Abstract
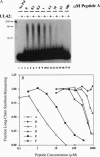
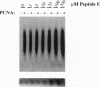
The herpes simplex virus DNA polymerase consists of two subunits--a catalytic subunit and an accessory subunit, UL42, that increases processivity. Mutations affecting the extreme C terminus of the catalytic subunit specifically disrupt subunit interactions and ablate virus replication, suggesting that new antiviral drugs could be rationally designed to interfere with polymerase heterodimerization. To aid design, we performed circular dichroism (CD) spectroscopy and analytical ultracentrifugation studies, which revealed that a 36-residue peptide corresponding to the C terminus of the catalytic subunit folds into a monomeric structure with partial alpha-helical character. CD studies of shorter peptides were consistent with a model where two separate regions of alpha-helix interact to form a hairpin-like structure. The 36-residue peptide and a shorter peptide corresponding to the C-terminal 18 residues blocked UL42-dependent long-chain DNA synthesis at concentrations that had no effect on synthesis by the catalytic subunit alone or by calf thymus DNA polymerase delta and its processivity factor. These peptides, therefore, represent a class of specific inhibitors of herpes simplex virus DNA polymerase that act by blocking accessory-subunit-dependent synthesis. These peptides or their structures may form the basis for the synthesis of clinically effective drugs.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ace C. I., McKee T. A., Ryan J. M., Cameron J. M., Preston C. M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989 May;63(5):2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulnick A. D., Thompson J. R., Iyengar S., Pearson G., Ablashi D., Ricciardi R. P. Identification of a DNA-binding protein of human herpesvirus 6, a putative DNA polymerase stimulatory factor. J Gen Virol. 1993 Jun;74(Pt 6):1003–1009. doi: 10.1099/0022-1317-74-6-1003. [DOI] [PubMed] [Google Scholar]
- Bravo R., Frank R., Blundell P. A., Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987 Apr 2;326(6112):515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- Brooks I., Watts D. G., Soneson K. K., Hensley P. Determining confidence intervals for parameters derived from analysis of equilibrium analytical ultracentrifugation data. Methods Enzymol. 1994;240:459–478. doi: 10.1016/s0076-6879(94)40060-1. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T., Chau K. H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974 Jul 30;13(16):3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- Cohen E. A., Gaudreau P., Brazeau P., Langelier Y. Specific inhibition of herpesvirus ribonucleotide reductase by a nonapeptide derived from the carboxy terminus of subunit 2. Nature. 1986 May 22;321(6068):441–443. doi: 10.1038/321441a0. [DOI] [PubMed] [Google Scholar]
- Digard P., Bebrin W. R., Weisshart K., Coen D. M. The extreme C terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J Virol. 1993 Jan;67(1):398–406. doi: 10.1128/jvi.67.1.398-406.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digard P., Chow C. S., Pirrit L., Coen D. M. Functional analysis of the herpes simplex virus UL42 protein. J Virol. 1993 Mar;67(3):1159–1168. doi: 10.1128/jvi.67.3.1159-1168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digard P., Coen D. M. A novel functional domain of an alpha-like DNA polymerase. The binding site on the herpes simplex virus polymerase for the viral UL42 protein. J Biol Chem. 1990 Oct 15;265(29):17393–17396. [PubMed] [Google Scholar]
- Dutia B. M., Frame M. C., Subak-Sharpe J. H., Clark W. N., Marsden H. S. Specific inhibition of herpesvirus ribonucleotide reductase by synthetic peptides. Nature. 1986 May 22;321(6068):439–441. doi: 10.1038/321439a0. [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Merutka G., Waltho J. P., Lerner R. A., Wright P. E. Folding of peptide fragments comprising the complete sequence of proteins. Models for initiation of protein folding. I. Myohemerythrin. J Mol Biol. 1992 Aug 5;226(3):795–817. doi: 10.1016/0022-2836(92)90633-u. [DOI] [PubMed] [Google Scholar]
- Eisenstein E., Osborne J. C., Jr, Chaiken I. M., Hensley P. Purification and characterization of ornithine transcarbamoylase from Saccharomyces cerevisiae. J Biol Chem. 1984 Apr 25;259(8):5139–5145. [PubMed] [Google Scholar]
- Ertl P. F., Powell K. L. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J Virol. 1992 Jul;66(7):4126–4133. doi: 10.1128/jvi.66.7.4126-4133.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Hall J. D., Mount D. W., Retondo M. J., Weller S. K., Coen D. M. Sequence and mapping analyses of the herpes simplex virus DNA polymerase gene predict a C-terminal substrate binding domain. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7969–7973. doi: 10.1073/pnas.82.23.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D. J., Weller S. K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J Virol. 1988 Jan;62(1):196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb J., Marcy A. I., Coen D. M., Challberg M. D. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990 Dec;64(12):5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh A., Greaves R., O'Hare P. Interference with the assembly of a virus-host transcription complex by peptide competition. Nature. 1990 Mar 15;344(6263):257–259. doi: 10.1038/344257a0. [DOI] [PubMed] [Google Scholar]
- Hamatake R. K., Bifano M., Tenney D. J., Hurlburt W. W., Cordingley M. G. The herpes simplex virus type 1 DNA polymerase accessory protein, UL42, contains a functional protease-resistant domain. J Gen Virol. 1993 Oct;74(Pt 10):2181–2189. doi: 10.1099/0022-1317-74-10-2181. [DOI] [PubMed] [Google Scholar]
- Hernandez T. R., Lehman I. R. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J Biol Chem. 1990 Jul 5;265(19):11227–11232. [PubMed] [Google Scholar]
- Hirsch M. S., Schooley R. T. Resistance to antiviral drugs: the end of innocence. N Engl J Med. 1989 Feb 2;320(5):313–314. doi: 10.1056/NEJM198902023200510. [DOI] [PubMed] [Google Scholar]
- Johnson M. L., Faunt L. M. Parameter estimation by least-squares methods. Methods Enzymol. 1992;210:1–37. doi: 10.1016/0076-6879(92)10003-v. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Jr Protein secondary structure and circular dichroism: a practical guide. Proteins. 1990;7(3):205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- Kost R. G., Hill E. L., Tigges M., Straus S. E. Brief report: recurrent acyclovir-resistant genital herpes in an immunocompetent patient. N Engl J Med. 1993 Dec 9;329(24):1777–1782. doi: 10.1056/NEJM199312093292405. [DOI] [PubMed] [Google Scholar]
- Lam P. Y., Jadhav P. K., Eyermann C. J., Hodge C. N., Ru Y., Bacheler L. T., Meek J. L., Otto M. J., Rayner M. M., Wong Y. N. Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science. 1994 Jan 21;263(5145):380–384. doi: 10.1126/science.8278812. [DOI] [PubMed] [Google Scholar]
- Li J. S., Zhou B. S., Dutschman G. E., Grill S. P., Tan R. S., Cheng Y. C. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol. 1987 Sep;61(9):2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi M., Déziel R., Moss N., Beaulieu P., Bonneau A. M., Bousquet C., Chafouleas J. G., Garneau M., Jaramillo J., Krogsrud R. L. A potent peptidomimetic inhibitor of HSV ribonucleotide reductase with antiviral activity in vivo. Nature. 1994 Dec 15;372(6507):695–698. doi: 10.1038/372695a0. [DOI] [PubMed] [Google Scholar]
- Marsden H. S., Murphy M., McVey G. L., MacEachran K. A., Owsianka A. M., Stow N. D. Role of the carboxy terminus of herpes simplex virus type 1 DNA polymerase in its interaction with UL42. J Gen Virol. 1994 Nov;75(Pt 11):3127–3135. doi: 10.1099/0022-1317-75-11-3127. [DOI] [PubMed] [Google Scholar]
- Monahan S. J., Barlam T. F., Crumpacker C. S., Parris D. S. Two regions of the herpes simplex virus type 1 UL42 protein are required for its functional interaction with the viral DNA polymerase. J Virol. 1993 Oct;67(10):5922–5931. doi: 10.1128/jvi.67.10.5922-5931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianka A. M., Hart G., Murphy M., Gottlieb J., Boehme R., Challberg M., Marsden H. S. Inhibition of herpes simplex virus type 1 DNA polymerase activity by peptides from the UL42 accessory protein is largely nonspecific. J Virol. 1993 Jan;67(1):258–264. doi: 10.1128/jvi.67.1.258-264.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G., Tan C. K., Kostura M., Mathews M. B., So A. G., Downey K. M., Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987 Apr 2;326(6112):517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- Scholtz J. M., Baldwin R. L. The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Sequences at the C-terminus of the herpes simplex virus type 1 UL30 protein are dispensable for DNA polymerase activity but not for viral origin-dependent DNA replication. Nucleic Acids Res. 1993 Jan 11;21(1):87–92. doi: 10.1093/nar/21.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney D. J., Hurlburt W. W., Bifano M., Stevens J. T., Micheletti P. A., Hamatake R. K., Cordingley M. G. Deletions of the carboxy terminus of herpes simplex virus type 1 UL42 define a conserved amino-terminal functional domain. J Virol. 1993 Apr;67(4):1959–1966. doi: 10.1128/jvi.67.4.1959-1966.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney D. J., Micheletti P. A., Stevens J. T., Hamatake R. K., Matthews J. T., Sanchez A. R., Hurlburt W. W., Bifano M., Cordingley M. G. Mutations in the C terminus of herpes simplex virus type 1 DNA polymerase can affect binding and stimulation by its accessory protein UL42 without affecting basal polymerase activity. J Virol. 1993 Jan;67(1):543–547. doi: 10.1128/jvi.67.1.543-547.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Itzstein M., Wu W. Y., Kok G. B., Pegg M. S., Dyason J. C., Jin B., Van Phan T., Smythe M. L., White H. F., Oliver S. W. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993 Jun 3;363(6428):418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]