Abstract
Natural products have been and continue to be the source and inspiration for a substantial fraction of human therapeutics. Although the pharmaceutical industry has largely turned its back on natural product discovery efforts, such efforts continue to flourish in academia with promising results. Natural products have traditionally been identified from a top-down perspective, but more recently genomics- and bioinformatics-guided bottom-up approaches have provided powerful alternative strategies. Here we review recent advances in natural product discovery from both angles, including diverse sampling and innovative culturing and screening approaches, as well as genomics-driven discovery and genetic manipulation techniques for both native and heterologous expression.
Introduction
It is no secret that historically, the human pharmaceutical armamentarium is significantly indebted to Nature, and in particular to the natural products of bacteria, fungi, and plants. To date, natural products and compounds derived there from and inspired thereby command a substantial market share, comprising 61% of anticancer compounds and 49% of anti-infectives approved in the past 30 years [1]. However, since the 1990s major pharmaceutical companies have largely turned away from natural product discovery efforts, citing difficulties in supply, screening, and characterization of natural products relative to completely synthetic libraries, and in particular the increasing rediscovery rates of known compounds [2]. Does this indicate that the diversity of extant natural products has been sufficiently sampled? With the advent of fast and inexpensive next generation genome sequencing technologies, the answer to the question has been shown to be a resounding “no.” Across all domains of life, the presence of multiple natural product gene clusters for which the cognate product has never been observed has become not the exception but the expectation in sequenced genomes. As a result, natural product discovery efforts continue to flourish at the academic level.
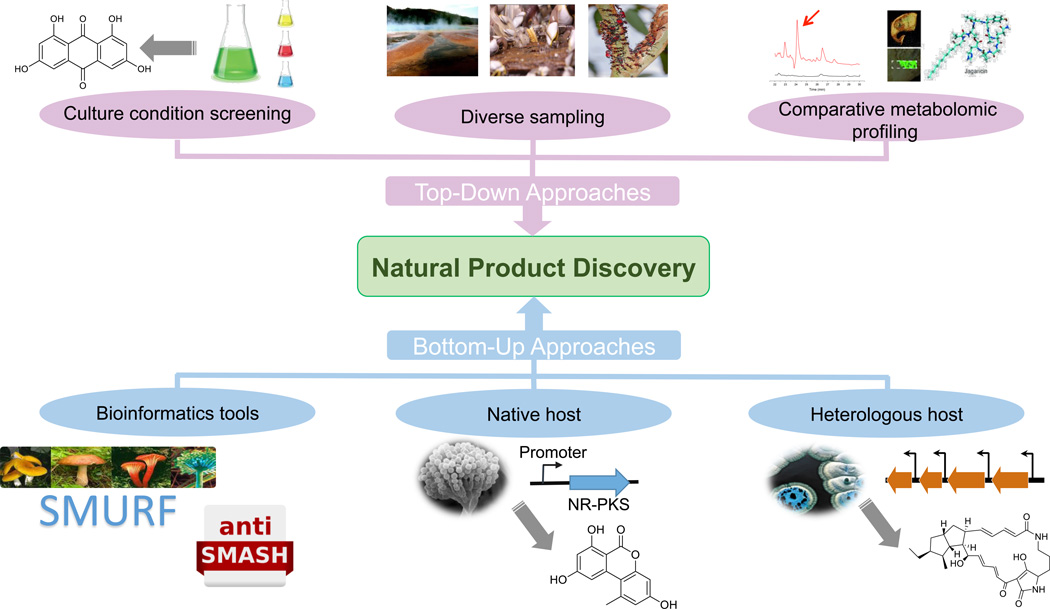
Here we broadly classify natural product discovery efforts into two categories, namely top-down and bottom-up approaches (Figure 1). Top-down approaches are defined as those that begin at the organism level, utilizing systems-level perturbations to elicit production of new natural products without prior knowledge of the genes and enzymes involved in their biosynthesis. Bottom-up approaches, in contrast, are those that first identify a gene cluster of interest and then utilize various gene manipulation techniques to drive transcription, translation, and eventual synthesis of the corresponding natural product. Though contrasting in style, both sets of techniques have enabled the discovery of many new and interesting natural products. As a result, new examples of their application continue to appear, and new techniques continue to be developed. In this review we highlight some of the most recent advances in natural product discovery from the past two years.
Figure 1.
Overview of the recent strategies applied in natural product discovery.
Top-Down Approaches
Top-down approaches have traditionally been the primary means of natural product discovery, as they do not require genome sequencing or sophisticated genetic manipulation. Such efforts begin with the collection of biological samples from diverse environments for either direct extraction or laboratory cultivation. Extracts are then screened for a desired bioactivity, with “hits” isolated for structural characterization. These methods are best suited to “low-hanging fruits,” i.e. compounds that are synthesized reproducibly and in relative abundance either in the native environment or under laboratory conditions. Nevertheless, continued innovation in sampling and screening methodologies has mitigated the risks of rediscovery of low-hanging fruit(s), allowing the top-down approach to remain a viable means of natural product discovery.
Diverse sampling and culturing
Natural products provide organisms with leverage against environmental stresses to survive and prosper. Often, these stresses come from interspecies interactions across all three domains of life, and so sampling particularly diverse environments and intimate interactions can lead to the discovery of new and interesting compounds. For example, isolation of two fungi from a microbial mat in an iron-rich spring led to the characterization of six novel natural products: clearanols A-E and disulochrin [3]. In particular, endosymbiotic bacteria represent a rich source of natural products [4], as many higher organisms rely on them to supplement their own defense systems. In a family of bivalve mollusks known as shipworms, boronated natural products with antibiotic activity were recently discovered from the cultivated symbiotic bacterium Teredinibacter turnerae [5]. Similarly, diverse pyrone polyketides have been discovered from Nocardiopsis alba CR167, an actinomycete endosymbiont of the cone snails Conus rolani and Conus tribblei [5]. Of course, marine organisms are not the only source of valuable endosymbionts, as several agriculturally relevant insects have also proven to be sources of antibiotic natural products [6].
To mimic in a laboratory setting the diverse environmental stresses an organism may experience, screening of different growth conditions is often employed. A recent example of this approach is the utilization of chemostat fermentation of Aspergillus nidulans to manipulate specific growth rate as well as carbon, nitrogen, and phosphorous levels [7]. Transcriptomic and metabolomic analyses revealed production of many polyketides under different conditions, including one novel compound. An alternate technique to introduce external stresses in laboratory culture is to challenge a target organism with other co-cultured species. This method has been demonstrated in many biotechnological applications, and has been recently reviewed elsewhere [8,9]. As a recent example, a novel prenylated polyketide was discovered from the fungal pathogen Aspergillus fumigatus only when cultured with the actinomycete bacterium Streptomyces rapamycinicus [10].
More unorthodox culture techniques have also been employed. For example, the Cichewicz group found that medium containing Cheerios breakfast cereal promoted growth and secondary metabolism of several isolated fungal strains, including the production of a novel diarylcyclopentendione metabolite and a novel biphenyl metabolite from Preussia typharum (Sacc.) Cain [11]. They also demonstrated an in vivo culture method to elicit the production of typically silent glidomycin/luminmycin metabolites from Photorhabdus asymbiotica by injecting the bacterium into live crickets and extracting the carcasses [12]. It should be noted, however, that the four compounds observed, including two new derivatives, were first identified in defined liquid medium culture.
Robust and high-throughput screening
Through decades of top-down screening for natural products, many of the low-hanging fruits have been discovered and characterized. However, a limiting factor in previous efforts has been the sensitivity and throughput of the screening techniques employed, such as traditional bioactivity assays. As a result, potentially interesting compounds can go undetected if they are present in low abundance or masked by the activity of more abundant known compounds. With continued advances in the large-scale acquisition and analysis of metabolomic data, it is possible to thoroughly process complex metabolite samples for the discovery of novel small molecules. For example, a statistical approach called principal component analysis was used to compare liquid chromatography/mass spectrometry (LC/MS) profiles of marine Streptomyces spp. to identify a novel peptide antibiotic, bottromycin D [13]. Alternatively, imaging MS was used to discover the lipopeptide jagaricin from Janthinobacterium agaricidamnosum, a bacterium responsible for soft-rot disease in fungi [14]. Here, slices of the button mushroom Agaricus bisporus were first infected with the bacterium and then scanned by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) to look for new metabolites within the damaged tissue.
Although MS-based metabolomic profiling is a very sensitive screening approach, it is limited in terms of the practical throughput that can be achieved. Thus, there is a continuing interest in developing “low-tech” high-throughput strategies with robust, quantifiable output. One such example is the use of indicators thymol blue and bromothymol blue to detect the pH drop concomitant with sugar fermentation in Vibrio cholerae cultures [15]. After screening of 39,000 crude extracts, 49 were found to block fermentation, and three were characterized as novel broad-spectrum antibiotics. An alternative colorimetric assay utilizes resazurin, a general indicator of metabolic activity, to detect antifungal activity against the human pathogen A. fumigatus [16]. High throughput screens can also be carried out against engineered assay strains, as exemplified in a screen for activators of the heat shock response in human cells [17]. Here, the reporter gene encoding enhanced green fluorescent protein (eGFP) was placed under the regulation of a promoter containing a heat-shock element, such that heat shock response would be accompanied by robust eGFP expression.
Bottom-Up Approaches
While traditional top-down approaches have been successful in identifying many bioactive natural products, the volume of genome sequence data now available has revised our view of the biosynthetic potential and metabolic capabilities of microorganisms, sparking a renaissance in natural product discovery. Genomics-based bottom-up approaches have been developed to unveil new natural products that were undetected under standard fermentation conditions [18]. These strategies leverage powerful functional genomics, bioinformatics, and genetic manipulation tools to identify and activate gene clusters of interest either in native or heterologous hosts.
Bioinformatics tools
Bioinformatics tools are indispensible in identification and characterization of potential natural product gene clusters from sequenced genomes and metagenomes. Recently, two powerful bioinformatics tools have been developed: Secondary Metabolite Unknown Regions Finder (SMURF) [19] and antibiotics & Secondary Metabolite Analysis Shell (antiSMASH) [20]. SMURF is the first comprehensive pipeline capable of predicting putative backbone genes in fungal genomes with high accuracy, and can identify gene clusters for nonribosomal peptides, polyketides, nonribosomal peptide-polyketide hybrids, indole alkaloids, and terpenes. AntiSMASH, on the other hand, expands the coverage beyond fungal genomes to the whole range of biosynthetic loci of known natural product classes from any input genome sequence. A highly improved antiSMASH2.0 that was recently released enables simultaneous processing of multiple contigs, analysis of protein sequences, and expanded structure prediction, making it the current state of the art for in silico natural product gene cluster analysis [21].
Native host expression
With genome sequence data in hand for tractable strains, it becomes possible to probe predicted natural product gene clusters in their native context, both to correlate in silico discovered clusters with known products and to discover previously undetected natural products. In the past decade, this has most often been achieved via gene knockout and subsequent comparative metabolomic profiling. Such techniques continue to be applied today, with recent examples including the identification of a phytotoxin cichorine gene cluster from A. nidulans [22], a strain not previously known to produce this compound; an endocrocin gene cluster from A. fumigatus [23]; and an echinocandin B gene cluster from Emericella rugulosa NRRL 11440 [24]. It is notable that this last example required development of a gene deletion method for the host of interest; in many cases, this can be impractical or impracticable. An alternative deletion-based strategy for activation of silent gene clusters in the native host is kinase knockout. Kinases play an important role in regulation and signal transduction. Screening of a genome-wide kinase knockout library of A. nidulans identified the production of aspernidine A from a mitogen-activated protein kinase gene, mpkA [25].
While gene knockout is useful in cases where gene clusters are expressed in the native host or pathway repressors can be identified, the fact remains that most natural product gene clusters remain silent under typical laboratory conditions. Thus, a “knock-in” promoter replacement strategy has recently been developed to activate these silent genes [26] (Figure 2). Through efficient gene targeting techniques, the promoters of nonreducing polyketide synthase (NR-PKS) genes, key genes in natural product gene clusters, and other genes necessary for NR-PKS product formation or release have been replaced in A. nidulans, which led to the discovery of seven novel compounds [27]. Similarly, the novel polyketide burkholderic acid was discovered from the pathogenic bacterium Burkholderia thailandensis via insertion of a constitutive promoter in front of the nonribosomal peptide synthetase (NRPS)-PKS gene burA [28]. However, when the native promoters of 13 NRPS-like genes in A. nidulans were replaced with the inducible alcohol dehydrogenase (alcA) promoter, the only discernable effect was enhanced production of a known compound, microperfuranone, with induction of the gene AN3396.4 [29]. Induction of the remaining 12 genes had no effect on the metabolite profile, highlighting the limitations of this approach.
Figure 2.
The “knock-in” strategy used in Aspergillus nidulans. A. Replacement of the promoters in front of the transcription activators identified three compounds from Aspergillus nidulans. B. Replacement of the promoters in front of the non-reducing PKSs identified eight compounds from Aspergillus nidulans.
Heterologous host expression
For the majority of organisms, genetic manipulation is either difficult or yet-to-be established. Therefore, heterologous expression of a single gene, a cassette of genes, or an entire biosynthetic gene cluster in a genetically tractable host is a practical alternative route for identifying the corresponding natural product. As an example, a terpene synthase gene from Streptomyces avermitilis was expressed in Escherichia coli, resulting in the synthesis of the novel tricyclic sesquiterpene, avermitilol [30]. Meanwhile, Saccharomyces cerevisiae has often been used as a heterologous host expressing genes from fungi. For example, a method was developed in Saccharomyces cerevisiae for cloning and heterologous expression of PKS and NRPS genes from various fungi, resulting in the identification of a novel product from an A. fumigatus NRPS [31]. Another popular heterologous host for expression of genes from fungi is A. nidulans. Recently an efficient approach for amplification, cloning, and expression of heterologous pathways in A. nidulans has been demonstrated, and was applied to express a silent asperfuranone pathway from Aspergillus terreus [32]. An analogous approach was used to express neosartoricin gene clusters from dermatophytic fungi in A. nidulans as well, resulting in discovery of the novel analog neosartoricin B [33]. Several other strategies for activating silent gene clusters in fungal genomes have previously been developed and were recently reviewed elsewhere [34,35].
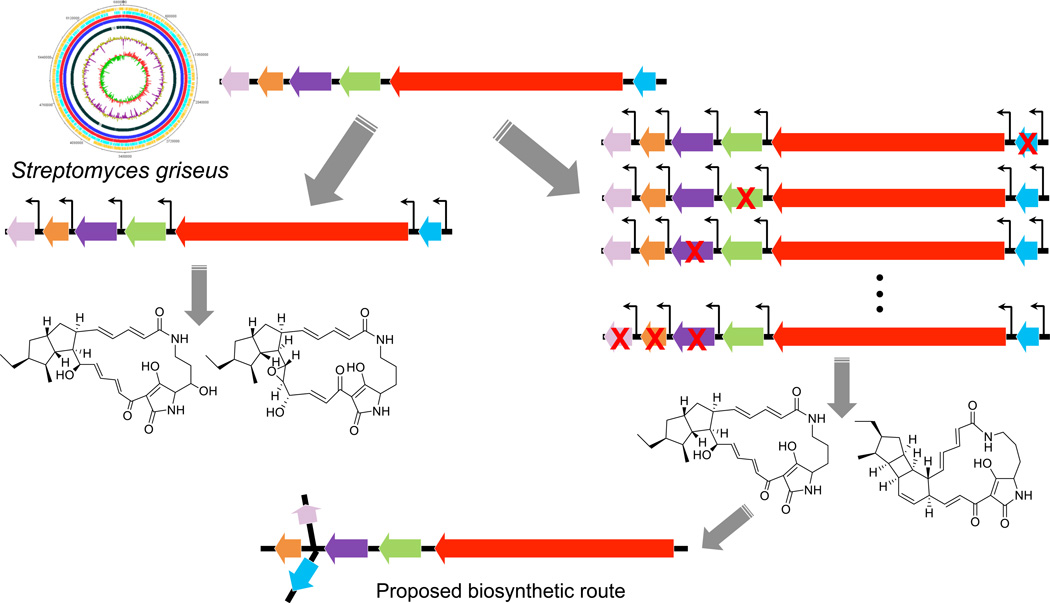
To facilitate the activation of cryptic pathways in a heterologous host, promoters were usually inserted into clusters to drive gene expressions. Recently, in the RecET direct cloning method, one strong promoter was added in front of the whole cluster to stimulate the production of natural products [36]. With the development of powerful DNA assembly methods such as DNA assembler [37], more comprehensive reconstruction of clusters has been achieved in a single-step manner, which results in the creation of novel compounds by combinatorial biosynthesis [38] and activation of a silent cluster for known compounds [39]. A similar strategy has been applied to discover novel polycyclic tetramate macrolactams (PTMs) [40]. The complete refactoring of a highly conserved PTM pathway by a plug-and-play scaffold revealed three novel PTMs and shed new light on the biosynthetic mechanisms (Figure 3).
Figure 3.
The plug-and-play scaffold developed for cryptic pathway activation and mechanistic study. The whole gene cluster was refactored by inserting constitutive promoters in front of each gene involved in the PTM biosynthesis pathway. Single-deletion and multiple-deletion constructs were assembled to elucidate the biosynthetic steps.
Heterologous hosts can further be engineered to facilitate natural product discovery by providing a clean secondary metabolite background, as previously demonstrated in the genome-minimized S. avermitilis strains. Recently, twenty diverse natural product gene clusters from a variety of organisms were successively cloned and introduced into genome-minimized strains SUKA17 or SUKA22, and production of the corresponding products was observed [41]. Notably, a few clusters required substitution of an alternative promoter or expression of a pathway-specific regulatory gene, and production levels did not always exceed those of the native hosts.
Apart from being used to activate cryptic gene clusters, heterologous expression is often employed for combinatorial biosynthesis to generate novel compounds as well. The macrolide antibiotic erythromycin A and its semisynthetic analogues are among the most useful antibacterial agents for the treatment of infectious diseases. Therefore, it is no surprise that the erythromycin biosynthetic pathway has been well-studied for combinatorial biosynthesis [42]. Recently, novel erythromycin analogs with different glycosylation patterns have been produced in E. coli by replacing the original deoxysugar pathways with the alternative D-mycaminose and D-olivose pathways. In addition to demonstrating the intrinsic flexibility of the biosynthetic system to accommodate alternative tailoring pathways, these results also offer an initial attempt to leverage the E. coli platform for erythromycin analogue production [43]. Another example of combinatorial biosynthesis of novel erythromycin analogs utilized precursor-directed biosynthesis, generating a new class of alkynyl- and alkenyl-substituted macrolides with activities comparable to that of the parent compound [44].
Beyond erythromycin, combinatorial biosynthetic approaches have been applied to a variety of other natural product gene clusters, and have been recently reviewed elsewhere [45,46]. As an example, the cyanobactin ribosomal peptide natural product pathway was manipulated to incorporate multiple tandem mutations and non-proteinogenic amino acids when expressed in E. coli [47]. This enabled the production of 22 compounds, including other known products from different pathways as well as novel derivatives. In A. nidulans, heterologous expression of a NR-PKS gene from Aspergillus niger triggered the production of two polyketides, and replacement of the terminal reducing domain with a phylogenetically related thioesterase domain facilitated the production of two metabolites with carboxyl groups replacing the corresponding aldehyde groups [48]. These results indicate that rational domain swapping may provide a route to engineer the functionalization of valuable chemicals. Finally, a fluoroacetate pathway was exploited as a source of fluorinated building blocks for introduction of fluorine into natural-product scaffolds [49]. The Chang group showed that fluoroacetate can be used to generate novel fluorinated polyketide products both in vitro and in vivo, highlighting the prospects for the production of complex fluorinated natural products using synthetic biology.
Conclusions and future perspectives
With continued progress in both top-down and bottom-up approaches, natural product discovery has remained an active field of academic research. Recent trends of sampling symbiotic microbes should continue to provide novel natural products, as these microbes synthesize many interesting compounds to interact with their hosts. Further diversity in sampling is also being introduced through “crowdsourcing” efforts, enabling collection of samples from all around the world [50]. As powerful analytical techniques (MS, NMR, and software for comparative data analysis) continue to evolve, we will continue to pull back the curtain of "silent" natural products by enabling detection and characterization of compounds present in minute quantities. Further, although it has primarily been demonstrated as a tool to increase production of known natural products or connect known natural products to their gene clusters, epigenetic perturbation via small molecules or genetic modifications can also be used to unlock unknown compounds [51].
Meanwhile, as whole-genome sequencing has become more economical, it will likely continue to be routinely applied in place of traditional library-based techniques for identification of gene clusters of interest [52]. The exponentially accumulating data from these genome sequencing projects have already shown that the metabolic potential of many microorganisms is severely underestimated, revealing a “silent majority” of as-yet-undiscovered natural products. As a result, more genomics-inspired strategies need to be developed in order to fully exploit this untapped biosynthetic potential. Among the most challenging goals concerning natural products is the robust activation of silent gene clusters. Careful manipulation of culture conditions and genetic manipulation of gene clusters in the native host have been and will continue to be highly effective for select cases. However, the uncultivable, intractable majority of microorganisms remains a significant obstacle to this approach. With the development of powerful DNA assembly techniques [53], the future will likely see an increase not only in the heterologous expression of full gene clusters, but also the complete redesign and refactoring of these clusters in well-studied, highly tractable hosts tailor-made for heterologous expression.
As genomics-driven natural product discovery efforts have revealed not only new compounds but also their cognate biosynthetic pathways, they have provided new insights into natural product biosynthesis. Discovery of a single pathway split between multiple genomic loci [16], for example, and multiple pathways intertwined at a single locus [54] have challenged the traditional idea of a single, co-localized "gene cluster." Further, isolation of new and unusual compounds has revealed novel biochemistry [55] and complex ecological interactions [56]. These discoveries not only satisfy scientific curiosity, but also can feed-forward to inform future discovery efforts. Such efforts will likely include not only further exploration of typical natural product “workhorses,” such as actinomycete bacteria and Aspergillus fungi, but also a wider array of underexplored or neglected organisms [57]. One can envision that new natural products possessing unprecedented structural features and novel modes of action can be discovered, fueling the next generation of therapeutic agents. This vision has recently begun to be embraced by major pharmaceutical companies, as evidenced by the disclosure of the natural products kibdelomycin by Merck, pyrrolamide by AstraZeneca, and MDN-0057 by researchers from Cubist Pharmaceuticals and coworkers [58]. GlaxoSmithKline has also showed a renewed commitment to natural products by developing an in-house natural products library, now 13,000 compounds strong [59]. Thus, natural product discovery efforts are poised to thrive in the years to come.
Highlights.
Natural products are discovered through top-down and bottom-up approaches.
Diverse sampling provides diverse natural products.
Powerful analytical techniques enable robust product detection.
Sophisticated genetic manipulation unlocks silent gene clusters.
Heterologous expression bypasses native host regulation of silent genes.
Acknowledgements
We gratefully acknowledge financial support from the National Institutes of Health (GM077596) and National Academies Keck Futures Initiative on Synthetic Biology.
Abbreviations
- antiSMASH
antibiotic & Secondary Metabolite Analysis Shell
- LC
liquid chromatography
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- NMR
nuclear magnetic resonance
- NR-PKS
nonreducing polyketide synthase
- NRPS
nonribosomal peptide synthetase
- PKS
polyketide synthase
- SMURF
Secondary Metabolite Unknown Regions Finder
- eGFP
enhanced green fluorescent protein
- PTM
polycyclic tetramate macrolactam
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li JW, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 3.Gerea AL, Branscum KM, King JB, You J, Powell DR, Miller AN, Spear JR, Cichewicz RH. Secondary metabolites produced by fungi derived from a microbial mat encountered in an iron-rich natural spring. Tetrahedron Lett. 2012;53:4202–4205. doi: 10.1016/j.tetlet.2012.05.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt EW, Donia MS, McIntosh JA, Fricke WF, Ravel J. Origin and variation of tunicate secondary metabolites. J Nat Prod. 2012;75:295–304. doi: 10.1021/np200665k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elshahawi SI, Trindade-Silva AE, Hanora A, Han AW, Flores MS, Vizzoni V, Schrago CG, Soares CA, Concepcion GP, Distel DL, et al. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc Natl Acad Sci U S A. 2013;110:E295–E304. doi: 10.1073/pnas.1213892110. This paper describes the identification of polyketide products and their cognate gene cluster from a cultivable endosymbiotic bacterium, with implications for their ecological role.
- 6.Ramadhar TR, Beemelmanns C, Currie CR, Clardy J. Bacterial symbionts in agricultural systems provide a strategic source for antibiotic discovery. J Antibiot (Tokyo) 2013 doi: 10.1038/ja.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar A, Funk AN, Scherlach K, Horn F, Schroeckh V, Chankhamjon P, Westermann M, Roth M, Brakhage AA, Hertweck C, et al. Differential expression of silent polyketide biosynthesis gene clusters in chemostat cultures of Aspergillus nidulans. J Biotechnol. 2012;160:64–71. doi: 10.1016/j.jbiotec.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Seyedsayamdost MR, Traxler MF, Clardy J, Kolter R. Old meets new: using interspecies interactions to detect secondary metabolite production in actinomycetes. Methods Enzymol. 2012;517:89–109. doi: 10.1016/B978-0-12-404634-4.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bader J, Mast-Gerlach E, Popovic MK, Bajpai R, Stahl U. Relevance of microbial coculture fermentations in biotechnology. J Appl Microbiol. 2010;109:371–387. doi: 10.1111/j.1365-2672.2009.04659.x. [DOI] [PubMed] [Google Scholar]
- 10.Konig CC, Scherlach K, Schroeckh V, Horn F, Nietzsche S, Brakhage AA, Hertweck C. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. Chembiochem. 2013;14:938–942. doi: 10.1002/cbic.201300070. [DOI] [PubMed] [Google Scholar]
- 11.Du L, King JB, Morrow BH, Shen JK, Miller AN, Cichewicz RH. Diarylcyclopentendione metabolite obtained from a Preussia typharum isolate procured using an unconventional cultivation approach. J Nat Prod. 2012;75:1819–1823. doi: 10.1021/np300473h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theodore CM, King JB, You J, Cichewicz RH. Production of cytotoxic glidobactins/luminmycins by Photorhabdus asymbiotica in liquid media and live crickets. J Nat Prod. 2012;75:2007–2011. doi: 10.1021/np300623x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou Y, Tianero MD, Kwan JC, Wyche TP, Michel CR, Ellis GA, Vazquez-Rivera E, Braun DR, Rose WE, Schmidt EW, et al. Structure and biosynthesis of the antibiotic bottromycin D. Org Lett. 2012;14:5050–5053. doi: 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graupner K, Scherlach K, Bretschneider T, Lackner G, Roth M, Gross H, Hertweck C. Imaging mass spectrometry and genome mining reveal highly antifungal virulence factor of mushroom soft rot pathogen. Angew Chem Int Ed Engl. 2012;51:13173–13177. doi: 10.1002/anie.201206658. Imaging mass spectrometry is applied to probe a host-pathogen interaction in situ, revealing production of a novel lipopeptide virulence factor.
- 15.Ymele-Leki P, Cao S, Sharp J, Lambert KG, McAdam AJ, Husson RN, Tamayo G, Clardy J, Watnick PI. A high-throughput screen identifies a new natural product with broad-spectrum antibacterial activity. PLoS One. 2012;7:e31307. doi: 10.1371/journal.pone.0031307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteiro MC, de la Cruz M, Cantizani J, Moreno C, Tormo JR, Mellado E, De Lucas JR, Asensio F, Valiante V, Brakhage AA, et al. A new approach to drug discovery: high-throughput screening of microbial natural extracts against Aspergillus fumigatus using resazurin. J Biomol Screen. 2012;17:542–549. doi: 10.1177/1087057111433459. [DOI] [PubMed] [Google Scholar]
- 17. Santagata S, Xu YM, Wijeratne EM, Kontnik R, Rooney C, Perley CC, Kwon H, Clardy J, Kesari S, Whitesell L, et al. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem Biol. 2012;7:340–349. doi: 10.1021/cb200353m. The authors present an innovative screening approach in which a cell line is engineered to enable in vivo screening for novel compounds that act via a known pathway.
- 18.Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. Here the authors present the most comprehensive, state-of-the-art tool for the in silico identification and analysis of natural product gene clusters.
- 22.Sanchez JF, Entwistle R, Corcoran D, Oakley BR, Wang CC. Identification and molecular genetic analysis of the cichorine gene cluster in Aspergillus nidulans. Medchemcomm. 2012;3 doi: 10.1039/C2MD20055D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim FY, Hou Y, Chen Y, Oh JH, Lee I, Bugni TS, Keller NP. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2012;78:4117–4125. doi: 10.1128/AEM.07710-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cacho RA, Jiang W, Chooi YH, Walsh CT, Tang Y. Identification and characterization of the echinocandin B biosynthetic gene cluster from Emericella rugulosa NRRL 11440. J Am Chem Soc. 2012;134:16781–16790. doi: 10.1021/ja307220z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaegashi J, Praseuth MB, Tyan SW, Sanchez JF, Entwistle R, Chiang YM, Oakley BR, Wang CC. Molecular genetic characterization of the biosynthesis cluster of a prenylated isoindolinone alkaloid aspernidine A in Aspergillus nidulans. Org Lett. 2013;15:2862–2865. doi: 10.1021/ol401187b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakley CE, Edgerton-Morgan H, Oakley BR. Tools for manipulation of secondary metabolism pathways: rapid promoter replacements and gene deletions in Aspergillus nidulans. Methods Mol Biol. 2012;944:143–161. doi: 10.1007/978-1-62703-122-6_10. [DOI] [PubMed] [Google Scholar]
- 27.Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, Lo HC, Yeh HH, Oakley BR, Wang CC. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Franke J, Ishida K, Hertweck C. Genomics-driven discovery of burkholderic acid, a noncanonical, cryptic polyketide from human pathogenic Burkholderia species. Angew Chem Int Ed Engl. 2012;51:11611–11615. doi: 10.1002/anie.201205566. Promoter replacement is applied in the native host to unlock an unusual, cryptic NRPS-PKS product, and specialized purification protocol is developed to overcome its inherent instability for structural characterization.
- 29.Yeh HH, Chiang YM, Entwistle R, Ahuja M, Lee KH, Bruno KS, Wu TK, Oakley BR, Wang CC. Molecular genetic analysis reveals that a nonribosomal peptide synthetase-like (NRPS-like) gene in Aspergillus nidulans is responsible for microperfuranone biosynthesis. Appl Microbiol Biotechnol. 2012;96:739–748. doi: 10.1007/s00253-012-4098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou WK, Fanizza I, Uchiyama T, Komatsu M, Ikeda H, Cane DE. Genome mining in Streptomyces avermitilis: cloning and characterization of SAV_76, the synthase for a new sesquiterpene, avermitilol. J Am Chem Soc. 2010;132:8850–8851. doi: 10.1021/ja103087w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishiuchi K, Nakazawa T, Ookuma T, Sugimoto S, Sato M, Tsunematsu Y, Ishikawa N, Noguchi H, Hotta K, Moriya H, et al. Establishing a new methodology for genome mining and biosynthesis of polyketides and peptides through yeast molecular genetics. Chembiochem. 2012;13:846–854. doi: 10.1002/cbic.201100798. [DOI] [PubMed] [Google Scholar]
- 32.Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, Sung CT, Wang CC, Oakley BR. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yin WB, Chooi YH, Smith AR, Cacho RA, Hu YC, White TC, Tang Y. Discovery of cryptic polyketide metabolites from Dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synthetic Biology. 2013;2:629–634. doi: 10.1021/sb400048b. In the two papers above, a new technique is presented to integrate heterologous gene clusters in A. nidulans and easily select the desired transformants.
- 34.Wiemann P, Keller NP. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol. 2014;41:301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 35.Scharf DH, Brakhage AA. Engineering fungal secondary metabolism: a roadmap to novel compounds. J Biotechnol. 2013;163:179–183. doi: 10.1016/j.jbiotec.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Cobb RE, Zhao H. Direct cloning of large genomic sequences. Nature Biotechnology. 2012;30:405–406. doi: 10.1038/nbt.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao Z, Zhao H. DNA assembler: a synthetic biology tool for characterizing and engineering natural product gene clusters. Methods Enzymol. 2012;517:203–224. doi: 10.1016/B978-0-12-404634-4.00010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao Z, Luo Y, Zhao H. Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol Biosyst. 2011;7:1056–1059. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao Z, Rao G, Li C, Abil Z, Luo Y, Zhao H. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol. 2013;2:662–669. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. The authors demonstrate how modern DNA assembly techniques can be used to construct and reconfigure a full natural product gene cluster for expression in a heterologous host.
- 41.Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-ya K, et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol. 2013;2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong FT, Khosla C. Combinatorial biosynthesis of polyketides--a perspective. Curr Opin Chem Biol. 2012;16:117–123. doi: 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang M, Zhang H, Park SH, Li Y, Pfeifer BA. Deoxysugar pathway interchange for erythromycin analogues heterologously produced through Escherichia coli. Metab Eng. 2013;20C:92–100. doi: 10.1016/j.ymben.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Harvey CJ, Puglisi JD, Pande VS, Cane DE, Khosla C. Precursor directed biosynthesis of an orthogonally functional erythromycin analogue: selectivity in the ribosome macrolide binding pocket. J Am Chem Soc. 2012;134:12259–12265. doi: 10.1021/ja304682q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winter JM, Tang Y. Synthetic biological approaches to natural product biosynthesis. Curr Opin Biotechnol. 2012;23:736–743. doi: 10.1016/j.copbio.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu MC, Law B, Wilkinson B, Micklefield J. Bioengineering natural product biosynthetic pathways for therapeutic applications. Curr Opin Biotechnol. 2012;23:931–940. doi: 10.1016/j.copbio.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal route to small-molecule diversity. J Am Chem Soc. 2012;134:418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh HH, Chang SL, Chiang YM, Bruno KS, Oakley BR, Wu TK, Wang CC. Engineering fungal nonreducing polyketide synthase by heterologous expression and domain swapping. Org Lett. 2013;15:756–759. doi: 10.1021/ol303328t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walker MC, Thuronyi BW, Charkoudian LK, Lowry B, Khosla C, Chang MC. Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science. 2013;341:1089–1094. doi: 10.1126/science.1242345. Here the authors demonstrate that fluorinated polyketides can be synthesized using a fluoroacetate pathway which could be applicable to many diverse polyketide scaffolds.
- 50.Du L, Robles AJ, King JB, Powell DR, Miller AN, Mooberry SL, Cichewicz RH. Crowdsourcing natural products discovery to access uncharted dimensions of fungal metabolite diversity. Angew Chem Int Ed Engl. 2014;53:804–809. doi: 10.1002/anie.201306549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cichewicz R. Epigenetic Regulation of Secondary Metabolite Biosynthetic Genes in Fungi. In: Witzany G, editor. Biocommunication of Fungi. Netherlands: Springer; 2012. pp. 57–69. [Google Scholar]
- 52.Gerken T, Walsh CT. Cloning and sequencing of the chaetocin biosynthetic gene cluster. Chembiochem. 2013;14:2256–2258. doi: 10.1002/cbic.201300513. [DOI] [PubMed] [Google Scholar]
- 53.Cobb RE, Ning JC, Zhao H. DNA assembly techniques for next-generation combinatorial biosynthesis of natural products. J Ind Microbiol Biotechnol. 2014;41:469–477. doi: 10.1007/s10295-013-1358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiemann P, Guo CJ, Palmer JM, Sekonyela R, Wang CC, Keller NP. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci U S A. 2013;110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peck SC, van der Donk WA. Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology. Curr Opin Chem Biol. 2013;17:580–588. doi: 10.1016/j.cbpa.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. Metabolic potential of endophytic bacteria. Curr Opin Biotechnol. 2014;27:30–37. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pidot SJ, Coyne S, Kloss F, Hertweck C. Antibiotics from neglected bacterial sources. Int J Med Microbiol. 2013 doi: 10.1016/j.ijmm.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 58.Brown DG, Lister T, May-Dracka TL. New natural products as new leads for antibacterial drug discovery. Bioorg Med Chem Lett. 2014;24:413–418. doi: 10.1016/j.bmcl.2013.12.059. [DOI] [PubMed] [Google Scholar]
- 59.Coma I, Bandyopadhyay D, Diez E, Ruiz EA, de Los Frailes MT, Colmenarejo G. Mining natural-products screening data for target-class chemical motifs. J Biomol Screen. 2014;19:749–757. doi: 10.1177/1087057114521463. [DOI] [PubMed] [Google Scholar]