Abstract
The genetic etiology of sporadic neuroblastoma is still largely obscure. In a genome-wide association study, we identified single nucleotide polymorphisms (SNP) associated with neuroblastoma at the LINC00340, BARD1, LMO1, DUSP12, HSD17B12, HACE1 and LIN28B gene loci, but these explain only a small fraction of neuroblastoma heritability. Other neuroblastoma susceptibility genes are likely hidden among signals discarded by the multiple testing corrections. In this study, we evaluated 8 additional genes selected as candidates for further study based on proven involvement in neuroblastoma differentiation. SNP at these candidate genes were tested for association with disease susceptibility in 2101 cases and 4202 controls, with the associations found replicated in an independent cohort of 459 cases and 809 controls. Replicated associations were further studied for cis-effect using gene expression, transient overexpression, silencing and cellular differentiation assays. The neurofilament gene NEFL harbored three SNP associated with neuroblastoma (rs11994014; Pcombined=0.0050; OR=0.88, rs2979704; Pcombined=0.0072; OR=0.87, rs105911; Pcombined=0.0049; OR=0.86). The protective allele of rs1059111 correlated with increased NEFL expression. Biological investigations showed that ectopic overexpression of NEFL inhibited cell growth specifically in neuroblastoma cells carrying the protective allele. NEFL overexpression also enhanced differentiation and impaired the proliferation and anchorage-independent growth of cells with protective allele and basal NEFL expression, while impairing invasiveness and proliferation of cells homozygous for the risk genotype. Clinically, high levels of NEFL expression in primary neuroblastoma specimens was associated with better overall survival (P=0.03; HR=0.68). Our results show that common variants of NEFL influence neuroblastoma susceptibility and they establish that NEFL expression influences disease initiation and progression.
Keywords: neuroblastoma, SNP, NEFL, genetic association, GWAS
Introduction
Neuroblastoma is a cancer of the sympathetic nervous system. It is the most frequent solid tumor of early childhood with a remarkable variation in clinical presentation ranging from favorable localized tumors that can spontaneously regress to metastatic disease that shows relentless progression (1). Despite intensive therapies, the survival rate for the patient sub-group with the most aggressive form remains approximately 40%. While familial neuroblastoma, which accounts for approximately 1% of cases, is in large proportion due to mutations in the ALK gene (2), the genetic bases of sporadic neuroblastoma remain largely unknown (3). Our ongoing genome-wide association study (GWAS) has demonstrated that genetic variants within the LINC00340, BARD1, LMO1, DUSP12, HSD17B12, HACE1 and LIN28B genes (4–8) are strongly associated with neuroblastoma in North American patients of European descent, and each of these associations have been replicated in an independent Italian population (9). However, these risk variants only explain a small proportion of neuroblastoma heritability and additional predisposing variants to neuroblastoma remain to be discovered.
The remaining genetic risk is probably made up by a combination of common variants with very modest effect sizes and by rare variants with stronger effects. Some of the common genetic risk variants are probably hidden among signals discarded by the multiple testing correction needed in the analysis of GWAS data. This multiple testing correction is necessary to exclude false-positive loci, but simultaneously it discards many true-positive risk loci. Different research strategies have been proposed (10) for extracting these hidden true-positive loci, such as increasing the GWAS sample size, performing a meta-analysis of GWAS data sets, replicating hundreds to thousands of GWAS signals in a larger cohort, performing imputation and epistasis analysis, using pathway-based and single nucleotide polymorphism (SNP)-set enrichment approaches, and others.
Here, in order to discover new common variants associated with neuroblastoma, we adopted a strategy based on the selection of genes with high relevance to neuroblastoma differentiation which were identified in our previous proteomic study (11). Arrested differentiation of neuroblasts at various stages is a crucial early event in neuroblastoma pathogenesis, especially in the more aggressive cases. Indeed a particular characteristic of neuroblastic tumors is their broad spectrum of cellular differentiation, ranging from undifferentiated cells that indicate a poor prognosis to those showing greater differentiation and predicting a generally favorable outcome (12). This heterogeneity suggests that dysregulated differentiation of sympathetic progenitor cells plays a key role in neuroblastoma pathogenesis. We thus hypothesized that variation in genes involved in neuroblastic differentiation might predispose to neuroblastoma development. This hypothesis is supported by recent research showing that functional alterations of key regulators of neuron development can induce neuroblastic malignant transformation (13, 14).
In the present study, we performed a gene-based association analysis of eight genes recently shown to be major regulators of neuroblastoma differentiation (11). A large cohort of 2101 patients and 4202 control subjects was used as discovery set whereas an independent population of 459 cases and 809 controls was used as replication set. The results showed a significant association of SNPs within NEFL with neuroblastoma risk. Additional in silico and in vitro analyses demonstrated a functional role of the neuroblastoma-associated SNPs and indicated that NEFL likely has a role in disease development and progression.
Materials and Methods
Study subjects
This study was approved by the Ethics Committee of the Medical University of Naples and the Children’s Hospital of Philadelphia.
This study included a GWAS dataset of 2101 neuroblastoma patients registered through the North American-based Children’s Oncology Group (COG) and 4202 cancer-free children of self-reported Caucasian ancestry who were recruited and genotyped by the Center for Applied Genomics at the Children’s Hospital of Philadelphia (CHOP). European American cases and controls have been described in detail in a previous publication (8). In addition, this study also included 459 neuroblastoma patients and 809 cancer-free controls of Italian origin. Additional details and eligibility criteria for genotyping of both populations are reported in Supplementary Information. This study was approved by the Ethics Committee of the Medical University of Naples and the Children’s Hospital of Philadelphia.
Selection of genes involved in neuroblastoma differentiation
Our previous study has demonstrated that eight proteins are differentially expressed during neuroblastoma differentiation by two-dimensional differential in-gel electrophoresis analysis of the cytosolic and nuclear protein expression patterns of LAN-5 cells following neuronal differentiating agent all-trans-retinoic acid treatment (11). The retinoic acid differential expression patterns of these eight proteins were further validated in three cell lines (LAN-5, SH-SY5Y and SK-N-BE) by western blotting and gene expression (11). This motivated us to select the genes (EEF1A, EEF2, GNB2, NEFL, PCNA, PRDX2, SCG2, VBP1) that encode the eight proteins to be tested for association with neuroblastoma development (Table 1).
Table 1.
Gene-based association analysis in neuroblastoma GWAS dataset (2101 cases and 4202 controls)
| GENE SETS | NSNP | NSIG | ISIG | EMP | FDR | SNPs |
|---|---|---|---|---|---|---|
| EEF1A1 | 4 | 0 | 0 | 1 | 1 | NS |
| EEF2 | 6 | 0 | 0 | 1 | 1 | NS |
| GNB2 | 3 | 0 | 0 | 1 | 1 | NS |
| NEFL | 8 | 4 | 2 | 0.0076 | 0.0304 | rs118727|rs169061 |
| PCNA | 6 | 0 | 0 | 1 | 1 | NS |
| PRDX2 | 2 | 0 | 0 | 1 | 1 | NS |
| SCG2 | 7 | 1 | 1 | 0.0037 | 0.0296 | rs4673067 |
| VBP1 | 0 | 0 | 0 | 1 | 1 | NS |
NSNP: Number of SNPs in gene and surrounding genomic region (+/− 20Kb)
NSIG: Total number of SNPs below p-value threshold
ISIG: Number of significant also passing LD-criterion
EMP: Empirical gene-based p-value
NS: not significant
FDR: False discovery rate correction method applied to EMP.
SNP genotyping
The European American DNA samples were genotyped using the Illumina Infinium II BeadChip HumanHap550 v1, v3, and Quad610 arrays according to methods detailed elsewhere (8). The quality control analyses for this GWAS dataset are described in detail elsewhere (8). We analyzed six SNPs of NEFL (rs196830, rs169061, rs11994014, rs2979704, rs1059111, rs3761) in the Italian cohort. The DNA samples were genotyped using SNP Genotyping Assay on 7900HT Real-time PCR system (Applied Biosystems). More details on SNP genotyping are reported in Supplementary Information.
Gene-based association analysis
The SNP genotypes of each selected gene and surrounding genomic region (+/− 20Kb) were extracted from the GWAS dataset. The analysis was carried out by using the set-based test implemented in PLINK software (15). Briefly, for each gene, a standard single SNP analysis (case/control association) is performed. SNPs with p-value > 0.05 and/or a Linkage Disequilibrium (LD) coefficient r2 > 0.5 with a significant SNP (defined as a p-value < 0.05) are filtered out. From the remaining independent SNPs, the statistic for each gene is calculated as the mean of the single SNP statistics. To correct for testing multiple SNPs within a gene, an empirical gene-based p-value is calculated based on 10,000 replicates of this procedure after random permutation of case/control status, as the proportion of replicates with a statistic larger than the one observed. To correct for testing multiple genes, we applied the false discovery rate method to the empirical gene-based p-values.
Genotype Imputation of putative functional SNPs at NEFL locus
As SNPs rs2979704, rs1059111, rs3761 were not included in the Illumina HumanHap550 array, genotype imputation was performed in the European American GWAS. Pre-phasing was performed first using SHAPEIT (16), followed by imputation using 1000 Genomes data (Phase I integrated release) and IMPUTE2 (17). SNPs with minor allele frequency (MAF) <1% and/or IMPUTE2-info quality score <0.8 were removed. To account for imputation uncertainty, the remaining SNPs were tested for association with neuroblastoma using the frequentist test under the additive model with the “score” method implemented in SNPTEST (18). A detailed description of genotype imputation is reported in Supplementary Information.
SNP-gene expression correlation analysis in tumor tissues and neuroblastoma cell lines
The influence of SNPs on NEFL gene expression was evaluated using data from genome-wide mRNA expression profiling (GSE3960) and SNP array genotyping (pha002845) in 51 neuroblastoma patients, and from qRT-PCR analysis in 8 neuroblastoma cell lines. A detailed description of the analysis is reported in Supplementary Information.
In vitro functional analysis
A detailed description of the luciferase assay and other experiments performed to evaluate the NEFL effect on neuroblastoma cell line phenotype is reported in Supplementary Information.
Cell lines
The human SH-SY5Y, SK-N-AS, IMR-32, SK-N-BE2c and HEK293T/17 cell lines were obtained from the American Type Culture Collection (respectively ATCC #CRL-2266, #CRL-2137, #CCL-127, #CRL-2268 and #CRL-11268). SH-SY5Y, SK-N-AS and HEK293T/17 cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM, Sigma); IMR32 cell line was grown in minimum essential medium eagle (MEM, Sigma) and SK-N-BE2c cell line was grown in DMEM/F12 medium (Sigma). The medium were supplemented with 10% heat-inactivated FBS (Sigma), 1mM L-glutamine, penicillin (100 U/ml) and streptomycin (100 μg/ml) (Invitrogen). The cells were cultured at 37°C, 5% CO2 in a humidified atmosphere. The cumulative culture length of the cells was fewer than 6 months after resuscitation. Early passage cells were used for all experiments and they were not re-authenticated.
Gene expression data for survival analysis and association with neuroblastoma stages
NEFL normalized gene expression array data of three independent sets of neuroblastoma patients (399 total) were downloaded from the website Oncogenomics (http://home.ccr.cancer.gov/oncology/oncogenomics/): 1) “Kahn dataset” composed of 46 samples (Affymetrix Human Exon 1.0 ST Array, GSE27608); 2) “Oberthuer dataset” composed of 251 samples (Agilent array Chip_10k_v1, E-TABM-38); 3) “Seeger dataset” composed of 102 samples (Affymetrix HG-U133A and HG-U133B array, GSE16254). Median centered normalization was used for all gene expression data. This allowed us to perform survival analysis on the three datasets combined. However, to overcome any biases due to the different platforms used in each dataset, we also performed a meta-analysis on the summary statistics (see below). Analysis of association between NEFL expression and neuroblastoma stages was performed using normalized microarray data freely available at GEO database (GSE45547, GSE14880). Data for tumor stages were available.
Statistical analysis
Hardy-Weinberg equilibrium was evaluated using the goodness-of-fit chi-square test in control subjects. For genotyped SNPs, two-sided chi-square tests were used to evaluate differences in the distributions of allele frequencies between all patients and controls. ORs and 95% CIs were calculated to assess the relative disease risk conferred by a specific allele. Combined analysis was performed using the weighted Z-score method in METAL based on sample size, P-value and direction of effect in each study (19). Student’s t test was used to compare the differences in the mRNA expression levels. LD and haplotype analyses were performed using the web-site tool SNAP v2.2 (http://www.broadinstitute.org/mpg/snap/index.php) (20) and Haploview v4.2 software (21). qRT-PCR data were analyzed by 2^−Δct method as described in our previous paper (22). To test association of gene expression levels with overall survival, individual gene expression profiles were dichotomized by median split into ‘high’ or ‘low’ expression groups, and Kaplan-Meier survival curves were plotted for each group. Cox regression analysis was applied to calculate HR for meta-analysis. A test of heterogeneity of combined HRs was carried out using Cochran’s Q test (significant at P<0.05) and Higgins I-squared statistic. I-squared >50% were considered to represent significant heterogeneity. Given the absence of heterogeneity among studies (P > 0.1), a fixed effects generic inverse variance model was used. Meta-analysis was performed using Review manager 5.0 (http://www.cochrane.org).
Results
Biologically driven gene-based association analysis
Eight genes shown to be associated with neuroblastoma differentiation (11) were tested for genetic associations with neuroblastoma risk. The gene-based test performed on the discovery set (2101 cases and 4202 controls) showed that the two genes NEFL and SCG2 were significantly associated with neuroblastoma susceptibility (Table 1). However, NEFL had 8 SNPs, 5 of which were significant, but only 2 of which were independently significant based on an r-squared threshold of 0.5; whereas SCG2 had 7 SNPs, but only 1 was significant.
Prioritization of SNPs in SCG2 and NEFL
To select putative functional SNPs to be further validated in the Italian cohort, we examined the genetic association and the predicted biological role of SNPs at both loci. At the SCG2 locus (surrounding region +/− 20Kb), only SNP rs4673067 resulted to be significant (data not shown). SNP function prediction analysis using the web tool SNPinfo Web Server (23) showed no relevant function for this SNP and those in LD with it (r2 > 0.60) (Supplementary Table S1). At the NEFL locus, three intergenic and two flanking (in the 3′ downstream and promoter regions) polymorphisms were significantly associated with neuroblastoma (Table 2, Fig. 1A). SNP function prediction analysis (23) showed that none of these five SNPs had putative biological function (data not shown). Thus, we carried out a prioritization functional analysis on SNPs in LD (r2 > 0.60) with the five typed SNPs (Supplementary Table S2, S3, S4, S5, S6). The results indicated that three polymorphisms within the NEFL 3′UTR (rs3761, rs2979704, and rs1059111) had a potential biological effect (Table 2, Fig. 1A). Each of these SNPs was located in evolutionary conserved regions and predicted to influence the binding of diverse transcription factors and microRNAs (Supplementary Table S7, S8, S9). In contrast, the analysis by UTRscan (http://itbtools.ba.itb.cnr.it/utrscan) showed that none of the above-mentioned SNPs altered sequence characteristics of the specific UTR motifs (such as: polyadenylation signal, AU rich element, Selenocysteine Insertion Sequence and others reported in the Supplementary Table S10). Taken together, in silico data suggest that the functional role of SNPs located in the 3′ UTR of NEFL are likely due to alteration of miRNA binding sites, enhancer elements or both. Notably, rs3761 was in LD with three significant typed tag-SNPs (rs118727, rs196830, rs17830286) while rs2979704 and rs1059111 (in strong LD with each other, r2=1, Supplementary Fig. 1) were in LD with only one typed tag-SNP (rs11994014). To further verify if other functional SNPs could be associated with neuroblastoma, we used the 1000 Genomes data to impute all of the common variants (MAF>0.05) across SCG2 and NEFL. No significant association was found at SCG2 locus (Supplementary Table S11). The rs3761 SNP in NEFL resulted to be the strongest significantly associated with neuroblastoma. Other two SNPs (rs62503767 and rs62503769) were significant (Supplementary Table S11) but no relevant biological function was predicted (data not shown). Together these data led us to exclude SCG2 SNPs from further analyses and focused our attention on NEFL SNPs.
Table 2.
Associations and functional prioritization of SNPs in NEFL and its surrounding region (+/−20Kb)
| SNP Association in GWAS dataset | SNP functional prioritization | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| SNP | Role | A1 | F_A | F_U | A2 | P | OR | L95 | U95 | Proxy SNP ID | Proxy SNP function | * r2 | RegPotential Score | Conservation Score |
| rs118727 | intergenic | C | 0.14 | 0.12 | T | 0.0026 | 1.18 | 1.06 | 1.32 | rs3761 | NEFL/3′-UTR | 1 | 0.21 | 0.132 |
| rs196830 | intergenic | C | 0.15 | 0.12 | A | 0.0012 | 1.19 | 1.07 | 1.33 | rs3761 | NEFL/3′-UTR | 1 | 0.21 | 0.132 |
| rs169061 | intergenic | C | 0.40 | 0.43 | T | 0.0008 | 0.88 | 0.81 | 0.95 | ^ no-LD | - | - | - | - |
| rs11994014 | Downstream | A | 0.20 | 0.22 | G | 0.0368 | 0.91 | 0.83 | 0.99 | ° rs2979704 and rs1059111 | NEFL/3′-UTR | 0.63 | 0.07 and 0.36 | 0.96 and 1.00 |
| rs2979685 | Promoter | T | 0.36 | 0.36 | G | 0.8723 | 1.01 | 0.93 | 1.09 | - | - | - | - | - |
| rs17830286 | Promoter | T | 0.16 | 0.15 | C | 0.0085 | 1.15 | 1.04 | 1.27 | rs3761 | NEFL/3′-UTR | 0.64 | 0.21 | 0.132 |
| rs13254844 | Promoter | T | 0.05 | 0.05 | C | 0.5270 | 0.95 | 0.80 | 1.12 | - | - | - | - | - |
| rs17830392 | Promoter | G | 0.08 | 0.07 | A | 0.2101 | 1.09 | 0.95 | 1.26 | - | - | - | - | - |
A1: minor allele; A2: major allele; F_A: minor allele frequency in cases; F_U: minor allele frequency in controls
OR: odds ratio; L95: lower bound of 95% confidence interval for odds ratio; U95: upper bound of 95% confidence interval for odds ratio
Cut-off = 0.60
rs2979704 and rs1059111 are in strong LD (r2=1)
No SNP was found in LD with r2>0.60
RegPotential: regulatory potential score (ESPERR regulatory potential based on 7 species) downloaded from UCSC genome bioinformatics web site
Conservation: vertebrate Multiz Alignment and Conservation score (17 Species) downloaded from UCSC genome website
Figure 1. NEFL genotype and gene expression association.
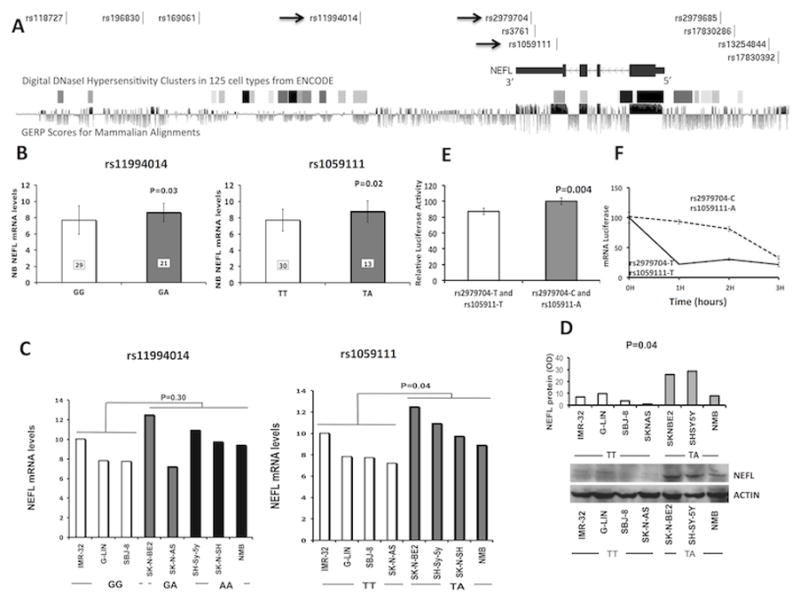
(A) Plot showing NEFL gene and analyzed SNPs. The arrows indicate the SNPs validated in the Italian population. DNase hypersensitive areas in 125 cell types from ENCODE data and GERP score for mammalian alignment (data retrieved from UCSC genome browser, https://genome.ucsc.edu/) are also plotted. (B) Microarray-based expression profiling on primary tumors demonstrates that NEFL expression correlates with rs11994014 (GG: 7.7±1.7 and GA: 8.6±1.1) and rs1059111 genotype (TT: 7.7±1.3 and TA: 8.8±1.3). The genotypes of rs1059111 have been imputed (see Supplementary Information). (C) qRT-PCR (rs11994014 GG: 8.5±1.3; rs11994014 GG/GA: 10.0±1.9; rs1059111 TT: 8.2±1.2; rs1059111 TA: 10.6±1.4) and (D) western blotting (rs1059111 TT: 5.3±3.7; rs1059111 TA: 20.8±11.2) analysis on neuroblastoma cell lines confirms the association between rs1059111 protective genotype and NEFL expression. Data are shown as mean ± standard deviation. (E) Luciferase report gene assay carried out in HEK293 cells confirms the same association (rs1059111 T: 87.1±3.8; rs1059111 A: 100±4.4). Data shown in percentage are the mean ± standard deviation from nine independent transfection experiments, each done in triplicate and compared with promoter less control. (F) Analysis of 3′UTR-allelic mRNA stability in HEK293 cells. The data are presented as the means value ± standard deviation from three independent experiments (P<0.05).
Replication study
We genotyped and tested for association in 459 neuroblastoma patients and 809 controls of Italian origin two tag-SNPs (rs196830, the most significant among the three tag-SNPs, and rs11994014), the three prioritized SNPs (rs3761, rs2979704, rs1059111), in addition to the significant typed SNP not in LD with other SNPs (rs169061) (Table 3). Only the tag-SNP rs11994014 and the linked functional SNPs rs2979704 and rs1059111 confirmed the associations with neuroblastoma (Table 3). Specifically, the minor alleles rs11994014-A, rs2979704-C, and rs1059111-A were associated with a decreased risk of neuroblastoma (Pcombined=0.0050; OR=0.88; 95% CI: 0.81–0.96, Pcombined=0.0072; OR=0.87; 95% CI: 0.78–0.96, Pcombined=0.0049; OR=0.86; 95% CI: 0.77–0.95). All of these SNP alleles were associated with the favorable clinical parameter age at diagnosis < 18 months in the European American cohort (Supplementary Table S12) but not in the Italian cohort (Supplementary Table S13). This may be due to the relatively low number of Italian cases in the clinical sub-groups. We found no significant correlation between the associated SNPs and other clinical covariates such as risk group, INSS stage, and MYCN status.
Table 3.
SNP associations at the NEFL locus
| European Americans | Italians | Combined | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||
| SNP | Role | A1 | F_A | F_U | A2 | P | OR | L95 | U95 | F_A | F_U | P | OR | L95 | U95 | Meta P | OR | L95 | U95 |
| rs196830 | intergenic | C | 0.146 | 0.125 | A | 0.0012 | 1.19 | 1.07 | 1.33 | 0.11 | 0.12 | 0.5189 | 0.92 | 0.70 | 1.20 | ||||
| rs169061 | intergenic | C | 0.396 | 0.427 | T | 0.0008 | 0.88 | 0.81 | 0.95 | 0.46 | 0.50 | 0.0351 | 1.20 | 1.01 | 1.41 | ||||
| rs11994014 | Downstream | A | 0.200 | 0.216 | G | 0.0368 | 0.91 | 0.83 | 0.99 | 0.26 | 0.30 | 0.0230 | 0.78 | 0.64 | 0.95 | 0.0050 | 0.88 | 0.81 | 0.96 |
| * rs2979704 | 3′UTR | C | 0.122 | 0.132 | T | 0.0877 | 0.91 | 0.81 | 1.02 | 0.15 | 0.19 | 0.0049 | 0.72 | 0.57 | 0.90 | 0.0072 | 0.87 | 0.78 | 0.96 |
| * rs1059111 | 3′UTR | A | 0.122 | 0.132 | T | 0.0876 | 0.91 | 0.81 | 1.02 | 0.14 | 0.19 | 0.0016 | 0.69 | 0.55 | 0.87 | 0.0049 | 0.86 | 0.77 | 0.95 |
| * rs3761 | 3′UTR | A | 0.139 | 0.121 | G | 0.0043 | 1.17 | 1.04 | 1.30 | 0.11 | 0.13 | 0.3286 | 0.87 | 0.67 | 1.15 | ||||
A1: minor allele; A2 major allele; F_A: minor allele frequency in cases; F_U: minor allele frequency in controls
OR: odds ratio; L95: lower bound of 95% confidence interval for odds ratio; U95: upper bound of 95% confidence interval for odds ratio
Meta-analysis P value calculated using METAL.
Imputed SNPs in the European American GWAS dataset
Functional analysis of significant SNPs
To investigate if the neuroblastoma-associated SNPs have a cis-effect on NEFL, we tested for SNP-gene expression associations at tag-SNP rs11994014 and prioritized SNP rs1059111 using different approaches. For these analyses, we chose the putative functional polymorphism rs1059111 because it showed the highest scores for potential regulation and conservation and it is predicted to alter miRNA binding sites (Table 2, Fig. 1A and Supplementary Table S8). Moreover, an additional analysis using HaploReg v2 (24) showed that rs1059111 resides within a DNase hypersensitive area in the SK-N-BE2c neuroblastoma cell line, the highest conserved region among the regions of other SNPs (Fig. 1A) and is predicted to alter the regulatory motifs of the transcriptional factor neuron restrictive silencer factor (NRSF) (Supplementary Table 9). Interestingly, the NEFL gene expression increased when we silenced NRSF in two neuroblastoma cell lines (SH-SY5Y and SK-N-BE2c) (Supplementary Fig. 2A–D). These findings strength the role of NRSF as oncogene and repressor of differentiation in neuroblastoma (25, 26) and provide evidences to support the role of NEFL in neuroblastoma differentiation. The analysis of gene expression variation using genome-wide expression and SNP arrays of neuroblastoma tumors demonstrated that both SNPs affect expression of NEFL. In particular, presence of the protective alleles (A and A) for SNPs rs11994014 and rs1059111 significantly correlated with increased NEFL mRNA expression (Fig. 1B). A qRT–PCR gene expression analysis was performed in neuroblastoma cell lines. The mRNA expression of NEFL was significantly higher in neuroblastoma cell lines heterozygous at rs1059111 (TA) (Fig. 1C). Only a trend toward association between high mRNA levels and presence of the protective allele A was observed for rs11994014 without reaching the threshold for statistical significance (Fig. 1C). These results were confirmed using freely available data (http://www.broadinstitute.org/ccle) on gene expression and SNP arrays for 16 neuroblastoma cell lines (Supplementary Fig. 3A) and for 198 lymphoblastoid cell lines (Supplementary Fig. 3B). The correlation of SNP rs1059111 with NEFL expression was validated by western blotting (Fig. 1D). Induction of 3′UTR activity of the construct containing rs2979704-C and rs1059111-A alleles was higher than that of the construct containing T alleles as assessed by luciferase report gene assay (Fig. 1E). Finally, NEFL mRNA abundance from clones containing the rs2979704-C and rs1059111-A alleles after actinomycin-D addition was significantly higher than expression from clones containing the T alleles (Fig. 1F). Together these data indicate that the A allele of SNP rs1059111 in 3′UTR of NEFL confers a decreased neuroblastoma risk and induces gene over-expression in neuroblastoma tumors compared to the alternative allele T. This is probably due to the alteration of miRNA binding site, enhancer elements or both. Further investigations are needed to address this issue. Moreover, transient over-expression of NEFL resulted in significant growth inhibition in neuroblastoma cell heterozygous for the rs1059111 protective A allele that had high NEFL expression (Fig. 2A). In the cell lines homozygous for the risk T allele that had low NEFL expression, the growth inhibition was less marked (Fig. 2B,C). The same results were confirmed in cell lines with stable expression of NEFL (Fig. 2E,D). These data suggest that the protective alleles are associated with growth disadvantage through increased NEFL expression. We have also evaluated cell growth and invasion after NEFL silencing in two TA genotype cell lines (SH-SY5Y and SK-N-BE2c). We observed no effect on cell growth, but increased cell invasion was evident in both cell lines (Supplementary Fig. 4A–C). We hypothesize that decrease of cell growth observed in cell lines with enforced expression of NEFL (Fig. 2A) is probably due to the effect of cell conversion into more differentiated status.
Figure 2. The protective allele A of the SNP rs1059111 is associated with cell growth disadvantage through increased NEFL expression.
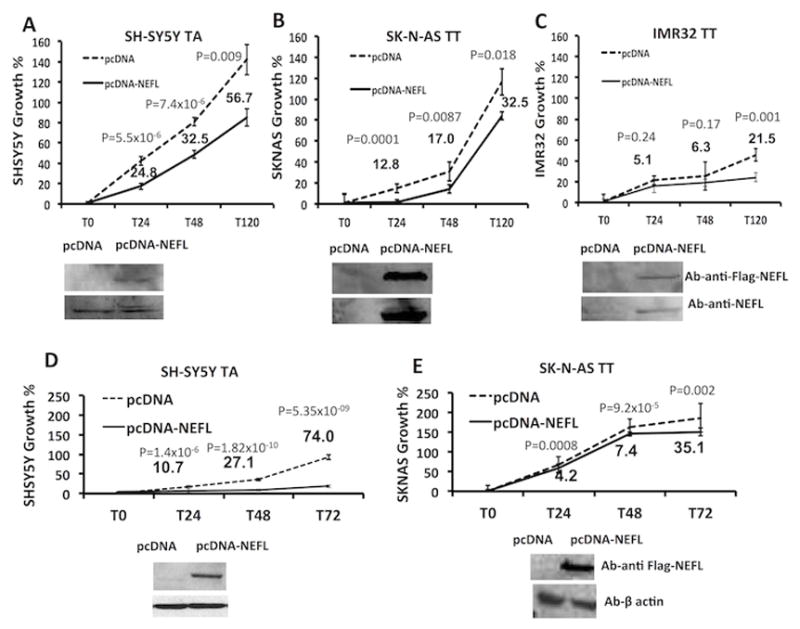
(A) In the cells with neuroblastoma protective allele and higher NEFL expression levels, NEFL transient over-expression leads to inhibition of cellular growth. (B,C) In cells homozygous for the risk allele that have low NEFL expression, the growth inhibition is less marked. Detection of NEFL protein by using antibody against Flag and NEFL assessed the NEFL transiently overexpression in SH-SY5Y, SK-N-AS, IMR32 cell lines. The variability of the forced NEFL expression may to be due to the different transfection efficiency of the three different cell lines. (D,E) We thus produced a stable over-expression of NEFL in SH-SY5Y and SK-N-AS cells to obtain a further validation. The experiments have been performed in three diverse clones for each experimental point and the shown results are representative of the mean among the different clones. The numbers represent the cell number difference between control and NEFL over-expression. Times (T) represent the hours.
Functional analysis of NEFL overexpression
To unravel NEFL contribution to neuroblastoma tumorigenesis and progression, we generated NEFL stable clones in two cell lines heterozygous for the protective minor allele (SH-SY5Y) and homozygous for the risk major allele (SK-N-AS). SH-SY5Y NEFL stable clones showed an enhanced differentiated phenotype as assessed by increased neurites length whereas SK-N-AS NEFL stable clones became flatter and larger than control pcDNA clones (Fig. 3A,B). Moreover, the expression of neuronal differentiation markers is also enhanced by NEFL overexpression in SH-SY5Y cells but not in SK-N-AS cells (Fig. 3C). The analysis using freely available data (http://www.broadinstitute.org/ccle) showed that three of these neuronal markers were up-regulated in neuroblastoma cell lines with the protective allele and high level of NEFL basal expression (Supplementary Fig. 5). In both NEFL overexpressing cell lines we observed a reduced proliferation as assessed by PCNA western blotting analysis (Fig. 3D) but not an increased caspase activity (data not shown). In an experiment of anchorage-independent growth in soft agar we observed NEFL overexpression in SH-SY5Y cells impaired the growth ability (Fig. 3E). In contrast SK-N-AS cells showed no or very little anchorage-independent growth in soft agar (data not shown). NEFL overexpression in SH-SY5Y (Fig. 3F) and SK-N-AS (Fig. 3G) cells impaired the migratory ability in invasion assay. These findings suggest that different phenotypic effect of the over-expression might be due to underlying genotype and basal NEFL expression in the two neuroblastoma cell lines.
Figure 3. Different phenotypic effect of the NEFL over-expression in two neuroblastoma cells with protective and risk genotype for the SNP rs1059111.
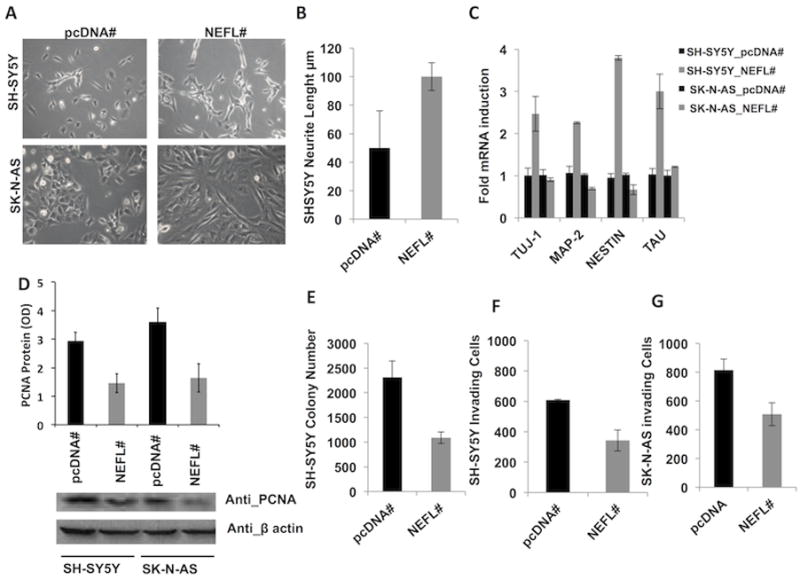
(A) SH-SY5Y (TA genotype) NEFL overexpressing stable clones show longer neurites processes than pcDNA control cells whereas SK-N-AS (TT genotype) NEFL overexpressing stable clones become flatter. (B) Neurites length in SH-SY5Y cell clones were measured from the cell body to the furthest tip of the process using LAS/AF tool (Leica Mycrosistem). (C) The expression of neuronal differentiating markers is enhanced in SH-SY5Y NEFL stable clones whereas it does not change in SK-N-AS NEFL stable clones, as shown by qRT-PCR. (D) The expression of PCNA is decreased in NEFL stable clones in both cell lines. (E) NEFL overexpression impairs the growth in soft agar of SH-SY5Y stable clones as shown by a decreased number of colonies with respect to pcDNA cells. NEFL impairs the number of invading cells in 2-dimentional invasion assay in (F) SH-SY5Y and (G) SK-N-AS stable clones. The experiments have been performed in three diverse clones for each experimental point and the shown results are representative of the mean among the different clones. Optical Density (OD).
Correlation analysis of NEFL gene expression with clinical outcome
To examine the relevance of NEFL in patients, we investigated if gene expression was associated with clinical outcome. Analysis of three publicly available gene expression array data of neuroblastoma showed that high NEFL expression is associated with better survival (Fig. 4A–D, Pcombined=0.04; Fig. 4E, Pmeta-analysis=0.03; HR=0.68, 95% CI: 0.48–0.97), well-differentiated tumors (Fig. 4F, P=0.002) and favorable stages (Fig. 4G, stage 1 vs. stage 4; P=2.3×10−4). No significant association was found between NEFL expression and the prognostic variables age at diagnosis, MYCN status and risk group (data not shown).
Figure 4. NEFL expression is associated with good outcome and favorable neuroblastoma stages.
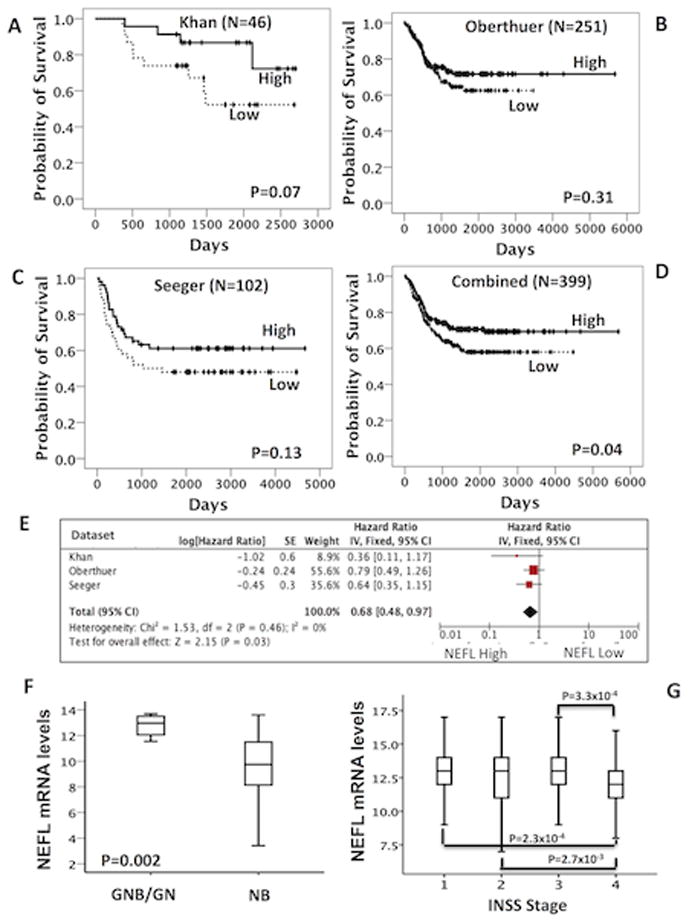
(A–C) Kaplan Meier analysis using published array data (Oncogenomics lab web tool). (D) Kaplan Meier analysis using combined array data from the three independent sets. (E) Meta-analysis of Hazard ratios from the Cox regression analysis of each dataset. (F) Changes in expression for NEFL in ganglioneuroblastoma (GNB) and ganglioneuroma (GN) and (G) in favorable stage using published array data (GEO database).
Discussion
Further investigation of 8 genes that we previously identified as being essential mediators of neuroblastoma differentiation has identified NEFL as a susceptibility gene. Particularly, the minor allele of SNP rs1059111 correlates with increased levels of NEFL and confers protection against neuroblastoma development, and is associated with a more benign phenotype in those that do develop. In silico analyses suggest that the SNP rs1059111 may alter the binding site of microRNAs and transcription factors. However, in vitro validation experiments of these data need to be performed in future studies. Forced over-expression of NEFL enhanced cellular differentiation in cells with the protective allele and basal NEFL expression, but had little effect on cells with the risk genotype and undetectable basal NEFL expression. Silencing of NEFL in cells with the protective allele did not affect cell proliferation but induced cell invasion. Here we present preliminary data providing evidence that NEFL expression sustains differentiated phenotype, impairs soft agar growth and proliferation in cells with the protective allele whereas impairs invasiveness and proliferation in cells homozygous for the risk major allele. Accordingly, high NEFL mRNA expression levels correlate with well-differentiated tumors and show association with a better clinical outcome. This suggests a dual function for NEFL: suppressing cancer initiation and also cancer progression. Taken together, these data provide direct evidence that a functional DNA variant in the 3′UTR region of NEFL influences neuroblastoma susceptibility and that low expression of NEFL plays a role in malignant neuroblastic transformation and disease progression, likely by disrupting a normal neuronal differentiation program.
The NEFL gene encodes a type IV intermediate filament heteropolymer that functionally maintains the neuronal caliber and plays an important role in the intracellular transport of neurotransmitters to axons and dendrites (27). Mutations in NEFL cause one of the most severe Charcot-Marie-Tooth disease phenotypes (28). Recently, several studies have proposed NEFL as a tumor suppressor gene. Indeed, deletions of chromosomal region 8p21, where NEFL is located, have been found in different cancer diseases such as prostate cancer (29–32), breast cancer (33–35), colorectal cancer (36, 37), hepatocellular carcinoma (38) and squamous cell carcinoma of the head and neck (HNSCC) (39, 40). Very recently, Chen and co-workers have demonstrated that in HNSCC, NEFL protein is physically associated with tuberous sclerosis 1 (TSC1), and NEFL down-regulation by methylation process leads to functional activation of mTOR pathway and consequentially confers cisplatin resistance (41). The same research group also showed that NEFL expression induces cancer cell apoptosis and inhibits invasion in HNSCC cell lines (42). In agreement with our findings, literature data strongly suggest that NEFL might play a critical role in suppressing cancer initiation and/or progression.
Neuroblastoma can be considered a malignancy due, at least in part, to a loss of normal differentiation pathways (12). Here, we suggest that down-regulation of NEFL expression through constitutional DNA variation can predispose to neuroblastoma development because low levels of NEFL can affect the normal differentiation of sympathetic neurons. As a consequence, the derivate immature cells may be susceptible to secondary mutations that could ultimately lead to neuroblastoma. In agreement with our hypothesis, two research groups have already reported that genetic events affecting the dosage of neuronal pro-differentiation genes might lead to the accumulation of a cell population that is unable to differentiate and can thus acquire the necessary transforming genetic or epigenetic aberrations (43, 44). Wu and colleagues have demonstrated that genes involved in brain development and neuronal differentiation, such as BMP4, POU4F3, GDNF, OTX2, NEFM, CNTN4, OTP, SIM1, FYN, EN1, CHAT, GSX2, NKX6-1, PAX6, RAX, and DLX2 are strongly enriched among genes frequently methylated in astrocytomas (44). Pei and colleagues showed that a reduced dosage of PHOX2B during development, through either a heterozygous deletion or dominant-negative mutation, imposes a block in the differentiation of sympathetic neuronal precursors, resulting in a cell population that is likely to be susceptible to secondary transforming events (43). Even if our findings suggest that low NEFL expression can play a role in tumor initiation by affecting neuronal differentiation, we do not exclude that NEFL can be involved in different molecular mechanisms whose alteration can contribute to tumor initiation and progression in neuroblastoma. Indeed, NEFL has been found to interact with proteins such as TRIM2 (45) and TSC1 (41) with high relevance in cancer and that participate in various important cellular processes.
Interestingly, another gene mutated in Charcot-Marie-Tooth disease, KIF1B (28), seems to act as a haploinsufficient tumor suppressor, and its down-regulation might potentially contribute to tumorigenesis of cancers, including neuroblastoma (46, 47). These data strengthen the hypothesis that alteration in normal expression of genes involved in neuronal development can induce neuroblastoma carcinogenesis.
In summary, we have identified NEFL as a novel susceptibility gene for neuroblastoma starting from the analysis of a set of genes involved in neuronal differentiation and using data from a neuroblastoma GWAS. Our results support the hypothesis that down-regulation of NEFL gene expression through functional hereditable DNA variation can contribute to malignant transformation of sympathetic progenitor cells. Moreover, we provide evidence that NEFL expression enhances differentiation and impairs cancer progression in neuroblastoma cell lines. This study has demonstrated that a hypothesis-driven GWAS follow-up study is a useful strategy for identifying novel disease susceptibility genes and that genetic and functional datasets can be merged to maximize discovery efforts. GWASs so far have led to substantial advances in our understanding of the role of common variation in complex disease and have identified previously unknown pathogenic pathways in neuroblastoma (4–8, 48) and other disorders (49). However, many disease-associated variants remain unknown or functionally uncharacterized and biological implications of risk-associated variants in pathogenesis are largely unknown (49). The methodological approach here presented might be a useful tool to overcome some of the limits emerged from GWAS studies.
Supplementary Material
Acknowledgments
Funding: This study was supported by grants from Associazione Italiana per la Ricerca sul Cancro 10537 (MC), MIUR- FIRB Ricerca in Futuro RBFR08DWQ3 (MC), Fondazione Italiana per la Lotta al Neuroblastoma (MC) and Associazione Oncologia Pediatrica e Neuroblastoma (MC). This work was also supported in part by NIH Grants R01-CA124709 (JMM), 5P30 CA016520 (JMM), K99-CA151869 (SJD), P30-HD026979 (MD), the Giulio D’Angio Endowed Chair (JMM), the Alex’s Lemonade Stand Foundation (JMM), Andrew’s Army Foundation (JMM), the PressOn Foundation (JMM), Ali’s Discovery Fund (JMM) and the Abramson Family Cancer Research Institute (JMM).
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capasso M, Diskin SJ. Genetics and genomics of neuroblastoma. Cancer Treat Res. 2010;155:65–84. doi: 10.1007/978-1-4419-6033-7_4. [DOI] [PubMed] [Google Scholar]
- 4.Maris JM, Mosse YP, Bradfield JP, Hou C, Monni S, Scott RH, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen le B, Diskin SJ, Capasso M, Wang K, Diamond MA, Glessner J, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–30. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capasso M, Diskin SJ, Totaro F, Longo L, De Mariano M, Russo R, et al. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis. 2013;34:605–11. doi: 10.1093/carcin/bgs380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimmino F, Spano D, Capasso M, Zambrano N, Russo R, Zollo M, et al. Comparative proteomic expression profile in all-trans retinoic acid differentiated neuroblastoma cell line. J Proteome Res. 2007;6:2550–64. doi: 10.1021/pr060701g. [DOI] [PubMed] [Google Scholar]
- 12.Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao L, Ding J, Zha Y, Yang L, McCarthy BA, King W, et al. HOXC9 links cell-cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. Cancer Res. 2011;71:4314–24. doi: 10.1158/0008-5472.CAN-11-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 17.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 19.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Capasso M, Avvisati RA, Piscopo C, Laforgia N, Raimondi F, de Angelis F, et al. Cytokine gene polymorphisms in Italian preterm infants: association between interleukin-10 -1082 G/A polymorphism and respiratory distress syndrome. Pediatr Res. 2007;61:313–7. doi: 10.1203/pdr.0b013e318030d108. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–5. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negrini S, Prada I, D’Alessandro R, Meldolesi J. REST: an oncogene or a tumor suppressor? Trends Cell Biol. 2013;23:289–95. doi: 10.1016/j.tcb.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Singh A, Rokes C, Gireud M, Fletcher S, Baumgartner J, Fuller G, et al. Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(beta-TRCP) in neuroblastoma cells. Cancer. 2011;117:5189–202. doi: 10.1002/cncr.26145. [DOI] [PubMed] [Google Scholar]
- 27.Leermakers FA, Zhulina EB. How the projection domains of NF-L and alpha-internexin determine the conformations of NF-M and NF-H in neurofilaments. Eur Biophys J. 2010;39:1323–34. doi: 10.1007/s00249-010-0585-z. [DOI] [PubMed] [Google Scholar]
- 28.Gentil BJ, Cooper L. Molecular basis of axonal dysfunction and traffic impairments in CMT. Brain Res Bull. 2012;88:444–53. doi: 10.1016/j.brainresbull.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Bova GS, Carter BS, Bussemakers MJ, Emi M, Fujiwara Y, Kyprianou N, et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res. 1993;53:3869–73. [PubMed] [Google Scholar]
- 30.MacGrogan D, Levy A, Bostwick D, Wagner M, Wells D, Bookstein R. Loss of chromosome arm 8p loci in prostate cancer: mapping by quantitative allelic imbalance. Genes Chromosomes Cancer. 1994;10:151–9. doi: 10.1002/gcc.2870100302. [DOI] [PubMed] [Google Scholar]
- 31.Kagan J, Stein J, Babaian RJ, Joe YS, Pisters LL, Glassman AB, et al. Homozygous deletions at 8p22 and 8p21 in prostate cancer implicate these regions as the sites for candidate tumor suppressor genes. Oncogene. 1995;11:2121–6. [PubMed] [Google Scholar]
- 32.Trapman J, Sleddens HF, van der Weiden MM, Dinjens WN, Konig JJ, Schroder FH, et al. Loss of heterozygosity of chromosome 8 microsatellite loci implicates a candidate tumor suppressor gene between the loci D8S87 and D8S133 in human prostate cancer. Cancer Res. 1994;54:6061–4. [PubMed] [Google Scholar]
- 33.Seitz S, Werner S, Fischer J, Nothnagel A, Schlag PM, Scherneck S. Refined deletion mapping in sporadic breast cancer at chromosomal region 8p12-p21 and association with clinicopathological parameters. Eur J Cancer. 2000;36:1507–13. doi: 10.1016/s0959-8049(00)00135-0. [DOI] [PubMed] [Google Scholar]
- 34.Yaremko ML, Kutza C, Lyzak J, Mick R, Recant WM, Westbrook CA. Loss of heterozygosity from the short arm of chromosome 8 is associated with invasive behavior in breast cancer. Genes Chromosomes Cancer. 1996;16:189–95. doi: 10.1002/(SICI)1098-2264(199607)16:3<189::AID-GCC6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 35.Yaremko ML, Recant WM, Westbrook CA. Loss of heterozygosity from the short arm of chromosome 8 is an early event in breast cancers. Genes Chromosomes Cancer. 1995;13:186–91. doi: 10.1002/gcc.2870130308. [DOI] [PubMed] [Google Scholar]
- 36.Yaremko ML, Wasylyshyn ML, Paulus KL, Michelassi F, Westbrook CA. Deletion mapping reveals two regions of chromosome 8 allele loss in colorectal carcinomas. Genes Chromosomes Cancer. 1994;10:1–6. doi: 10.1002/gcc.2870100102. [DOI] [PubMed] [Google Scholar]
- 37.Lerebours F, Olschwang S, Thuille B, Schmitz A, Fouchet P, Laurent-Puig P, et al. Deletion mapping of the tumor suppressor locus involved in colorectal cancer on chromosome band 8p21. Genes Chromosomes Cancer. 1999;25:147–53. doi: 10.1002/(sici)1098-2264(199906)25:2<147::aid-gcc10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 38.Becker SA, Zhou YZ, Slagle BL. Frequent loss of chromosome 8p in hepatitis B virus-positive hepatocellular carcinomas from China. Cancer Res. 1996;56:5092–7. [PubMed] [Google Scholar]
- 39.Coon SW, Savera AT, Zarbo RJ, Benninger MS, Chase GA, Rybicki BA, et al. Prognostic implications of loss of heterozygosity at 8p21 and 9p21 in head and neck squamous cell carcinoma. Int J Cancer. 2004;111:206–12. doi: 10.1002/ijc.20254. [DOI] [PubMed] [Google Scholar]
- 40.el-Naggar AK, Hurr K, Batsakis JG, Luna MA, Goepfert H, Huff V. Sequential loss of heterozygosity at microsatellite motifs in preinvasive and invasive head and neck squamous carcinoma. Cancer Res. 1995;55:2656–9. [PubMed] [Google Scholar]
- 41.Chen B, Chen J, House MG, Cullen KJ, Nephew KP, Guo Z. Role of neurofilament light polypeptide in head and neck cancer chemoresistance. Mol Cancer Res. 2012;10:305–15. doi: 10.1158/1541-7786.MCR-11-0300. [DOI] [PubMed] [Google Scholar]
- 42.Huang Z, Zhuo Y, Shen Z, Wang Y, Wang L, Li H, et al. The role of NEFL in cell growth and invasion in head and neck squamous cell carcinoma cell lines. J Oral Pathol Med. 2013 doi: 10.1111/jop.12109. [DOI] [PubMed] [Google Scholar]
- 43.Pei D, Luther W, Wang W, Paw BH, Stewart RA, George RE. Distinct neuroblastoma-associated alterations of PHOX2B impair sympathetic neuronal differentiation in zebrafish models. PLoS Genet. 2013;9:e1003533. doi: 10.1371/journal.pgen.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X, Rauch TA, Zhong X, Bennett WP, Latif F, Krex D, et al. CpG island hypermethylation in human astrocytomas. Cancer Res. 2010;70:2718–27. doi: 10.1158/0008-5472.CAN-09-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balastik M, Ferraguti F, Pires-da Silva A, Lee TH, Alvarez-Bolado G, Lu KP, et al. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:12016–21. doi: 10.1073/pnas.0802261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munirajan AK, Ando K, Mukai A, Takahashi M, Suenaga Y, Ohira M, et al. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem. 2008;283:24426–34. doi: 10.1074/jbc.M802316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlisio S, Kenchappa RS, Vredeveld LC, George RE, Stewart R, Greulich H, et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884–93. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosse KR, Diskin SJ, Cole KA, Wood AC, Schnepp RW, Norris G, et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72:2068–78. doi: 10.1158/0008-5472.CAN-11-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–58. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


