Abstract
Agrobacterium tumefaciens has a unique ability to transfer genes into plant genomes. This ability has been utilized for plant genetic engineering. However, the efficiency is not sufficient for all plant species. Several studies have shown that ethylene decreased the Agrobacterium-mediated transformation frequency. Thus, A. tumefaciens with an ability to suppress ethylene evolution would increase the efficiency of Agrobacterium-mediated transformation. Some studies showed that plant growth-promoting rhizobacteria (PGPR) can reduce ethylene levels in plants through 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, which cleaves the ethylene precursor ACC into α-ketobutyrate and ammonia, resulting in reduced ethylene production. The whole genome sequence data showed that A. tumefaciens does not possess an ACC deaminase gene in its genome. Therefore, providing ACC deaminase activity to the bacteria would improve gene transfer. As expected, A. tumefaciens with ACC deaminase activity, designated as super-Agrobacterium, could suppress ethylene evolution and increase the gene transfer efficiency in several plant species. In this review, we summarize plant–Agrobacterium interactions and their applications for improving Agrobacterium-mediated genetic engineering techniques via super-Agrobacterium.
Keywords: Agrobacterium-mediated gene transfer, Agrobacterium tumefaciens, ACC deaminase, ethylene, Agrobacterium—plant interaction
INTRODUCTION
Agrobacterium tumefaciens is a soil-borne bacterium known to cause crown gall disease in plants. A. tumefaciens strains that induce crown gall have a large plasmid (tumor-inducing plasmid: Ti plasmid), which is essential for the establishment of crown gall disease (van Larebeke et al., 1974, 1975; Zaenen et al., 1974; Watson et al., 1975; Currier and Nester, 1976). Chilton et al. (1977) detected part of the Ti plasmid in the crown gall genome, showing that plant cell conversion resulted in genetic transformation. This work clearly showed that A. tumefaciens has the ability to transfer T-DNA into plant genomes (Chilton et al., 1977).
Since this discovery in 1977, many investigators have attempted to generate A. tumefaciens suitable for plant genetic engineering. In early techniques, the oncogenic T-DNA region of the Ti plasmid was replaced with genes of interest by single or double homologous recombination (Zambryski et al., 1983). Although the recombination steps are easy, they are limited by the potential recombination of repetitive sequences during or after recombination in the Ti plasmid replicon in A. tumefaciens. Hoekema et al. (1983) showed that A. tumefaciens is able to deliver the T-DNA even if the Ti plasmid is divided into two plasmids (the T-DNA region and the vir region). This finding made it possible to extract only the T-DNA region from A. tumefaciens, and the modification was easier without the sequence limitation. After this finding, a binary plant vector strategy became the standard method in plant genetic engineering (Hoekema et al., 1983; Bevan, 1984). Increasing the virulence of A. tumefaciens is a useful strategy for higher transformation efficiency. The identification and application of vir gene inducers, such as plant phenolic compounds (Stachel et al., 1985, 1986) and sugars (Cangelosi et al., 1990; He et al., 2009; Hu et al., 2013), enabled this technique to be adapted for a wide range of plant species, especially monocot crops. The selection of hyper-virulent strains also spread the adaptation throughout the host range. Super-binary vectors, which have shown higher vir gene expression, have provided critical improvements in transformation efficiency (Komari et al., 2006). Agrobacterium-mediated gene transfer has been well established through many modifications, and it is a basic technique in plant science, but the gene transfer efficiencies are not sufficient for many plant species, especially crops and biomass plants. Therefore, improvements in these techniques are required.
Some plant metabolites and phytohormones inhibit Agrobacterium-mediated transformation. In particular, ethylene showed a negative effect on Agrobacterium-mediated T-DNA transformation in many plant species. In this review, we focus on the effect of ethylene on Agrobacterium-mediated gene transfer into plant cells and introduce an engineering strategy by which to increase the transformation efficiency of A. tumefaciens via ethylene removal. The established A. tumefaciens strain is designated as super-Agrobacterium.
ETHYLENE SUPPRESSES Agrobacterium-MEDIATED T-DNA TRANSFER
The gaseous phytohormone ethylene regulates multiple physiological and developmental processes in plants, such as seedling emergence, leaf and flower senescence, ripening, organ abscission, growth-phase transitions, rhizobium–legume interaction, and plant–pathogen interaction (Beyer, 1981; Yang and Hoffman, 1984; Yang, 1985; Abeles, 1992; Ogawara et al., 2003). Ethylene synthesis is stimulated by biotic or abiotic stress. Ethylene also modulates A. tumefaciens–plant interactions. Ethylene is a crucial determinant of crown gall development. Plants treated with inhibitors of ethylene synthesis or perception, such as aminoethoxyvinylglycine (AVG), and ethylene-insensitive Never-ripe mutants (tomato) suppress crown gall growth (Aloni et al., 1998). Because vascularization is essential for efficient assimilate import from the host plant into the tumor cells, if the vascularization is suppressed, nutrient supply stops (Malsy et al., 1992; Pradel et al., 1996, 1999), resulting in the inhibition of crown gall development. Ethylene stimulates crown gall development by inducing vascular development (Wächter et al., 2003), and it induces crown gall development but inhibits Agrobacterium-mediated genetic transformation of plant cells. The enhancement of ethylene production by supplying its immediate precursor, 1-aminocyclopropane-1-carboxylic acid (ACC), suppresses gene transfer in tomatoes and melons (Davis et al., 1992; Ezura et al., 2000). Ethylene is increased by wounding and A. tumefaciens infection stress during co-cultivation (Ezura et al., 2000). Reducing ethylene production during co-cultivation using ethylene biosynthesis inhibitors such as AVG or suppressing plant ethylene perception by adding silver ions to the tissue culture medium has improved the transformation efficiency in melons (Ezura et al., 2000), cauliflowers (Chakrabarty et al., 2002), apricots (Burgos and Alburquerque, 2003), apple trees (Petri et al., 2005; Seong et al., 2005), and bottle gourds (Han et al., 2005). The stable transformation frequency was also increased in the Arabidopsis thaliana ethylene-insensitive mutants etr1-1 and etr1-2 (Nonaka et al., 2008a).
These results indicate that ethylene inhibits gene transfer in plants. One possible explanation for this phenomenon might involve the plant defense response via ethylene signaling. Previous studies have found that ethylene regulates several genes that are involved in the defense response, including those that encode the PR proteins chitinase, β-1, 3-glucanase, and PR1 (Deikman, 1997) in addition to phytoalexin synthetic enzymes (Ecker and Davis, 1987), defensins (Penninckx et al., 1996), and hydroxy-Pro-rich glycoproteins (Toppan et al., 1982). These compounds suppress bacterial growth because of their antibacterial activity. Indeed, in tomatoes, the reduced expression of LeETR4, which encodes a tomato ethylene receptor, resulted in increased sensitivity to ethylene, an enhanced hypersensitive response, and the reduced growth of Xanthomonas campestris pv. vesicatoria compared to wild-type (WT) plants (Ciardi et al., 2000). Ethylene insensitivity results in reduced resistance to the potato soft rot agent Erwinia carotovora subsp. carotovora in A. thaliana etr1-1 and ein2-1 mutants (Norman-Setterblad et al., 2000).
In general, ethylene levels increase in plant defense responses; however, the inhibitory effect of ethylene on Agrobacterium-mediated gene transfer does not occur through plant defense. The model scheme is described in Figure 1. Microarray and differential display analysis showed that A. tumefaciens infection induces the plant genes necessary for transformation while simultaneously repressing host defense response genes (Veena et al., 2003). Ethylene evolution was induced by A. tumefaciens inoculation in the early stage of the infection (Nonaka et al., 2008b; Lee et al., 2009). Although A. tumefaciens infection increased ethylene levels, the plant genes that encode ethylene receptors and their downstream signaling components, including defense response genes, are not induced (Lee et al., 2009). These results support the hypothesis that the inhibitory effect of ethylene on Agrobacterium-mediated gene transfer is independent of the plant defense response. Indeed, the plant ethylene response did not affect A. tumefaciens growth during co-cultivation (Nonaka et al., 2008a). The plant ethylene response inhibits T-DNA transfer through the suppression of vir gene expression (Nonaka et al., 2008a). The application of exudate from plants showing an ethylene response to A. tumefaciens reduced vir gene expression, which decreased T-DNA transfer. Such an inhibitory effect was partially overcome by the application of acetosyringone, a vir gene inducer, and in an A. tumefaciens strain constitutively expressing the vir gene. From this result, two possibilities were considered: in plants showing an ethylene response, the amount of the vir gene inducer would be reduced or the antagonist of the vir gene inducer would be produced. This deficient recovery indicates that vir gene suppression is one of the reasons for a reduction in T-DNA transfer via the ethylene response. The partial restoration upon the application of the vir gene inducer suggests that there is another inhibitory mechanism of ethylene. Therefore, the suppression of the vir gene is not sufficient to explain the negative effect of ethylene on Agrobacterium-mediated gene transfer. Because the inhibitory mechanism of ethylene on Agrobacterium-mediated gene transfer still needs to be clarified, to improve the transformation frequency, the introduction of the ability to reduce ethylene in A. tumefaciens would be effective.
FIGURE 1.
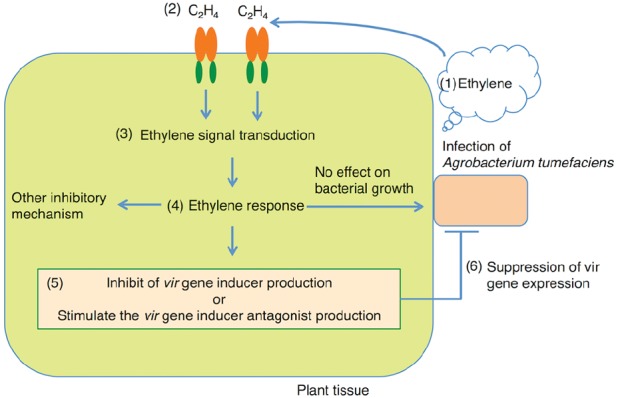
Schematic drawing of the potential mechanisms of inhibition through ethylene signaling and vir gene expression. (1) The infection of A. tumefaciens induces ethylene evolution from plant tissue. (2) Ethylene is received by the ethylene receptor. (3) Ethylene signal transduction is induced and, (4) subsequently, the plant shows the ethylene response. (5) In plants that show the ethylene response, vir gene inducer production would be suppressed or an antagonist of the inducer would be produced, (6) resulting in a reduction of vir gene expression in A. tumefaciens.
STRATEGY TO REDUCE ETHYLENE PRODUCTION IN PLANT CELLS INOCULATED WITH Agrobacterium
Ethylene is generated through the ethylene biosynthetic pathway, which was elucidated largely by the pioneering work of Yang and co-workers in the 1970s and 1980s (Wang et al., 2002). Ethylene is synthesized from the amino acid methionine, which is converted to S-adenosyl-L-methionine (SAM) by SAM synthase (ADS). SAM is the major methyl donor in plants and is involved in the methylation of lipids, proteins, and nucleic acids. SAM is converted by the enzyme ACC synthase (ACS) to 5′-methylthioadenosine (MTA), which is converted back to methionine via the Yang Cycle and to ACC, the precursor of ethylene. ACC is finally oxidized by ACC oxidase (ACO) to form ethylene, cyanide, and carbon dioxide. The conversion of SAM to ACC is considered to be the rate-limiting step in ethylene biosynthesis and consequently has been studied intensively. Therefore, the degradation of ACC, the immediate precursor of ethylene, effectively reduces ethylene production in plants. The reduction of ethylene via the degradation of ACC is found in some soil bacteria.
The pyridoxal 5-phosphate-dependent enzyme ACC deaminase catalyzes the decomposition of ACC to α-ketobutyrate and ammonia (Honma and Shimomura, 1978; Minami et al., 1998; Hontzeas et al., 2004; McDonnell et al., 2009). Although ACC deaminase genes have been isolated in a wide range of organisms, the ACC deaminase gene was isolated from many plant growth-promoting rhizobacteria (PGPR; Table 1). As shown in. PGPR with ACC deaminase activity can lower the ethylene level in plant cells through the degradation of ACC.
Table 1.
Organisms with ACC deaminase activity.
| Source organisms | Reference |
|---|---|
| Bacteria (Gram-negative) | |
| Enterobacter cloacae | Holguin and Glick (2001) |
| Achromobacter xylosoxidans | Belimov et al. (2001) |
| Rhizobium leguminosarum | Ma et al. (2003) |
| Pseudomonas putida | Hontzeas et al. (2004) |
| Burkholderia phytofirmans | Sessitsch et al. (2005) |
| Variovorax paradoxus | Madhaiyan et al. (2006) |
| Methylobacterium fujisawaense | Belimov et al. (2009) |
| Cronobacter sakazakii | Jha et al. (2012) |
| Mesorhizobium sp. | Gontia-Mishra et al. (2014) |
| Haererehalobacter sp. | Gontia-Mishra et al. (2014) |
| Halomonas sp. | Gontia-Mishra et al. (2014) |
| Bacteria (Gram-positive) | |
| Rhodococcus sp. | Belimov et al. (2001) |
| Brevibacterium iodinum | Dastager et al. (2010) |
| Bacillus licheniformis | Siddikee et al. (2011) |
| Zhihengliuela alba | Gontia et al. (2011) |
| Micrococcus sp. | Jha et al. (2012) |
| Brachybacterium saurashtrense | Gontia-Mishra et al. (2014) |
| Brevibacterium casei | Gontia-Mishra et al. (2014) |
| Archaebacteria | |
| Pyrococcus horikoshii | Fujino et al. (2004) |
| Yeasts | |
| Hansenula saturnus | Minami et al. (1998) |
| Issatchenkia occidentalis | Palmer et al. (2007) |
| Fungi | |
| Penicillium citrinum | Jia et al. (1999) |
| Trichoderma asperellum | Viterbo et al. (2010) |
| Phytophthora sojae | Singh and Kashyap (2012) |
| Plants | |
| Arabidopsis thaliana | McDonnell et al. (2009) |
| Populus tremula | Plett et al. (2009) |
| Solanum lycopersicum | Gontia-Mishra et al. (2014) |
In response to tryptophan and other small molecules present in root exudates, IAA is synthesized and secreted by a PGPR that is bound to the surface of the root (Whipps, 1990; Hong et al., 1991; Fallik et al., 1994; Dimkpa et al., 2012). Some of the newly synthesized IAA is taken up by the plant and can stimulate ACS to convert SAM to ACC (Kende, 1993). The uptake and subsequent cleavage of ACC by the PGPR decrease the amount of ACC outside the plant (Penrose and Glick, 2001).
The Km of ACC deaminase is lower than ACC oxidase. The various plant ACC oxidase for ACC ranges from approximately 8 μM (for ripening apples) to 120 μM (for etiolated beans; Smith et al., 1992). The Km of ACC deaminase for ACC ranges from approximately 1.5 to 3.4 mM (Honma and Shimomura, 1978; Hontzeas et al., 2004). This means that ACC oxidase has about a 100-fold greater affinity for ACC than does ACC deaminase. Despite the fact that ACC oxidase has a much higher affinity for ACC, the kinetic calculations indicate that ACC deaminase can be more effective in lowering ACC than ACC oxidase when the amount of ACC deaminase is much greater than the amount of ACC oxidase (Glick et al., 1998). These results indicate that the ACC metabolite reaction in PGPR was more effective than that in plant cells and that the ACC level are lower in inner plant cells than in external cells. Therefore, to maintain equilibrium between internal and external ACC levels, the plant increases the level of ACC exudate. PGPRs have the ability to utilize ACC as a sole source of nitrogen in plant roots (Glick et al., 1994; Jacobson et al., 1994) and to proliferate under conditions in which other soil bacteria cannot grow. The reduction of the inner ACC level caused by the utilization of ACC in bacteria causes the plant to synthesize more ACC and to effectively exude ACC from the plant.
A significant correlation was found between in vitro bacterial ACC deaminase activity and the growth-promoting activity of these bacteria under pot and field trial conditions (Shaharoona et al., 2006a,b). In fact, many types of PGPR containing the ACC deaminase gene reduced ethylene production, resulting in physiological changes in many types of plants. The rhizobacterium Variovorax paradoxus 5C-2, which possesses ACC deaminase, promotes the growth (leaf area and shoot biomass) and development of WT A. thaliana and the ethylene-overproducing mutant eto1-1 but does not have these effects in ethylene-insensitive mutants (etr1-1 and ein2-1; Chen et al., 2013). Furthermore, V. paradoxus 5C-2 decreased the ACC concentrations in the rosette leaves of WT plants by 59% and foliar ethylene emission in both WT plants and eto1-1 mutants by 42 and 37%, respectively, (Chen et al., 2013). Rhizobacteria possessing ACC deaminase induced maximum waterlogging tolerance in Ocimum sanctum, as treated waterlogged plants exhibited the maximum growth and biomass yield with minimal ethylene levels (Barnawal et al., 2012). Bacterial endophytes expressing ACC deaminase delay flower senescence (Ali et al., 2012). Mesorhizobium ciceri LMS-1 expressing an exogenous ACC deaminase increases nodulation abilities and chickpea plant resistance to soil constraints (Nascimento et al., 2012).
SUPER-Agrobacterium WITH ACC DEAMINASE ACTIVITY INCREASES T-DNA TRANSFER EFFICIENCY VIA A. tumefaciens
The introduction of ACC deaminase activity into A. tumefaciens may reduce ethylene levels during co-cultivation and increase A. tumefaciens-mediated transformation efficiency. Whole-genome sequence analysis revealed that the A. tumefaciens strain C58 does not have an ACC deaminase gene (Wood et al., 2001); therefore, this strategy seems to be effective in this species. The ACC deaminase gene was amplified from Pseudomonas sp. via PCR. This amplified fragment was inserted into pBBR1MCS-5 (pBBRacdS). The pBBR1MCS-5 plasmid is compatible with IncP, IncQ, and IncW plasmids, and it has a different origin of the RK2 vector, which is used as a binary vector (Kovach et al., 1994). Because their origins are different, pBBRacdS and the binary vector are able to coexist in A. tumefaciens. The expression of ACC deaminase was controlled by the lacZ promoter, which constitutively and strongly expresses genes in A. tumefaciens. The A. tumefaciens strain harboring ACC deaminase genes showed ACC deaminase activity and effectively inhibited ethylene production in melons (Figure 2A), resulting in increased transient T-DNA transformation in melons (Figure 2B; Nonaka et al., 2008b).
FIGURE 2.
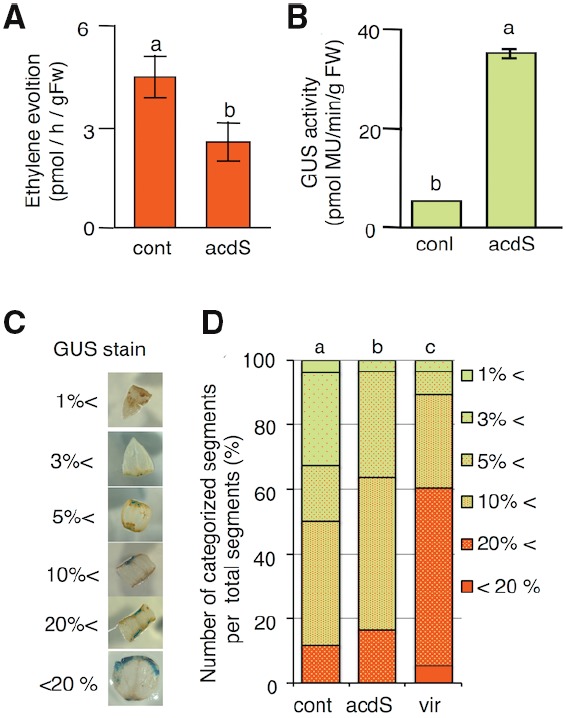
Agrobacterium tumefaciens with ACC deaminase activity inhibits ethylene evolution during co-cultivation, increasing gene transformation frequency in tomatoes and melons. (A) Measurement of ethylene evolution. The accumulation of ethylene in the headspace was measured on a gas chromatograph. Bars represent standard deviations (n = 3). The characters a and b show significant differences (t-test; P < 0.05). fw, fresh weight. (B) Quantification of gene transfer by a GUS assay. Melon cotyledon segments were co-cultured with three different A. tumefaciens strains for 3 days. (C) Classification of GUS-stained cotyledon explants. GUS-stained tomato cotyledons were categorized based on the stained area: less than 1, 1–3, 3–5, 5–10, 10–20, and more than 20%. (D) The frequency of each GUS staining category in tomato explants. Bacterial strains with significant differences (Student’s t-test and Kruskal–Wallis test; P < 0.01) are indicated by different letters. cont: A. tumefaciens GV2260 (pBBR1MCS-5, pIG121); acdS: A. tumefaciens GV2260 (pBBRacdS, pIG121-Hm) acdS gene driven by lac promoter; virD1acdS: A. tumefaciens GV2260 (pvirD1acdS, pIG121-Hm) acdS gene driven by the virD1 promoter. Panels (A,B) are cited from Nonaka et al. (2008b). Panels (C,D) are cited from Someya et al. (2013).
For the further improvement of Agrobacterium-mediated transformation, we attempted to increase the expression level of ACC deaminase. Klüsener et al. (2010) showed that virB1, virD1, and virE1 of A. tumefaciens were among the most highly expressed genes in acidic medium (AB medium at pH 5.5) containing 100 mol/L acetosyringone (Klüsener et al., 2010). We compared the activities of these three promoters to select the highest one, and the virD1 promoter showed the strongest transcription activity. The virD1 promoter conferred fourfold increased transcriptional activity compared with the lacZ promoter. The time course analysis (0–72 h) showed that ACC deaminase expression was induced at 6 h by adding acetosyringone to MS medium, and the high expression level was maintained until 72 h. By contrast, the expression of the ACC deaminase gene driven by the lacZ promoter started to decrease at 6 h. Compared with the lacZ promoter, the virD1 promoter maintained ACC deaminase gene expression at a higher level for a long time, increasing the ACC deaminase activity in A. tumefaciens. Generally, the co-cultivation period is 72 to 96 h, so the virD1 promoter seems to be suitable as the ACC deaminase gene activator. This newly developed A. tumefaciens strain showed higher T-DNA transformation efficiency in tomatoes (Figures 2C,D) and higher biomass production than Erianthus ravennae, which shows very low transformation frequency. Therefore, A. tumefaciens with higher ACC deaminase activity is a powerful tool for the Agrobacterium-mediated genetic engineering of plants (Someya et al., 2013). Introducing ACC deaminase into A. tumefaciens is effective at increasing the stable transformation frequency. A. tumefaciens with ACC deaminase succeeded in increasing the stable transformation frequency in Egusi melons (Ntui et al., 2009) and three canola cultivars (Brassica napus cv. Westar, B. napus cv. Hyola 401 and B. napus cv. 4414RR; Hao et al., 2010). We herein designate A. tumefaciens with ACC deaminase activity as super-Agrobacterium, which has the potential to improve the transformation efficiency of recalcitrant plant species.
CONCLUSION
Agrobacterium-mediated transformation is an important tool for plant genetic engineering. Although a sophisticated protocol has been established for model plants, such as A. thaliana, tobacco, and rice, the transformation efficiency was not sufficiently high for commercially important crops such as maize, sorghum, soybean, barley, and E. ravennae. There has therefore been a need for the improvement of this methodology. Among the various negative factors contributing to low Agrobacterium-mediated transformation rates, ethylene has been well known as an inhibitor of transformation efficiency for a long time. There are chemicals to reduce ethylene production or ethylene perception. However, they are hard to use because they are expensive, in a gaseous form or toxic to bacteria; they are thus not suitable for improving Agrobacterium-mediated transformation. Therefore, we attempted to reduce ethylene in A. tumefaciens. Some PGPRs have an enzyme that degrades the ethylene precursor ACC. This strategy is not toxic for bacteria, and it is very easy to perform. We introduced the enzyme ACC deaminase into A. tumefaciens. The strain has been designated as super-Agrobacterium. It showed increased transient gene delivery into melon cotyledons (Nonaka et al., 2008b), tomatoes, and E. ravennae (Someya et al., 2013). In addition, the super-Agrobacterium also increased stable transformation in Egusi melon and three canola species (Ntui et al., 2009; Hao et al., 2010). To improve the “super-Agrobacterium,” a thorough analysis will be required in the future. One of the negative effects of ethylene is the suppression of vir gene expression in A. tumefaciens. However, the constitutive vir gene expression strain or the addition of the vir gene expression inducer could only partially overcome the inhibitory effect of ethylene on vir gene expression. This result showed the possibility of a different inhibitory mechanism that is caused by ethylene; therefore, a thorough analysis will be required in the future.
Our study showed that there are two points that should be considered when improving the efficiency of Agrobacterium-mediated gene transfer: first, the transient transformation frequency is important for increasing the stable transformation efficiency. Second, the regulation of plant-derived signals is an effective method for increasing the A. tumefaciens-mediated gene transfer frequency. For a more sophisticated strategy by which to increase transformation, it is effective to analyze the plant-derived signals in Agrobacterium–plant interactions. The removal of these signals would further increase the A. tumefaciens-mediated transformation frequency in recalcitrant plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGMENTS
We thank Prof. M. Mii (Chiba University, Japan) and Dr. I. Nakamura (Chiba University, Japan) for kindly providing the E. ravennae calli and binary plasmid, respectively. This research was supported in part by the New Energy and Industrial Technology Development Organization (NEDO) to Hiroshi Ezura This work was also supported by JSPS KAKENHI, a Grant-in-Aid for Young Scientists (B); (Grant Number 24780001) to Satoko Nonaka, and the Cooperative Research Grant of Gene Research Center, the University of Tsukuba to Hiroshi Ezura and Satoko Nonaka.
REFERENCES
- Abeles F. B. (1992). “Regulation of ethylene production by internal, environmental, and stress factors,” in Ethylene in Plant Biology, 2nd Edn, eds Abeles F. B., Morgan P. W., II, Saltveit M. E. (San Diego: Academic Press; ), 56–119. [Google Scholar]
- Ali S., Charles T. C., Glick B. R. (2012). Delay of flower senescence by bacterial endophytes expressing 1-aminocyclopropane-1-carboxylate deaminase. J. Appl. Microbiol. 113, 1139–1144. 10.1111/j.1365-2672.2012.05409.x [DOI] [PubMed] [Google Scholar]
- Aloni R., Wolf A., Feigenbaum P., Avni A., Klee H. J. (1998). The never ripe mutant provides evidence that tumor-induced ethylene controls the morphogenesis of Agrobacterium tumefaciens-induced crown galls on tomato stems. Plant Physiol. 117, 841–849. 10.1104/pp.117.3.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnawal D., Bharti N., Maji D., Chanotiya C. S., Kalra A. (2012). 1-Aminocyclopropane-1-carboxylic acid (ACC) deaminase-containing rhizobacteria protect Ocimum sanctum plants during waterlogging stress via reduced ethylene generation. Plant Physiol. Biochem. 58, 227–235. 10.1016/j.plaphy.2012.07.008 [DOI] [PubMed] [Google Scholar]
- Belimov A., Dodd I. C., Safronova V. I., Davies W. J. (2009). ACC deaminase-containing rhizobacteria improve vegetative development and yield of potato plants grown under water-limited conditions. Asp. Appl. Biol. 98, 163–169. [Google Scholar]
- Belimov A. A., Safronova V. I., Sergeyeva T. A., Egorova T. N., Matveyeva V. A., Tsyganov V. E., et al. (2001). Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 47, 642–652. 10.1139/cjm-47-7-642 [DOI] [PubMed] [Google Scholar]
- Bevan M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. 10.1093/nar/12.22.8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer E. M. (1981). “Ethylene action and metabolism,” in Recent Advances in the Biochemistry of Fruits and Vegetables, eds Friend J., Rhodes M. J. C. (New York: Academic Press; ), 107–121. [Google Scholar]
- Burgos L., Alburquerque N. (2003). Ethylene inhibitors and low kanamycin concentrations improve adventitious regeneration from apricot leaves. Plant Cell Rep. 21, 1167–1174. 10.1007/s00299-003-0625-6 [DOI] [PubMed] [Google Scholar]
- Cangelosi G. A., Ankenbauer R. G., Nester E. W. (1990). Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natil. Acad. Sci. U.S.A. 87, 6708–6712 10.1073/pnas.87.17.6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty R., Viswakarma N., Bhat S. R., Kirti P. B., Singh B. D., Chopra V. L. (2002). Agrobacterium-mediated transformation of cauliflower: optimization of protocol and development of Bt-transgenic cauliflower. J. Biosci. 27, 495–502. 10.1007/BF02705046 [DOI] [PubMed] [Google Scholar]
- Chen L., Dodd I. C., Theobald J. C., Belimov A. A., Davies W. J. (2013). The rhizobacterium Variovorax paradoxus 5C-2, containing ACC deaminase, promotes growth and development of Arabidopsis thaliana via an ethylene-dependent pathway. J. Exp. Bot. 64, 1565–1573. 10.1093/jxb/ert031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton M. D., Drummond M. H., Merio D. J., Sciaky D., Montoya A. L., Gordon M. P., et al. (1977). Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11, 263–271 10.1016/0092-8674(77)90043-5 [DOI] [PubMed] [Google Scholar]
- Ciardi J. A., Tieman D. M., Lund S. T., Jones J. B., Stall R. E., Klee H. J. (2000). Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol. 123, 81–92. 10.1104/pp.123.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. (1976). Evidence for diverse types of large plasmids in tumor-inducing strains of Agrobacterium. J. Bacteriol. 126, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastager S. G., Deepa C. K., Pandey A. (2010). Isolation and characterization of novel plant growth promoting Micrococcus sp NII-0909 and its interaction with cowpea. Plant Physiol. Biochem. 48, 987–992. 10.1016/j.plaphy.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Davis M. E., Miller A. R., Lineberger R. D. (1992). Studies on the effects of ethylene on transformation of tomato cotyledons (Lycopersicon esculentum Mill.) by Agrobacterium tumefaciens. J. Plant Physiol. 139, 309–312 10.1016/S0176-1617(11)80343-3 [DOI] [Google Scholar]
- Deikman J. (1997). Molecular mechanisms of ethylene regulation of gene transcription. Physiol. Plant. 100, 561–566 10.1111/j.1399-3054.1997.tb03061.x [DOI] [Google Scholar]
- Dimkpa C. O., Zeng J., McLean J. E., Britt D. W., Zhan J., Anderson A. J. (2012). Production of indole-3-acetic acid via the indole-3-acetamide pathway in the plant-beneficial bacterium Pseudomonas chlororaphis O6 is inhibited by ZnO nanoparticles but enhanced by CuO nanoparticles. Appl. Environ. Microbiol. 78, 1404–1410. 10.1128/AEM.07424-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker J. R., Davis R. W. (1987). Plant defense genes are regulated by ethylene. Proc. Natl. Acad. Sci. U.S.A. 84, 5202–5206. 10.1073/pnas.84.15.5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezura H., Yuhashi K., Yasuta T., Minamisawa K. (2000). Effect of ethylene on Agrobacterium tumefaciens-mediated gene transfer to melon. Plant Breed. 119, 75–79 10.1046/j.1439-0523.2000.00438.x [DOI] [Google Scholar]
- Fallik E., Sarig S., Okon Y. (1994). “Morphology and physiology of plant roots associated with Azospirillum,” in Azospirillum–Plant Associations, ed. Okon Y. (Boca Raton: CRC Press; ), 77–85. [Google Scholar]
- Fujino A., Ose T., Yao M., Tokiwano T., Honma M., Watanabe N., et al. (2004). Structural and enzymatic properties of 1-aminocyclopropane-1-carboxylate deaminase homologue from Pyrococcus horikoshii. J. Mol. Biol. 341, 999–1013. 10.1016/j.jmb.2004.06.062 [DOI] [PubMed] [Google Scholar]
- Gontia I., Kavita K., Schmid M., Hartmann A., Jha B. (2011). Brachybacterium saurashtrense sp. nov., a halotolerant root associated bacterium with plant growth-promoting potential. Int. J. Syst. Evol. Microbiol. 61, 2799–2804. 10.1099/ijs.0.023176-0 [DOI] [PubMed] [Google Scholar]
- Gontia-Mishra I., Sasidharan S., Tiwari S. (2014). Recent developments in use of 1-aminocyclopropane-1-carboxylate (ACC) deaminase for conferring tolerance to biotic and abiotic stress. Biotechnol. Lett. 36, 889–898. 10.1007/s10529-014-1458-9 [DOI] [PubMed] [Google Scholar]
- Glick B. R., Jacobson C. B., Schwarze M. M. K., Pasternak J. J. (1994). 1-Aminocyclopropane-1-carboxylic acid deaminase mutants of the plant growth promoting rhizobacterium Pseudomonas putida GR12-2 do not stimulate canola root elongation. Can. J. Microbiol. 40, 911–915 10.1139/m94-146 [DOI] [Google Scholar]
- Glick B. R., Penrose D. M., Li J. (1998). A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theor. Biol. 190, 63–68. 10.1111/j.1574-6968.2010.01977.x [DOI] [PubMed] [Google Scholar]
- Han J. S., Kim C. K., Park S. H., Hirschi K. D., Mok I. G. (2005). Agrobacterium-mediated transformation of bottle gourd (Lagenaria siceraria Standl.). Plant Cell Rep. 23, 692–698. 10.1007/s00299-004-0874-z [DOI] [PubMed] [Google Scholar]
- Hao Y., Charles T. C., Glick B. R. (2010). ACC deaminase increases the Agrobacterium tumefaciens-mediated transformation frequency of commercial canola cultivars. FEMS Microbiol. Lett. 7, 185–190. 10.1111/j.1574-6968.2010.01977.x [DOI] [PubMed] [Google Scholar]
- He F., Nair G. R., Soto C. S., Chang Y., Hsu L., Ronzone E., et al. (2009). Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens. J. Bacteriol. 191, 5802–5813. 10.1128/jb.00451-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A., Hirsch P. R., Hooykaas P. J. J., Schilperoort R. A. (1983). A binary plant vector strategy based on separation of the Vir-and T-region on the Agrobacterium tumefaciens Ti plasmid. Nature 303, 179–180 10.1038/303179a0 [DOI] [Google Scholar]
- Holguin G., Glick B. R. (2001). Expression of the ACC deaminase gene from Enterobacter cloacae UW4 in Azospirillum brasilense. Microb. Ecol. 41, 281–288. 10.1007/s002480000040 [DOI] [PubMed] [Google Scholar]
- Hong Y., Glick B. R., Pasternak J. J. (1991). Plant–microbial interactions under gnotobiotic conditions: a scanning electron microscope study. Curr. Microbiol. 23, 111–114 10.1007/BF02092259 [DOI] [Google Scholar]
- Honma S., Shimomura T. (1978). Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42, 1825–1831 10.1271/bbb1961.42.1825 [DOI] [Google Scholar]
- Hontzeas N., Zoidakis J., Glick B. R., Abu-Omar M. M. (2004). Expression and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the rhizobacterium Pseudomonas putida UW4: a key enzyme in bacterial plant growth promotion. Biochim. Biophys. Acta 1703, 11–19. 10.1016/j.bbapap.2004.09.015 [DOI] [PubMed] [Google Scholar]
- Hu X., Zhao J., Degrado W. F., Vinns A. N. (2013). Agrobacterium tumefaciens recognizes its host environment using ChvE to bind divers plant sugars as virulence signals. Proc. Natil. Acad. Sci. U.S.A. 110, 678–683. 10.1073/pnas.1215033110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson C. B., Pasternak J. J., Glick B. R. (1994). Partial purification and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Can. J. Microbiol. 40, 1019–1025 10.1139/m94-162 [DOI] [Google Scholar]
- Jha B., Gontia I., Hartmann A. (2012). The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil 356, 265–277 10.1007/s11104-011-0877-9 [DOI] [Google Scholar]
- Jia Y. J., Kakuta Y., Sugawara M., Igarashi T., Oki N., Kisaki M., et al. (1999). Synthesis and degradation of 1-aminocyclopropane-1-carboxylic acid by Penicillium citrinum. Biosci. Biotechnol. Biochem. 63, 542–549. 10.1271/bbb.63.542 [DOI] [PubMed] [Google Scholar]
- Kende H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 283–307 10.1146/annurev.pp.44.060193.001435 [DOI] [Google Scholar]
- Komari T., Takakura Y., Ueki J., Kato N., Ishida Y., Hiei Y. (2006). Binary vectors and super-binary vectors. Methods Mol. Biol. 343, 15–41. 10.1385/1-59745-130-4:15 [DOI] [PubMed] [Google Scholar]
- Kovach M. E., Phillips R. W., Elzer P. H., Roop R. M., II, Peterson K. M. (1994). pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16, 800–802. [PubMed] [Google Scholar]
- Klüsener S., Hacker S., Tsai Y. L., Bandow J. E., Gust R., Lai E. M., et al. (2010). Proteomic and transcriptomic characterization of a virulence-deficient phosphatidylcholine-negative Agrobacterium tumefaciens mutant. Mol. Genet. Genomics 283, 575–589. 10.1007/s00438-010-0542-7 [DOI] [PubMed] [Google Scholar]
- Lee C. W., Efetova M., Engelmann J. C., Kramell R., Wasternack C., Ludwig-Muller J., et al. (2009). Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21, 2948–2962. 10.1105/tpc.108.064576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Guinel F. C., Glick B. R. (2003). Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 69, 4396–4402. 10.1128/AEM.69.8.4396-4402.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhaiyan M., Poonguzhali S., Ryu J., Sa T. (2006). Regulation of ethylene levels in canola (Brassica campestris) by 1-aminocyclopropane-1-carboxylate deaminase-containing Methylobacterium fujisawaense. Planta 224, 268–278. 10.1007/s00425-005-0211-y [DOI] [PubMed] [Google Scholar]
- Malsy S., van Bel A. J. E., Kluge M., Hartung W., Ullrich C. I. (1992). Induction of crown galls by Agrobacterium tumefaciens (strain C–58) reverses assimilate translocation and accumulation in Kalanchoë daigremontiana. Plant Cell Environ. 15, 519–529 10.1111/j.1365-3040.1992.tb01485.x [DOI] [Google Scholar]
- McDonnell L., Plett J. M., Andersson-Gunnerås S., Kozela C., Dugardeyn J., Van Der Straeten D., et al. (2009). Ethylene levels are regulated by a plant encoded 1-aminocyclopropane-1-carboxylic acid deaminase. Physiol. Plant. 136, 94–109. 10.1111/j.1399-3054.2009.01208.x [DOI] [PubMed] [Google Scholar]
- Minami R., Uchiyama K., Murakami T., Kawai J., Mikami K., Yamada T., et al. (1998). Properties, sequence, and synthesis in Escherichia coli of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus. J. Biochem. 123, 1112–1118. 10.1093/oxfordjournals.jbchem.a022050 [DOI] [PubMed] [Google Scholar]
- Nascimento F. X., Brigido C., Glick B. R., Oliveira S., Alho L. (2012). Mesorhizobium ciceri LMS-1 expressing an exogenous 1-aminocyclopropane-1-carboxylate (ACC) deaminase increases its nodulation abilities and chickpea plant resistance to soil constraints. Lett. Appl. Microbiol. 55, 15–21. 10.1111/j.1472-765X.2012.03251.x [DOI] [PubMed] [Google Scholar]
- Nonaka S., Yuhashi K., Takada K., Sugaware M., Minamisawa K., Ezura H. (2008a). Ethylene production in plants during transformation suppresses vir gene expression in Agrobacterium tumefaciens. New Phytol. 178, 647–656. 10.1111/j.1469-8137.2008.02400.x [DOI] [PubMed] [Google Scholar]
- Nonaka S., Sugawara M., Minamisawa K., Yuhashi K., Ezura H. (2008b). 1-Aminocyclopropane-1-carboxylate deaminase enhances Agrobacterium tumefaciens-mediated gene transfer into plant cells. Appl. Environ. Microbiol. 74, 2526–2528. 10.1128/AEM.02253-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman-Setterblad C., Vidal S., Palva E. T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall degrading enzymes from Erwinia carotovora. Mol. Plant Microbe Interact. 13, 430–438. 10.1094/MPMI.2000.13.4.430 [DOI] [PubMed] [Google Scholar]
- Ntui V. O., Thirukkumaran G., Iioka S., Mii M. (2009). Efficient plant regeneration via organogenesis in “Egusi” melon (Colocynthis citrullus L.). Sci. Hortic. 119, 397–402 10.1016/j.scienta.2008.08.031 [DOI] [Google Scholar]
- Ogawara T., Higashi K., Kamada H., Ezura H. (2003). Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J. Plant Physiol. 160, 1335–1340. 10.1078/0176-1617-01129 [DOI] [PubMed] [Google Scholar]
- Palmer C., Golden K., Danniels L., Ahmad H. (2007). ACC deaminase from Issatchenkia occidentalis. J. Biol. Sci. 7, 188–193 10.3923/jbs.2007.188.193 [DOI] [Google Scholar]
- Pradel K. S., Ullrich C. I., Santa Cruz S., Oparka K. J. (1999). Symplastic continuity in Agrobacterium tumefaciens-induced tumours. J. Exp. Bot. 50, 183–192 10.1093/jxb/50.331.183 [DOI] [Google Scholar]
- Pradel S. K., Rezmer C., Krausgrill S., Rausch T., Ullrichm C. I. (1996). Evidence for symplastic phloem unloading with concomitant high activity of acid cell wall invertase in Agrobacterium tumefaciens-induced plant tumors. Bot. Acta 109, 397–404 10.1111/j.1438-8677.1996.tb00589.x [DOI] [Google Scholar]
- Penninckx I., Eggermont K., Terras F. R. G., Thomma B. P. H. J., Samblanx G. W. D., Buchala A., et al. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. 10.1105/tpc.8.12.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose D. M., Glick B. R. (2001). Levels of ACC and related compounds in exudate and extracts of canola seeds treated with ACC deaminase-containing plant growth-promoting bacteria. Can. J. Microbiol. 47, 368–372. 10.1139/w01-014 [DOI] [PubMed] [Google Scholar]
- Petri C., Alburquerque N., Perez-Tornero O., Burgos L. (2005). Auxin pulses and a synergistic interaction between polyamines and ethylene inhibitors improve adventitious regeneration from apricot leaves and Agrobacterium-mediated transformation of leaf tissues. Plant Cell Tissue Organ Cult. 82, 105–111 10.1007/s11240-004-7013-y [DOI] [Google Scholar]
- Plett J. M., McDonnell L., Regan S. (2009). Plant encoded 1-aminocyclopropane-1-carboxylic acid deaminase activity implicated in different aspects of plant development. Plant Signal. Behav. 4, 1186–1189 10.1111/j.1399-3054.2009.01208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong E. S., Song K. J., Jegal S., Yu C. Y., Chung I. M. (2005). Silver nitrate and aminoethoxyvinylglycine affect Agrobacterium-mediated apple transformation. Plant Growth Regul. 45, 75–82 10.1007/s10725-004-6126-y [DOI] [Google Scholar]
- Sessitsch A., Coenye T., Sturz A. V., Vandamme P., Barka E., Wang-Pruski G., et al. (2005). Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 55, 1187–1192. 10.1099/ijs.0.63149-0 [DOI] [PubMed] [Google Scholar]
- Shaharoona B., Arshad M., Zahir Z. A. (2006a). Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett Appl. Microbiol. 42, 155–159. 10.1111/j.1472-765X.2005.01827.x [DOI] [PubMed] [Google Scholar]
- Shaharoona B., Arshad M., Zahir Z. A., Khalid A. (2006b). Performance of Pseudomonas spp. containing ACC-deaminase for improving growth and yield of maize (Zea mays L.) in the presence of nitrogenous fertilizer. Soil Biol. Biochem. 38, 2971–2975 10.1016/j.soilbio.2006.03.024 [DOI] [Google Scholar]
- Stachel S. E., Messens E., van Montagu M., Zambryski P. (1985). Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318, 624–629 10.1038/318624a0 [DOI] [Google Scholar]
- Stachel S. E., Nester E. W., Zambryski P. C. (1986). A plant cell factor induces Agrobacterium tumefaciens vir gene expression. Proc. Natil. Acad. Sci. U.S.A. 83, 379–383 10.1073/pnas.83.2.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddikee M. A., Glick B. R., Chauhan P. S., Yim W. J., Sa T. (2011). Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol. Biochem. 49, 427–434. 10.1016/j.plaphy.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Singh N., Kashyap S. (2012). In silico identification and characterization of 1-aminocyclopropane-1-carboxylate deaminase from Phytophthora sojae. J. Mol. Model. 18, 4101–4111. 10.1007/s00894-012-1389-0 [DOI] [PubMed] [Google Scholar]
- Smith J. J., Ververidis P., John P. (1992). Characterization of the ethylene-forming enzyme partially purified from melon. Phytochemistry 31, 1485–1494 10.1016/0031-9422(92)83092-D [DOI] [Google Scholar]
- Someya T., Nonaka S., Nakamura K., Ezura H. (2013). Increased 1-aminocyclopropane-1-carboxylate deaminase activity enhances Agrobacterium tumefaciens-mediated gene delivery into plant cells. Microbiologyopen 2, 873–880. 10.1002/mbo3.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppan A., Roby D., Esquerre-Tugaye M. T. (1982). Cell surfaces in plant–microorganism interactions: III. In vivo effect of ethylene on hydroxyproline-rich glycoprotein accumulation in the cell wall of diseased plants. Plant Physiol. 70, 82–86. 10.1104/pp.70.1.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Larebeke N., Engler G., Holsters M., van den Elsacker S., Schelperoort R. A., Schell J. (1974). Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252, 169–170. 10.1038/252169a0 [DOI] [PubMed] [Google Scholar]
- van Larebeke N., Genetello C., Schell J., Schilperoort R. A., Hermans A. K., van Montagu M., et al. (1975). Acquisition of tumour-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature 255, 742–743. 10.1038/255742a0 [DOI] [PubMed] [Google Scholar]
- Veena, Jiang, H., Doerge, R. W., Gelvin S. B. (2003). Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 35, 219–236. 10.1046/j.1365-313X.2003.01796.x [DOI] [PubMed] [Google Scholar]
- Viterbo A., Landau U., Kim S., Chernin L., Chet I. (2010). Characterization of ACC deaminase from the biocontrol and plant growth-promoting agent Trichoderma asperellum T203. FEMS Microbiol. Lett. 305, 42–48. 10.1111/j.1574-6968.2010.01910.x [DOI] [PubMed] [Google Scholar]
- Wang K. L., Li H., Ecker J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl.), S131–S151. 10.1105/tpc.001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächter R., Langhans M., Aloni R., Götz S., Weilmünster A., Koops A., et al. (2003). Vascularization, high-volume solution flow, and localized roles for enzymes of sucrose metabolism during tumorigenesis by Agrobacterium tumefaciens. Plant Physiol. 133, 1024–1037. 10.1104/pp.103.028142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson B., Currier T. C., Gordon M. P., Chilton M. D., Nester E. W. (1975). Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 123, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipps J. M. (1990). “Carbon utilization,” in The Rhizosphere, ed. Lynch J. M. (Chichester: Wiley; ), 59–97. [Google Scholar]
- Wood D. W., Setubal J. C., Kaul R., Monks D. E., Kitajima J. P., Okura V. K., et al. (2001). The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323. 10.1126/science.1066804 [DOI] [PubMed] [Google Scholar]
- Yang S. F. (1985). Biosynthesis and action of ethylene. HortScience 20, 41–45. [Google Scholar]
- Yang S. F., Hoffman N. E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189 10.1146/annurev.pp.35.060184.001103 [DOI] [Google Scholar]
- Zaenen I., van Larebeke N., Van Montagu M., Schell J. (1974). Supercoiled circular DNA in crown-gall inducing Agrobacterium strains. J. Mol. Biol. 86, 109–127 10.1016/S0022-2836(74)80011-2 [DOI] [PubMed] [Google Scholar]
- Zambryski P., Joos H., Genetello C., Leemans J., Montagu M. V., Schell J. (1983). Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 2, 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]


