Abstract
Rice BGlu1 (Os3BGlu7) is a glycoside hydrolase family 1 β-glucosidase that hydrolyzes cellooligosaccharides with increasing efficiency as the degree of polymerization (DP) increases from 2 to 6, indicating six subsites for glucosyl residue binding. Five subsites have been identified in X-ray crystal structures of cellooligosaccharide complexes with its E176Q acid-base and E386G nucleophile mutants. X-ray crystal structures indicate that cellotetraose binds in a similar mode in BGlu1 E176Q and E386G, but in a different mode in the BGlu1 E386G/Y341A variant, in which glucosyl residue 4 (Glc4) interacts with Q187 instead of the eliminated phenolic group of Y341. Here, we found that the Q187A mutation has little effect on BGlu1 cellooligosaccharide hydrolysis activity or oligosaccharide binding in BGlu1 E176Q, and only slight effects on BGlu1 E386G glycosynthase activity. X-ray crystal structures showed that cellotetraose binds in a different position in BGlu1 E176Q/Y341A, in which it interacts directly with R178 and W337, and the Q187A mutation had little effect on cellotetraose binding. Mutations of R178 and W337 to A had significant and nonadditive effects on oligosaccharide hydrolysis by BGlu1, pNPGlc cleavage and cellooligosaccharide inhibition of BGlu1 E176Q and BGlu1 E386G glycosynthase activity. Hydrolysis activity was partially rescued by Y341 for longer substrates, suggesting stacking of Glc4 on Y341 stabilizes binding of cellooligosaccharides in the optimal position for hydrolysis. This analysis indicates that complex interactions between active site cleft residues modulate substrate binding and hydrolysis.
Keywords: rice, β-glucosidase, glycosynthase, hydrolysis, oligosaccharide binding, active site cleft
Introduction
Plant derived cellulose and related β-glucans comprise the largest source of biomass on earth, and breakdown of these β-glucans is of great interest for biomass conversion and biofuel production.1 Bacteria and fungi make use of the abundant β-glucan biomass as a nutritional source, while plants need to recycle their cell walls during development, and each of these phyla has developed their own set of enzymes for β-glucan breakdown. The enzymes involved in β-glucan breakdown include endoglucanases, exoglucanases, and β-glucosidases (β-d-glucoside glucohydrolases; E.C. 3.2.1.21), and in microorganisms polysaccharide lyases and polysaccharide oxidases,1,2 which have been catagorized into families based on protein sequence similarity in the Carbohydrate-Active enZYme (CAZY) database (www.cazy.org).3 The binding of the glycoside hydrolases to cellooligosaccharide substrates at their active site must be strong enough to position and stabilize the substrate for hydrolysis, yet weak enough to allow release of the products.4,5 A more thorough understanding of this binding is needed for engineering of oligosaccharide-active β-glucosidases and transglucosidases.
β-glucosidases hydrolyze β-glycosidic linkages of oligosaccharides and glycosides to release nonreducing terminal glucosyl residues.6 Henrissat et al. have categorized β-glucosidases into glycoside hydrolase (GH) families GH1, GH3, GH5, GH9, GH30, and GH116, with GH1 containing the most β-glucosidases that have been described.3,7 β-glucosidases can have varying levels of specificity, from enzymes that appear to hydrolyze a single substrate, such as Sorghum bicolor dhurrinase, to enzymes that act on a variety of alkyl and aryl glycosides, as well as oligosaccharides.8–11 Most β-glucosidases act through a two-step retaining mechanism, in which the catalytic nucleophile (a glutamate residue for GH1 enzymes) displaces the aglycone with acid-assistance from the catalytic acid-base (a glutamic acid residue for GH1) in the first step to form an α-linked glucosyl-enzyme intermediate. A water molecule, which is deprotonated by the catalytic acid-base, displaces the enzyme to release β-d-glucose in the second step.
Mutations of the catalytic acid-base and nucleophile have been developed to allow transglycosylation to proceed with minimal hydrolysis. Glycosynthases are nucleophile mutants of glycosidases that can be used for synthesis of oligosaccharides and glycoconjugates from glycosyl fluoride donors and suitable acceptors without hydrolysis of the products.12–15 Thioglycoligases are mutants in which the acid–base residue is replaced with a nonionizable amino acid residue to allow transfer of a glycosyl residue to the enzyme from substrates with good leaving groups that do not require acid-assistance and transfer to nucleophiles that do not require basic-assistance, such as thiols.16–18 Both of these types of mutations result in transglycosylation, with the glycosynthases acting through an inverting mechanism, while the thoiglycoligases maintain a retaining mechanism.
Rice BGlu1 β-glucosidase, also known as Os3BGlu7, acts as an exo-β-glucosidase on β-1,3- and β-1,4-linked gluco-oligosaccharides and belongs to glycoside hydrolase family GH110,19 BGlu1 also exhibits transglucosylation activity toward oligosaccharide substrates. Kinetic subsite analysis of cellooligosaccharide hydrolysis indicated that rice BGlu1 has at least six subsites for binding of β-1,4-linked d-glucosyl residues. The BGlu1 nucleophile mutant E386G is a glycosynthase that can synthesize long para-nitrophenyl (pNP)-cellooligosaccharides of up to 11 β-1,4-linked glucosyl residues.20 The BGlu1 E176Q acid-base mutant can cleave aryl glycosides with good leaving groups, such as pNPGlc, but cannot cleave oligosaccharides, such as cellotriose, cellotetraose, cellopentaose, cellohexaose and laminaribiose.4 These oligosaccharides nonetheless bind in the active site cleft and act as competitive inhibitors.
The X-ray crystal structures of BGlu1 E176Q acid-base mutant and BGlu1 E386G glycosynthase in complexes with cellotetraose and cellopentaose show that the sidechains of N245, S334 and Y341 interact with glucosyl residues in cellooligosaccharide binding subsites +2, +3 and +4.4,21 In complexes with cellotetraose and cellopentaose, the N245 sidechain forms direct hydrogen bonds to the glucosyl residue in the +2 subsite in BGlu1 E176Q and to the glucosyl residues in both the +1 and +2 subsites in BGlu1 E386G. The BGlu1 N245V mutant had higher Km values for hydrolysis of pNP-β-d-glucopyranoside (pNPGlc) and cellobiose than does wild type BGlu1,22 and it also had a 10-fold lower catalytic efficiency value (kcat/Km) than wild type for cellotriose, but only 3-fold lower for cellotetraose and cellopentaose.21 The Y341 residue interacts via a water-mediated hydrogen bond to the glycosidic oxygen between subsites +2 and +3 and aromatic-sugar stacking interactions at subsites +3 and +4 in the active sites of the BGlu1 E176Q acid-base mutant and BGlu1 E386G glycosynthase complexes with cellotetraose and cellopentaose.4,21,22 However, the BGlu1 Y341A variant had only slightly higher Km values and slightly lower kcat/Km values for cellotriose, cellotetraose and cellopentaose compared to wild type BGlu1. When the same mutation was made in the BGlu1 E386G glycosynthase, the E386G/Y341 variant synthesized shorter pNP-cellooligosaccarides compared to the original BGlu1 E386G glycosynthase. The structure of BGlu1 E386G/Y431A in complex with cellotetraose revealed that the Glc2, Glc3, and Glc4 residues of cellotetraose at subsites +1, +2, and +3 were rotated nearly 180° and moved across the active site groove compared to the structure of BGlu1 E386G with cellotetraose. The cellotetraose position in BGlu1 E386G/Y341A allowed the O6 of Glc4 to form direct hydrogen bonds to Q187. The alternative binding mode observed for cellotetraose in the structures of the BGlu1 E386G/Y341A glycosynthase variant was suggested to explain the fact that BGlu1 Y341A had similar hydrolysis activity to wild type BGlu1, but BGlu1 E386G/Y341A exhibited poorer synthesis of long oligosaccharides than the BGlu1 E386 glycosynthase.
To further investigate the roles of active site cleft residues of BGlu1 in cellooligosaccharide binding and hydrolysis and in glycosynthase production of long cellooligosaccharides, the glucose binding sites of the wild type BGlu1, BGlu1 E176Q acid-base mutant and BGlu1 E386G glycosynthase were disrupted by site-directed mutagenesis and the functional and structural effects of these mutations characterized.
Results
BGlu1 variants with the Q187A mutation
Hydrolysis
Based on the observed interactions between Q187 and cellotetraose in the BGlu1 E386G/Y431A complex with cellotetraose,21 BGlu1 variants with the mutations Q187A, Y341A/Q187A, and N245V/Q187A were generated to investigate how the loss of the Q187 side chain would affect oligosaccharide binding, hydrolysis and synthesis of cellooligosaccharides. As shown in Table1, the BGlu1 Q187A variant had similar Km values and slightly lower kcat/Km values for hydrolysis of cellotriose, cellotetraose and cellopentaose compared to wild type BGlu1. The Y341A/Q187A mutant had similar kcat/Km values for cellotriose and cellotetraose and a slightly lower kcat/Km value for cellopentaose compared to BGlu1 Y341A. Moreover, the N245V/Q187A variant had slightly lower kcat/Km values for cellotriose, cellotetraose, and cellopentaose than BGlu1 N245V. Kinetic subsite analysis by the method of Hiromi23 verified that the Q187A, Y341A/Q187A and N245V/Q187A mutations had little effect on the apparent affinity at the +3 and +4 subsites, as illustrated in Figure 1. The only significant change was that the Y341A/Q187A mutant gave lower subsite +4 affinity than BGlu1 Y341A, suggesting that the Q187 residue only affects binding at the +4 subsite when the phenolic group of Y341 is missing.
Table I.
Kinetic Parameters of BGlu1 Wild Type and its Variants for Hydrolysis of Cellotriose, Cellotetraose, and Cellopentaose
| Substrate | Kinetic parameters | BGlu121 | Y341A21 | N245V21 | Q187A | Y341A; Q187A | N245V; Q187A | R178A | W337A | Y341A; R178A | Y341A; W337A | Y341A; R178A; W337A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellotriose | Km (mM) | 0.50 ± 0.05 | 0.97 ± 0.07 | 11.2 ± 0.8 | 0.48 ± 0.04 | 0.75 ± 0.06 | 15.2 ± 0.9 | 1.91 ± 0.11 | 1.14 ± 0.07 | 6.4 ± 0.5 | 2.12 ± 0.16 | 12.0 ± 0.7 |
| kcat (s−1) | 15.6 ± 0.6 | 26.5 ± 0.7 | 36.1 ± 0.9 | 13.6 ± 0.4 | 19.9 ± 0.6 | 34.0 ± 0.8 | 19.0 ± 0.4 | 15.1 ± 0.3 | 34.8 ± 1.0 | 27.8 ± 0.7 | 44.0 ± 0.9 | |
| kcat/Km (s−1 M−1) | 31,000 | 27,000 | 3200 | 28,000 | 27,000 | 2200 | 9900 | 13,000 | 5400 | 13,000 | 3,700 | |
| Cellotetraose | Km (mM) | 0.37 ± 0.04 | 0.70 ± 0.05 | 5.3 ± 0.2 | 0.42 ± 0.03 | 0.57 ± 0.04 | 6.66 ± 0.4 | 1.03 ± 0.07 | 0.52 ± 0.04 | 3.87 ± 0.27 | 1.09 ± 0.05 | 11.1 ± 0.60 |
| kcat (s−1) | 23.7 ± 1.1 | 35.5 ± 1.0 | 103 ± 1.6 | 24.0 ± 1.0 | 29.5 ± 0.7 | 96.0 ± 2.5 | 43.5 ± 1.2 | 22.8 ± 0.6 | 37.1 ± 0.9 | 33.9 ± 0.6 | 63.0 ± 1.2 | |
| kcat/Km (s−1M−1) | 64,000 | 51,000 | 20,000 | 57,000 | 52,000 | 14,000 | 42,000 | 44,000 | 9600 | 31,000 | 5700 | |
| Cellopentaose | Km (mM) | 0.28 ± 0.02 | 0.44 ± 0.02 | 3.83 ± 0.18 | 0.28 ± 0.03 | 0.43 ± 0.03 | 4.5 ± 0.4 | 0.59 ± 0.03 | 0.26 ± 0.02 | 3.80 ± 0.34 | 1.18 ± 0.06 | 11.8 ± 0.4 |
| kcat (s−1) | 27.5 ± 1.2 | 32.3 ± 0.6 | 139 ± 3 | 25.8 ± 1.7 | 25.1 ± 0.6 | 118 ± 5 | 45.6 ± 0.9 | 21.9 ± 0.6 | 25.5 ± 1.1 | 31.1 ± 0.6 | 40 ± 3 | |
| kcat/Km (s−1M−1) | 98,000 | 73,000 | 36,000 | 92,000 | 58,000 | 26,000 | 77,000 | 84,000 | 6700 | 26,000 | 3400 |
Figure 1.
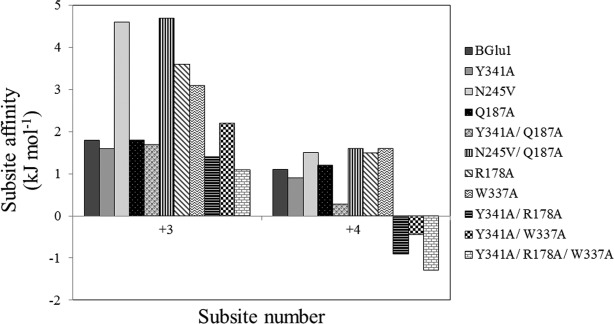
Subsite affinities for subsites +3 and +4 in BGlu1 and its mutants determined by kinetic subsite mapping. Subsite affinities were calculated by the method of Hiromi et al.23 as applied to cellooligosaccharides by Hrmova et al.24: Ai = −ΔG = RTln[(kcat/Km)n/(kcat/Km)n − 1] as a means of estimating the effect of each successive sugar residue.
Oligosaccharide binding by BGlu1 E176Q mutants
Previously, we used this rescue of the BGlu1 E176Q acid–base mutant for kinetic investigation of the binding of oligosaccharides to the BGlu1 E176Q active site by inhibition studies.4 Cellooligosaccharides are not hydrolyzed by the BGlu1 E176Q and its variants, but instead display competitive inhibition of the release of pNP from pNPGlc in the presence of azide. Although cellobiose displays mixed inhibition, the competitive inhibition constant, Ki, of 44 mM is much lower than the uncompetitive inhibition constant, Ki,u, of 649 mM, so the contribution from binding of the substrate to an enzyme-inhibitor complex is negligible and the competitive Ki serves as a reasonable estimate of the KD for cellobiose as well. Applying this method to the active site cleft mutations made in the BGlu1 E176Q background allowed us to calculate binding affinities for different length oligosaccharides.
First, we assessed the effects of the mutations on pNPGlc hydrolysis. The BGlu1 E176Q/Q187A, BGlu1 E176Q/Y341A and BGlu1 E176Q/Y341A/Q187A variants had slightly lower kcat/Km values than BGlu1 E176Q for pNPGlc hydrolysis (Table2). The BGlu1 E176Q/Q187A variant had similar apparent competitive inhibition constants (Ki) compared to the BGlu1 E176Q for cellobiose, cellotriose, cellotetraose, cellopentaose and cellohexaose (Table3). Moreover, the binding free energy changes for glucosyl residue addition (ΔGb,Cn→Cn+1, where n is the number of β-1,4-linked glucosyl residues in the cellooligosaccharide C) were similar to those for BGlu1 E176Q. The BGlu1 E176Q/Y341A variant had a slightly higher Ki value compared to BGlu1 E176Q for cellobiose, whereas, it had 5-, 11-, 46-, and 111-fold higher Ki values for cellotriose, cellotetraose, cellopentaose and cellohexaose, respectively, thereby verifying that Y341 is involved in binding longer oligosaccharides. The BGlu1 E176Q/Y341A/Q187A variant had similar Ki values to the BGlu1 E176Q/Y341A variant for cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose, and consequently, their binding energies were similar to those of BGlu1 E176Q/Y341A. However, the ΔGC4→C5, which was unfavorable for BGlu1 E176Q/Y341A (+1.7 kJ mol−1), was more unfavorable for BGlu1 E176Q/Y341A/Q187A (+3.0 kJ mol−1). Therefore, despite the observation of cellotetraose binding to Q187 in the crystal structure of the BGlu1 E386G/Y341A variant complex with cellotetraose,21 the Q187A mutation has little effect on cellooligosaccharide binding in the active sites of BGlu1 and its E176Q acid-base mutant.
Table II.
Kinetic Parameters of BGlu1 E176Q and its Variants for Hydrolysis of pNPGlc
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (s−1M−1) |
|---|---|---|---|
| E176Q | 0.188 ± 0.007 | 3.68 ± 0.06 | 20,000 |
| E176Q/Y341A | 0.225 ± 0.004 | 3.71 ± 0.03 | 17,000 |
| E176Q/Q187A | 0.188 ± 0.005 | 3.11 ± 0.03 | 17,000 |
| E176Q/Y341A/Q187A | 0.249 ± 0.005 | 3.86 ± 0.03 | 16,000 |
| E176Q/R178A | 0.108 ± 0.004 | 1.05 ± 0.01 | 9800 |
| E176Q/W337A | 0.144 ± 0.009 | 2.25 ± 0.04 | 17,000 |
| E176Q/Y341A/R178A | 0.135 ± 0.006 | 1.28 ± 0.02 | 9500 |
| E176Q/Y341A/W337A | 0.204 ± 0.012 | 3.03 ± 0.06 | 15,000 |
| E176Q/Y341A/R178A/W337A | 0.333 ± 0.006 | 1.98 ± 0.01 | 6000 |
Table III.
Inhibition of Cleavage of pNPGlc by BGlu1 E176Q and its Variants by Cellooligosaccharides
| BGlu1 E176Q4 | BGlu1 E176Q Y341A | BGlu1 E176Q Q187A | BGlu1 E176Q Y341A Q187A | BGlu1 E176Q R178A | BGlu1 E176Q; W337A | BGlu1 E176Q Y341A R178A | BGlu1 E176Q Y341A W337A | BGlu1 E176Q Y341A R178A; W337A | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Cellobiose | Ki (mM) | 44 ± 3 | 50 ± 1 | 36 ± 2.2 | 59 ± 0.8 | 61 ± 3 | 64 ± 4 | 116 ± 3 | 103 ± 3 | 118 ± 1 |
| KA (M−1)ab | 23 | 20 | 28 | 17 | 16 | 16 | 8.6 | 9.7 | 8.5 | |
| ΔGbinding (kJ mol−1)a | −7.9 | −7.5 | −8.4 | −7.1 | −7.0 | −7.0 | −5.4 | −5.7 | −5.4 | |
| Cellotriose | Ki (mM) | 0.48 ± 0.03 | 2.4 ± 0.09 | 1.4 ± 0.07 | 2.3 ± 0.04 | 6.7 ± 0.2 | 10 ± 0.1 | 14 ± 0.2 | 9.7 ± 0.14 | 24 ± 0.2 |
| KA (M−1) | 2100 | 420 | 710 | 430 | 150 | 100 | 71 | 100 | 42 | |
| ΔGbinding (kJ mol−1) | −19.3 | −15.2 | −16.5 | −15.3 | −12.6 | −11.6 | −10.7 | −11.6 | −9.4 | |
| ΔGbinding, C2→C3 (kJ mol−1) | −11.4 | −7.7 | −8.1 | −8.2 | −5.6 | −4.6 | −5.3 | −5.9 | −4.0 | |
| Cellotetraose | Ki (mM) | 0.052 ± 0.006 | 0.57 ± 0.03 | 0.081 ± 0.007 | 0.42 ± 0.05 | 1.1 ± 0.05 | 0.68 ± 0.03 | 9.2 ± 0.18 | 2.7 ± 0.02 | 13 ± 0.4 |
| KA (M−1) | 19,200 | 1800 | 12,300 | 2400 | 910 | 1500 | 110 | 370 | 77 | |
| ΔGbinding (kJ mol−1) | −24.8 | −18.9 | −23.7 | −19.5 | −17.2 | −18.4 | −11.8 | −14.9 | −10.9 | |
| ΔGbinding, C3→C4 (kJ mol−1) | −5.5 | −3.7 | −7.2 | −4.2 | −4.6 | −6.8 | −1.1 | −3.3 | −1.5 | |
| Cellopentaose | Ki (mM) | 0.024 ± 0.002 | 1.1 ± 0.03 | 0.030 ± 0.002 | 1.4 ± 0.04 | 0.61 ± 0.04 | 0.31 ± 0.02 | 11 ± 0.1 | 4.8 ± 0.08 | 18 ± 0.4 |
| KA (M−1) | 41,700 | 910 | 33,300 | 710 | 1600 | 3200 | 91 | 210 | 56 | |
| ΔGbinding (kJ mol−1) | −26.8 | −17.2 | −26.2 | −16.5 | −18.6 | −20.3 | −11.4 | −13.5 | −10.1 | |
| ΔGbinding, C4→C5 (kJ mol−1) | −2 | 1.7 | −2.5 | 3.0 | −1.4 | −1.9 | 0.4 | 1.4 | −0.8 | |
| Cellohexaose | Ki (mM) | 0.018 ± 0.001 | 2.0 ± 0.01 | 0.037 ± 0.003 | 3.0 ± 0.08 | 0.61 ± 0.03 | 0.38 ± 0.02 | 11 ± 0.5 | 11 ± 0.2 | 11 ± 0.2 |
| KA (M−1) | 55,600 | 500 | 27,000 | 330 | 1600 | 2600 | 91 | 91 | 91 | |
| ΔGbinding (kJ mol−1) | −27.5 | −15.7 | −25.7 | −14.6 | −18.6 | −19.8 | −11.4 | −11.4 | −11.4 | |
| ΔGbinding, C5→C6 (kJ mol−1) | −0.7 | 1.5 | 0.5 | 1.9 | 0 | 0.5 | 0 | 2.1 | −1.3 |
The apparent KA and ΔGbinding are calculated based on the assumption of purely competitive inhibition. The equations used to calculate these values were: KA = 1/KD = 1/Ki; ΔGbinding = −RTln KA, where R is the gas constant (8.314 J K−1 mol−1) and T is the absolute temperature (303 K). ΔGCn-1→Cn = ΔGbinding, n − ΔGbinding, n-1. These are equivalent to subsite binding affinities: Ai = − ΔGCn-1→Cn, i = n − 1, when the cellooligosaccharides are binding nearly exclusively with the nonreducing residue in the −1 subsite and length n and n − 1 cellooligosaccharides bind in the same tract (utilize the same subsites other than subsite i).
Since cellobiose gave mixed inhibition, the KA is not strictly the inverse of Ki. However, since the uncompetitive inhibition constants were much larger than the competitive constants, they should provide reasonable estimates.
Transglucosylation
Previously, the BGlu1 E386G/Y341A and BGlu1 E386G/N245V glycosynthases were shown to synthesize shorter pNP-cellooligosaccharides than BGlu1 E386G glycosynthase.21 BGlu1 E386G/Q187A was found to have slightly higher synthesis activity, producing approximately twofold as much pNP-cellooligosacchairdes with DP of 7–9, compared to the BGlu1 E386G glycosynthase (Table4). In contrast, BGlu1 E386G/Y341A/Q187A produced 2 to 7-fold less DP 3–5 pNP-cellooligosaccharides than BGlu1 E386G/Y341A, and no detectable DP6 product, which was clearly detectable for BGlu1 E386G/Y341A. Thus, the Q187A mutation had differential effects on cellooligosaccharide synthesis activity of BGlu1 E386G glycosynthase, which depended on the presence of Y341.
Table IV.
Products of Glycosynthase Reactions of BGlu1 E386G Glycosynthase and its Variants with Equal Amounts of α-glucosyl Fluoride Donor and pNPC2 Acceptor
| Glycosynthase | Percent of pNP in each oligo-glycoside producta | |||||||
|---|---|---|---|---|---|---|---|---|
| DP 2 | DP 3 | DP 4 | DP 5 | DP 6 | DP 7 | DP 8 | DP 9 | |
| E386G | 56.7 ± 0.07 | 26.8 ± 0.03 | 9.8 ± 0.04 | 4.75 ± 0.014 | 1.52 ± 0.005 | 0.37 ± 0.005 | 0.07 ± 0.001 | 0.009 ± 0.0001 |
| E386G/Y341A | 70.6 ± 0.32 | 24.1 ± 0.26 | 4.94 ± 0.06 | 0.35 ± 0.009 | 0.021 ± 0.0017 | — | — | — |
| E386G/Q187A | 49.0 ± 0.07 | 28.2 ± 0.10 | 12.8 ± 0.02 | 6.70 ± 0.016 | 2.42 ± 0.02 | 0.67 ± 0.006 | 0.14 ± 0.002 | 0.022 ± 0.0004 |
| E386G/Y341A/Q187A | 89.0 ± 0.24 | 10.3 ± 0.22 | 0.68 ± 0.025 | 0.009 ± 0.0004 | — | — | — | — |
| E386G/R178A | 98.1 ± 0.01 | 1.91 ± 0.01 | 0.009 ± 0.0001 | — | — | — | — | — |
| E386G/W337A | 83.1 ± 0.22 | 14.8 ± 0.23 | 1.51 ± 0.036 | 0.09 ± 0.004 | 0.005 ± 0.0001 | — | — | — |
| E386G/Y341A/R178A | 98.6 ± 0.01 | 1.38 ± 0.01 | — | — | — | — | — | — |
| E386G/Y341A/W337A | 96.3 ± 0.12 | 3.59 ± 0.12 | 0.066 ± 0.0007 | — | — | — | — | — |
| E386G/Y341A/R178A/W337A | 96.8 ± 0.24 | 3.21 ± 0.24 | — | — | — | — | — | — |
Reactions in 150 mM ammonium bicarbonate buffer, pH 7.0, 30°C for 24 h, at a 1:1 molar ratio of α-GlcF donor: pNPC2 acceptor, and 40 μM enzyme concentration. The relative percentages are in terms of peak area per total 300 nm absorbance in pNP-glycoside products and substrate peaks separated by HPLC on a ZORBAX carbohydrate column. The molecular mass [Mass + 35Cl]- of each eluted compound was confirmed by mass spectrometry: pNPC2 (498 m/z), pNPC3 (660 m/z), pNPC4 (822 m/z), pNPC5 (984 m/z), pNPC6 (1146 m/z), pNPC7 (1308 m/z), pNPC8 (1471 m/z), and pNPC9 (1633 m/z).
Structures of BGlu1 Y341A variants with cellotetraose
Despite the previous observation that cellotetraose forms hydrogen bonds to Q187 in the E386G/Y341A variant,21 mutation of Q187 to A had only minor effects on hydrolysis by BGlu1/Y341A and on BGlu1 E386G glycosynthase activity. To elucidate the structural basis for these results, BGlu1 E176Q/Y341A, BGlu1 E176Q/Y341A/Q187A and E386G/Y341A/Q187A were crystallized, cellotetraose was soaked into these crystals, and the structures were determined.
The X-ray diffraction statistics are shown in Table5. The observed electron densities, shown in Figure 2, indicated that cellotetraose bound with the nonreducing residue in the −1 subsite as a hydrolysis substrate or transglycosylation product, as previously seen for the E176Q acid-base mutant4 and E386G glycosynthase.21 To obtain sufficient ligand occupancies in the active site, the crystals were soaked in cryoprotectant saturated with the cellotetraose, thus an extra cellotetraose was bound on the surface of the protein, as was previously found in the structures of cellotetraose complexes with BGlu1 E386G, BGlu1 E386G/S334A and BGlu1 E386G/Y341A.21 Aside from the mutated residues, the amino acid residues in the active sites of the BGlu1 E176Q/Y341A and BGlu1 E176Q/Y341A/Q187A acid-base variants and BGlu1 E386G/Y341A/Q187A glycosynthase variant were similar to those in the active sites of BGlu1 E176Q and BGlu1 E386G in complex with cellotetraose (Fig. 3). The Glc2, Glc3, and Glc4 residues at subsites +1, +2, and +3 in the structure of the BGlu1 E386G/Y341A/Q187A glycosynthase variant were rotated nearly 180° compared to the structure of BGlu1 E386G with cellotetraose, and the Glc3 and Glc4 residues in the active site of the BGlu1 E386G/Y341A/Q187A variant moved to positions similar to their positions in the complex of cellotetraose with BGlu1 E386G/Y341A glycosynthase [Fig. 3(A)].21 Hydrogen bonding interactions between the glucosyl residues of cellotetraose, ordered water molecules and amino acid residues in the active site of BGlu1 E386G/Y341A/Q187A are shown in Figure 4(A). The hydrogen bonds and aromatic-sugar stacking interactions of the Glc2, Glc3, and Glc4 residues at subsites +1, +2, and +3 are similar to those in the BGlu1 E386G/Y341A complex with cellotetraose. A new ordered water molecule hydrogen bonded to O6 of Glc4 in place of the Q187 sidechain [Figs. 3(A) and 4(A)].
Table V.
Data Collection and Refinement Statistics
| PDB code | E386G/Y341A/Q187A +C4; 4QLJ | E176Q/Y341A + C4; 4QLK | E176Q/Y341A/Q187A + C4; 4QLL |
|---|---|---|---|
| Beamline | BL13B1 | BL13B1 | BL13B1 |
| Wavelength (Å) | 1 | 1 | 1 |
| Space group | P212121 | P21 | P212121 |
| Unit cell parameters (Å, ˚) | a = 79.5 | a = 63.7 | a = 79.6 |
| b = 101.2 | b = 79.5 | b = 101.3 | |
| c = 127.3 | c = 102.0 | c = 127.4 | |
| α = β = γ = 90˚ | α = γ = 90˚; β =98˚ | α = β = γ = 90˚ | |
| No. of molecules per ASU | 2 | 2 | 2 |
| Resolution range (Å) | 30–1.95 (2.02–1.95) | 30-1.83 (1.90–1.83) | 30–1.85 (1.92–1.85) |
| Completeness (%) | 100.0 (100.0) | 98.7 (92.8) | 97.5 (96.4) |
| I/σ(I) | 18.3 (4.4) | 13.7 (2.0) | 25.5 (4.9) |
| Rsym (%)a | 10.3 (45.6) | 8.4 (46.5) | 7.7 (41.6) |
| Rfactor (%)b | 17.5 | 15.3 | 16.2 |
| Rfree (%)c | 20.2 | 16.9 | 18.5 |
| No. of amino-acid residues | 944 | 944 | 944 |
| No. of protein atoms | 7586 | 7604 | 7596 |
| No. of water molecules | 835 | 839 | 839 |
| Refined carbohydrate | Cellotetraose | Cellotetraose | Cellotetraose |
| No. of carbohydrate atoms | 135 | 135 | 135 |
| No. of other hetero atoms | 35 | 35 | 35 |
| Ramachandran statisticsd | |||
| Most favored regions (%) | 89.1 | 89.3 | 90.0 |
| R.m.s.d. bond distances (Å) | 0.010 | 0.008 | 0.008 |
| R.m.s.d. bond angle (˚) | 1.28 | 1.28 | 1.31 |
| Mean B factors (Å2) | |||
| All protein atoms | 16.17 | 18.12 | 16.62 |
| Waters | 29.12 | 31.52 | 29.65 |
| Hetero atoms | 32.42 | 36.13 | 31.31 |
| Carbohydrate atoms | 36.78 | 39.74 | 33.05 |
| Subsite −1 (A/B) | 25.71/25.72 | 31.77/30.55 | 25.57/24.73 |
| Subsite +1 (A/B) | 26.97/27.81 | 34.36/33.89 | 29.69/31.15 |
| Subsite +2 (A/B) | 30.29/31.44 | 33.75/35.55 | 29.76/30.29 |
| Subsite +3 (A/B) | 42.05/40.81 | 34.20/37.59 | 30.97/32.63 |
Numbers in parentheses are outer shell parameters.
Rsym= Σhkl Σi |Ii(hkl) − <I(hkl)>|/ Σhkl Σi I(hkl).
Rfactor=(Σ|Fo| − |Fc|/Σ|Fo|).
Based on 5% of the unique observations not included in the refinement.
Ramachandran values were determined from PROCHECK.25
Figure 2.
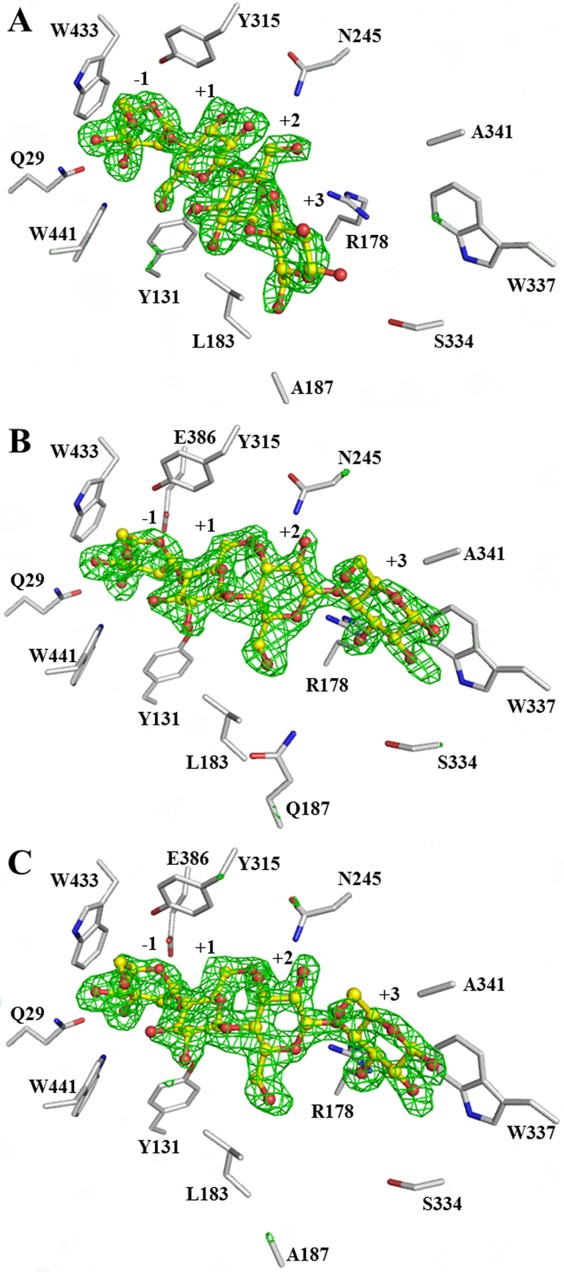
The Fo − Fc electron density maps for cellotetraose in the active sites of BGlu1 E386G/Y341A/Q187A (A), BGlu1 E176Q/Y341A (B), and BGlu1 E176Q/Y341A/Q187A (C) calculated with the ligand omitted. The omit maps are shown in green mesh at +3σ. The cellotetraose from the final structures are shown in ball and stick representation with carbons colored in yellow and oxygen atoms in red. The sidechains of amino acid residues in the active site are shown with gray carbons. Use this link to access the interactive version of this figure
Figure 3.
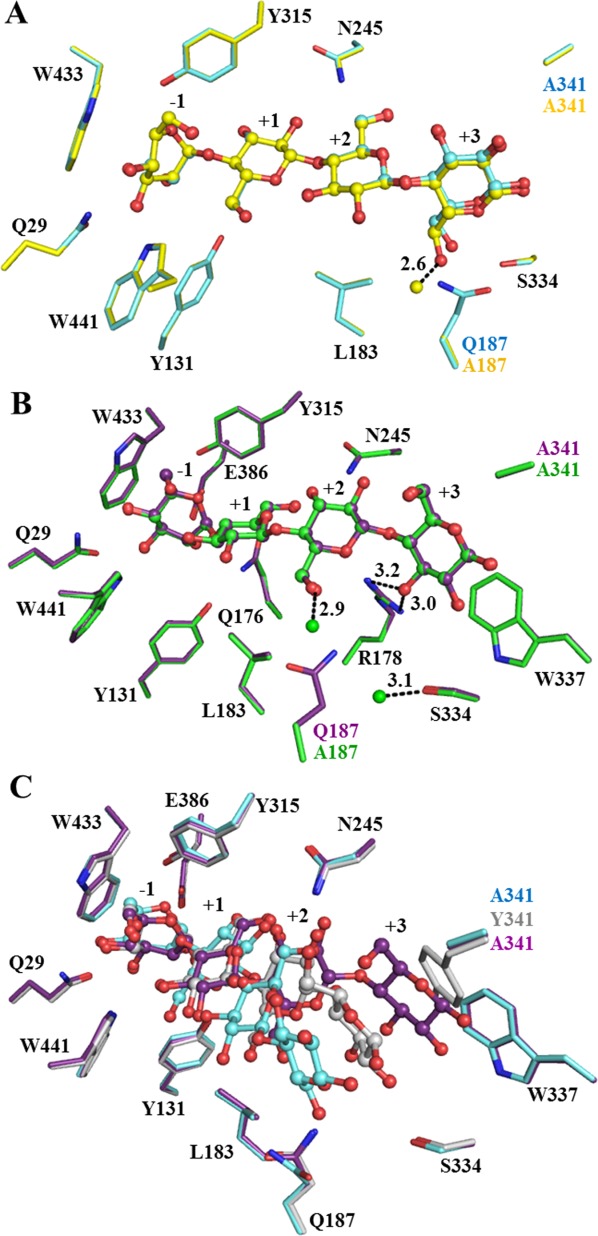
Cellotetraose binding in the active site of BGlu1 E386G/Y341A/Q187A, BGlu1 E176Q/Y341A and BGlu1 E176Q/Y341A/Q187A. (A) Comparison of cellotetraose binding in active sites of BGlu1 E386G/Y341A (cyan carbons) and BGlu1 E386G/Y341A/Q187A (yellow carbons). The fixed water molecule seen in E386G/Y341A/Q187A, but not in E386G/Y341A is shown as a yellow ball. (B) Cellotetraose binding in the active sites of BGlu1 E176Q/Y341A (violet carbons) and BGlu1 E176Q/Y341A/Q187A (green carbons), which has new fixed water molecules that are shown as green balls. In both BGlu1 E176Q/Y341A and E176Q/Y341A/Q187A, O3 of Glc4 forms direct hydrogen bonds with R178 (shown as dashed lines) and Glc4 was stacked upon W337. (C) Cellotetraose in the active sites of BGlu1 E176Q (gray carbons), BGlu1 E386G/Y341A (cyan carbons) and BGlu1 E176Q/Y341A (violet carbons). The cellotetraose is represented as balls and sticks.
Figure 4.
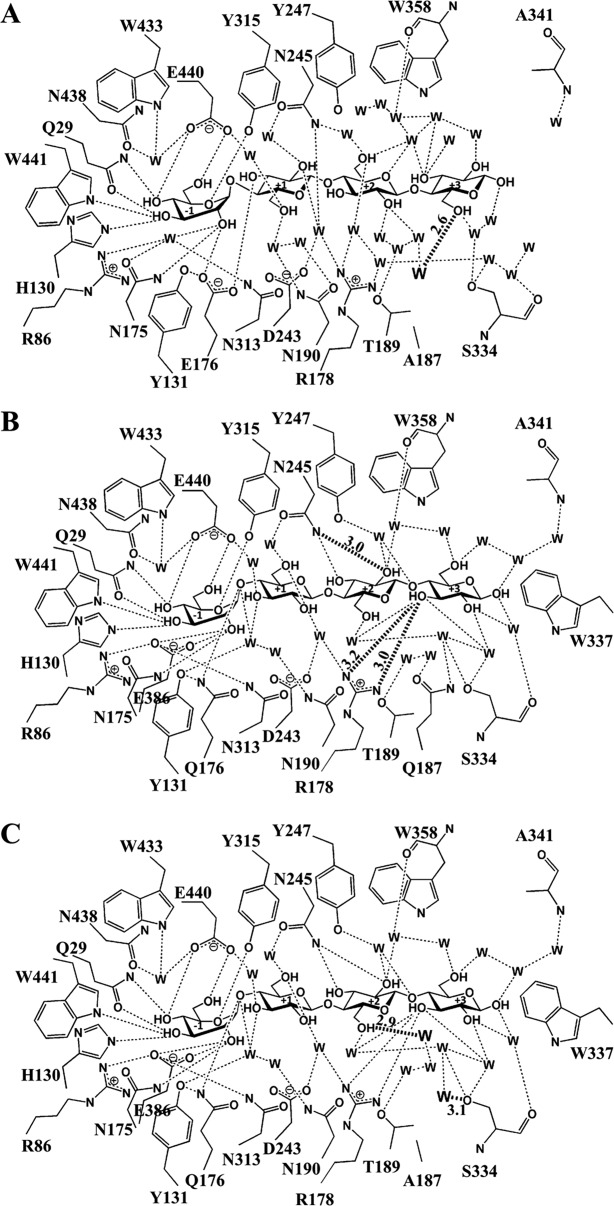
Hydrogen bonds between the glucose residues of cellotetraose, surrounding amino acid residues and the network of water molecules (W) in the active sites of BGlu1 E386G/Y341A/Q187A (A), BGlu1 E176Q/Y341A (B) and BGlu1 E176Q/Y341A/Q187A (C). Hydrogen bonds with measured distances of 3.2 Å or less are shown as dashed lines to indicate the hydrophilic interactions. The diagram in A shows the flip of the glucosyl residues in subsites +1 to +3 that results in a completely different network than to those in the active sites of the BGlu1 E386G and E176Q complexes with cellotetraose. The hydrogen bond between O6 of Glc4 and a water found in the place of the Q187 amide (W) is shown as a darkened dashed line. The diagram in B shows the direct hydrogen bonds with R178 and hydrogen bond from N245 that are shorter than those in the BGlu1 E176Q complex with cellotetraose as darkened dashed lines with their distances in Å marked on the lines. The diagram of BGlu1 E176Q/Y341A/Q187A in C shows similar hydrogen bonds as in B, except for those of the two waters bound in place of Q187 with O6 of Glc3 and S334 Oγ, which are shown as darkened dashed lines.
In contrast, the Glc2, Glc3 and Glc4 residues at subsites +1, +2, and +3 in the structures of the BGlu1 E176Q/Y341A and BGlu1 E176Q/Y341A/Q187A acid-base mutant variants were found in positions that were different from both those in the cellotetraose complexes with BGlu1 E176Q4 and BGlu1 E386G,21 and the BGlu1 E386G/Y341A variants [Fig. 3(B,C)]. In this new position, the O3 of Glc4 directly hydrogen bonds to R178 (3.0 Å) and Glc4 stacks onto the indole ring of W337 (3.9 Å). Moreover, the shift in the positions of the oligosaccharide glucosyl moieties allowed the N245 Nδ to form hydrogen bonds with Glc3 O2 [Fig. 4(B)]. The positions of the amino acid residues and cellotetraose in the active sites of BGlu1 E176Q/Y341A and BGlu1 E176Q/Y341A/Q187A were similar and the Q187 amide was replaced by two water molecules that formed hydrogen bonds to Glc3 O6 and S334 Oγ in the latter [Figs. 3(B) and 4(C)].
The BGlu1 variant active site cleft structures indicate that the position of binding of cellotetraose in the presence of the Y341A mutation is dependent on the nature of the inactivating mutation at the −1 subsite, despite the fact that the positions of cellotetraose in its complexes with BGlu1 E176Q and E386G are quite similar. The only significant difference noted in the BGlu1 E176Q and E386G complexes was that N245 moved to interact with Glc2 and Glc3 in BGlu1 E386G, whereas N245 remains in a similar position as in the native structure in the BGlu1 E176Q cellotetraose complex and only interacts with Glc3.21
Effects of R178 and W337 mutations on oligosaccharide binding with and without Y341
Hydrolysis
On the basis of the observed interactions of R178 and W337 with cellotetraose in the BGlu1 E176Q/Y341A and BGlu1 E176Q/Y341A/Q187A structures (Figs. 3 and 4), we hypothesized that these residues should play roles in the binding of cellotetraose to BGlu1 and BGlu1 E176Q in the absence of the Y341 phenolic group. To test this hypothesis, BGlu1 variants with the mutations R178A, W337A, Y341A/R178A, Y341A/W337A and Y341A/R178A/W337A were characterized. Table1 shows that the BGlu1 R178A variant had a nearly fourfold higher Km value and threefold lower kcat/Km value for hydrolysis of cellotriose compared to wild type BGlu1, with smaller effects on cellotetraose, and cellopentaose hydrolysis. The BGlu1 Y341A/R178A variant had 5-, 5-, and 11-fold lower kcat/Km values than the BGlu1 Y341A variant for cellotriose, cellotetraose, and cellopentaose, respectively. The W337A variant was similar to the R178A variant, with a 2.4-fold lower kcat/Km value for cellotriose and slightly lower kcat/Km values for cellotetraose and cellopentaose compared to wild type BGlu1, while the Y341A/W337A variant had 2- to 3-fold lower kcat/Km values than BGlu1 Y341A for cellotriose, cellotetraose, and cellopentaose. The BGlu1 Y341A/R178A/W337A kcat/Km values for cellotriose, cellotetraose, and cellopentaose were about 7-, 9-, and 21-fold lower than those of BGlu1 Y341A variant, and it had about twofold lower kcat/Km values than those of BGlu1 Y341A/R178A. The results suggested that R178 and W337 play important role in cellooligosaccharide hydrolysis in the absence of Y341.
The kinetic subsite analysis displayed in Figure 1 verified that the W337A and R178A mutations had significant effects on the apparent affinities at the +3 and +4 subsites, giving higher apparent subsite +3 and +4 affinities than wild type BGlu1. In contrast, the BGlu1 Y341A/R178A, BGlu1 Y341A/W337A and BGlu1 Y341A/R178A/W337A variants had lower apparent subsite +4 affinities than BGlu1 Y341A.
Oligosaccharide binding by BGlu1 E176Q variants
The BGlu1 E176Q/W337A and BGlu1 E176Q/Y341A/W337A variants had approximately 10% lower kcat/Km values than BGlu1 E176Q and BGlu1 E176Q/Y341A for pNPGlc hydrolysis, while the kcat/Km values were twofold lower for BGlu1 E176Q/R178A and BGlu1 E176Q/Y341A/R178A, and threefold lower for BGlu1 E176Q/Y341A/R178A/W337A (Table2). Thus, these mutations could affect even short substrates that bind out to the +1 to +2 subsite, as also seen in the R178A mutation effects on cellotriose hydrolysis.
BGlu1 E176Q/R178A had 1.4-, 14-, 21-, 25-, and 34-fold higher Ki values than BGlu1 E176Q for cellobiose, cellotriose, cellotetraose, cellopentaose and cellohexaose, whereas BGlu1 E176Q/Y341A/R178A had 2-, 6-, 16-, 10-, and 6-fold higher Ki values than BGlu1 E176Q/Y341A, respectively. Based on these inhibition constants, the binding energy increments for addition of glucosyl residues 3 to 6 in BGlu1 E176Q/R178A and E176Q/R178A/Y341A were all less favorable than in BGlu1 E176Q and BGlu1 E176Q/Y341A. The BGlu1 E176Q/W337A variant had 1.5-, 21-, 13-, 13-, and 21-fold higher Ki values than BGlu1 E176Q for cellobiose, cellotriose, cellotetraose, cellopentaose and cellohexaose, respectively, indicating less favorable binding energies. The BGlu1 E176Q/Y341A/W337A variant had 2-, 4-, 5-, 4-, and 6-fold higher Ki values than BGlu1 E176Q/Y341A for cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose, respectively, and correspondingly less favorable binding energies than BGlu1 E176Q/Y341A. Thus, the effects of both the R178A and W337A mutations on cellooligosaccharide binding appeared stronger in the absence of the Y341A mutation, and were stronger than those of the Q187A mutation in either the presence or absence of the Y341A mutation. BGlu1 E176Q/Y341A/W337A had lower Ki values, and correspondingly higher binding energies, than those of BGlu1 E176Q/Y341A/R178A for all oligosaccharides, except cellohexaose, although their overall effects were similar. The BGlu1 E176Q/Y341A/R178A/W337A variant had higher Ki values and less favorable binding energies than the BGlu1 E176Q/Y341A/R178A and BGlu1 E176Q/Y341A/W337A variants for cellotriose, cellotetraose and cellopentaose, but these effects were small in comparison to either mutation alone. Thus, mutations of the R178 and W337 residues in active site of BGlu1 affected binding of all lengths of cellooligosaccharide both in the presence and absence of Y341, and both residues have stronger influence than Q187.
Transglucosylation
Because the R178A and W337A mutations were in the oligosaccharide binding site and had effects on cellooligosaccharide hydrolysis, the same mutations of E386G glycosynthase were tested to see whether they affected the production of long oligosaccharides in transglycosylation reactions with α-GlcF donor and pNPC2 acceptor. The synthesis activities of the E386G glycosynthase26 and E386G glycosynthase variants containing the mutations R178A, W337A, Y341A/R178A, Y341A/W337A, and Y341A/R178A/W337A were compared. BGlu1 E386G/R178A and BGlu1 E386G/W337A were found to have lower synthesis activity and synthesize shorter cellooligosaccharides compared to the BGlu1 E386G glycosynthase (Table4). The R178A variant could synthesize cellooligosaccharides up to DP 4 (pNPC4), whereas, the variant with the W337A mutation could synthesize cellooligosaccharides up to DP 6 (pNPC6). BGlu1 E386G/Y341A/R178A, BGlu1 E386G/Y341A/W337A and BGlu1 E386G/Y341A/R178A/W337A had lower synthesis activity compared to BGlu1 E386G/Y341A and synthesized shorter pNP-cellooligosaccharides up to only DP 3 for the E386G/Y341A/R178A and E386G/Y341A/R178A/W337A variants and DP 4 for the E386G/Y341A/W337A variant. Therefore, the R178A and W337A mutations had significant effects on synthesis activity of BGlu1 E386G glycosynthase.
Discussion and Conclusion
Assessment of cellooligosaccharide binding
In this article, we have assessed the cellooligosaccharide binding in BGlu1 variants by X-ray crystallography and two different kinetic methods, the kinetic subsite analysis developed by Hiromi and colleagues23,27 and inhibition of pNPGlc cleavage by cellooligosaccharides in BGlu1 E176Q acid-base mutant variants.4 Although these methods generally show similar patterns of free energy changes upon addition of glucosyl residues, the values are not the same and there are significant differences in the effects of the mutations for the outer subsites. Both the methods assume that all cellooligosaccharide binding modes in the same tract (defined here as a set of binding modes that can be obtained by moving the nonreducing end glucosyl residue from subsite −1 to outer subsites while maintaining the glucosyl residue positions in the occupied subsites), with simply an extension on the reducing end as each glucosyl residue is added (illustrated in Supporting Information Fig. S1). This assumption may not be completely valid given the three binding modes of cellotetraose observed in the BGlu1 E176Q, E386G, E176Q/Y341A, and E386G/Y341A variants by X-ray crystallography, which fell in three different tracts.
The Hiromi kinetic subsite computation method has the additional assumption of an intrinsic kcat for hydrolysis,23,27 which may not be valid due to a slight shift in the hydrolyzed bond when the cellooligosaccharide moves from one tract to another. The structural subsites and kinetics based subsites do not strictly coincide, since there the alternative binding modes in the different tracts (with alternative subsites) and multiple productive binding interactions may contribute to the observed hydrolysis or inhibition kinetics. However, as long as one binding mode is predominant, they should correlate to each other. The advantage of the kinetic subsite analysis is that, as long as the productive state for all lengths of oligosaccharides have the same intrinsic kcat, the contribution of the nonproductive binding modes will be equal for the kcat and Km and the kcat/Km will be equivalent to that for binding only in the productive mode.23
In contrast, in cellooligosaccharide inhibition the difference in binding free energy between oligosaccharides of length n and n-1 only approximates the subsite affinity when each oligosaccharide binds with its nonreducing end in the −1 subsite (equivalent to the productive mode for hydrolysis) to the extent that other binding modes can be ignored. In general, the observed association constant for binding is the sum of the association constants for all binding modes:
| 1 |
where KAn,i is the association constant of a cellooligosaccharide of length n with its nonreducing end glucosyl residue in subsite i, and
| 2 |
where the free energy of binding is the sum of the loss of mixing energy, ΔGmix (entropy of mixing with water), and the free energy of binding of each subsite23,27:
| 3 |
Subtracting the binding energy of oligosaccharide with a DP of n-1 from that of the oligosaccharide with a DP of n provides an assessment of the contribution of the nth glucosyl residue to binding, ΔGb,Cn-1→Cn, which is a measure of binding energy at subsite n-1, if the predominant binding mode is that with the nonreducing end glucosyl residue in the −1 subsite (i.e., KAn,−1 is much greater than the sum of all other binding modes, so KA = KAn,−1). The fractional occupancy of each subsite can be calculated as:
| 4 |
By Hiromi subsite analysis,23 Opassiri et al.19 calculated subsite affinities for the wild type BGlu1 of 13.5 kJ mol−1 for subsite −1 and −0.7, 16.7, 2.2, 0.4, and 1.9 kJ mol−1 for subsites +1 to +5, respectively. By adding the subsite affinities for each binding mode and calculating the KA values from KA = e−ΔG/RT, it can be calculated from Eq. (4) that cellobiose should be predominantly span subsites +2 to +3 (71%), while all longer cellooligosaccharides should bind with the nonreducing end in the −1 subsite (97.6% in this “productive” mode for cellotriose). Therefore, for wild type BGlu1 cellooligosaccharide inhibition could not readily be used to measure subsite +2 affinity, but could provide a means to estimate affinities at subsites +3 to +5. The E176Q mutation may modulate these subsite affinities slightly and other mutations that change hydrolysis and binding of cellooligosaccharides, like N245V, R187A, and W337A, may also have significant effects. Nonetheless, the electron densities observed in all crystal structures of BGlu1 variants with cellotetraose and cellopentaose were consistent with the cellooligosaccharides binding nearly exclusively with their nonreducing ends in the −1 subsite.4,21 This discussion above implies that ΔGb,C4→C5 should be a good estimate of subsite +4, but when considering the effects of active site cleft mutations, the effects of different binding modes must be considered, as discussed below.
Assessment of the roles of the active site cleft residues in hydrolysis and transglycosylation
Despite our previous observation that cellotetraose moved to hydrogen bond to Q187 in the E386G/Y341A variant of rice BGlu1,21 no effects were seen on cellooligosaccharide binding and hydrolysis in BGlu1 and BGlu1 E176Q when the Q187A mutation was made in these enzymes, except for the binding of cellopentaose and cellohexaose in the presence of the Y341A mutation. In contrast, the Q187 mutation caused a significant increase in glycosynthase activity when introduced to the E386G glycosynthase alone, but a significant decrease when combined with Y341A in the E386G/Q187A/Y341A variant. The differential effects of the Q178A mutation may indicate that Q187 stabilizes an oligosaccharide binding mode that competes with the mode with binding of the oligosaccharides upon Y341, which is optimal for cellooligosaccharide elongation. In the absence of Y341, the binding mode stabilized by Q187 could help compensate for its loss.
The crystal structure of E176Q/Y341A with cellotetraose provided an explanation for the lack of effect of the Q187A mutation on oligosaccharide binding and hydrolysis, since the oligosaccharide had moved to interact more closely with R178 and W337, rather than interacting with Q187. This difference in cellotetraose binding was in contrast to the similarity of the cellotetraose complexes with the BGlu1 E176Q and E386G variants, in which the only obvious difference was that N245 stayed in a similar position as in the native wild type structure in BGlu1 E176Q to interact with Glc3 in the +2 subsite,4 while it swung inward to interact with Glc2 in the +1 subsite, as well as Glc3, in BGlu1 E386G.21 The shift in N245 in BGlu1 E386G was accompanied by a very small shift in the cellooligosaccharide position that was considered insignificant until the differential effects of adding the Y341A mutation on cellotetraose binding to the acid-base and nucleophile mutants were observed in the current work. This difference corresponds to the differences in the sensitivity of the BGlu1 hydrolase, BGlu1 E176Q transglucosidase and BGlu1 E386G glycosynthase activities to the active site cleft mutations. Thus, when investigating active site cleft binding, different inactivating mutations cannot be considered equivalent, since each has its own effect on substrate (in this case oligosaccharide) binding, however subtle these differences may appear at first glance.
Although the Q187A mutation for the most part only affected glycosynthase activity, the R178A and W337A mutations had significant effects on the activities of BGlu1, BGlu1 E176Q, and BGlu1 E386G, either alone or in combination with the Y341A mutation. R178 extends into the active site and makes water mediated hydrogen bonds with the +2 and +3 subsites in the BGlu1 E176Q and E386G complexes with cellooligosaccharides,4,21 which may explain why mutating R178 would affect binding of cellooligosaccharides of 3 or more glucosyl residues. The fact that the R178A mutation even affected pNPGlc hydrolysis and cellobiose binding suggests it has effects on the water network all the way to the +1 subsite. The extensive effect of the W337A mutation is more difficult to rationalize, as it forms a hydrophobic surface over the active site wall between Y341 and R178, which would have little interaction with cellooligosaccharides in the presence of Y341 (Fig. 5). However, it may affect the positions of R178 and Y341 and the hydrophobic surface may affect the active site water matrix structure as well. It has previously been noted that residues outside the active site may affect GH1 glycosidase specificity via amino acid interaction networks to active site residues,28 but in this case an interaction through the active site cleft water network seems more plausible.
Figure 5.
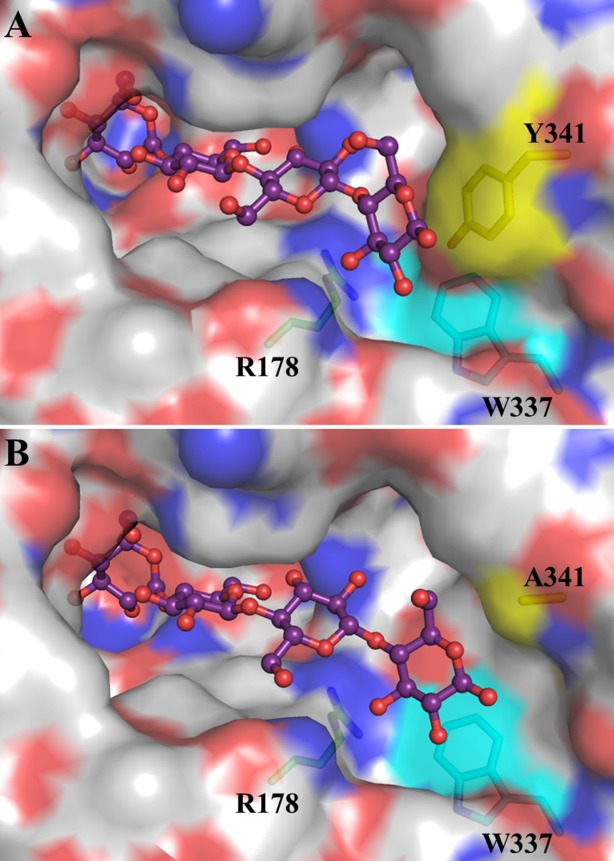
Comparison active site cleft of BGlu1 E386G (A) and BGlu1 E176Q/Y341A (B) in complex with cellotetraose. The sidechains of R178 (green carbons), W337 (cyan carbons) and Y341 (yellow carbons) residues noted to have significant effects on the activities of BGlu1, BGlu1 E176Q and BGlu1 E386G are shown in stick representation behind the active site surface. The cellotetraose is represented as balls and sticks. The surface representation was created in Pymol (Schrödinger LLC) and colored with the color of the underlying atoms, where carbon is grey in non-highlighted amino acid residues, oxygen is red, and nitrogen is blue.
The effects of the Y341A, R178A, and W337A mutations were nonadditive, consistent with the R178A and W337A acting at the same point of catalysis, that is, in binding the same cellooligosaccharide glucosyl residues, and sitting next to one another in the active site cleft wall, as shown in Figure 5.29 In the kinetic subsite analysis, the R178A and W337A mutations resulted in increases in the apparent affinity at subsites +3 and +4 (Fig. 1). These increased subsite affinities appear to result from a compensation at outer subsites, since these mutations cause decreased hydrolysis of shorter substrates, as seen by the 3.5-fold decrease in the kcat/Km for pNPGlc hydrolysis by BGlu1 E176Q/R178A compared to BGlu1 E176Q. A similar phenomenon is seen with the BGlu1 N245V, which has a nearly 10-fold decrease in the kcat/Km for cellotriose, but only approximately threefold decreases for cellotetraose and cellopentaose (Table1 and Ref.21). A plausible explanation for these observations is that these mutations destabilize the binding of the substrates in the most productive binding mode (i.e., the binding mode with the nonreducing glucosyl residue in subsite −1 in the tract with the glycosidic bond position that leads to the highest intrinsic kcat). Once the cellooligosaccharides are long enough to stack onto the Y341 phenol ring, this stacking stabilized the binding in the productive position, resulting in a jump in the kcat/Km that translates to a high apparent subsite affinity. The Y341A mutation suppresses the high apparent subsite +3 and +4 affinities because stabilization of the product position by Y341 is lost, consistent with the binding mode observed for cellotetraose in the BGlu1 E176Q/Y341A and E386G/Y341A variants.
In contrast, the subsite affinities calculated from cellooligosaccharide inhibition of pNPGlc cleavage by the BGlu1 E176Q acid/base mutant variants were all similar to or weaker than those for BGlu1 E176Q, except that ΔGC3→C4 of the E176Q/Q187A variant was −7.2 kJ mol−1 and that of the E176Q/W337A variant was −6.8 kJ mol−1 compared to −5.5 kJ mol−1 for BGlu1 E176Q. This lower sensitivity to the postulated tract change may be due to the fact that any binding position will affect the KD of binding of the oligosaccharides in BGlu1 E176Q and as long as the primary binding modes block pNPGlc access to the active site, complete inhibition will be observed. Thus, glycosidic bond position, as reflected in the intrinsic kcat, does not affect the cellooligosaccharide inhibition. However, for cellopentaose and cellohexaose, significant effects were observed in the presence of the Y341A mutation, most likely because in the binding mode observed for cellotetraose in BGlu1 E176Q/Y341A in Figures 3(B) and 4(B), there does not appear to be enough space to accommodate the 5th glucosyl residue in cellopentaose in the same tract. Although it has been noted that small movements can relieve steric clashes between substrates and active site residues,30 it is likely the longer oligosaccharide will adopt a different binding tract, since the binding modes observed with BGlu1 E176Q and BGlu1 E386G/Y341A are readily accessible. This is reflected in the unfavorable ΔGb,C4→C5 and ΔGb,C5→C6 for BGlu1 E176AQ/Y341A variants (Y341A, Y341A/Q187A, Y341A/R178A, Y341A/W337A, and Y341/R178A/W337A) in Table3. This effect is also seen in the lower subsite +4 affinities for the BGlu1 Y341A variants (Y341A/Q187A, Y341A/R178A, Y341A/W341A, and Y341A/R178A/W337A) compared to wild type in Table1.
As seen in our previous work,21 the glycosynthase activity appears to be the most sensitive to loss of active site functional groups. The R178 residue appears to be especially critical to glycosynthase activity, since all glycosynthase variants with the R178A mutation converted <3.3% of the pNPC2 acceptor substrate to product. No direct hydrogen bonds were seen between R178 and the ligand in the structures of BGlu1 E386G with cellotetraose and cellopentaose and E386G/Y341A and E386G/Y341A/Q187A with cellotetraose, but it did coordinate water-mediated hydrogen bonds with the ligand in each case. However, each of these structures was a product complex and may not represent the exact position of the acceptor substrate before glucosyl transfer. In both BGlu1 E376G and Streptomyces β-glucosidase E383A glycosynthases, it has been noted that longer acceptor substrates with more active site cleft interactions lead to greater regioselectivity,26,31 which also depends on the donor substrate and may not correspond precisely to that seen in the parent hydrolase.32
In conclusion, we have shown that rice BGlu1 and its E176Q transglucosidase and E386G glycosynthase show remarkable plasticity in their active site cleft interactions with cellooligosaccharides, with three binding modes observed depending on the combination of inactivating and active site cleft mutations. The mutation made in the catalytic acid-base or nucleophile affects the binding mode of the oligosaccharide at the outer subsites, while mutations made in residues located at the +3 to +4 subsites, R178A and W337A, affect the activity of the enzymes toward shorter substrates that are not expected to reach this far, likely due to their effects on the active site cleft shape, environment and water network. Stacking of Glc4 in longer cellooligosaccharides on Y341 appears to compensate for the disruption caused by these mutations. Nonetheless, Y341 appears to be dispensable in the absence of these disruptive mutations, since alternative binding modes can result in effective hydrolysis as well. Thus, binding of cellooligosaccharides is mediated by complex interactions between the residues in the active site cleft, the cellooligosaccharides and the surrounding water molecules.
Materials and Methods
Mutation of rice BGlu1, BGlu1 E176Q, and BGlu1 E386G
Here, we used the pET32a/BGlu1 E386G-2 construct26 for all glycosynthase mutations and designate these protein variants containing this mutation as simply BGlu1 E386G. The mutations for BGlu1 E176Q, BGlu1 N245V, BGlu1 Y341A, and BGlu1 E386G have been described previously.21,22,26 The mutations for the variants Q187A, Y341A/Q187A, N245V/Q187A, R178A, Y341A/R178A, W337A, Y341A/W337A, Y341A/R178A/W337A, E176Q/Q187A, E176Q/Y341A, E176Q/Y341A/Q187A, E176Q/R178A, E176Q/Y341A/R178A, E176Q/W337A, E176Q/Y341A/W337A, E176Q/Y341A/R178A/W337A, E386G/Q187A, E386G/Y341A/Q187A, E386G/R178A, E386G/Y341A/R178A, E386G/W337A, E386G/Y341A/W337A, and E386G/Y341A/R178A/W337A were made in the rice bglu1, rice bglu1 E176Q and rice bglu1 E386G-2 cDNA in the pET32a expression vector10,22,26 by Quikchange (Stratagene, Agilent, La Jolla, CA) site-directed mutagenesis. The primers used for the previously unpublished mutations were as follows: 5′-GTA GCA CTT CTT GGT TAT GAC GCG GGA ACA AAT CCT CCT AAA AGG TG-3′ and its reverse complement for Q187A; 5′-cac tgg ttt aca ttt aat gag cca GCT ata gta gca ctt ctt ggt tat gac-3′ and its reverse complement for R178A; 5′-cac tgg ttt aca ttt aat cag cca GCT ata gta gca ctt ctt ggt tat gac-3′ and its reverse complement for E176Q/R178A; 5′-cg acg agt tac tca gcc gat GCT cag gtt acc tat gtt ttt gcg-3′ and its reverse complement for W337A and 5′-g acg agt tac tca gcc gat GCT cag gtt acc gct gtt ttt gc-3′ and its reverse complement for Y341A/W337A (the mutated nucleotides are underlined).
Protein expression and purification
The BGlu1, BGlu1 E386G glycosynthase and BGlu1 E176Q acid-base variants described above were expressed in Escherichia coli strain Origami (DE3) and purified by immobilized metal affinity chromatography (IMAC), enterokinase digestion and IMAC, as previously described.10,22 The purified protein concentration was determined by measuring absorbance at 280 nm. Extinction coefficients, ε280 of 113,560 M−1 cm−1 for most of the BGlu1 proteins and ε280 of 112,280 M−1 cm−1 for BGlu1 Y341A-containing variants, were calculated by the method of Gill and von Hippel.33 Proteins were checked by SDS-PAGE with Coomassie brilliant blue staining, in which all BGlu1 variants had purities >90%, as shown in Supporting Information Figures S2, S3, and S4.
Oligosaccharide hydrolysis
The activities of the BGlu1 mutants toward cellotriose, cellotetraose and cellopentaose were assayed as release of glucose in 50 mM sodium acetate buffer, pH 5.0, at 30°C, as previously described.10 The kinetic parameters Vmax and Km were calculated by nonlinear regression of Michaelis–Menten plots with the Grafit 5.0 computer program (Erithacus Software, Horley, UK) and divided by the protein concentrations to determine the apparent kcat and kcat/Km. Relative subsite affinities were determined by the method of Hiromi.23
Oligosaccharide binding by BGlu1 E176Q mutants
Enzyme rates were determined in triplicate reactions, as previously described,4 with a time course to ensure linearity. Enzyme activities of BGlu1 E176Q mutants were determined in 50 mM MES buffer, pH 5.0, and 50 mM sodium azide with pNPGlc as a substrate. Kinetic parameters (Km, Vmax and apparent kcat) of BGlu1 E176Q variants in the reactions were calculated from nonlinear regression of Michaelis–Menten curves with Grafit 5.0. The competitive inhibition constants (Ki) were determined from the plots of the apparent Km/Vmax versus the inhibitor (oligosaccharide) concentration, and were taken to be the apparent dissociation constants (KD) of the inhibitors. The association constants were calculated as KA = 1/KD = 1/Ki; and Gibbs free energies of binding were calculated as ΔGbinding = −RTlnKA, where R is the gas constant (8.314 J K−1 mol−1) and T is the absolute temperature (303 K).
Oligosaccharide synthesis
The BGlu1 E386G glycosynthase and its mutants, BGlu1 E386G/Q187A, BGlu1 E386G/Y341A, BGlu1 E386G/Y341A/Q187A, BGlu1 E386G/R178A, BGlu1 E386G/W337A, and BGlu1 E386G/R178A/W337A, were purified as described above and the buffer was exchanged with 50 mM phosphate buffer, pH 6.0. The proteins were incubated with 10 mM α-GlcF donor and 10 mM pNPC2 acceptor in 150 mM ammonium bicarbonate buffer, pH 7.0, at 30°C for 24 h.26 The enzymes in the reaction mixtures were removed by boiling and centrifugation at 13,000 g for 5 min. Five microliters of the products from the reactions were loaded onto a ZORBAX carbohydrate column (4.6 mm × 250 mm, Agilent, USA) connected to an Agilent 1100 series LC-MSD. The column was eluted with a linear gradient from 90 to 50% acetonitrile in water over 30 min at a flow rate of 1 mL min−1. The eluted peaks were detected at 300 nm with a UV–visible diode array detector and the product masses determined by mass spectrometry.21
X-ray crystallography
The BGlu1 E176Q/Y341A, BGlu1 E176Q/Y341A/Q187A, and BGlu1 E386G/Y341A/Q187A variant proteins were crystallized with 2 mM cellotetraose by hanging drop vapor diffusion with microseeding, optimized in 16–26%, polyethylene glycol monomethyl ether (PEG MME) 5000, 0.12–0.26 M (NH4)2SO4, and 2–5 mg mL−1 protein in 0.1M MES (pH 6.7) at 288 K, as previously described.22 Before flash cooling in liquid nitrogen, the crystals were soaked in cryo solution saturated with cellotetraose (∼75–100 mM).
Datasets were collected with 1.0 Å wavelength X-rays and an ADSC Quantum 315 CCD detector on the BL13B1 beamline at the National Synchrotron Radiation Research Center (NSRRC in Hsinschu, Taiwan). The crystals were maintained at 105 K during data collection with a nitrogen cold stream (Oxford Instruments). Data were processed and scaled with the HKL-2000 package.34
The crystals of BGlu1 E176Q/Y341A/Q187A and BGlu1 E386G/Y341A/Q187A with cellotetrase were isomorphous with wild type BGlu1 crystals,22 allowing the structures to be solved by rigid body refinement with the free BGlu1 structure (PDB: 2RGL) from which solvent and heteroatoms were deleted in REFMAC535 with the two molecules in the asymmetric unit refined as independent domains. However, the crystal of BGlu1 E176Q/Y341A with cellotetraose, which had P21 space group symmetry, was solved by molecular replacement using the MolRep program36 and free BGlu1 E176Q as a search model (PDB: 3F4V with solvent and heteroatoms deleted).4 The refinement was executed with REFMAC5 with tight noncrystallographic symmetry (NCS) restraints and model building with Coot.37 Water molecules were added with the Coot and ARP/wARP programs in the CCP4 suite. Glucosyl residues were built into the electron densities in the shapes that fit the densities best (4C1 relaxed chairs or 1S3 skew boats) and refined. The refined sugar residue coordinates were assigned their final conformation designation according to their Cremer–Pople parameters,38 calculated by the Cremer–Pople parameter calculator of Prof. Shinya Fushinobu (University of Tokyo, http://www.ric.hi-ho.ne.jp/asfushi/). In all cases, the nonreducing end glucosyl residue in the −1 subsite was in a shape between a 1S3 skew boat and a 4E envelope, while other Glc residues were in relaxed 4C1 chair conformations. The final models were analyzed with PROCHECK25 and MolProbity.39 The figures of protein structures were generated in PyMol (Schrödinger LLC).
Acknowledgments
The BGlu1 E176Q acid–base mutant vector was originally generated by Watchalee Chuenchor. Stephen G. Withers is thanked for providing α-fluoroglucoside and helpful comments, Jisnuson Svasti for discussions and support and Jitrayut Jitonnom for helpful analysis. Portions of this research were conducted at the National Synchrotron Radiation Research Center.
Glossary
- α-GlcF
α-d-glucosyl fluoride
- pNP
para-nitrophenyl
- pNPC2
pNP-β-cellobioside
- pNPC3
pNP-β-cellotrioside
- pNPC4
pNP-β-cellotetraoside
- pNPC5
pNP-β-cellopentaoside
- pNPC6
pNP-β-cellohexaoside
- pNPC7
pNP-β-celloheptaoside
- pNPC8
pNP-β-cellooctaoside
- pNPC9
pNP-β-cellononaoside
- pNPGlc
pNP-β-d-glucopyranoside
- DP
degree of polymerization
- IMAC
immobilized metal affinity chromatography
- LC-MS
liquid chromatography-mass spectrometry
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- Gilbert HJ. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010;153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert HJ, Knox JP, Boraston AB. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate binding modules. Curr Opin Struct Biol. 2013;23:669–677. doi: 10.1016/j.sbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucl Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuenchor W, Pengthaisong S, Robinson RC, Yuvaniyama J, Svasti J, Ketudat Cairns JR. The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase. J Struct Biol. 2011;173:169–179. doi: 10.1016/j.jsb.2010.09.021. [DOI] [PubMed] [Google Scholar]
- Davies GJ, Planas A, Rovira C. Conformational analyses of the reaction coordinate of glycosidases. Acc Chem Res. 2012;2:308–316. doi: 10.1021/ar2001765. [DOI] [PubMed] [Google Scholar]
- Ketudat Cairns JR, Esen A. β-glucosidases. Cell Mol Life Sci. 2010;67:3389–3405. doi: 10.1007/s00018-010-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DG, Withers SG. The purification and characterization of a β-glucosidase from Alcaligenes faecalis. Biochem Cell Biol. 1986;64:914–922. doi: 10.1139/o86-122. [DOI] [PubMed] [Google Scholar]
- Cicek M, Blanchard D, Bevan DR, Esen A. The aglycone specificity determining sites are different in 2,4-dihydroxy-7-methoxy-1,4-benzoazin-3-one (DIMBOA) glucosidase (maize β-glucosidase and dhurrinase (sorghum β-glucosidase) J Biol Chem. 2000;275:20002–20011. doi: 10.1074/jbc.M001609200. [DOI] [PubMed] [Google Scholar]
- Opassiri R, Ketudat Cairns JR, Akiyama T, Wara-Aswapati O, Svasti J, Esen A. Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 2003;165:627–638. [Google Scholar]
- Opassiri R, Maneesan J, Akiyama T, Pomthong B, Jin S, Kimura A, Ketudat Cairns JR. Rice Os4BGlu12 is a wound-induced β-glucosidase that hydrolyzes cell wall-β-glucan-derived oligosaccharides and glycosides. Plant Sci. 2010;179:273–280. [Google Scholar]
- Mackenzie LF, Wang Q, Warren RAJ, Withers SG. Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J Am Chem Soc. 1998;120:5583–5584. [Google Scholar]
- Williams, Withers SG. Glycosyl fluorides in enzymatic reactions. Carbohydr Res. 2000;327:27–46. doi: 10.1016/s0008-6215(00)00041-0. [DOI] [PubMed] [Google Scholar]
- Perugino G, Trincone A, Rossi M, Moracci M. Oligosaccharide synthesis by glycosynthases. Trends Biotechnol. 2004;22:31–37. doi: 10.1016/j.tibtech.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Faijes M, Planas A. In vitro synthesis of artificial polysaccharides by glycosidases and glycosynthases. Carbohydr Res. 2007;342:1581–1594. doi: 10.1016/j.carres.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Wang Q, Trimbur D, Graham R, Warren RAJ, Withers SG. Identification of the acid/base catalyst in Agrobacterium faecalis beta-glucosidase by kinetic analysis of mutants. Biochemistry. 1995;34:14554–14562. doi: 10.1021/bi00044a034. [DOI] [PubMed] [Google Scholar]
- Jahn M, Marles J, Warren RAJ, Withers SG. Thioglycoligases: mutant glycosidases for thioglycoside synthesis. Angew Chem Int Ed. 2003;42:352–354. doi: 10.1002/anie.200390114. [DOI] [PubMed] [Google Scholar]
- Müllegger J, Jahn M, Chen HM, Warren RAJ, Withers SG. Engineering of a thioglycoligase: randomized mutagenesis of the acid-base residue leads to identification of improved catalysts. Prot Eng Des Sel. 2005;18:33–40. doi: 10.1093/protein/gzi003. [DOI] [PubMed] [Google Scholar]
- Opassiri R, Hua Y, Wara-Aswapati O, Akiyama T, Svasti J, Esen A, Ketudat Cairns JR. β-Glucosidase, exo-β-glucanase and pyridoxine transglucosylase activities of rice BGlu1. Biochem J. 2004;379:125–131. doi: 10.1042/BJ20031485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommalai G, Withers SG, Chuenchor W, Ketudat Cairns JR, Svasti J. Enzymatic synthesis of cellooligosaccharides by mutated rice β-glucosidases. Glycobiology. 2007;17:744–753. doi: 10.1093/glycob/cwm039. [DOI] [PubMed] [Google Scholar]
- Pengthaisong S, Withers SG, Kuaprasert B, Svasti J, Ketudat Cairns JR. The role of the oligosaccharide binding cleft of rice BGlu1 in hydrolysis of cellooligosaccharides and in their synthesis by rice BGlu1 glycosynthase. Protein Sci. 2012;21:362–372. doi: 10.1002/pro.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuenchor W, Pengthaisong S, Robinson RC, Yuvaniyama J, Oonanant W, Bevan DR, Esen A, Chen CJ, Opassiri R, Svasti J, Ketudat Cairns JR. Structural insights into rice BGlu1 beta-glucosidase oligosaccharide hydrolysis and transglycosylation. J Mol Biol. 2008;377:1200–1215. doi: 10.1016/j.jmb.2008.01.076. [DOI] [PubMed] [Google Scholar]
- Hiromi K, Nitta Y, Namura C, Ono S. Subsite affinities of glucoamylase, examination of the validity of the subsite theory. Biochim Biophys Acta. 1973;302:362–375. doi: 10.1016/0005-2744(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Hrmova M, MacGregor AM, Biely P, Stewart RJ, Fincher GB. Substrate binding and catalytic mechanism of a barley β-d-glucosidase/(1,4)-β-d-glucan exohydrolase. J Biol Chem. 1998;273:11134–11143. doi: 10.1074/jbc.273.18.11134. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK, a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Pengthaisong S, Chen C-F, Withers SG, Kuaprasert B, Svasti J, Ketudat Cairns JR. Rice BGlu1 glycosynthase and wild type transglycosylation activities distinguished by cyclophellitol inhibition. Carbohydr Res. 2012;352:51–59. doi: 10.1016/j.carres.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Hiromi K. Interpretation of dependency of rate parameters on the degree of polymerization of substrate in enzyme-catalyzed reactions. Evaluation of subsite affinities of exo-enzyme. Biochem Biophys Res Commun. 1970;40:1–6. doi: 10.1016/0006-291x(70)91037-5. [DOI] [PubMed] [Google Scholar]
- Mendonça LMF, Marana SR. Single mutations outside the active site affect the substrate specificity in a β-glycosidase. Biochim Biophys Acta Proteins Proteom. 2011;1814:1616–1623. doi: 10.1016/j.bbapap.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Mildvan AS, Weber DJ, Kuiopulos A. Quantitative interpretations of double mutations of enzymes. Arch Biochem Biophys. 1992;294:327–340. doi: 10.1016/0003-9861(92)90692-p. [DOI] [PubMed] [Google Scholar]
- Warshel A, Levitt M. Theoretical studies of enzyme reactions: dielectric, electrostatic, and steric stabilization of carbonium ion in the reaction of lysozyme. J Mol Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- Faijes M, Saura-Valls M, Perez X, Conti M, Planas A. Acceptor dependent regioselectivietty of glycosynthase reactions by Steptomyces E383A β-glucosidase. Carbohydr Res. 2006;341:2055–2065. doi: 10.1016/j.carres.2006.04.049. [DOI] [PubMed] [Google Scholar]
- Pozzo T, Plaza M, Romero-Garcia J, Faijes M, Nordberg Karlsson E, Planas A. Glycosynthases from Thermotoga neapolitana β-glucosidase 1A: a comparison of α-glucosyl fluoride and in situ-generated α-glycosyl formate donors. J Mol Catal B Enzym. 2014;107:132–139. [Google Scholar]
- Gill, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Lebedev A, Vagin AA, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- Emsley P, Cowtan K. Coot, model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Cremer D, Pople JA. A general definition of ring puckering coordinates. J Am Chem Soc. 1975;97:1354–1358. [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity, all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


