Abstract
Sulfated low molecular weight lignins (LMWLs) have been found to bind in the heparin binding sites of coagulation proteinases. LMWLs represent a library of diverse non-carbohydrate, aromatic molecules which are structures different from heparin, but still potently inhibit thrombin and factor Xa. To better understand their mechanism of action, we studied the effects of three sulfated LMWLs (CDSO3, FDSO3, and SDSO3) on the active sites of thrombin and factor Xa. LMWLs were found to uniformly inhibit the catalytic activity of thrombin and factor Xa, regardless of the substrate used. Michaelis-Menten kinetic studies indicate that maximal velocity of hydrolysis of each chromogenic substrate decreases significantly in the presence of sulfated LMWLs, while the effect on Michaelis constant is dependent on the nature of the substrate. These studies indicate that LMWLs inhibit thrombin and factor Xa through allosteric disruption of the catalytic apparatus, specifically through the catalytic step. As opposed to heparin, LMWLs significantly alter the binding of the active site fluorescent ligand p-aminobenzamidine. LMWLs also had a greater effect on the molecular orientation of fluorescein-labeled His 57 than heparin. The molecular geometry surrounding the most important catalytic amino acid, Ser 195, was significantly altered by the binding of LMWLs while heparin had no measurable effect on Ser 195. These results further advance the concept of sulfated LMWLs as heparin mimics and will aid the design of anticoagulants based on their novel scaffold.
Keywords: Sulfated lignins, heparin, anticoagulation, allosteric mechanism, exosite II, thrombin
Introduction
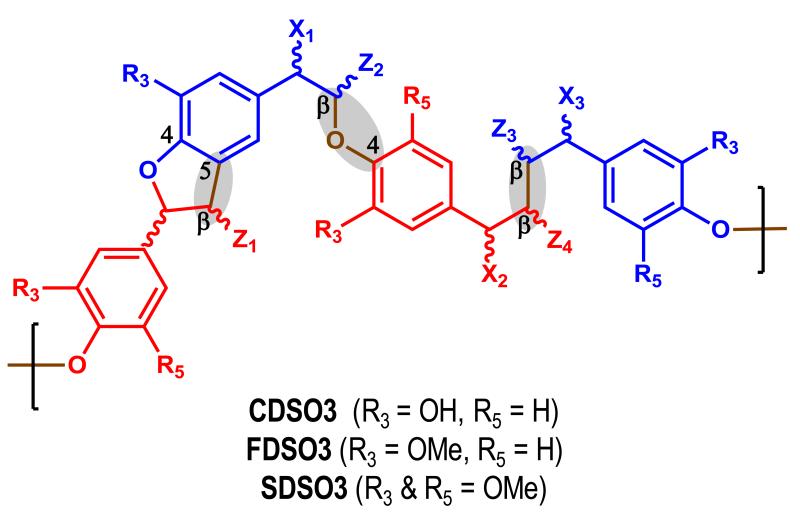
With a goal of developing better anticoagulants that are less polyanionic and more hydrophobic than heparins, we recently prepared sulfated low molecular weight lignins (LMWLs), as functional mimetics of heparin. These designed molecules, CDSO3, FDSO3 and SDSO3 – were prepared in a simple, two-step chemo-enzymatic process involving horseradish peroxidase catalyzed oligomerization of 4-hydroxycinnamic acid monomers followed by the chemical sulfation of the resulting LMWLs with triethylamine-sulfur trioxide complex [1]. In terms of structural polydispersity and heterogeneity, sulfated LMWLs are similar to low molecular weight heparins (LMWHs). Sulfated LMWLs are composed of many oligomeric chains of varying lengths and contain different inter-monomeric linkages such as β-O-4 and β-5 (Figure 1). Yet, sulfated LMWLs are completely unlike LMWHs with respect to the nature of their chemical backbone. In contrast to the highly anionic scaffold of the heparins, sulfated LMWLs possess a highly aromatic scaffold with fewer anionic groups. In fact, from a structural perspective, sulfated LMWLs are unlike any other class of anticoagulant investigated to-date, including the heparins, the coumarins, the hirudins, the peptidomimetics and the small molecule direct inhibitors.
Figure 1.
Sulfated low molecular weight lignins (LMWLs) are complex three-dimensional oligomers obtained from enzymatic condensation of 4-hydroxycinnamic acid monomers using horseradish peroxidase followed by chemical sulfation using sulfur trioxide. The oligomers primarily contain β-O-4, β -5 and β - β inter-residue linkages (shown shaded). Other less common linkages, e.g., 5-5, are also present (not shown). X1, X2 and X3 are substituents at the a-position and may be H, -OH, or -OSO3Na. Z1, Z2 and Z3 may be -H or -COONa. R3 and R5 may be either -H or -OMe depending upon the starting monomer, as shown. These variations generate a large number of sequences.
Functionally, the sulfated LMWLs display an anticoagulation profile in human plasma and blood similar to that of LMWHs [2]. Studies also suggested that CDSO3, FDSO3 and SDSO3 are capable of binding to antithrombin and inhibiting factor Xa and thrombin in an antithrombin–dependent as well as independent manner suggesting a potentially novel mode of inhibition [1,3]. Subsequent work showed that CDSO3, FDSO3, and SDSO3 possess high selectivity for inhibiting thrombin and factor Xa over other enzymes of the coagulation cascade [4, 5]. CDSO3 inhibits thrombin through allosteric disruption of its catalytic apparatus and preferentially binds in or near the region formed by anion-binding exosite II [6], which is also known as the heparin binding domain of thrombin [7]. Although heparin binds to exosite II, this is a non-inhibitory interaction [8]. Sulfated LMWLs appear to be the first molecules that induce allosteric inhibition of pro-coagulant proteinases, especially thrombin and factor Xa, through the heparin binding site. These prior studies suggest that sulfated LMWLs are structurally, functionally and mechanistically a very interesting class of molecules that may lead to novel anticoagulants.
Based on the observations of our previous work, it became apparent that our sulfated LMWLs have a predilection for interacting with heparin binding proteins. Given the large number of heparin binding proteins within the coagulation cascade, we attempted to understand the interaction of our heparin mimics with these proteins. Sulfated LMWLs potently and selectively inhibit four coagulation serine proteases by specifically recognizing unique structural motifs [4]. In general, the sulfated LMWLs bind thrombin, FIXa, FXa and FXIa through recognition of a heparin binding site that contains a cysteine-constrained loop. However, these prior studies have only looked at their ability to inhibit each serine protease with one substrate. It is well established in the literature that the same ligand-enzyme interaction can have different allosteric effects depending on the substrate used in the reaction [9, 10]. Studies with hirugen, the C-terminal dodecapeptide of hirudin that is known to bind in exosite I of thrombin, show that the kCAT/KM value increases or decreases approximately 2-fold depending on the nature of chromogenic substrate [9, 10]. This phenomenon raised the question of whether our exosite II binding LMWLs would exhibit similar mixed inhibitory – enhancing effects on serine protease catalysis depending on the substrate used or if our molecules behave as strict allosteric inhibitors. A corollary to this question would be to understand what changes occur to the molecular geometry and structure of catalytic active site of serine proteases during binding of LMWLs. By understanding how the active structure changes during binding, it may be possible to better understand the inhibitory mechanism of LMWLs.
In this paper, we report that our sulfated LMWLs allosterically inhibit thrombin and factor Xa regardless of the structure of the chromogenic substrate used. Additionally, we examined the effects of the binding of sulfated LMWLs on the active site of thrombin by using three complimentary fluorescent probes. Although both sulfated LMWLs and heparin occupy exosite II [4, 6], fluorescence studies show a significant difference in the way in which LMWLs alter the active site of thrombin compared to heparin. This implies that sulfated LMWL binding realigns amino acids critical to thrombin’s activity in manner deleterious to efficient catalysis. By using covalent and non-covalent fluorescent active site probes, we show that sulfated LMWLs are able to alter the structure of the active site and more specifically, alter Ser195 and His57, members of thrombin’s catalytic triad.
Methods
Proteins and Chemicals
Sulfated dehydropolymers CDSO3, FDSO3 and SDSO3 (Fig. 1B) were prepared in two steps from 4-hydroxycinnamic acid monomers, caffeic acid (CA), ferulic acid (FA) and sinapic acid (SA) using chemo-enzymatic synthesis described by Monien et al [1]. The human plasma proteinases, factor Xa, α-thrombin and α-thrombin-FFPRCK (fluorescein-labeled thrombin), were purchased from Haematologic Technologies (Essex Junction, VT). Dansyl-labeled thrombin was prepared by the method described by Berliner et al [11]. Stock solutions of proteins were prepared in 20 mM sodium phosphate buffer, pH 7.4, containing 100 mM NaCl and 2.5 mM CaCl2 (thrombin) or 5 mM MES buffer, pH 5.45 containing 100 mM NaCl (factor Xa). Chromogenic substrates Spectrozyme TH (H-D-hexahydrotyrosol-Ala-Arg-p-nitroanilide), Spectrozyme FXa (Methoxycarbonyl-D-cyclohexylglycyl-Gyl-Arg-p-nitroanilide), Spectrozyme Pro (H-D- cyclohexylglycyl-propyl-arginine-p-nitroanilide) and Spectrozyme PCa (H-D-γ-carbobenzoxyl-lysyl-prolyl-arginine-p-nitroanilide) were purchased from American Diagnostica (Greenwich, CT). S-2238 (H-D-Phenylalanyl-L-pipecolyl-L-arginine-p-nitroaniline dihydrochloride) and S-2772 (N-α-Acetyl-D-arginyl-L-glycyl-L-arginine-p-nitroaniline dihydrochloride) were purchased from DiaPharm Group (West Chester, OH). Dansyl fluoride was purchased from TCI chemicals. Unfractionated heparin (MW ~15,000 Da) and the active site fluorophore p-aminobenzamidine (PABA) were purchased from Sigma Chemicals (St. Louis, MO). All other chemicals were analytical reagent grade from either Sigma Chemicals (St. Louis, MO) or Fisher (Pittsburgh, PA) and used without further purification.
Michaelis-Menten Kinetics of Chromogenic Substrate Hydrolysis by Thrombin or Factor Xa in the Presence of CDSO3
The initial rate of chromogenic substrate hydrolysis by thrombin or factor Xa was monitored from the linear increase in absorbance at 405 nm corresponding to less than 10% consumption of the substrate. The initial rate was measured as a function of various concentrations of the substrate (0.2 to 1000 μM) in the presence of fixed concentration of CDSO3 (10–300 nM) in either 20 mM Tris-HCl buffer, pH 7.4, containing 100 mM NaCl, 2.5 mM CaCl2 and 0.1 % PEG8000 at 25 °C for thrombin assays or 20 mM Tris-HCl buffer, pH 7.4, containing 100 mM NaCl, 2.5 mM CaCl2 and 0.1 % PEG8000 at 25 °C for factor Xa assays. The concentration of thrombin used for each substrate was as follows: 1 nM thrombin for Spectrozyme TH, 4 nM thrombin for Spectrozyme FXa, 4.5 nM thrombin for Spectrozyme Pro, 4.5 nM thrombin for Spectrozyme PCa and 0.2 nM thrombin for S-2238. The concentration of factor Xa used for each substrate was as follows: 4 nM FXa for Spectrozyme TH, 1 nM FXa for Spectrozyme FXa, 2.1 nM FXa for Spectrozyme Pro and 2 nM FXa for S-2772. The data were fitted using the standard Michaelis-Menten Eq. 1 to determine the KM and VMAX.
| Eq. 1 |
Fluorescence Spectroscopic Studies
Fluorescence spectroscopy was used to measure the affinity of sulfated LMWLs for thrombin, which was either covalently (fluorescein-FPRK or dansyl) or non-covalently (PABA) complexed to a fluorophore. The buffer was 20 mM sodium phosphate pH 7.4, 100 mM NaCl, 0.1 mM EDTA, 0.1% PEG 8000 and the temperature was 25 °C [8, 12]. Thrombin-FFPRCK has a fluorescein covalently attached to the active site His57 of thrombin through a D-Phe-Pro-Arg linkage using a method developed by Bock [13]. Human thrombin was labeled at the active site (Ser195) by reacting thrombin with Dansyl fluoride to yield Dansyl-labeled thrombin by the method described by Berliner et al [11]. Fluorescence experiments were performed using a QM4 fluorometer (Photon Technology International, Birmingham, NJ). Equilibrium dissociation constants (KD) for the sulfated LMWLs - Thrombin complexes were determined by titrating the LMWLs into a solution of thrombin-fluorophore probe complex (60 nM FFPRCK complex (λEX = 490 nm); 360 nM dansyl thrombin complex (λEX = 280 nm); 1.2 uM thrombin - PABA complex (λEX = 345 nm)) and monitoring the decrease in fluorescence at 520 nm, or 550 nm or 370 nm, respectively [8,9,11]. Inner filter effects were calculated from A280 values of sulfated LMWLs and the geometry of fluorescence cuvette using the equation developed by Parker and Barnes [14]. These effects were found to be close to zero for thrombin-FFPRCK complex, ~1 – 18% for thrombin-dansyl complex and 1 – 53% for thrombin – PABA complex. These effects were subtracted from the fluorescence of the complex to obtain the change in fluorescence due to thrombin binding (ΔFOBSt). The slit widths on the excitation and emission side were both 2 mm. The decrease in fluorescence signal due to the formation of the complex was fit to the quadratic equilibrium binding equation 2 to obtain the KD of interaction. In this equation, ΔF represents the change in fluorescence due to the formation of the complex following each addition of the ligand ([LMWL]O) from the initial fluorescence FO and ΔFMAX represents the maximal change in fluorescence observed on saturation of thrombin ([TH]O). A binding stoichiometry of 1:1 was assumed for the sulfated LMWL – thrombin interaction.
| Eq. 2 |
Results
Effects of CDSO3 on the Michaelis–Menten Kinetics of Thrombin Hydrolysis of Various Chromogenic Substrates
Previous work on the allosteric modulation of thrombin catalysis has shown that some exosite I ligands, e.g., hirugen or thrombomodulin fragments, decrease the rate of hydrolysis for some substrates (S2266, SPXa, and BzVGR) but increase the rate for other (S2238, S2288 and SPTH) [15]. This suggests that structural changes within the active site allosterically initiated by certain exosite I ligands create a new binding pocket for small chromogenic substrates. Depending on the structure of the chromogenic substrate, the new active site molecular geometry may improve substrate binding resulting in more efficient catalysis or reduced substrate binding resulting in inhibition. To investigate whether sulfated LMWLs also introduce such variable effects, we studied the kinetics of thrombin hydrolysis of Spectrozyme FXa, Spectrozyme TH, Spectrozyme Pro, Spectrozyme PCa and S-2338 in the presence of CDSO3.
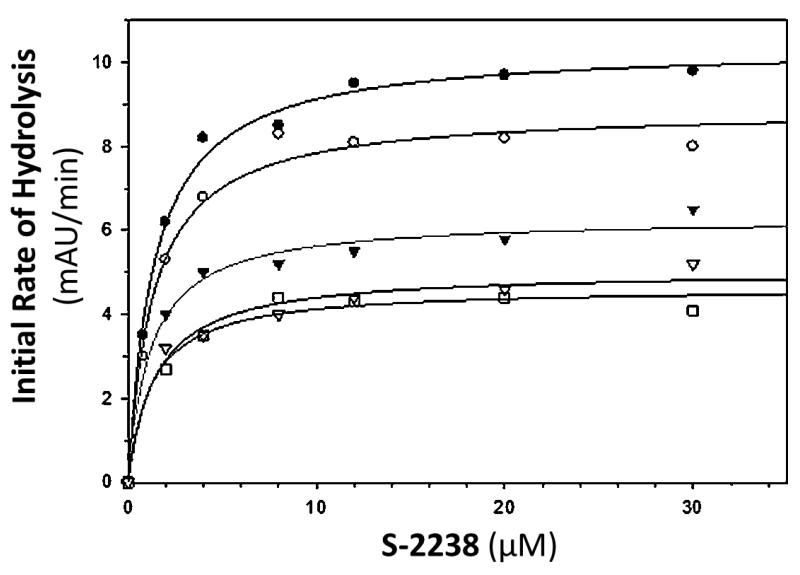
Table 1 shows the apparent KM and VMAX values for five different chromogenic substrates. In every case, the VMAX was observed to decrease in a concentration dependent manner, indicating that regardless of substrate used, CDSO3 was capable of making thrombin catalysis dysfunctional. For the hydrolysis of S-2238 by thrombin (figure 2), there was a concentration dependent decrease in VMAX without change in KM. This is representative of noncompetitive inhibition, because CDSO3 has no significant difference in affinity for thrombin or the thrombin:S-2238 complex. At the highest concentration tested, CDSO3 decreased the maximum rate of the reaction by 2.2 fold. For the thrombin hydrolysis of Spectrozyme TH, FXa and Pro, both KM and VMAX were found to decrease as a function of CDSO3 concentration. This is indicative of mixed inhibition where CDSO3 appears to function as an allosteric binding inhibitor (decreased VMAX) but has a greater affinity for the thrombin:substrate complex than thrombin alone (decreased KM). At the highest concentration tested, CDSO3 decreased the maximum rate of the reaction by 1.6 – 3 fold, while the substrate concentration at which the reaction rate is half VMAX decreased by 1.6 – 2.6 fold. Conversely, in the case of thrombin hydrolysis of Spectrozyme PCa, there was an increase in KM and a decrease in VMAX. This also suggests mixed inhibition, but in this case, CDSO3 has a greater affinity for thrombin alone then for the thrombin:Spectrozyme PCa complex. At the highest concentration tested, CDSO3 decreased the maximum rate of the reaction by 1.6 fold while the Michaelis constant increased by 2 fold. Overall, these results suggest that the five chromogenic substrates are sensing similar, deleterious changes in the active site conformation by CDSO3, but because of structural differences in the substrates, the affinity of enzyme substrate complex differs.
Table 1.
Effects of CDSO3 on the Michaelis–Menten Parameters of Thrombin and Various Substratesa
| [CDS03] (nM) | Substrate | Km (μM)b | Vmax (mAU/min)b |
|---|---|---|---|
| 0 | Spectrozyme TH | 2.2 ± 0.2 | 21 ± 0.7 |
| 30 | ” | 1.3 ± 0.1 | 15 ± 0.3 |
| 105 | ” | 1.2 ± 0.1 | 11 ± 0.3 |
| 300 | ” | 0.85 ± 0.1 | 9.5 ± 0.2 |
| 0 | S-2238 | 1.3 ± 0.1 | 10.4 ± 0.2 |
| 1 | ” | 1.3 ± 0.2 | 8.8 ± 0.2 |
| 10.5 | ” | 1.1 ± 0.2 | 6.2 ± 0.2 |
| 30 | ” | 1.4 ± 0.3 | 5.0 ± 0.2 |
| 105 | ” | 1.2 ± 0.3 | 4.6 ± 0.2 |
| 0 | Spectrozyme FXa | 61 ± 3 | 36 ± 0.6 |
| 10.5 | ” | 42 ± 4 | 31 ± 0.8 |
| 30 | ” | 40 ± 5 | 23 ± 0.9 |
| 105 | ” | 40 ± 5 | 18 ± 0.7 |
| 203 | ” | 39 ± 10 | 12 ± 1 |
| 0 | Spectrozyme Pro | 1.3 ± 0.2 | 36 ± 1 |
| 30 | ” | 0.5 ± 0.05 | 23 ± 0.3 |
| 120 | ” | 0.7 ± 0.1 | 21 ± 0.5 |
| 0 | Spectrozyme PCa | 4.5 ± 0.5 | 107 ± 3 |
| 30 | ” | 7.3 ± 2.0 | 82 ± 4 |
| 90 | ” | 9.1 ± 3.2 | 67 ± 5 |
KM and VMAX values of substrate hydrolysis by human thrombin were measured as described in Experimental Procedures. mAU indicates milliabsorbance units.
Error represents ±1 SE.
Figure 2.
Michaelis–Menten kinetics of S-2238 hydrolysis by thrombin in the presence of CDSO3. The initial rate of hydrolysis at various substrate concentrations was measured in pH 7.4 buffer as described in Experimental Procedures. The concentrations of CDSO3 chosen in the study were 0 (●), 1 nM (○), 10.5 nM (▼), 30 nM (△), 105 nM (□). Solid lines represent nonlinear regressional fits to the data by the Michaelis–Menten eq 1.
Effects of CDSO3 on the Michaelis–Menten Kinetics of Factor Xa Hydrolysis of Various Chromogenic Substrates
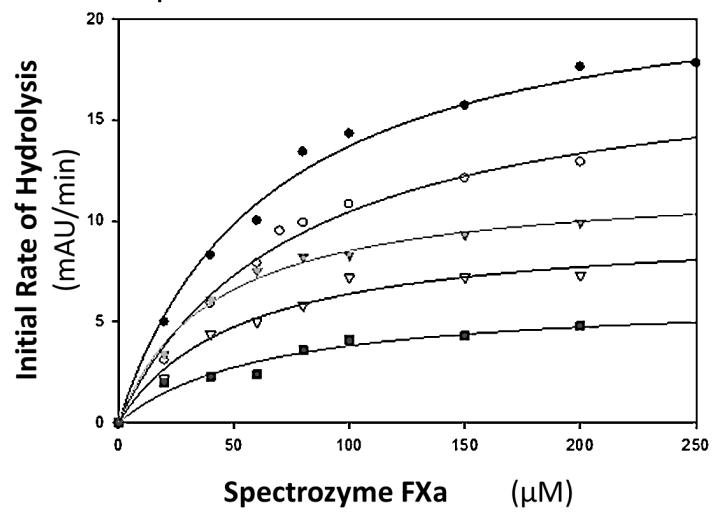
Table 2 shows the apparent KM and VMAX values for the hydrolysis of four different chromogenic substrates in the presence of factor Xa. As was seen in the case with thrombin, the VMAX of factor Xa was observed to decrease in a concentration dependent manner, indicating that regardless of substrate used, CDSO3 was capable of making the catalytic step dysfunctional. For the hydrolysis of Spectrozyme FXa by factor Xa (figure 3), there was a concentration dependent decrease in VMAX without change in KM. This is representative of noncompetitive inhibition, because CDSO3 has no significant difference in affinity for factor Xa or the factor Xa:substrate complex. At the highest concentration tested, CDSO3 decreased the maximum rate of the reaction by 3.5 fold. For factor Xa hydrolysis of S-2772, Spectrozyme TH and Pro, both KM and VMAX were found to decrease as a function of CDSO3 concentration. This is indicative of mixed inhibition where CDSO3 appears to function as an allosteric binding inhibitor but has a greater affinity for the factor Xa:substrate complex than factor Xa alone. At the highest concentration tested, CDSO3 decreased the maximum rate of the reaction by 2.2 – 10 fold, while KM decreased by 2.4 – 9.1 fold.
Table 2.
Effects of CDSO3 on the Michaelis–Menten Parameters of Factor Xa and Various Substrates
| [CDS03] (nM) |
Substrate |
Km (μM)b |
Vmax (mAU/min)b |
|---|---|---|---|
| 0 | Spectrozyme FXa | 64 ± 7 | 22 ± 0.8 |
| 10.5 | ” | 76 ± 11 | 18 ± 1 |
| 30 | ” | 41 ± 5 | 12 ± 0.5 |
| 105 | ” | 52 ± 12 | 9.7 ± 0.8 |
| 203 | ” | 63 ± 19 | 6.2 ± 0.7 |
| 0 | S-2772 | 212 ± 21 | 34 ± 1 |
| 10.5 | ” | 60 ± 5 | 12 ± 0.2 |
| 105 | ” | 42 ± 15 | 3.4 ± 0.3 |
| 0 | Spectrozyme TH | 597 ± 180 | 80 ± 16 |
| 10.5 | ” | 390 ± 105 | 50 ± 8 |
| 30 | ” | 168 ± 75 | 13 ± 2 |
| 83 | ” | 65 ± 30 | 6.6 ± 0.7 |
| 0 | Spectrozyme Pro | 188 ± 28 | 119 ±7 |
| 7.5 | ” | 112 ± 8 | 72 ± 2 |
| 30 | ” | 77 ± 9 | 53 ± 2 |
KM and VMAX values of substrate hydrolysis by human factor Xa were measured as described in Experimental Procedures. mAU indicates milliabsorbance units.
Error represents ±1 SE.
Figure 3.
Michaelis–Menten kinetics of Spectrozyme FXa hydrolysis by factor Xa in the presence of CDSO3. The initial rate of hydrolysis at various substrate concentrations was measured in pH 7.4 buffer as described in Experimental Procedures. The concentrations of CDSO3 chosen in the study were 0 (●), 10.5 nM (○), 30 nM (▼), 105 nM (△), 203 nM (■). Solid lines represent nonlinear regressional fits to the data by the Michaelis–Menten eq 1.
Effects of Sulfated Low Molecular Weight Lignins and Unfractionated Heparin upon Binding to the Reversible Thrombin:PABA complex
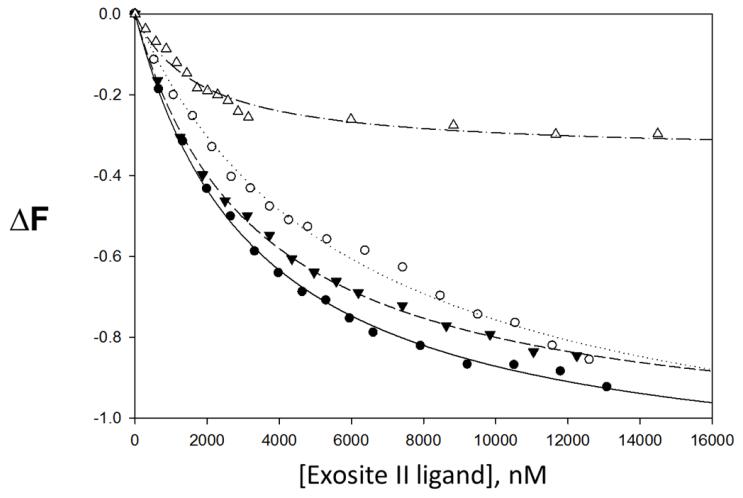
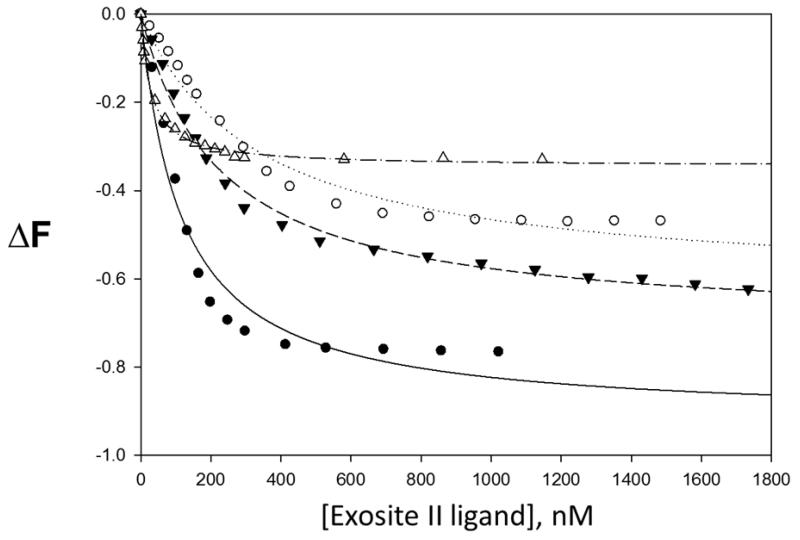
The fluorescent organic small molecule, para-amionbenzamidine (PABA) is a non-selective serine protease active site binding ligand. When dissolved alone in solution, it emits fluorescent energy after excitation. However, when bound to the active site of a serine protease like thrombin, the fluorescent emission signal is significantly greater. Since PABA fits into the active site in manner similar the chromogenic substrates discussed above, PABA was used to test the effects of sulfated LMWLs and heparin on the gross structural conformation of the thrombin active site. In this study, we found that the interaction of our sulfated LMWLs with thrombin resulted in a significant decrease in PABA fluorescence compared to heparin (see Table 3). Binding of sulfated LMWLs to thrombin resulted in ~100% decrease in PABA fluorescence (fluorescent emission signal similar to background signal of PABA alone in solution), which could be fitted by a standard quadratic binding equation to obtain the equilibrium dissociation constant KD(figure 4). The KD for sulfated LMWLs binding to thrombin in the presence of PABA ranges from 3.3-5.9 μM. In contrast to sulfated LMWLs, heparin is only able to decrease the fluorescence of PABA by 34%. The resulting KD is 1.7 μM, which is 1.9 – 3.4 times greater than sulfated LMWLs. It is important to note that the affinities measured using PABA are much weaker than those measured using fluorescein or dansyl probes (see below). The exact reason for this difference is not clear but may arise from several factors including the poor sensitivity of the probe, the significantly higher inner filter correction, and/or the weak affinity of the reversible probe for thrombin. Despite this anomaly, the allosteric effects, or lack thereof, observed for both heparin and LMWLs are likely to be accurate representation of the system.
Table 3.
Interaction of Sulfated LMWLs and Heparin with Thrombin Using PABA Probesa
| Ligand |
KD(μM)b |
ΔF
MAX
b
|
|---|---|---|
| CDS03 | 3.33 ± 0.12 | −1.16 ± 0.01 |
| FDS03 | 5.95 ± 0.44 | −1.20 ± 0.04 |
| SDS03 | 3.36 ± 0.11 | −1.07 ± 0.01 |
| UFH | 1.74 ± 0.21 | −0.34 ± 0.01 |
KD and ΔFMAX values of LMWLs and heparin were measured as described in Experimental Procedures. PABA reversibly binds to the active site of thrombin.
Error represents ±1 SE.
Figure 4.
Change in thrombin↔p-aminobenzamidine (PABA) complex fluorescence with increasing concentrations of exosite II ligand. • CDSO3, ○ FDSO3, ▼ SDSO3, △ heparin. ΔF is the relative change in fluorescence. Line of best fit for CDSO3 (—), FDSO3 (----), SDSO3 (---) and heparin (-----). PABA reversibly binds to the active site of thrombin.
Effects of Sulfated Low Molecular Weight Lignins and Unfractionated Heparin upon Binding to the Covalent Thrombin:Fluorescein complex
H57 is a component of the catalytic triad of thrombin and all serine proteases [16]. Since the proper alignment of H57 is crucial for effective catalysis, it stands to reason that the allosteric inhibitors, sulfated LMWLs, change the molecular orientation of residues like H57, while non-inhibitory molecules like heparin should not affect H57 to the same extent. By covalently attaching a fluorescent probe to H57, changes in the chemical environment around the probe and H57 as a result of being repositioned by allosteric binding from a ligand would be detected. A fluorescein probe was attached to H57 as previously described [13].
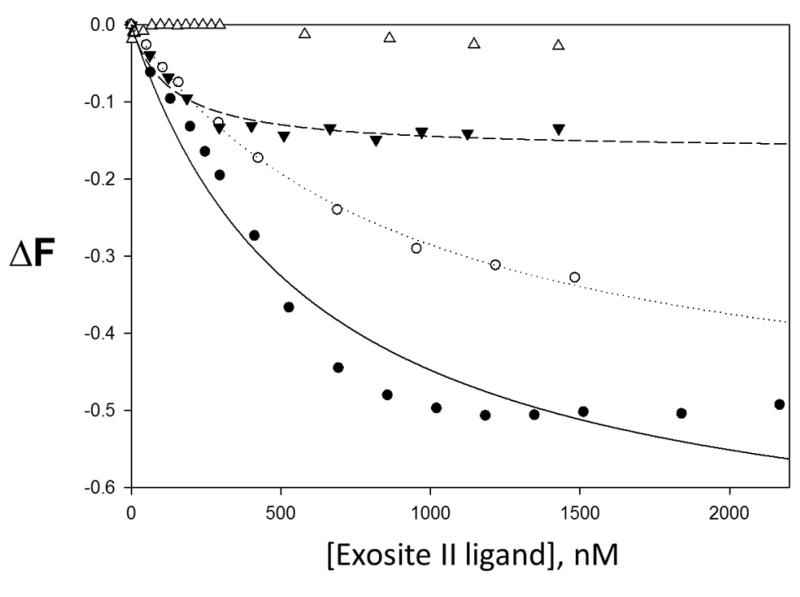
It was observed that as the concentration of sulfated LMWLs and heparin where increased, the fluorescent signal of fluorescein decreased (see Table 4). The binding of sulfated LMWLs to fluorescein-labeled thrombin resulted in 62-91% decrease in fluorescein fluorescence, which could be fitted by a standard quadratic binding equation to obtain the equilibrium dissociation constant KD(figure 5). The KD for sulfated LMWLs binding to fluorescein-labeled thrombin ranges from 115 - 332 nM. In contrast to sulfated LMWLs, heparin decreased the fluorescence of fluorescein by 34%. The resulting KD is 28 nM, which is 4.1 - 11.8 times greater than sulfated LMWLs.
Table 4.
Interaction of Sulfated LMWLs and Heparin with Fluorescein labeled Thrombina
| Ligand |
KD(nM)b |
ΔF
MAX
b
|
|---|---|---|
| CDS03 | 115 ± 20 | −0.91± 0.04 |
| FDS03 | 332 ± 43 | −0.62 ± 0.02 |
| SDS03 | 225 ± 15 | −0.70 ± 0.01 |
| UFH | 28 ± 1 | −0.34 ± 0.01 |
KM and ΔFMAX values of LMWLs and heparin were measured as described in Experimental Procedures. His 57 is the active site amino acid labeled by fluorescein.
Error represents ±1 SE.
Figure 5.
Change in thrombin:fluorescein complex fluorescence with increasing concentrations of exosite II ligand. ● CDSO3, ○ FDSO3, ▼ SDSO3, △ heparin. ΔF is the relative change in fluorescence. Line of best fit for CDSO3 (—), FDSO3 (----), SDSO3 (---) and heparin (-----). His 57 is the active site amino acid labeled by the fluorescein probe.
Effects of Sulfated Low Molecular Weight Lignins and Unfractionated Heparin upon Binding to the Covalent Thrombin:Dansyl complex
S195 is the most important component of the catalytic triad of thrombin, as it is directly responsible for initiating proteolytic cleavage of the peptide bond [16]. Since the proper alignment of S195 is crucial for effective catalysis, it stands to reason that the allosteric inhibitors, like sulfated LMWLs, change the molecular orientation of residues like S195, while non-inhibitory molecules like heparin should not affect S195 significantly, if at all. By covalently attaching a fluorescent probe to S195, changes in the chemical environment around the probe and S195 would be detected. A dansyl probe was attached to S195 as previously described [11].
It was observed that as the concentrations of sulfated LMWLs was increased, the fluorescence signal of dansyl group decreased (see Table 5). The binding of sulfated LMWLs to dansyl-labeled thrombin resulted in 16 - 71% decrease in dansyl fluorescence, which could be fitted by a standard quadratic binding equation to obtain the equilibrium dissociation constant KD(figure 6). The KD for sulfated LMWLs binding to dansyl-labeled thrombin ranges from 130 – 919 nM. In contrast to sulfated LMWLs, heparin had no detectable effect on the fluorescence of dansyl-labeled thrombin.
Table 5.
Interaction of Sulfated LMWLs and Heparin with Dansyl labeled Thrombina
| Ligand | KD(nM)b | ΔF MAX b |
|---|---|---|
| CDS03 | 601 ± 122 | −0.71 ± 0.05 |
| FDS03 | 919 ± 77 | −0.54 ± 0.02 |
| SDS03 | 130 ± 29 | −0.16 ± 0.01 |
| UFH | No interaction detected | |
KD and ΔFMAX values of LMWLs and heparin were measured as described in Experimental Procedures. Ser 195 is the active site amino acid labeled by the dansyl probe.
Error represents ±1 SE.
Figure 6.
Change in thrombin:dansyl complex fluorescence with increasing concentrations of exosite II ligand. ● CDSO3, ○ FDSO3, ▼ SDSO3, △ heparin. ΔF is the relative change in fluorescence. Line of best fit for CDSO3 (—), FDSO3 (----), SDSO3 (---) and heparin (-----). Ser 195 is the active site amino acid labeled by the dansyl probe.
Discussion
Earlier work on the allosteric modulation of thrombin catalysis has shown that some exosite I ligands, e.g., hirugen or thrombomodulin fragments, decrease the rate of hydrolysis for some substrates (S2266, SPXa, and BzVGR) but increase the rate for other (S2238, S2288 and SPTH) [10]. Since our sulfated LMWLs represent a novel allosteric mechanism of action, we studied the effect of sulfated LMWLs on thrombin and FXa catalysis of various small non-physiologic substrates to determine if certain substrates are not affected by sulfated LMWLs. Furthermore, the type of allosteric inhibition (non-competitive vs. mixed) had not previously been determined. CDSO3 showed a wide spectrum of inhibitory mechanisms against thrombin when various substrates were used. With respect to the substrate S-2238, CDSO3 acted as a traditional non-competitive inhibitor indicating an equal binding preference for thrombin and the thrombin:S-2238 complex. For three of the substrates used (Spectrozyme TH/FXa/Pro), CDSO3 preferentially bound to the thrombin:substrate complex. In the case of Spectrozyme PCa, the data indicated that CDSO3 bound more favorably to thrombin alone as opposed to the thrombin:substrate complex. Although not every known thrombin substrate was studied, these experiments continue to reinforce that sulfated LMWLs are uniformly exosite II – directed allosteric inhibitors. Interestingly, mixed inhibition was observed for some substrates, which is also a form of allosteric modulation and probably indicates some level of cooperativity arising from substrate binding in the active site on the conformation of exosite II. This aspect will have to be investigated more rigorously in future studies.
As was seen for thrombin, CDSO3 inhibits factor Xa regardless of the substrate used. CDSO3 only functions as a non-competitive inhibitor for one of the four substrates studied while the rest of the substrates show mixed allosteric inhibition. Therefore, like thrombin, the heparin binding site of factor Xa also appears to be altered by substrate binding in the factor Xa active site. In spite of these structural changes to the heparin binding site, CDSO3 is still capable of disrupting the factor Xa active site. It is important to note that these results for thrombin and factor Xa were obtained using small non-physiologic substrates and not endogenous substrates such as fibrinogen, factor V, factor II, etc. It remains to be seen whether the potency and mode of inhibition with endogenous substrates is maintained. However, our earlier whole blood and plasma studies have shown strong anticoagulation potential of these sulfated LMWLs alluding to a strong possibility of allosteric disruption of catalysis even when macromolecular substrates are present.
Despite having extensive indirect evidence that sulfated LMWLs where capable of altering the active site of thrombin, there was no direct evidence that sulfated LMWLs alter the conformation of the active site. Therefore, fluorescence binding assays were designed to probe the thrombin active site. The PABA assay showed that LMWL binding displaces PABA from the thrombin active site. This implies that the critical interactions necessary for PABA binding are being lost by allosteric alterations in the thrombin active site. Interestingly, heparin is also capable of displacing PABA, albeit to a significantly lesser extent. Similarly, in the thrombin-fluorescein model, both heparin and LMWL binding changed the fluorescence in a dose dependent manner but LMWL did so to a significantly greater extent. The thrombin:dansyl fluorescence assay probes Ser 195, the most critical amino acid in the thrombin active site. LMWLs were shown to alter dansyl emission intensity indicating that LMWLs alter the orientation of Ser 195. Interestingly, Ser195 is not affected by the binding of heparin. Heparin does appear to induce some small conformational changes in the active site suggesting that there is some degree of permissive flexibility within the active site, yet it is clear that the presence of a bound heparin does not induce catalytic dysfunction. Thus, there appears to be a threshold at which point exosite II allosteric forces disrupt optimal active site geometry causing allosteric inhibition. Sulfated LMWLs are capable of breaching this threshold.
The question of why LMWLs, which bind to the same exosite as heparin, are capable of inhibiting thrombin with nanomolar potency while heparin is catalytically neutral, is likely best explained by structure. The predominantly hydrophobic interactions that sulfated LMWLs make with exosite II as opposed to the predominately electrostatic interactions made by heparin, have a more profound and far reaching effect on the active site, most specifically Ser195 and His57. Although heparin is able to some extent alter the conformation of the active site enough to affect PABA binding and the chemical environment surrounding the fluorescein probe bound to His57, it does not fundamentally change the geometry of the catalytic triad. Sulfated LMWLs clearly have a greater effect on the gross structure of the thrombin active site as well as two of its most important amino acids, Ser195 and His57.
Sulfated LMWLs represent a completely novel class of anticoagulants which utilize the heparin binding domain to induce allosteric inhibition of a variety of serine coagulation proteases. Although other ligands have been described to bind to the heparin binding site of serine coagulation proteases, none have exhibited evidence a direct anticoagulant effect. Given this novel mechanism of action, we describe in this paper the molecular mechanism by which sulfated LMWLs are able to impart their inhibitory activity. Furthermore, we demonstrate why a catalytically neutral ligand like heparin is unable to function as an allosteric inhibitor. As we move forward with the development of more homogenous derivatives of sulfated LMWLs, these assays will provide vital information about their mechanism of action and ability to disrupt the catalytic active site. Future experiments will focus on identifying homogenous sulfated LMWLs with high selectivity for various coagulation serine proteases. It will be important to be able to accurately describe their mechanism of action and selectivity in detail. The assays developed and described in this manuscript represent some of the first steps towards advancing the development of sulfated LMWLs into therapeutic anticoagulants.
Highlights for Thrombosis Research paper.
Sulfated low molecular weight lignins represent a novel class of anticoagulants Bind to heparin binding site and allosterically inhibit thrombin and factor Xa Uniformly inhibit thrombin and factor Xa hydrolysis of chromogenic substrates LMWLs exhibit different effects on the thrombin active site compared to heaprin
Acknowledgemets
This work was supported by grants HL090586 and HL107152 from the National Institutes of Health.
Abbreviations
- UFH
unfractionated heparin
- LMWH
low molecular weight heparin
- fIIa
factor IIa (thrombin)
- fXa
factor Xa
- LMWL
low molecular weight lignin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature
- 1.Monien BH, Henry BL, Raghuraman A, Hindle M, Desai UR. Novel chemo-enzymatic oligomers of cinnamic acids as direct and indirect inhibitors of coagulation proteinases. Bioorg Med Chem. 2006;14:7988–98. doi: 10.1016/j.bmc.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 2.Henry BL, Thakkar JN, Martin EJ, Brophy DF, Desai UR. Characterization of the plasma and blood anticoagulant potential of structurally and mechanistically novel oligomers of 4-hydroxycinnamic acids. Blood Coagul Fibrinolysis. 2009;20:27–34. doi: 10.1097/MBC.0b013e328304e077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry BL, Connell J, Liang A, Krishnasamy C, Desai UR. Interaction of antithrombin with sulfated, low molecular weight lignins: opportunities for potent, selective modulation of antithrombin function. J Biol Chem. 2009;284:20897–908. doi: 10.1074/jbc.M109.013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry BL, Thakkar JN, Liang A, Desai UR. Sulfated, low molecular weight lignins inhibit a select group of heparin-binding serine proteases. Biochem Biophys Res Commun. 2012;417:382–6. doi: 10.1016/j.bbrc.2011.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henry BL, Abdel Aziz M, Zhou Q, Desai UR. Sulfated, low-molecular-weight lignins are potent inhibitorsof plasmin, in addition to thrombin and factor Xa: Novel opportunity for controlling complex pathologies. Thromb Haemost. 2010;103:507–15. doi: 10.1160/TH09-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry BL, Monien BH, Bock PE, Desai UR. A novel allosteric pathway of thrombin inhibition: Exosite II mediated potent inhibition of thrombin by chemo-enzymatic, sulfated dehydropolymers of 4-hydroxycinnamic acids. J Biol Chem. 2007;282:31891–9. doi: 10.1074/jbc.M704257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davie EW, Kulman JD. An overview of the structure and function of thrombin. Semin Thromb Hemost. 2006;32(Suppl 1):3–15. doi: 10.1055/s-2006-939550. [DOI] [PubMed] [Google Scholar]
- 8.Olson ST, Halvorson HR, Björk I. Quantitative characterization of the thrombin-heparin interaction. Discrimination between specific and nonspecific binding models. J Biol Chem. 1991;266:6342–52. [PubMed] [Google Scholar]
- 9.Ye J, Liu LW, Esmon CT, Johnson AE. The fifth and sixth growth factor-like domains of thrombomodulin bind to the anion-binding exosite of thrombin and alter its specificity. J. Biol. Chem. 1992;267:11023–11028. [PubMed] [Google Scholar]
- 10.Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J. Biol. Chem. 1991;266:16977–16980. [PubMed] [Google Scholar]
- 11.Berliner LJ, Shen YY. Active site fluorescent labeled dansyl and anthraniloyl human thrombins. Thromb Res. 1978;12:15–25. doi: 10.1016/0049-3848(78)90081-6. [DOI] [PubMed] [Google Scholar]
- 12.Olson ST. Transient kinetics of heparin-catalyzed protease inactivation by antithrombin III. Linkage of protease-inhibitor-heparin interactions in the reaction with thrombin. J Biol Chem. 1988;263:1698–708. [PubMed] [Google Scholar]
- 13.Bock PE. Active site selective labeling of serine proteases with spectroscopic probes using thioester peptide chloromethyl ketones: demonstration of thrombin labeling using N alpha-[(acetylthio)acetyl]-D-Phe-Pro-Arg-CH2Cl. Biochemistry. 1988;27:6633–9. doi: 10.1021/bi00417a063. [DOI] [PubMed] [Google Scholar]
- 14.Parker CA, Barnes WJ. Analyst. 1957;82:606. [Google Scholar]
- 15.Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J. Biol. Chem. 1991;266:16977–16980. [PubMed] [Google Scholar]
- 16.Higaki JN, Evnin LB, Craik CS. Introduction of a cysteine protease active site into trypsin. Biochemistry. 1989;28:9256–63. doi: 10.1021/bi00450a004. [DOI] [PubMed] [Google Scholar]