Abstract
The kinase mTOR (mechanistic target of rapamycin) integrates diverse environmental signals and translates these cues into appropriate cellular responses. mTOR forms the catalytic core of at least two functionally distinct signaling complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). mTORC1 promotes anabolic cellular metabolism in response to growth factors, nutrients, and energy and functions as a master controller of cell growth. While significantly less well understood than mTORC1, mTORC2 responds to growth factors and controls cell metabolism, cell survival, and the organization of the actin cytoskeleton. mTOR plays critical roles in cellular processes related to tumorigenesis, metabolism, immune function, and aging. Consequently, aberrant mTOR signaling contributes to myriad disease states, and physicians employ mTORC1 inhibitors (rapamycin and analogs) for several pathological conditions. The clinical utility of mTOR inhibition underscores the important role of mTOR in organismal physiology. Here we review our growing knowledge of cellular mTOR regulation by diverse upstream signals (e.g. growth factors; amino acids; energy) and how mTORC1 integrates these signals to effect appropriate downstream signaling, with a greater emphasis on mTORC1 over mTORC2. We highlight dynamic subcellular localization of mTORC1 and associated factors as an important mechanism for control of mTORC1 activity and function. We will cover major cellular functions controlled by mTORC1 broadly. While significant advances have been made in the last decade regarding the regulation and function of mTOR within complex cell signaling networks, many important findings remain to be discovered.
Keywords: mTOR, mTORC1, mTORC2, insulin, amino acids, energy
Introduction
All cells from single-celled organisms to those comprising multicellular organisms sense and respond rapidly to fluctuations in their nutritional and energetic environments in order to modulate cell metabolism appropriately and maintain cellular homeostasis. Consequently, cells coordinate nutritional and energetic supply and demand tightly to prevent engagement in ATP-consuming anabolic processes when environmental resources become limited. During evolution, multicellular organisms acquired the additional ability to sense and respond to long-range systemic signals (i.e. hormones; growth factors; mitogens; cytokines) (referred to collectively as “growth factors” here) to enable communication between tissues and organ systems. mTOR, the mechanistic target of rapamycin, functions as a critical integrator of these diverse environmental cues by integrating them into appropriate cellular responses.
mTOR, an evolutionarily conserved serine/threonine protein kinase, belongs to the phosphatidylinositol-3 kinase (PI3K)-related kinase (PIKK) superfamily. mTOR represents the functional target of a natural macrolide antibiotic called rapamycin (clinically known as sirolimus). Rapamycin, produced by the bacterium Streptomyces hygroscopicus, was discovered in soil samples from Easter Island (known as Rapa Nui to the native population) in the 1970s (1; 2). Rapamycin reduces eukaryotic cell proliferation to various degrees, with immune cells showing strong sensitivity. To identify the target of rapamycin, Hall and colleagues performed an elegant genetic screen in 1991 in the budding yeast Saccharomyces cerevisiae. Mutations in three genes, Fpr1 (an orthologue of FKBP12 [FK506-binding protein 12]), Tor1 and Tor2, conferred resistance to rapamycin (3) (and reviewed in (2)). Today we understand that rapamycin associates with an endogenous cellular protein, FKBP12, and this complex docks to the FRB (FKBP12 rapamycin binding) domain located immediately N-terminal to the C-terminal mTOR kinase domain (see the accompanying article for greater detail regarding mTOR structure), resulting in allosteric inhibition of mTOR activity and signaling (4–6). By affinity purification of FKPB12-rapamycin binding proteins, several groups identified the mammalian orthologue of budding yeast Tor1/2 in 1994–95 (7–9).
mTOR constitutes the catalytic core of two known signaling complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (10; 11). These mTOR complexes (mTORCs) possess distinct substrates, cellular functions, and sensitivity to rapamycin. Acute rapamycin treatment inhibits cellular mTORC1 but not mTORC2 signaling while longer-term rapamycin treatment suppresses mTORC2 function by compromising complex integrity to variable degrees depending on cell type (12). Rapamycin fails to inhibit the phosphorylation of all mTORC1 substrates equally (13; 14). It completely inhibits phosphorylation of S6K1 (ribosomal protein S6 kinase1) but only partially inhibits phosphorylation of 4EBP1 (eukaryotic initiation factor 4E binding protein 1). The development of ATP-competitive mTOR catalytic inhibitors (i.e. Torin1; Ku-0063794) revealed that mTORC1 phosphorylates substrates in both rapamycin-sensitive and -insensitive manners (15–18), possibly due to differential substrate access to the kinase active site controlled by the mTOR FRB domain (19) and/or due to differential substrate quality conferred by phosphorylation site consensus sequence (20; 21).
The exclusive mTOR partner raptor (regulatory-associated protein of mTOR) defines mTORC1 (22; 23) while the exclusive mTOR partner rictor (rapamycin-insensitive companion of mTOR) defines mTORC2 (24; 25) (Figure 1). In addition to raptor, mTORC1 contains mLST8/GβL (mammalian lethal with Sec 13 protein 8/G-protein β-protein subunit like) (26), PRAS40 (Akt/PKB substrate 40 kDa) (27; 28), and deptor (DEP-domain-containing mTOR interacting protein) (29). Raptor serves a scaffolding role, functioning to recruit substrates to the mTOR kinase through their TOS (TOR signaling) motifs (30; 31). Global deletion of raptor in mice results in early embryonic lethality (e5.5) (32), similar to the global knockout of mTOR (33). PRAS40 and deptor function as both suppressors and targets of mTORC1, likely by acting as competitive substrates (29; 34), while mLST8/ GβL is not essential for mTORC1 function (32). In addition to rictor (24; 25), mTORC2 contains mSIN1 (mammalian stress-activated protein kinase interacting protein 1) (35; 36), protor1/2 (protein observed with rictor 1/2) (aka PRR5) (37), mLST8/GβL (24; 25), and deptor (29). Thus, mTORC1 and mTORC2 contain distinct and shared partner proteins. Similar to raptor within mTORC1, rictor and mSin1 serve as critical scaffolds that control mTORC2 integrity, regulation by upstream signals, and substrate choice (24; 25; 35; 36). Unlike mTORC1, mLST8/GβL is required for mTORC2 function; like rictor, its deletion in mice causes embryonic lethality (e10.5) (32). The role of protor remains unclear. It is important to note that mTOR also assembles into relatively homologous TORC1 and TORC2 complexes in budding and fission yeast, underscoring the ancestral origin of the TORCs.
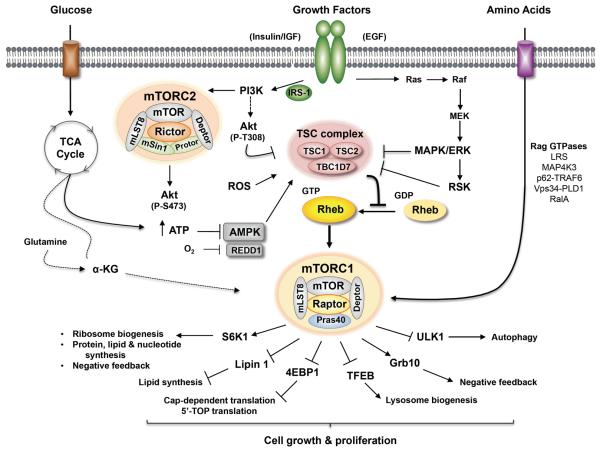
Figure 1. Regulation of the mTORC1 and mTORC2 signaling network by diverse upstream inputs.
Growth factors such as insulin or EGF activate mTORC1 through either the PI3K-Akt or the Ras-MAPK (ERK)-RSK axes, respectively. Growth factor-mediated activation of mTORC1 absolutely requires sufficient levels of amino acids, which are sensed through a variety of factors, as indicated. mTORC1 action also requires sufficient levels of energy (i.e. ATP) and/or oxygen, which are sensed by AMPK, REDD1, and TCA cycle metabolites (i.e. αKG). The TSC complex integrates diverse upstream signals to regulate mTORC1 action. TSC suppresses the conversion of Rheb-GDP to Rheb-GTP, a small GTPase that activates mTORC1. mTORC1 phosphorylates a limited known set of bona fide substrates to drive anabolic and suppress catabolic cellular processes and to mediate negative feedback toward PI3K. Growth factors (i.e. insulin) also activate mTORC2 through poorly defined signaling intermediates.
While TORC1 in yeast responds simply to environmental nutrients and energy, mTORC1 in higher eukaryotes responds to a broader array of upstream signals, integrating cues from growth factors (i.e. insulin; IGF; EGF; cytokines) to modulate cellular functions appropriately (Figure 1) (10; 11). mTORC1 function absolutely requires sufficient levels of amino acids such that their withdrawal inactivates mTORC1 signaling rapidly and renders mTORC1 activation refractory to virtually all other inputs, including growth factors. To limit cellular engagement in energy costly anabolic processes, nutrient and growth factor withdrawal as well as diverse types of cell stress (i.e. low energy; hypoxia; ER stress; ROS (reactive oxygen species)) downregulate mTORC1 signaling (38). Upon activation, mTORC1 signaling drives cap-dependent protein synthesis, cell growth, and cell proliferation through phosphorylation of the ribosomal protein S6 kinases (S6K1/2) and the eukaryotic initiation factor 4E (eIF4E) binding proteins 1–3 (4EBP1–3) at least in part (10; 11; 39). While the current set of direct mTORC1 substrates remains somewhat limited (e.g. S6Ks; 4EBPs; IRS-1; ULK1; Lipin1; TFEB; Grb10), quantitative phosphoproteomic screens identified a large number of downstream mTORC1 effectors, many of which likely represent bona fide mTORC1 substrates (40; 41) (Figure 1). mTORC1 promotes other anabolic processes including lipid and nucleotide synthesis and suppresses autophagy, a degradative process in which autophagosomes break down macromolecules and organelles during nutrient and energy starvation. Thus, mTORC1 drives anabolic and suppresses catabolic cellular processes. Our understanding of the regulation and function of mTORC2 lags far behind that of mTORC1 due to its more recent discovery (24; 25) and the lack of mTORC2-specific inhibitors. Growth factors activate mTORC2, which phosphorylates a limited set of known substrates including Akt (aka PKB), PKCα (protein kinase Cα), and SGK1 (serum and glucocorticoid-induced protein kinase). mTORC2 modulates cell metabolism and the organization of the actin cytoskeleton and enhances cell survival, due to its activation of the survival kinase Akt (42; 43) (Figure 1).
Aberrant mTORC1 function contributes to myriad pathologic conditions including cancer and benign tumor syndromes, metabolic disorders (e.g. type II diabetes; obesity), cardiovascular disorders, inflammatory disorders, and neurological disorders (10; 11). Consequently, clinicians employ rapamycin (aka sirolimus) and rapamycin analogs (rapalogs) (i.e. everolimus; temsirolimus) for immunosuppression following renal transplantation and for treatment of renal cell carcinoma, neuroendocrine tumors of pancreatic origin, tuberous sclerosis complex (TSC, a benign tumor syndrome), and cardiac restenosis following angioplasty (6). The role of mTOR in pathophysiology of disease combined with the utility of mTORC1 inhibitors in clinical medicine underscores the importance of elucidating the regulation and function of mTORC1 at the cellular level (44).
Growth factor sensing by the TSC-Rheb axis
Growth factors, in particular insulin/ IGF (insulin-like growth factor) and EGF (epidermal growth factor), represent the best understood inputs that lead to activation of mTORC1 upon converging on the TSC (tuberous sclerosis complex) / Rheb axis (45). TSC, composed of TSC1 (aka hamartin), TSC2 (aka tuberin), and a more recently discovered third subunit, TBC1D7 (TBC [Tre2-Bub2-Cdc16] 1 domain family member 7), functions as a tumor suppressor that inhibits mTORC1 (46) (Figure 1). Inactivating mutations in either TSC1 or TSC2 increases mTORC1 signaling and causes an autosomal dominant disease in which benign tumors form in various organs including brain, kidney, and heart (47). TSC inhibits mTORC1 by inhibiting Rheb, (Ras homolog enriched in brain), a small Ras-like GTPase essential for mTORC1 activation by all upstream signals.
Insulin-PI3K-Akt signaling
Insulin/IGF binding to its cognate cell surface tyrosine kinase receptor leads to tyrosine phosphorylation of IRS (insulin receptor substrate) proteins, which recruits and activates PI3K (phosphatidylinositol 3-kinase) (48) (Figure 1). Increased production of the phospho-lipid PI(3,4,5)P3 on lipid membranes by PI3K recruits Akt via its PH (pleckstrin homology) domain, leading to PDK1-mediated phosphorylation of Akt on its activation loop (T308) (49). It is important to note that additional phosphorylation of Akt on its hydrophobic motif (S473) by PI3K-controlled mTORC2 boosts Akt activity several fold further (49; 50). Activated Akt then phosphorylates TSC2 on several sites (S939; T1462) to suppress the inhibitory effect of the TSC complex toward mTORC1, thus leading to increased mTORC1 signaling (51; 52). TSC2 possesses a GAP (GTPase activating protein) domain that hydrolyzes active Rheb-GTP to inactive Rheb-GDP. Thus, in response to insulin/PI3K signaling, Akt phosphorylates and inactivates TSC2, which increases Rheb-GTP loading and mTORC1 kinase activity (27). While Rheb-GTP interacts with the mTOR kinase domain (27; 53), the underlying molecular mechanism by which Rheb activates mTORC1 remains unclear.
Other parallel mechanisms contribute to activation of mTORC1 by insulin/IGF. Akt and mTORC1 phosphorylate PRAS40, an mTORC1 inhibitory partner (on T246 and S183/S212/S221, respectively), inducing the dissociation of PRAS40 from mTORC1 and thus relieving PRAS40-mediated substrate competition (27; 28; 34; 54). Insulin/PI3K signaling also leads to mTORC1-mediated phosphorylation of raptor (on S863) to promote mTORC1 signaling (55; 56). Moreover, phosphorylation of mTOR itself (on S1261, S2159, and T2164) by unknown kinases (57; 58) and on S1415 by IKKα (59) contributes to increased mTORC1 signaling. While mTOR autophosphorylation on S2481 plays no known role in mTORC1 function, it serves as a biomarker for mTORC1 and mTORC2 catalytic activity in intact cells (5). Many phospho-proteomic studies agree that mTOR and its partner proteins undergo phosphorylation on many sites (40; 41; 60). Consequently, a challenge for the future will be to identify the kinases that act on these sites directly and to decipher the regulation and functional significance of complex mTORC phosphorylation.
EGF-Ras-MAPK signaling
EGF activates mTORC1 signaling independently of the PI3K/Akt axis. EGF binding to its cell surface tyrosine kinase receptor activates the Ras GTPase, which leads to activation of c-Raf, MEK (MAPK/ERK kinase), MAPK (mitogen activated protein kinase) (aka ERK) and RSK (p90 ribosomal S6 kinase (Figure 1). Similar to Akt, MAPK and RSK phosphorylate TSC2 on different sites (S540/S644 and S1798, respectively) to suppress the inhibitory action of TSC2 toward Rheb (61; 62). By a parallel pathway, the Ras/MAPK pathway converges on raptor. MAPK phosphorylates raptor (on S8/S696/S863) and RSK phosphorylates raptor (on S719/S721/S722)(63; 64) to promote mTORC1 signaling.
mTORC2 regulation
Insulin/PI3K signaling leads to mTORC2-mediated phosphorylation of Akt on its hydrophobic motif (HM) site, S473 (Figure 1) as well as the HM sites of other AGC kinases, PKCα (on S657), and SGK1 (on S422) (25; 50; 65). Insulin increases the kinase activity of mTORC2 in vitro in a manner sensitive to cellular treatment with the PI3K inhibitor wortmannin (66). Thus, insulin/PI3K signaling activates mTORC2; it is important to note, however, that the signaling intermediates that link PI3K to mTORC2 remain virtually unknown. Interestingly, while TSC inhibits mTORC1, TSC activates mTORC2 (66). MEFs lacking TSC2 display reduced mTORC2 kinase activity toward Akt in vitro and decreased Akt S473 phosphorylation in intact cells in a manner independent of the well-known mTORC1-mediated negative feedback loop that attenuates PI3K signaling (discussed below) (66). In addition, TSC associates with mTORC2. These data suggest quite different regulation of mTORC2 compared to mTORC1. On the other hand, the Rac1 GTPase interacts with mTOR and provides an activating signal to both mTORC1 and mTORC2 in response to growth factors in a PI3K/Akt-independent manner, suggesting that common upstream inputs co-regulate both mTORCs (67). There is no question that important discoveries await regarding mTORC2 regulation.
As mTORC2 mediates Akt S473 phosphorylation, it would seem that mTORC2 lies upstream of mTORC1. While such epistasis may hold true in certain cellular contexts, genetic knockout or knockdown of core mTORC2 components (i.e. rictor; mSin1) in many cultured cell types has no effect on TSC2 phosphorylation and mTORC1 signaling (32; 68). Thus, mTORC2 function is not required for mTORC1 action. On the other hand, mTORC2 function is required for Akt-mediated phosphorylation of other substrates (i.e. FoxO1/3a) (32; 68). These data can be explained by the known essential requirement for activation loop site (T308) but not HM site (S473) phosphorylation for Akt kinase activity; Akt S473 phosphorylation boosts Akt activity further and may modulate substrate specificity (32; 49; 68). Thus, in many cellular contexts Akt phosphorylation on its activation loop without HM-site phosphorylation provides sufficient activity to mediate downstream signaling to mTORC1.
Interestingly, mTORC2 associates with ribosomes in a growth factor sensitive manner (69; 70). Structurally intact ribosomes, but not protein synthesis itself, are required for mTORC2 kinase activity in vitro and signaling in intact cells (70). Thus, a direct interaction of mTORC2 with ribosomes may play a role in insulin/PI3K-meditated mTORC2 activation. mTORC2 also promotes turn-motif (TM) site phosphorylation of Akt (T450) and several conventional PKCs (PKCα T638 and PKCβ T641) in a co-translational manner independently of growth factor status, functioning to increase protein stability and folding (71; 72). In addition to interacting with ribosomes (likely those associated with ER engaged in protein translation), mTORC2 associates with an ER sub-compartment called MAM (mitochondrial-associated ER membrane) in a growth factor stimulated manner. mTORC2 inactivation decreases MAM integrity, mitochondrial metabolism, and cell survival (73).
Negative feedback signaling
Several negative feedback mechanisms modulate the mTOR signaling network, as signal attenuation limits signal amplitude and duration critical for homeostatic control of complex biological systems. Cellular TSC loss leads to elevated and constitutive mTORC1 signaling independent of growth factor status and attenuates PI3K signaling, thus producing a state of cellular insulin resistance (74; 75). S6K1 and mTORC1 phosphorylate IRS-1 directly to induce its degradation, thus uncoupling the insulin receptor from PI3K. mTORC2 also limits PI3K signaling by inducing IRS-1 degradation (76). mTORC2 phosphorylates and stabilizes Fbw7, an ubiquitin ligase subunit that targets IRS-1 for degradation (76). Grb10 was identified as a direct mTORC1 substrate in phosphoproteomic screens (40; 76). mTORC1-mediated phosphorylation of Grb10, a growth factor receptor-bound adaptor that limits growth factor signaling, stabilizes Grb10 and attenuates both PI3K and MAPK/ERK signaling. Depending on cellular context, either S6K1 or Akt phosphorylate mSin1 directly (on T86 and T398), a critical mTORC2 partner, dissociating mSin1 from the complex and decreasing mTORC2 signaling (40). Along similar lines, several groups reported that S6K1 phosphorylates rictor directly (T1135) (77–80), which may reduce mTORC2 signaling to Akt (78; 79). These data reveal that both mTORC1 and mTORC2 engage in negative feedback to maintain proper signaling by growth factor receptors and the mTORCs.
Amino acid sensing by the Rag-Ragulator axis and other emerging factors
Sufficient levels of amino acids, particularly leucine, are essential for basal mTORC1 signaling from yeast to mammals and for robust activation of mTORC1 in response to growth factor signals in higher eukaryotes (81; 82). How cells sense amino acid levels remains poorly defined, but great progress has been made in recent years identifying the machinery that propagates amino acid sensing proximal to mTORC1. While several signaling molecules that link amino acid sensing to mTORC1 have been identified (see text below), the Rag GTPases, the Ragulator complex, and the v-ATPase represent the best-characterized links between amino acid sensing and mTORC1.
The Rag GTPases
The evolutionarily conserved family of Rag GTPases function as obligate heterodimers in which Rag A or Rag B dimerizes with RagC or RagD (Figure 2). Upon amino acid stimulation, RagA/B loads with GTP and binds raptor while RagC/D loads with GDP (83; 84). Expression of dominant-active RagA/B mutants (loaded constitutively with GTP) promote mTORC1 signaling in the absence of cellular amino acids while expression of dominant-negative RagA/B mutants (nucleotide-free) suppress mTORC1 signaling in the presence of amino acids. Thus, heterodimers composed of RagA/BGTP-RagC/DGDP form during amino acid sufficiency to promote mTORC1 signaling and heterodimers composed of RagA/BGDP-RagC/DGTP form during amino acid withdrawal (83; 84). As exogenous expression of RagA/BGTP-RagC/DGDP heterodimers more strongly activate mTORC1 in amino acid deprived cells than exogenous expression of dominant-active RagA/B alone, these data suggest that the nucleotide-bound state of RagC/D as well as that of RagA/B indeed contributes to mTORC1 signaling in response to amino acids.
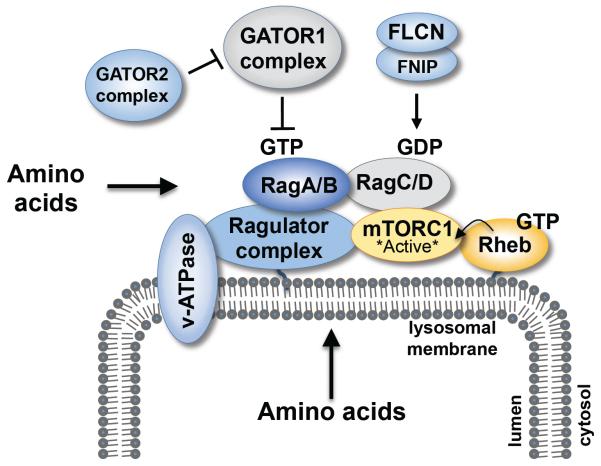
Figure 2. Rag heterodimers recruit mTORC1 to lysosomal membranes for Rheb-mediated activation.
Activation of mTORC1 by amino acids through Rag GTPase heterodimers involves the v-ATPase, ragulator complex, and Rag regulatory factors. The ragulator complex, which acts as a GEF toward RagA/B GTPases, induces formation of active RagA/BGTP-RagC/DGDP heterodimers. The GATOR1 complex functions as a GAP (GTPase activating protein) for RagA/B (thus inhibiting Rag heterodimers) while folliculin (FLCN) and its associated proteins (FNIP1/2) functions as a GAP for Rag C/D (thus promoting a Rag heterodimer active state). The GATOR2 complex suppresses GATOR1. Active RagA/BGTP-RagC/DGDP heterodimers bind mTORC1 through raptor to recruit mTORC1 to the lysosomal surface where Rheb resides. When loaded with GTP, Rheb activates mTORC1 through a poorly defined mechanism. An “inside-out” model proposes that the v-ATPase and ragulator respond to amino acid levels inside the lysosomal lumen to control Rag nucleotide binding state.
How do Rags control amino acid-mediated activation of mTORC1? While Rheb-GTP provided in vitro increases mTORC1 kinase activity directly (27), active RagA/BGTP-RagC/DGDP heterodimers provided in vitro are insufficient (83). Cellular amino acid stimulation induces the translocation of mTOR and raptor from a poorly defined cytoplasmic compartment to LAMP1/2-positive lysosomal membranes, a site to which the Rags and Rheb also localize (83; 85). Importantly, mTORC1 translocation requires the Rags (83). These data suggest a model in which the amino acid-Rag axis activates mTORC1 by controlling mTORC1 subcellular localization: Amino acid signaling drives the formation of active RagA/BGTP-RagC/DGDP heterodimers, which bind raptor to recruit mTORC1 to the lysosomal surface where mTORC1 receives an essential activating input from Rheb (and possibly from other inputs) (Figure 3). While Rheb docks to internal membranes by a farnesyl lipid moiety, Rag GTPases do not possess lipid-anchoring motifs.
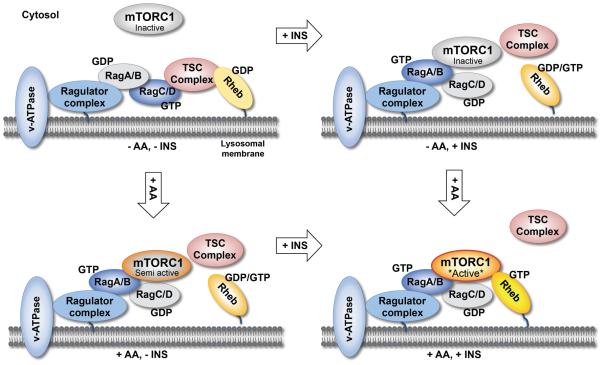
Figure 3. Integration of amino acid and insulin sensing at the lysosome by spatial-temporal regulation of mTORC1 and TSC.
Recent data suggest a model in which amino acids and insulin control mTORC1 activity at the lysosome by spatial-temporal regulation of not only mTORC1 but also its inhibitor TSC. In the absence of amino acids and insulin, inactive mTORC1 localizes to the cytosol while active TSC localizes to lysosomal membranes in a Rag-dependent manner. Upon stimulation with amino acids, active RagA/BGTP-RagC/DGDP heterodimers recruit mTORC1 to lysosomal membranes where Rheb resides to mediate basal mTORC1 activity; simultaneously, amino acids induce TSC dissociation from lysosomal membranes partially. Similar to amino acids, insulin stimulation induces TSC dissociation from lysosomal membranes. mTORC1 dissociates from lysosomes completely only when both Rag heterodimers and Rheb exist in their inactive states. Conversely, mTORC1 associates with lysosomal membranes fully only when Rag heterodimers and Rheb exist in their active states in the presence of both amino acids and insulin.
The Ragulator complex
A pentameric complex called the Ragulator, consisting of p18 (LAMTOR1), p14 (LAMTOR2), MP1 (LAMTOR3), c7orf59 (LAMTOR4), and HBXIP (hepatitis B virus X interacting protein) (LAMTOR5), was found to anchor the Rags to lysosomal membranes through p18 myristoylation (85; 86) (Figure 3). The Ragulator complex not only tethers the Rags to lysosomal membranes, it possesses GEF activity toward RagA/B to enable exchange of GDP for GTP, thus converting Rag heterodimers to an active state (85). Conversely, a complex of proteins termed GATOR1, which contains proteins DEPDC5, Nprl2, and Nprl3, binds Rag heterodimers and possesses GAP activity for RagA/B, thus converting Rag heterodimers to an inactive state (87). A complex called GATOR2, which contains proteins Mios, Wdr24, Wdr59, Seh1L, and Sec13, suppresses GATOR1. Indeed, inactivation of GATOR1 subunits renders mTORC1 resistant to amino acid deprivation while inactivation of GATOR2 subunits suppresses mTORC1 signaling (87).
RagC/D not only function as obligate binding partners for RagA/B, their nucleotide binding state also participates in amino acid controlled mTORC1 function. Folliculin (FLCN) and its interacting partners FNIP1/2 are required for amino acid-stimulated translocation of mTORC1 to lysosomes, where they dock to Rag GTPases in the absence of amino acids, poised to convert RagC/DGTP to RagC/DGDP upon docking of mTORC1 to the lysosomal surface upon amino acid addition (88; 89) (Figure 2). FLCN possess GAP activity toward RagC/D but not RagA/B and thus converts Rag heterodimers from an inactive RagA/BGDP-RagC/DGTP to an active RagA/BGTP-RagC/DGDP nucleotide-bound state that stabilizes mTORC1 docking (89). The discovery that a spatial-temporal mechanism governs amino acid-mediated mTORC1 signaling explained a long-standing mystery in the field regarding why growth factors fail to activate mTORC1 in the absence of amino acids: If mTORC1 localizes within the cell to the wrong place at the wrong time, mTORC1 cannot be activated by upstream inputs.
The v-ATPase
How do cells sense amino acid levels? The discovery that the v-ATPase (vacuolar H+-adenosine triphosphatase) interacts with the Ragulator complex and Rag GTPases on the lysosomal surface and senses amino acids levels, possibly from within the lysosomal lumen, begins to elucidate these important but poorly resolved questions (90). v-ATPase subunits and its ATP hydrolyzing catalytic activity are required for generation of active RagA/BGTP-RagC/DGDP heterodimers, for amino acid-mediated localization of mTORC1 to lysosomes, and for activation of mTORC1 (Figure 2). Its classical function as a proton pump that lowers luminal pH of lysosomes is not required for mTORC1 activation, however (90). Amino acids modulate interactions between the v-ATPase, Ragulator, and Rags (90), and amino acid accumulation within the lysosomal lumen correlates with recruitment of mTORC1 to lysosomal membranes. Thus, mTORC1 appears to sense intra-lysosomal amino acids by an “inside-out” mechanism. It is important to note, however, that the overall importance of intra-lysosomal amino acids for mTORC1 recruitment remains unclear. In this proposed model, the membrane-spanning v-ATPase senses luminal amino acids and undergoes conformational changes to relay the amino acid signal to the Ragulator-Rag complex on the cytosolic lysosomal face and ultimately to mTORC1 (90).
Emerging amino acid sensing factors
Other signaling factors linked to amino acid sensing by the Rag GTPases include leucyl-tRNA synthetase (LRS), the signaling adaptor p62 (aka sequestasome-1; SQSTM1),TRAF6 (TNF receptor associated factor 6), MAP4K3 (mitogen activated protein kinase kinase kinase kinase 3), and SH3BP4 (SH3 binding protein 4). Whether these Rag-linked signaling factors operate independently of or in conjunction with the Ragulator and v-ATPase remains unknown at this time. Other amino acid sensing factors not yet linked to the Rag GTPases include the Vps34 (vacuolar protein sorting 34)-PLD1 (phospholipase D1) pathway (91–93), the RalA GTPase (94) and IPMK (inositol polyphosphate multikinase) (95) (Figure 2).
Leucyl-tRNA synthetase (LRS), the enzyme that ligates leucine to its cognate tRNA, functions as an intracellular leucine sensor important for amino acid stimulated mTORC1 activation (96; 97). Amino acid stimulation induces the translocation of LRS to lysosomal membranes, where LRS associates with RagD and mTORC1. LRS possesses GAP activity for RagD and thus facilitates the generation of active Rag heterodimers (96). An LRS-dependent mechanism also functions in amino acid sensing in budding yeast, although some mechanistic details differ (97). The signaling adaptor p62 (aka sequestasome-1; SQSTM1), which targets proteins for autophagic degradation, co-localizes with Rags on lysosomal membranes and interacts with raptor and mTOR in an amino acid-dependent manner (98). p62 is required for amino acid-induced mTORC1 translocation to lysosomes and appears to stabilize active Rag heterodimers. p62 also interacts with TRAF6, an enzyme that catalyzes K63-linked ubiquitination (98; 99). In amino acid stimulated cells, p62 recruits TRAF6 to mTORC1, which catalyzes K63-linked ubiquitination of mTOR, an event required for mTORC1 signaling (99). As the p62-TRAF6 axis is not required for growth factor signaling, it likely provides a non-essential, parallel input that converges on the core Rag machinery to modify amino acid sensing. The Ste20 family member MAP4K3 (mitogen activated protein kinase kinase kinase kinase 3) and SH3BP4 (SH3 binding protein 4) also link to the Rag GTPases, although many molecular details remain unresolved. Amino acids but not insulin activate MAP4K3, which interacts with Rag GTPases and is required for amino acid-induced mTORC1 activation (100; 101). Consistently, PP2A/PR61ε, a phosphatase that inhibits MAP4K3, suppresses mTORC1 signaling upon amino acid deprivation (102). SH3BP4 binds to inactive Rag heterodimers during amino acid deprivation via its SH3 domain to inhibit formation of active Rag heterodimers, thus suppressing mTORC1 signaling (103). By a Rag GTPase independent pathway, the class III phosphatidylinositol 3-kinase Vps34 (91–93), the RalA GTPase (94) and IPMK (inositol polyphosphate multikinase) (95) are also required for amino acid-induced mTORC1 signaling.
Sufficient levels of leucine and glutamine are particularly important for amino acid-induced mTORC1 signaling (81; 82). The export of glutamine across the plasma out of the cell by the bidirectional amino acid permease SLC7A5-SLC3A2 drives leucine into the cell (104). Thus, cellular glutamine indirectly influences leucine-induced mTORC1 signaling. By a more direct mechanism, hydrolysis of glutamine itself by glutaminolysis into glutamate (by glutaminase (GLS)) and αKG (ketoglutarate) (by glutamate dehydrogenase (GDH)) activates mTORC1 (105). Glutaminolysis and its product αKG promote RagB GTP-loading, mTORC1 translocation to lysosomes, and activation of mTORC1 (105). Consistently, glutaminolysis promotes cell growth and suppresses autophagy, two important mTORC1 controlled processes. As leucine functions as an allosteric cofactor for GDH, leucine and glutamine cooperate to activate mTORC1. This work explains in part the glutamine addiction that many cancer cells demonstrate. Not only does glutamine metabolism promote mTORC1 signaling via production of αKG (105), mTORC1 signaling feeds forward to promote glutamine metabolism and the production of αKG as an anaplerotic source to fuel the TCA cycle (106). mTORC1 signaling represses transcription of SIRT4, a mitochondrial-localized sirtuin that inhibits GDH (106). Thus, mTORC1 augments GDH activity to increase levels of αKG to drive ATP production.
The lysosome as a critical platform for upstream signal integration and mTORC1 activation
Discovery of the mTORC1 proximal amino acid sensing machinery- composed of the Rags, Ragulator complex and v-ATPase- identified the lysosome as a critical platform for mTORC1 docking and activation. Moreover, it revealed subcellular localization as an important molecular mechanism for regulation of mTORC1. Absence of mTORC1 from an activating site- the lysosomal surface-explains why diverse growth factors fail to activate mTORC1 in the absence of amino acids. Consistent with this model, amino acid depletion inhibits mTORC1 in the absence of TSC complex function, indicating that the amino acid sensing Rag pathway functions in a parallel and dominant manner to the growth factor sensing Akt-TSC pathway (107; 108). Inconsistent with this model, loss of TSC was also reported to attenuate the ability of amino acid depletion to inhibit mTORC1 signaling (109; 110). Recent work resolves these seemingly discrepant results by revealing that amino acids and insulin each modulate the subcellular localization of the TSC complex in a dynamic manner (110; 111). Upon amino acid withdrawal, inactive Rag GTPases (i.e. RagA/BGDP-RagC/DGTP) dissociate mTORC1 but recruit the TSC complex (via TSC2) to the lysosomal surface; due to spatial proximity, the GAP activity of TSC2 engages Rheb to hydrolyze active Rheb-GTP to inactive Rheb-GDP (110) (Figure 3). Thus, the Rag GTPases recruit both mTORC1 and the TSC complex in an opposite fashion depending on nucleotide binding state. Upon insulin stimulation, PI3K signaling mediates Akt-dependent phosphorylation of TSC2, which induces TSC complex dissociation from the lysosomal surface (111); due to spatial restriction, the GAP activity of TSC2 cannot hydrolyze active Rheb-GTP into inactive Rheb-GDP. The finding that insulin-PI3K-Akt signaling induces dynamic TSC movement off lysosomal membranes provided a molecular mechanism for how TSC2 phosphorylation suppresses its GAP activity toward Rheb, a longstanding question in the field, as little experimental evidence supported the hypothesis that TSC2 phosphorylation by Akt modulates the intrinsic GAP activity of TSC2. Taken together, amino acid and growth factor signals converge on the TSC complex by controlling its dynamic subcellular localization on and off the lysosomal surface, thus effecting the integration of these signals (Figure 3). The dynamic localization of TSC in response to amino acids and insulin, combined with variable cellular sensitivity to these signals depending on cell type and culture conditions, likely explains the seemingly variable role of the TSC complex in amino acid responsiveness in various studies.
Other work reinforces the concept of the lysosome as a critical platform for mTORC1 activation. In response to amino acids, PLD1 (phospholipase D1) translocates to lysosomal membranes where it participates in activation of mTORC1 (93) (Figure 1). The amino acid sensing lipid kinase Vps34 mediates activation of PLD1 (phospholipase D1) by generating PI(3)P on endomembranes and creating a docking site for the PX-domain of PLD1 (93). The activation of mTORC1 in response to amino acids by this Vps34-PLD1 axis occurs in parallel to the Rag pathway and thus represents an independent mechanism for amino acid sensing by mTORC1 (93). Interestingly, PLD1 also functions downstream of Rheb, suggesting that a Vps34-PLD1 axis senses amino acids while a Rheb-PLD1 axis senses growth factors (112). In response to growth factors, activation of PLD1 and generation of its product phosphatidic acid (PA), a lipid second messenger, activates mTORC1 (113–115). PA binds mTOR near the FRB (FKBP12-rapamycin binding)-domain to activate mTORC1 by an allosteric mechanism and by displacing FKBP38 (FK506 binding protein 38), an mTORC1 inhibitory protein (116). Thus, similar to the TSC complex, PLD1 integrates amino acid and growth factor signals (117).
Not only does the lysosome function as a signaling platform that integrates upstream signals to effect proper regulation mTORC1, mTORC1 action on the lysosomal surface also controls the biogenesis of the organelle itself. Lysosomes function in various degradative events including autophagy as well as energy metabolism and cell signaling (118). TFEB (transcription factor EB), a master regulator of gene expression that controls lysosome biogenesis, co-localizes with TORC1 on lysosomal membranes (119–121). During amino acid sufficiency, active Rag heterodimers recruit both mTORC1 and TFEB to the lysosome where mTORC1 interacts with and phosphorylates TFEB (on S142; S211); mTORC1-mediated TFEB phosphorylation inhibits TFEB action by inducing its cytosolic retention and thus nuclear exclusion to reduce lysosome biogenesis (119–121). During amino acid withdrawal, hypo-phosphorylated TFEB translocates into the nucleus to drive gene expression and lysosome biogenesis. Consistently, FLCN is required for mTORC1-mediated phosphorylation and cytosolic sequestration of TFEB (88).
Do other endomembranes function as mTORC1 docking platforms to control mTORC1 activation? In response to ROS (reactive oxygen species), the TSC complex localizes to peroxisomes by binding to PEX19 (peroxisomal biogenesis factor 19) and PEX5 where it inactivates Rheb and mTORC1 to drive autophagy (122; 123). Phospho-lipids also control mTORC1 subcellular localization, reinforcing the emerging concept of endomembranes as important sites for mTORC1 activation (124; 125). The class III lipid kinase Vps34 and the class II lipid kinase PI3K-C2alpha, which generate the phospholipid PI(3)P on endomembranes, functions in an mTORC1 amino acid sensing pathway (91; 92; 124). In addition, the lipid kinase that phosphorylates PI(3)P to generate PI(3,5)P2 on endomembranes, PIKFYVE (a phosphatidylinositol-3-phosphate-5-kinase), controls mTORC1 activation in both mammalian cells and yeast, indicating an evolutionarily conserved function for this signaling lipid (124; 125). Moreover, PI(3,5)P2 binds to raptor, and in response to insulin in 3T3-L1 adipocytes, generation of PI(3,5)P2 contributes to mTORC1 activation and mediates mTORC1 translocation to the plasma membrane (124). Altered function of Rab5, a small GTPase that controls early to late endosome maturation and that regulates PI(3)P synthesis on endosomes, impairs both amino acid- and insulin-induced activation and subcellular localization of mTORC1 (126; 127). Taken together, these data point to an emerging role for phospholipids in endocytic trafficking, subcellular localization, and activation of mTORC1, possibly on other endomembrane sites in addition to lysosomes.
Energy and stress sensing
Cellular energy produced by sufficient levels of nutrients (i.e. amino acids, glucose, and oxygen) enables cells to engage in ATP-consuming anabolic metabolism. Energy deprivation caused by nutrient insufficiency rapidly alters cell metabolism to shut down anabolic cellular processes and turn up catabolic cellular processes (i.e. autophagy) capable of providing emergency energy sources. mTORC1 functions not only as a critical integrator of amino acid signals, it functions as a critical integrator of energy signals. In addition, diverse forms of cell stress suppress mTORC1.
Energy stress caused by glucose deprivation, hypoxia, or pharmacologic inhibition of glycolysis or mitochondrial function reduces cellular ATP levels (128). The resulting rise in AMP/ATP and ADP/ATP ratios activates AMPK (AMP-activated protein kinase), which phosphorylates TSC2 (on S1345) to augment inhibitory action of the TSC complex toward mTORC1 (129) (Figure 1). In TSC null cells, energy stress partially decreases mTORC1 signaling, indicating that an AMPK-independent pathway downregulates mTORC1. Indeed, AMPK directly phosphorylates raptor (on S792) to inhibit mTORC1 signaling (130). AMPK also stimulates autophagy by phosphorylating ULK1 (on S317) to ramp up catabolic metabolism (131). Severe energy stress caused by withdrawal of glucose and glutamine disassembles the TTT-RUVBL1/2 complex (composed of the proteins Tel2, Tti1, and Tti2), which normally interacts with mTOR and facilitates mTORC1 dimer formation (106). Impaired mTORC1 dimer formation reduces mTORC1 interaction with active Rag GTPases on lysosomal membranes and thus reduces mTORC1 signaling (106). Independent of cellular ATP levels, hypoxia also stabilizes and activates HIF-1 (hypoxia-inducible factor 1), a transcription factor that drives expression of REDD1 (regulated in development and DNA damage responses 1) (aka RPT801) (132–134). REDD1 converges on the TSC complex to facilitate TSC-mediated suppression of mTORC1 signaling through an incompletely defined mechanism that may involve shuttling of 14-3-3 proteins from TSC to REDD1 (135; 136). Glucose and oxygen deprivation also impair protein maturation in the ER, leading to accumulation of misfolded proteins, ER stress, and induction of the unfolded protein response (UPR), a set of integrated signal transduction pathways that slow protein synthesis (137). Along one signaling branch of the UPR, ER stress induces REDD1 expression by the transcription factor ATF4 (138) (139) and thus suppresses mTORC1. ER stress not only modulates mTORC1 function but mTORC1 also modulates the ER stress response. Aberrantly high mTORC1 signaling, as occurs during loss of TSC, induces a state of ER stress presumably caused by aberrantly high protein synthesis (140). Consequently, the UPR acts to restrain protein synthesis by attenuating mTORC1 signaling and mTORC1-mediated protein synthesis, at least in part. In response to DNA damage and genotoxic stress, stabilization and activation of p53, a transcription factor and tumor suppressor, induces expression of Sestrins 1 and 2 (141). The sestrins bind to and activate AMPK (by a poorly defined mechanism), which suppresses mTORC1 signaling by AMPK-mediated TSC2 phosphorylation, as described above. p53 may also transcriptionally induce REDD1, thus suppressing mTORC1 by an AMPK-independent mechanism (142).
Major cellular functions controlled by mTORC1
Cell growth (an increase in cell mass and size) and cell proliferation (an increase in cell number through cell division) represent major cellular functions controlled by mTORC1. Deletion of mTOR in flies or suppression of mTORC1 function in cultured cells with rapamycin causes a cell autonomous decrease in cell size with reduced cell cycle progression and cell proliferation (143–146). mTORC1 is reported to preferentially drive cell growth through S6K1 and cell proliferation through 4EBP1 (147). Commitment to these processes requires sufficient levels of cellular components such as proteins, lipids, and nucleotides. It is not surprising, therefore, that mTORC1 functions as a master controller of anabolic metabolism by promoting protein synthesis, lipid synthesis, and nucleotide synthesis.
Protein synthesis represents the best-understood cellular function of mTORC1. Indeed, the best-characterized substrates of mTORC1 (4EBP1 and S6K1) control different aspects of protein synthesis (Figure 1). mTORC1-mediated phosphorylation of the translation repressor protein 4EBP1 on several sites (T37/46; T70; S65) induces dissociation of 4EBP1 from eIF4E, a translation initiation factor that binds the m7-GTP (7-methylguanosine) cap structure at the 5' end of mRNAs (148; 149). eIF4E then interacts with other translation initiation factors (i.e. eIF4G (a modular scaffold); eIF4A (a helicase)) to form the eIF4F complex, which recruits the 40S ribosome and other factors (see accompanying review article for greater detail). Thus, mTORC1-mediated phosphorylation of 4EBP1 drives cap-dependent translation and thus augments global translation rates. mTORC1 also induces the selective translation of specific types of mRNA transcripts, those with 5'-TOP (terminal oligopyrimidine) or 5'-TOP-like motifs, which tend to encode components of the translation apparatus such as ribosomal proteins and translation factors (150–152). Indeed, deletion of 4EBPs renders 5'-TOP translation (152) and cell proliferation (147) resistant to inhibition with mTORC1 inhibitors. These data indicate that mTORC1-mediated signaling to 4EBP1 promotes protein synthesis by driving translation initiation and biosynthesis of the translational apparatus itself, events important for mTORC1-driven cell proliferation. It is important to note that mTORC1 appears to promote 5'-TOP translation through additional factors under certain cellular contexts (153). Indeed, mTORC1 was shown recently to promote 5'-TOP translation through LARP1 (La-related protein 1) (154), a raptor-associated protein that binds 5'-TOP mRNAs. LARP1 associates with the translation initiation apparatus in an mTORC1-dependent manner to promote efficient 5'-TOP translation as well as cell cycle progression and cell proliferation. While significantly less well understood than 4EBP1, S6K1 also promotes protein synthesis downstream of mTORC1. mTORC1-mediated phosphorylation of S6K1 on its HM site (T389) (155; 156) induces the ribosome biogenesis (RiBi) transcriptional program, which increases ribosome number and augments overall protein biosynthetic capacity (157). In addition, S6K1 associates with eIF3 and phosphorylates directly many proteins linked to protein synthesis including eIF4B, PDCD4, eEF2K, and ribosomal protein S6 (39).
More recently, mTORC1 was shown to promote lipid and nucleotide synthesis (158) (Figure 1). mTORC1 signaling through S6K1 promotes the activation of SREBPs (sterol regulatory element-binding proteins 1 and 2), transcription factors that induce the expression of genes involved in de novo synthesis of fatty acids and sterols (159; 160). In addition, mTORC1-mediated phosphorylation of lipin 1, a phosphatidic acid phosphatase, promotes SREBP1/2-mediated transcription by blunting nuclear entry of lipin 1, a repressor of SREB1/2 function (161). mTORC1 signaling through SREBPs also transcriptionally upregulates genes involved in the pentose phosphate pathway (PPP), which produces ribose for nucleotide synthesis and the co-factor NADPH critical for many metabolic reactions including lipid synthesis (160). S6K1 also phosphorylates CAD (on S1859) (carbamoyl-phosphate synthetase 2; aspartate transcarbamylase; dihydro-orotase), which catalyzes the first several steps of de novo pyrimidine synthesis (162). Thus, through both transcriptional and phosphorylation-mediated mechanisms, mTORC1 signaling drives the synthesis of new nucleotides essential for an anabolic response.
While mTORC1 drives anabolic metabolism, it coordinately suppresses catabolic metabolism. During nutrient and energy abundance, mTORC1 suppresses autophagy, the cellular process by which double-membrane structures called autophagosomes form and engulf organelles and macromolecules, and after fusion with lysosomes, degrade the engulfed material into core building blocks (Figure 1). Mechanistically, mTORC1 phosphorylates and inhibits ULK1 (on S758) (an autophagy initiating kinase), as well as ATG13 (a positive regulator of ULK1) and AMBRA1 (on S52)(autophagy/beclin-1 regulator 1) to suppress initiation of autophagy (163–166). During energy starvation, AMPK inactivates mTORC1, thus reducing inhibitory phosphorylation on ULK1, which enables AMPK to bind, phosphorylate (S317), and activate ULK1 to initiate autophagy (163; 164) (see accompanying review article for greater detail).
Future directions
While great progress has been made in elucidating the complex regulation and function of mTOR signaling networks, many questions remain. Understanding the direct molecular mechanisms that govern regulation of mTORC1 and mTORC2 and identifying the signaling intermediates that relay diverse mTOR-regulatory signals to the mTORCs represents an important area of future investigation. It is important to note that upstream regulation of mTORC2 remains a large black box. The clinical utility of mTOR inhibition to treat organ transplant rejection, kidney cancer, tuberous sclerosis complex (TSC), and coronary artery stent restenosis underscores the important role of mTOR in organismal physiology and highlights the importance of elucidating the regulation and function of mTOR signaling networks at the cellular and molecular levels (6). Results obtained in cell culture need to be validated in vivo in more physiologic contexts to understand their significance for organismal physiology and pathophysiology. For example, experiments in cell culture employing complete amino deprivation followed by amino acid re-addition to elucidate amino acid regulation of mTORC1 represents a quite non-physiological approach. To date, a fairly limited set of bona fide mTOR substrates have been identified; it will be important in the future to identify the complete repertoire of mTORC1 and mTORC2 substrates and to link these substrates to cellular functions controlled by the mTORCs, both in cell culture and in animal models.
The identification of lysosomal and peroxisomal membranes as critical mTORC1-regulatory platforms and the realization that dynamic subcellular localization of mTOR and TSC (and possibly other mTOR pathway regulatory molecules) mediate mTORC1 regulation suggests the possibility that other cellular endomembranes may also serve critical mTORC1 regulatory roles (123; 167). Emerging data on the importance of phospholipids and vesicular trafficking in mTORC1 regulation (91; 92; 124; 125) support the idea that specific membrane sub-compartments may control mTORC1 (and mTORC2) differentially in response to different types of upstream signals and may translate these cues into different types of cellular responses. At this point, we understand mTORC2 subcellular localization poorly. The recent discovery that mTORC2 localization to mitochondrial-associated ER membranes (MAMs) (73) controls mTORC2 function suggests that specific endomembrane platforms may participate in mTORC2 as well as mTORC1 regulation and function. The recent discovery that K63-linked mTOR ubiquitination promotes mTORC1 function (99) indicates that this and other PTMs (post-translational modifications) in addition to phosphorylation may serve regulatory roles in mTORC1 an/or mTORC2 function. A major challenge for the future will be to decipher the role of complex phosphorylation on the mTORCs, which has potential to identify novel upstream regulatory molecules and pathways. Another emerging area for future research will be to elucidate crosstalk between mTORC pathways and other established signaling systems; indeed, emerging data link mTOR to the Wnt, Notch, Hippo, and innate immunity signaling pathways (for review see (168)).
Interdisciplinary approaches in which researchers engage in basic research in cultured cells and animal models together with translational approaches will enable improved understanding of the role of mTOR in human physiology and disease. Complete understanding of mTOR signaling networks may enable identification of novel drug targets that can be exploited to treat the myriad diseases linked to aberrant mTOR function, which include cancer, benign tumor syndromes, diabetes, aging, cardiovascular disorders, neurodegenerative disorders, and inflammatory disorders (6; 10; 11).
Acknowledgments
We thank Fingar lab members for review of the manuscript and Andrew Tee for editorial assistance. KH has been supported by a postdoctoral fellowship from the AHA; Fingar lab research is supported by NIH-R01 DK-100722, NIH-R01 DK-103877, and ADA #1-12-BS-49. Importantly, we apologize to those researchers whose important work could not be cited due to space restrictions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abraham RT, Wiederrecht GJ. Immunopharmacology of rapamycin. Annu Rev Immunol. 1996;14:483–510. doi: 10.1146/annurev.immunol.14.1.483. PMID. [DOI] [PubMed] [Google Scholar]
- 2.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. PMID: 15094765. [DOI] [PubMed] [Google Scholar]
- 3.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. PMID: 1715094. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, Fingar DC. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. PMID: 20022946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. PMID: 24508508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. PMID: 7518356. [DOI] [PubMed] [Google Scholar]
- 8.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. PMID: 8008069. [DOI] [PubMed] [Google Scholar]
- 9.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. PMID: 7822316. [DOI] [PubMed] [Google Scholar]
- 10.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. PMID: 22500797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013 doi: 10.1016/j.gde.2012.12.005. PMID: 23317514. [DOI] [PubMed] [Google Scholar]
- 12.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. PMID: 16603397. [DOI] [PubMed] [Google Scholar]
- 13.Choo AY, Blenis J. Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle. 2009;8:567–572. doi: 10.4161/cc.8.4.7659. PMID: 19197153. [DOI] [PubMed] [Google Scholar]
- 14.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4EBP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. PMID: 18955708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci. Signal. 2009;2:pe24. doi: 10.1126/scisignal.267pe24. PMID. [DOI] [PubMed] [Google Scholar]
- 16.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. PMID: 19150980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoreen CC, Sabatini DM. Rapamycin inhibits mTORC1, but not completely. Autophagy. 2009;5:725–726. doi: 10.4161/auto.5.5.8504. PMID: 19395872. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. PMID: 19402821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. PMID: 23636326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. PMID: 23888043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon SO, Roux PP. Rapamycin resistance: mTORC1 substrates hold some of the answers. Curr Biol. 2013;23:R880–883. doi: 10.1016/j.cub.2013.08.030. PMID: 24112984. [DOI] [PubMed] [Google Scholar]
- 22.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. PMID: 12150925. [DOI] [PubMed] [Google Scholar]
- 23.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. PMID: 12150926. [DOI] [PubMed] [Google Scholar]
- 24.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. PMID: 15467718. [DOI] [PubMed] [Google Scholar]
- 25.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. PMID: 15268862. [DOI] [PubMed] [Google Scholar]
- 26.Kim DH, Sarbassov dos D, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. PMID: 12718876. [DOI] [PubMed] [Google Scholar]
- 27.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. PMID: 17386266. [DOI] [PubMed] [Google Scholar]
- 28.van der Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signaling to mTOR mediated by the AKT/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. PMID. [DOI] [PubMed] [Google Scholar]
- 29.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. PMID: 19446321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schalm SS, Blenis J. Identification of a conserved motif required for mTOR signaling. Curr Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. PMID: 11967149. [DOI] [PubMed] [Google Scholar]
- 31.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS Motif-Mediated Raptor Binding Regulates 4E-BP1 Multisite Phosphorylation and Function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. PMID: 12747827. [DOI] [PubMed] [Google Scholar]
- 32.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. PMID: 17141160. [DOI] [PubMed] [Google Scholar]
- 33.Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, Kozma SC. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. PMID: 15485918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Harris TE, Roth RA, Lawrence JC., Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. PMID: 17510057. [DOI] [PubMed] [Google Scholar]
- 35.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. PMID: 16919458. [DOI] [PubMed] [Google Scholar]
- 36.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. PMID: 17043309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. PMID: 17461779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. PMID: 17041623. [DOI] [PubMed] [Google Scholar]
- 39.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. PMID: 22168436. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. PMID: 21659605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. PMID: 21659604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. PMID: 17604717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–2316. doi: 10.4161/cc.10.14.16586. PMID: 21670596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest. 2011;121:1231–1241. doi: 10.1172/JCI44145. PMID: 21490404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. PMID: 21531565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. PMID: 22795129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. PMID: 18466115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers MG, Jr., White MF. Insulin signal transduction and the IRS proteins. Annu Rev Pharmacol Toxicol. 1996;36:615–658. doi: 10.1146/annurev.pa.36.040196.003151. PMID: 8725404. [DOI] [PubMed] [Google Scholar]
- 49.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. PMID: 8978681. [PMC free article] [PubMed] [Google Scholar]
- 50.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. PMID: 15718470. [DOI] [PubMed] [Google Scholar]
- 51.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. PMID: 12150915. [DOI] [PubMed] [Google Scholar]
- 52.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. PMID: 12271141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. PMID: 15854902. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Harris TE, Lawrence JC., Jr. Regulation of Proline-rich Akt Substrate of 40 kDa (PRAS40) Function by Mammalian Target of Rapamycin Complex 1 (mTORC1)-mediated Phosphorylation. J Biol Chem. 2008;283:15619–15627. doi: 10.1074/jbc.M800723200. PMID: 18372248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Lawrence JC, Jr., Sturgill TW, Harris TE. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J Biol Chem. 2009;284:14693–14697. doi: 10.1074/jbc.C109.002907. PMID: 19346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foster KG, Acosta-Jaquez HA, Romeo Y, Ekim B, Soliman GA, Carriere A, Roux PP, Ballif BA, Fingar DC. Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. J Biol Chem. 2010;285:80–94. doi: 10.1074/jbc.M109.029637. PMID: 19864431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Acosta-Jaquez HA, Keller JA, Foster KG, Ekim B, Soliman GA, Feener EP, Ballif BA, Fingar DC. Site-specific mTOR phosphorylation promotes mTORC1-mediated signaling and cell growth. Mol Cell Biol. 2009;29:4308–4324. doi: 10.1128/MCB.01665-08. PMID: 19487463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ekim B, Magnuson B, Acosta-Jaquez HA, Keller JA, Feener EP, Fingar DC. mTOR Kinase Domain Phosphorylation Promotes mTORC1 Signaling, Cell Growth, and Cell Cycle Progression. Mol Cell Biol. 2011;31:2787–2801. doi: 10.1128/MCB.05437-11. PMID: 21576368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dan HC, Ebbs A, Pasparakis M, Van Dyke T, Basseres DS, Baldwin AS. Akt-dependent activation of mTORC1 involves phosphorylation of mTOR by IKKa. J. Biol. Chem. 2014 doi: 10.1074/jbc.M114.554881. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12:260–271. doi: 10.1021/pr300630k. PMID: 23186163. [DOI] [PubMed] [Google Scholar]
- 61.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. PMID: 15851026. [DOI] [PubMed] [Google Scholar]
- 62.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. PMID: 15342917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carriere A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, Roux PP. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. PMID: 18722121. [DOI] [PubMed] [Google Scholar]
- 64.Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1) J Biol Chem. 2011;286:567–577. doi: 10.1074/jbc.M110.159046. PMID: 21071439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. PMID: 18925875. [DOI] [PubMed] [Google Scholar]
- 66.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. PMID: 18411301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saci A, Cantley LC, Carpenter CL. Rac1 Regulates the Activity of mTORC1 and mTORC2 and Controls Cellular Size. Mol Cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. PMID: 21474067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. PMID: 16962653. [DOI] [PubMed] [Google Scholar]
- 69.Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. PMID: 21045808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. PMID: 21376236. [DOI] [PubMed] [Google Scholar]
- 71.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, Sessa WC, Qin J, Zhang P, Su B, Jacinto E. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. Embo J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. PMID: 18566586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. Embo J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. PMID: 18566587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci U S A. 2013;110:12526–12534. doi: 10.1073/pnas.1302455110. PMID: 23852728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. PMID: 15380067. [DOI] [PubMed] [Google Scholar]
- 75.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. PMID: 15249583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim SJ, DeStefano MA, Oh WJ, Wu CC, Vega-Cotto NM, Finlan M, Liu D, Su B, Jacinto E. mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Mol Cell. 2012;48:875–887. doi: 10.1016/j.molcel.2012.09.029. PMID: 23142081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boulbes D, Chen CH, Shaikenov T, Agarwal NK, Peterson TR, Addona TA, Keshishian H, Carr SA, Magnuson MA, Sabatini DM, Sarbassov dos D. Rictor phosphorylation on the Thr-1135 site does not require mammalian target of rapamycin complex 2. Mol Cancer Res. 2010;8:896–906. doi: 10.1158/1541-7786.MCR-09-0409. PMID: 20501647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009;29:5657–5670. doi: 10.1128/MCB.00735-09. PMID: 19720745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Julien LA, Carriere A, Moreau J, Roux PP. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol. 2010;30:908–921. doi: 10.1128/MCB.00601-09. PMID: 19995915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Treins C, Warne PH, Magnuson MA, Pende M, Downward J. Rictor is a novel target of p70 S6 kinase-1. Oncogene. 2010;29:1003–1016. doi: 10.1038/onc.2009.401. PMID: 19935711. [DOI] [PubMed] [Google Scholar]
- 81.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. PMID. [DOI] [PubMed] [Google Scholar]
- 83.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. PMID: 18497260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. PMID: 18604198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. PMID: 20381137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. PMID: 22980980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. PMID: 23723238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202:1107–1122. doi: 10.1083/jcb.201307084. PMID: 24081491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. PMID: 24095279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. PMID: 22053050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. PMID: 16049009. [DOI] [PubMed] [Google Scholar]
- 92.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. PMID: 16176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoon MS, Du G, Backer JM, Frohman MA, Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol. 2011;195:435–447. doi: 10.1083/jcb.201107033. PMID: 22024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 2008;283:35053–35059. doi: 10.1074/jbc.M805822200. PMID: 18948269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, Chakraborty A, Koldobskiy MA, Cha SH, Barrow R, Snowman AM, Snyder SH. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–221. doi: 10.1016/j.cmet.2011.01.007. PMID: 21284988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. PMID: 22424946. [DOI] [PubMed] [Google Scholar]
- 97.Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. PMID: 22424774. [DOI] [PubMed] [Google Scholar]
- 98.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. PMID: 21981924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. PMID: 23911927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. PMID: 17253963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bryk B, Hahn K, Cohen SM, Teleman AA. MAP4K3 regulates body size and metabolism in Drosophila. Dev Biol. 2010;344:150–157. doi: 10.1016/j.ydbio.2010.04.027. PMID: 20457147. [DOI] [PubMed] [Google Scholar]
- 102.Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, Lamb RF. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. PMID: 20227368. [DOI] [PubMed] [Google Scholar]
- 103.Kim YM, Stone M, Hwang TH, Kim YG, Dunlevy JR, Griffin TJ, Kim DH. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol Cell. 2012;46:833–846. doi: 10.1016/j.molcel.2012.04.007. PMID: 22575674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. PMID: 19203585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. PMID: 22749528. [DOI] [PubMed] [Google Scholar]
- 106.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. PMID: 23663782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. PMID: 15772076. [DOI] [PubMed] [Google Scholar]
- 108.Roccio M, Bos JL, Zwartkruis FJ. Regulation of the small GTPase Rheb by amino acids. Oncogene. 2006;25:657–664. doi: 10.1038/sj.onc.1209106. PMID: 16170341. [DOI] [PubMed] [Google Scholar]
- 109.Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. PMID: 12172555. [DOI] [PubMed] [Google Scholar]
- 110.Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell. 2014;156:786–799. doi: 10.1016/j.cell.2014.01.024. PMID: 24529380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. PMID: 24529379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci U S A. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. PMID: 18550814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. PMID. [DOI] [PubMed] [Google Scholar]
- 114.Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. PMID: 17210675. [DOI] [PubMed] [Google Scholar]
- 115.Sun Y, Chen J. mTOR signaling: PLD takes center stage. Cell Cycle. 2008;7:3118–3123. doi: 10.4161/cc.7.20.6881. PMID: 18927511. [DOI] [PubMed] [Google Scholar]
- 116.Yoon MS, Sun Y, Arauz E, Jiang Y, Chen J. Phosphatidic acid activates mammalian target of rapamycin complex 1 (mTORC1) kinase by displacing FK506 binding protein 38 (FKBP38) and exerting an allosteric effect. J Biol Chem. 2011;286:29568–29574. doi: 10.1074/jbc.M111.262816. PMID: 21737445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Foster DA, Salloum D, Menon D, Frias M. Phospholipase D and the Maintenance of Phosphatidic Acid Levels for Regulation of mTOR. J Biol Chem. 2014 doi: 10.1074/jbc.R114.566091. PMID: 24990952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. PMID: 23609508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. PMID: 22343943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. PMID: 22692423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–491. doi: 10.1083/jcb.201209135. PMID: 23401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, Kwiatkowski E, Dunlop EA, Dodd KM, Folkerth RD, Faust PL, Kastan MB, Sahin M, Walker CL. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. PMID: 23955302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Benjamin D, Hall MN. TSC on the peroxisome controls mTORC1. Nat Cell Biol. 2013;15:1135–1136. doi: 10.1038/ncb2849. PMID: 24084861. [DOI] [PubMed] [Google Scholar]
- 124.Bridges D, Ma JT, Park S, Inoki K, Weisman LS, Saltiel AR. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–2962. doi: 10.1091/mbc.E11-12-1034. PMID: 22696681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jin N, Mao K, Jin Y, Tevzadze G, Kauffman EJ, Park S, Bridges D, Loewith R, Saltiel AR, Klionsky DJ, Weisman LS. Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol Biol Cell. 2014;25:1171–1185. doi: 10.1091/mbc.E14-01-0021. PMID: 24478451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Flinn RJ, Yan Y, Goswami S, Parker PJ, Backer JM. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21:833–841. doi: 10.1091/mbc.E09-09-0756. PMID: 20053679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bridges D, Fisher K, Zolov SN, Xiong T, Inoki K, Weisman LS, Saltiel AR. Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J Biol Chem. 2012;287:20913–20921. doi: 10.1074/jbc.M111.334060. PMID: 22547071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. PMID: 22436748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. PMID: 14651849. [DOI] [PubMed] [Google Scholar]
- 130.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. PMID: 18439900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. PMID: 21892142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, Kalinski H, Kamer I, Rozen A, Mor O, Keshet E, Leshkowitz D, Einat P, Skaliter R, Feinstein E. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. PMID: 11884613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. PMID: 15545626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. PMID: 22562152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. PMID: 15545625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. PMID: 18198340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov. 2013;12:703–719. doi: 10.1038/nrd3976. PMID: 23989796. [DOI] [PubMed] [Google Scholar]
- 138.Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem Biophys Res Commun. 2009;379:451–455. doi: 10.1016/j.bbrc.2008.12.079. PMID: 19114033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jin HO, Seo SK, Woo SH, Kim ES, Lee HC, Yoo DH, An S, Choe TB, Lee SJ, Hong SI, Rhee CH, Kim JI, Park IC. Activating transcription factor 4 and CCAAT/enhancer-binding protein-beta negatively regulate the mammalian target of rapamycin via Redd1 expression in response to oxidative and endoplasmic reticulum stress. Free Radic Biol Med. 2009;46:1158–1167. doi: 10.1016/j.freeradbiomed.2009.01.015. PMID: 19439225. [DOI] [PubMed] [Google Scholar]
- 140.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. PMID: 18342602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. PMID: 18692468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, Tanti JF, Giorgetti-Peraldi S, Bost F. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res. 2011;71:4366–4372. doi: 10.1158/0008-5472.CAN-10-1769. PMID: 21540236. [DOI] [PubMed] [Google Scholar]
- 143.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. PMID: 12080086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR Controls Cell Cycle Progression through Its Cell Growth Effectors S6K1 and 4E-BP1/Eukaryotic Translation Initiation Factor 4E. Mol Cell Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. PMID: 14673156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. PMID: 20508131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. PMID: 19339977. [DOI] [PubMed] [Google Scholar]
- 150.Jefferies HBJ, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypryimidine tract” nRNA family. P.N.A.S. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. PMID: 11029573. [DOI] [PubMed] [Google Scholar]
- 152.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. PMID: 22552098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miloslavski R, Cohen E, Avraham A, Iluz Y, Hayouka Z, Kasir J, Mudhasani R, Jones SN, Cybulski N, Ruegg MA, Larsson O, Gandin V, Rajakumar A, Topisirovic I, Meyuhas O. Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J Mol Cell Biol. 2014;6:255–266. doi: 10.1093/jmcb/mju008. PMID: 24627160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation. Genes Dev. 2014;28:357–371. doi: 10.1101/gad.231407.113. PMID: 24532714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Isotani S, Hara K, Tokunaga C, Inoue H, Avruch J, Yonezawa K. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J Biol Chem. 1999;274:34493–34498. doi: 10.1074/jbc.274.48.34493. PMID: 10567431. [DOI] [PubMed] [Google Scholar]
- 157.Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, Pende M. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene. 2014;33:474–483. doi: 10.1038/onc.2012.606. PMID: 23318442. [DOI] [PubMed] [Google Scholar]
- 158.Ricoult SJ, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013;14:242–251. doi: 10.1038/embor.2013.5. PMID: 23399656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. PMID: 18762023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. PMID: 20670887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. PMID: 21816276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. PMID: 23429704. [DOI] [PubMed] [Google Scholar]
- 163.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. PMID: 21258367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–644. doi: 10.4161/auto.7.6.15123. PMID: 21460621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. PMID: 23524951. [DOI] [PubMed] [Google Scholar]
- 166.Nazio F, Cecconi F. mTOR, AMBRA1, and autophagy: an intricate relationship. Cell Cycle. 2013;12:2524–2525. doi: 10.4161/cc.25835. PMID: 23907135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Benjamin D, Hall MN. mTORC1: turning off is just as important as turning on. Cell. 2014;156:627–628. doi: 10.1016/j.cell.2014.01.057. PMID: 24529368. [DOI] [PubMed] [Google Scholar]
- 168.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. PMID: 24556838. [DOI] [PubMed] [Google Scholar]