;2:620–633. doi: 10.1364/BOE.2.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H, Knöpfel T. Probing neuronal activities with genetically encoded optical indicators: from a historical to a forward-looking perspective. Pflügers Archiv - European Journal of Physiology. 2013;465:361–371. doi: 10.1007/s00424-012-1202-z. [DOI] [PubMed] [Google Scholar]
- Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–84. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden I, Wang J, Lu Y, May T, Lee J, Goo W, O’Shea DJ, Kalanithi P, Diester I, Diagne M, Deisseroth K, Shenoy KV, Nurmikko AV. A coaxial optrode as multifunction write-read probe for optogenetic studies in non-human primates. Journal of Neuroscience Methods. 2013;219:142–154. doi: 10.1016/j.jneumeth.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak CA, Sakai Y, Thattaliyath BD, Mah CS, Byrne BJ. Tissue specific promoters improve specificity of AAV9 mediated transgene expression following intra-vascular gene delivery in neonatal mice. Genet Vaccines Ther. 2008;6:13. doi: 10.1186/1479-0556-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Nagayama T, Mukhopadhyay P, Batkai S, Kass DA. Measurement of cardiac function using pressure-volume conductance catheter technique in mice and rats. Nat Protocols. 2008;3:1422–1434. doi: 10.1038/nprot.2008.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Roska B, Hausser M. Targeting neurons and photons for optogenetics. Nat Neurosci. 2013;16:805–815. doi: 10.1038/nn.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante BA, Giovannone S, Fang-Yu L, Zhang J, Liu N, Kang G, Dun W, Boyden PA, Fishman GI. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–94. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KM, Smith RS, Xu Y, French BA. A single direct injection into the left ventricular wall of an adeno-associated virus 9 (AAV9) vector expressing extracellular superoxide dismutase from the cardiac troponin-T promoter protects mice against myocardial infarction. J Gene Med. 2011a;13:333–41. doi: 10.1002/jgm.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KM, Xu Y, Yang Z, Acton ST, French BA. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther. 2011b;18:43–52. doi: 10.1038/gt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr S. Myofibroblasts in diseased hearts: new players in cardiac arrhythmias? Heart rhythm : the official journal of the Heart Rhythm Society. 2009;6:848–56. doi: 10.1016/j.hrthm.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Rohr S. Arrhythmogenic Implications of Fibroblast-Myocyte Interactions. Circulation: Arrhythmia and Electrophysiology. 2012;5:442–452. doi: 10.1161/CIRCEP.110.957647. [DOI] [PubMed] [Google Scholar]
- Rosen AB, Kelly DJ, Schuldt AJ, Lu J, Potapova IA, Doronin SV, Robichaud KJ, Robinson RB, Rosen MR, Brink PR, Gaudette GR, Cohen IS. Finding fluorescent needles in the cardiac haystack: tracking human mesenchymal stem cells labeled with quantum dots for quantitative in vivo three-dimensional fluorescence analysis. Stem Cells. 2007;25:2128–2138. doi: 10.1634/stemcells.2006-0722. [DOI] [PubMed] [Google Scholar]
- Ruiz O, Lustig BR, Nassi JJ, Cetin A, Reynolds JH, Albright TD, Callaway EM, Stoner GR, Roe AW. Optogenetics through windows on the brain in the nonhuman primate. Journal of Neurophysiology. 2013;110:1455–1467. doi: 10.1152/jn.00153.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E, Bushong EA, Haustein MD, Tong X, Jackson-Weaver O, Kracun S, Xu J, Sofroniew MV, Ellisman MH, Khakh BS. Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J Gen Physiol. 2013;141:633–47. doi: 10.1085/jgp.201210949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedemark-Margulies N, Trapani JG. Tools, methods, and applications for optophysiology in neuroscience. Frontiers in Molecular Neuroscience. 2013:6. doi: 10.3389/fnmol.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Koos T, Buzsáki G. Diode probes for spatiotemporal optical control of multiple neurons in freely moving animals. Journal of Neurophysiology. 2012;108:349–363. doi: 10.1152/jn.00153.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu K, Snider P, Conway SJ, Nagai R. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120:254–65. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez C, Morley GE. The origin and arrhythmogenic potential of fibroblasts in cardiac disease. J Cardiovasc Transl Res. 2012;5:760–7. doi: 10.1007/s12265-012-9408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigts J, Siegle JH, Pritchett DL, Moore CI. The flexDrive: An ultra-light implant for optical control and highly parallel chronic recording of neuronal ensembles in freely moving mice. Frontiers in Systems Neuroscience. 2013:7. doi: 10.3389/fnsys.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wagner F, Borton DA, Zhang J, Ozden I, Burwell RD, Nurmikko AV, van Wagenen R, Diester I, Deisseroth K. Integrated device for combined optical neuromodulation and electrical recording for chronic in vivo applications. Journal of Neural Engineering. 2012;9:016001. doi: 10.1088/1741-2560/9/1/016001. [DOI] [PubMed] [Google Scholar]
- Williams JC, Xu J, Lu Z, Klimas A, Chen X, Ambrosi CM, Cohen IS, Entcheva E. Computational Optogenetics: Empirically-Derived Voltage- and Light-Sensitive Channelrhodopsin-2 Model. PLoS Comput Biol. 2013;9:e1003220. doi: 10.1371/journal.pcbi.1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PD, Ranjzad P, Kakar SJ, Kingston PA. Development of viral vectors for use in cardiovascular gene therapy. Viruses. 2010;2:334–71. doi: 10.3390/v2020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Abilez OJ, Kuhl E. Computational Optogenetics: A Novel Continuum Framework for the Photoelectrochemistry of Living Systems. J Mech Phys Solids. 2012;60:1158–1178. doi: 10.1016/j.jmps.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Gutbrod SR, Bonifas AP, Su Y, Sulkin MS, Lu N, Chung HJ, Jang KI, Liu Z, Ying M, Lu C, Webb RC, Kim JS, Laughner JI, Cheng H, Liu Y, Ameen A, Jeong JW, Kim GT, Huang Y, Efimov IR, Rogers JA. 3D multifunctional integumentary membranes for spatiotemporal cardiac measurements and stimulation across the entire epicardium. Nat Commun. 2014;5:3329. doi: 10.1038/ncomms4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–8. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Yu J, Boyle PM, Ambrosi CM, Trayanova N, Entcheva E. High-throughput contactless optogenetic assay for cellular coupling: Illustration by ChR2-light-sensitized cardiac fibroblasts and cardiomyocytes. Circulation. 2013;128:A14943. [Google Scholar]
- Yue Y, Ghosh A, Long C, Bostick B, Smith BF, Kornegay JN, Duan D. A single intravenous injection of adeno-associated virus serotype-9 leads to whole body skeletal muscle transduction in dogs. Mol Ther. 2008;16:1944–52. doi: 10.1038/mt.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–9. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zlochiver S, Munoz V, Vikstrom KL, Taffet SM, Berenfeld O, Jalife J. Electrotonic myofibroblast-to-myocyte coupling increases propensity to reentrant arrhythmias in two-dimensional cardiac monolayers. Biophysical journal. 2008;95:4469–80. doi: 10.1529/biophysj.108.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsebo K, Yaroshinsky A, Rudy JJ, Wagner K, Greenberg B, Jessup M, Hajjar RJ. Long-term effects of AAV1/SERCA2a gene transfer in patients with severe heart failure: analysis of recurrent cardiovascular events and mortality. Circ Res. 2014;114:101–8. doi: 10.1161/CIRCRESAHA.113.302421. [DOI] [PubMed] [Google Scholar]