Abstract
Nucleic acid can catalyze the conversion of α-helical cellular prion protein to β-sheet rich Proteinase K resistant prion protein oligomers and amyloid polymers in vitro and in solution. Because unfolding of a protein molecule from its ordered α-helical structure is considered to be a necessary step for the structural conversion to its β-sheet rich isoform, we have studied the unfolding of the α-helical globular 121–231 fragment of mouse recombinant prion protein in the presence of different nucleic acids at neutral and acid pH. Nucleic acids, either single or double stranded, do not have any significant effect on the secondary structure of the protein fragment at neutral pH; however the protein secondary structure is modified by the nucleic acids at pH 5. Nucleic acids do not show any significant effect on the temperature induced unfolding of the globular prion protein domain at neutral pH which, however, undergoes a gross conformational change at pH 5 as evidenced from the lowering of the midpoint of thermal denaturation temperatures, Tm, of the protein. The extent of Tm decrease shows a dependence on the nature of nucleic acid. The interaction of nucleic acid with the nonpolar groups exposed from the protein interior at pH 5 probably contributes substantially to the unfolding process of the protein.
Keywords: prion protein, nucleic acids, pH 5, PloyA, denaturation temperature (Tm)
Introduction
The fatal neurodegenerative prion diseases are both genetic and infectious and can inflict on humans and other animals. Structural change in the α-helix rich normal cellular prion protein, PrPC to its β-sheet rich scrapie isoform, PrPSc, has been considered as the obligatory step to the occurrence and propagation of the prion disease.1–3 PrPSc can exist as a monomer or a multimer.1 PrPSc can catalyse the conversion PrPC to PrPSc and on the basis of genetic experiments, the process of conversion has been postulated to require the binding of PrPC to a still unidentified “protein X” (a cofactor) before its conversion by PrPSc can occur.1
Synthetic nucleic acids, in solution and in vitro, can catalyze conversion of recombinant and cellular PrPC to PrPSc as evidenced from secondary structural studies of the protein and proteinase K (PK) resistance properties.4–7 Our studies also indicated that bending and unwinding of nucleic acid occurred by prion protein and natural poly amines (spermine and spermidine) can inhibit this interaction.8,9 Highly structured small RNA binds to PrPC at neutral pH and yields Proteinase K resistant component in the presence of cellular cofactors.6
Studies using brain tissues have shown that an endogenous 300 nucleotide long RNA (100 kDa) can convert endogenous PrPC to Proteinase K resistant form in vitro.7,10,11 These results indicate that nucleic acid can act as a cofactor for the conversion of PrPC to PrPSc and can be the active transmissible spongiform encephalopathy (TSE) agent. In addition, the interaction between recombinant PrPC and nucleic acids (DNAs, tRNA, and PolyA) simultaneously produces a mixture of condensed and functionally active nucleoprotein complex, PrPSc oligomers and linear and spherical amyloids.8–12 A specific nucleic acid as a cofactor for the propagation of prion infection has not been identified.13 PrPC is a cell surface protein and nucleic acids in extracellular circulation can interact with it.14 However it has been considered that the relevant nucleic acid for PrPC conversion would be of cytoplasmic origin.4,5,7,14,15 Besides, we have also found that osmolyte can induce recombinant alpha-helical prion protein to its soluble beta-structured form at high temperature.16
The presence of prion protein in cytoplasm of cells including neurons has been shown and although the exact biological role of prion protein–nucleic acid interaction is not known at present, it is possible that structural conversion of PrPC to PrPSc can be catalyzed by cytoplasmic nucleic acids that can play a role in the prion diseases.12,17,18 A separate study indicates the neurotoxic effect found on the cultured rat cortical neurons of the complex of the ovine prion protein (OvPrP(C)) and RNA.19,20 Besides, unique quadruplex structure and selective interaction of an RNA aptamer against bovine prion protein and may inhibits the prion disease propagation.21
Understanding the processes of PrPC structural changes, viz. its unfolding and refolding to polymeric scrapie isoform are of paramount importance.1 Thermal denaturation of proteins is one of the methods used to study the unfolding processes of protein. We encountered experimental difficulty to study the unfolding properties of the full-length protein in nucleic acid solutions due to the appearance of turbidity and coagulation of protein during thermal unfolding of the protein.
The full-length recombinant prion proteins corresponding to different species show the presence of three α-helices and two short α-strands in the globular segment spanning 121–231 residues [Fig. 1(A)], and it has been shown that, the three-dimensional fold of the full-length protein is fully preserved in this C-terminal 121–231 protein fragment.22,23 Previously we have shown that the α-helical moPrP 121–231 fragment in the presence of double stranded nucleic acids is converted to a protein rich in β-sheet at pH 5 which after incubation resulted in linear amyloids determined from electron microscopy, Congo Red and Thiaflavin T binding.24,25 In this present study, we determine the thermal unfolding of the moPrP 121–231 fragment in the presence of different nucleic acids at neutral and pH5 since the protein fragment did not coagulate on heating. We considered that the information obtained from the study of this globular fragment would be relevant to understand the unfolding processes of the full-length prion protein in nucleic acid solutions.
Figure 1.
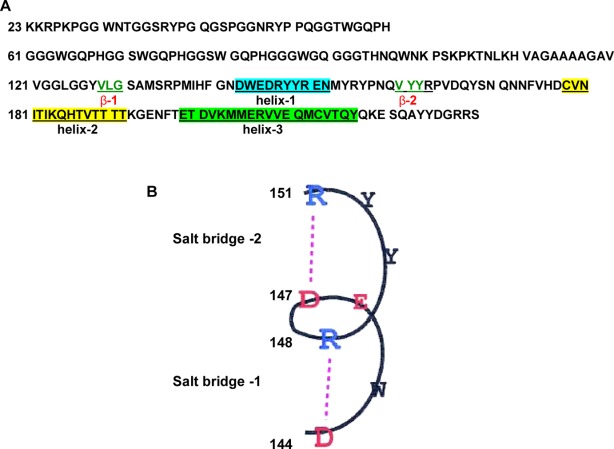
A. Sequence of the mouse recombinant full-length prion protein (moPrP 23–231). The N-terminal 23–125 segment is highly basic and unstructured and rich in glycine and tryptophan groups. The amino acid residues forming three α-helices have been marked. β-Sheets are represented by green letters. B. The two salt bridges in the prion protein helix 1 (DWEDRYYR) can be perturbed by nucleic acids inducing unfolding of the prion protein fragment 121–231. (Adapted from Morrissey and Shakhnovich, 1999, with permission from Proc. Natl. Acad. Sci. USA.)
Results
Secondary structure of the protein in nucleic acid solutions
The circular dichroism (CD) experiments of the mouse prion protein fragment moPrP (121–231) showed characteristic of double pick (minima) at 208 and 222 nm (Fig. 2), which indicated a significant α-helical content of the protein fragment at neutral pH (lower spectrum) which did not change at pH 5 (upper trace). The CD spectra of the protein in nucleic acid solutions were corrected after subtracting the corresponding free nucleic acid spectra from the CD spectra of the moPrP (121–231)—DNA mixture.24,26 Nucleic acids even at their maximum concentrations used in this study do not have significant CD values between 200 and 250 nm compared to the CD values of 20 µM protein (Supporting Information Fig. 1).
Figure 2.
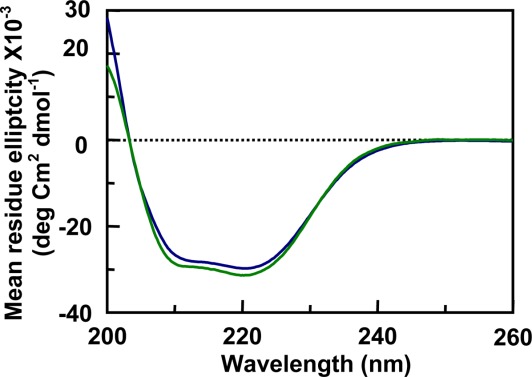
Circular dichroism spectra of moPrP 121–231 fragment (25 µM) inTris HCl pH 7.2 (bottom) and acetate buffer pH 5 (top) were using the same concentration of the protein. The result shows that the secondary structures of the C-terminal domain of prion protein and the globular fragment of the protein do not change to any significant extent from neutral to pH 5.
The CD spectra of the 121–231 protein fragment in the presence of nucleic acids, tRNA and gcDNA (80 µM nucleotide concentrations), at pHs 7.2 and 5 have been presented in Figures 3 and 4, respectively. At pH 7.2, the ratio of the intensity of the two peaks changed in the presence of 1 µM tRNA [Fig. 3(A)]. However, gcDNA did not induce any similar change under similar experimental conditions [Fig. 4(A)]. The positive CD values at 265 nm represent the contribution of tRNA to the CD spectra. The intensities of the CD spectrum decreased and the peaks in the spectrum were not pronounced in the presence of tRNA at pH 5 [Fig. 3(B)]. However, gcDNA decreased the CD intensity of the 240–200 nm region of the spectrum and the CD peak at 220 nm became more pronounced comppared to the peak at 208 nm in buffer which appeared as a shoulder at pH 5 [Fig. 4(B)]. The CD intensities of the protein peaks decreased (∼20%) in the presence of both the nucleic acids at pH 5 suggesting a decrease in the secondary structural content of the protein at pH 5 [Figs. 3(B) and 4(B)].
Figure 3.
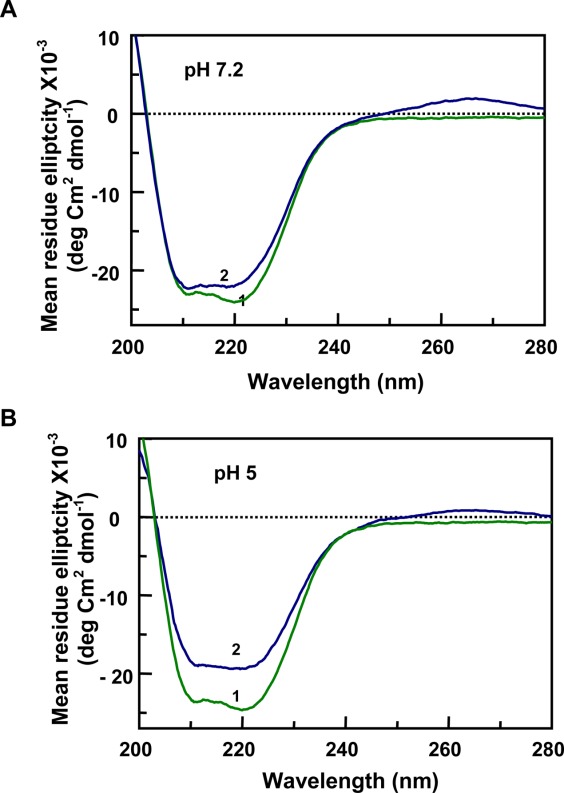
Secondary structural changes observed by CD spectra of the moPrP 121–231 fragment in the presence of tRNA. In A and B (pH 7.2 and pH 5, respectively), trace-1 represents the protein in buffer (moPrP 121–231; 20 µM), showing two characteristic peaks for α-helix at 222 and 208 nm. In the presence of 1 µM tRNA ([Nucleotide]/[PrP]) = 4), there is no significant effect on the protein secondary structure at neutral pH (A, trace-2,). However there is a substantial reduction of the CD intensities at the peaks observed at pH 5 (B, trace-2).
Figure 4.
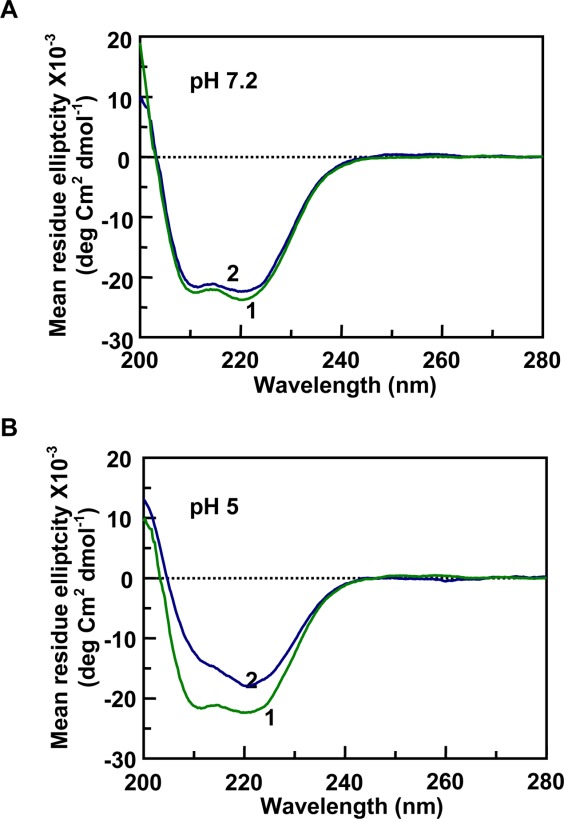
Secondary structural changes observed by CD spectra of the moPrP 121–231 fragment in the presence of gcDNA. In A and B (pH 7.2 and pH 5, respectively), trace-1 represents the protein in buffer (moPrP 121–231; 20 µM), showing two characteristic peaks for α-helix at 222 and 208 nm. In the presence of the nucleic acid ([Nucleotide]/[PrP]) = 4), there is no significant effect on the protein secondary structure at neutral pH (A, trace-2,). However there is a substantial reduction of the CD intensities at the peaks observed at pH5 (B, trace-2) where reduction in the CD intensity at 222 nm reduced to a lesser extent than the peak at 208 nm.
To characterize the effect of nucleic acids on the structural stability of moPrP (121–231) fragment, we studied the thermal denaturation of the protein (20 µM) in the presence of increasing nucleic acid concentrations. The mid points of the structural transitions (Tm) for moPrP (121–231) in pH 7.2 and pH 5 buffers were 66.5°C ± 0.4°C and 65.4°C ± 0.2°C, respectively indicating that changes in the pH do not perturb the structural stability of the protein (traces 1, Figs. 5 and 6). The stability of the protein, as determined from the Tm values, in the presence of either gcDNA or tRNA, does not change at neutral pH [traces 2; Figs. 5(A) and 6(A)]. However all the nucleic acids studied here decreased the Tm values of the protein at pH 5 indicating nucleic acid induced structural destabilization of the protein at pH 5 [Figs. 5(B) and 6(B)]. The variation of Tm values as a function of nucleic acid concentration (plotted as nucleotide to protein mole ratio) has been presented in Figure 7. It can be seen that the decrease in the Tm values remained constant after nucleotide to protein ratio of 2 for the nucleic acids studied, although the extent of decrease in the Tm values at saturation varied on the type of the nucleic acids (Fig. 7 and Table1). The data in Table1 also shows that maximum decrease (nucleotide to protein ratio of 4) in the Tm values occurred with tRNA and mDNA (Tm ∼45°C) and the effect was minimum with PolyC (Tm, 53°C).
Figure 5.
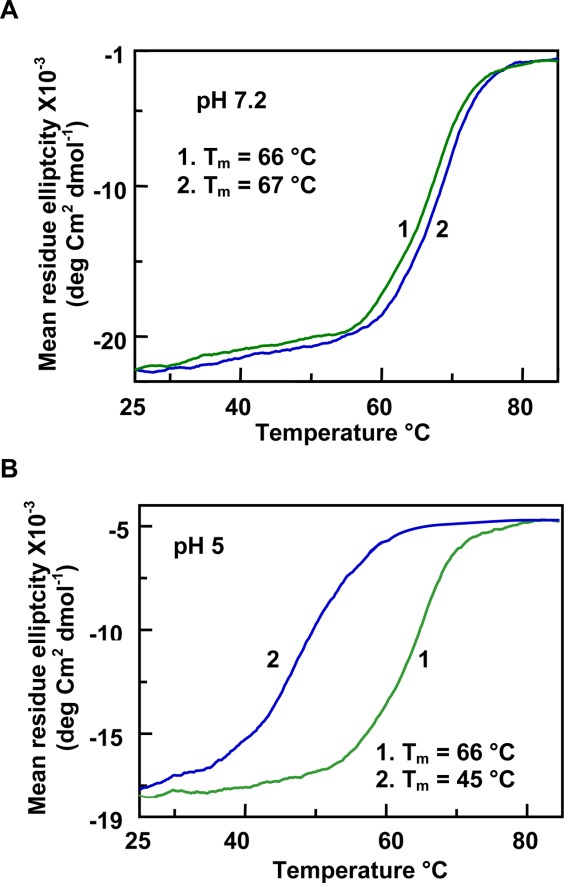
Effect of tRNA on the thermal unfolding of moPrP (121–231) monitored at pH 7.2 (A) and pH 5 (B). The change in the CD intensity was measured at 222 nm. In both figures, trace 1 represents the protein in buffer and trace 2 represents the protein in the presence of 1 µM tRNA. Nucleotide to protein concentration ratio was 4. It can be seen that the structural transition of the protein fragment is broader at pH 5.
Figure 6.
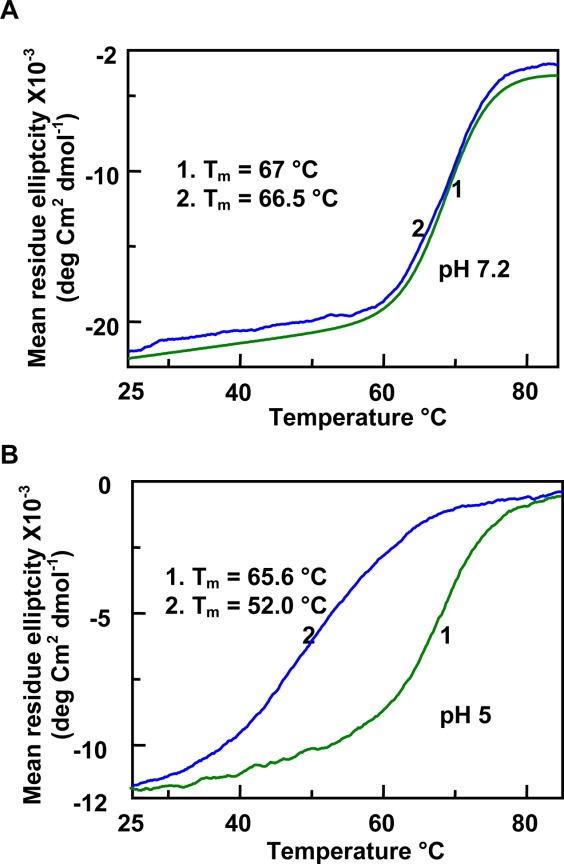
Effect of gcDNA on the thermal unfolding of moPrP (121–231) monitored at pH 7.2 (A) and pH 5 (B). The change in the CD intensity was measured at 222 nm. In both figures, trace 1 represents the protein in buffer and trace 2 represents the protein in the presence of gcDNA. Nucleotide to protein concentration ratio was 4. It can be seen that the structural transition of the protein fragment is broader at pH 5.
Figure 7.
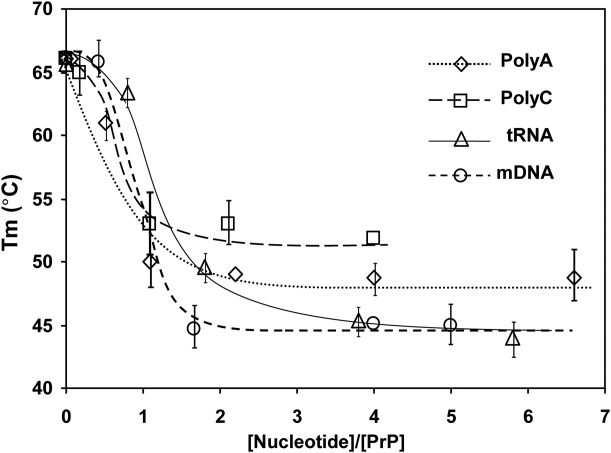
The decrease in the temperature of the thermally induced midpoint of structural unfolding, Tm, of moPrP (121–231) induced by increasing concentration of different nucleic acids at pH 5. The protein concentration was kept constant (20 µM) and nucleic acid concentrations were varied. The results show increased destabilization with increasing concentrations of the nucleic acids at pH 5. The maximum extent of the decrease in the Tm values depends upon the composition of the nucleic acids.
Table I.
Comparison of the Tm Values for the Unfolding of moPrP 121–231 Fragment in the Presence of Different Nucleic Acids at a Constant [Nucleotide]/[PrP] Ratio of 4 at pH 5
| Nucleic acids | No. of nucleotides (nt) | Tm (°C) at a fixed [nucleotide]/[PrP] = 4 ratios |
|---|---|---|
| MoPrP(121–231) in Buffer, pH5 | — | 65.4 ± 0.2 |
| MoPrP(121–231) in Buffer, pH6 | — | 65.8 ± 0.3 |
| MoPrP(121–231) in Buffer, pH7.2 | — | 66.5 ± 0.4 |
| Polyadenylic acid [PolyA] | 910 | 49.0 ± 0.5 |
| Polycytidylic acid [PolyC] | 2500 | 53.0 ± 0.6 |
| Polyuradylic acid [PolyU] | 2727 | 48.0 ± 0.4 |
| ds poly (dA-dC).(dG-dT) [mDNA] | 5316 | 45.0 ± 0.3 |
| Transfer RNA [tRNA] | 80 | 45.0 ± 0.5 |
Nature of interaction between nucleic acid and moPrP (121–231) fragment
Next, we sought to characterize the nature of interaction between nucleic acid and moPrP 121–231. Unlike the full-length prion protein where we have used intrinsic fluorescence polarization values of tryptophan (numbering 7) to study the interaction with nucleic acids, we have used the intrinsic fluorescence intensity of the single tryptophan group present in the 145 position of the first α-helix of the protein [see the protein sequence, Fig. 1(A)]. The emission at 350 nm arises from the single tryptophan 145 of the protein fragment, which might reflect either a partial exposure of this Trp (W) to the surrounding solvent or the influence of a local charged environment around it [DWEDR in the helix-1, Fig. 1(B)] or a combination of both of these factors.27
The titration results with tRNA showed that its addition quenched the fluorescence intensity of the tryptophan group of moPrP121–231 and the titration curve did not show a saturation effect at higher tRNA concentrations (mole ratio of protein to nucleotide >15) shown in Figure 8. In increasing concentrations of NaCl, the extent of quenching of the tryptophan fluorescence in the presence of tRNA decreased showing that its interaction with the prion protein fragment decreased in the presence of salt. This suggests that electrostatic interaction contributes substantially to the interaction between nucleic acid and moPrP fragment 121–231 (pI, 7.2).
Figure 8.
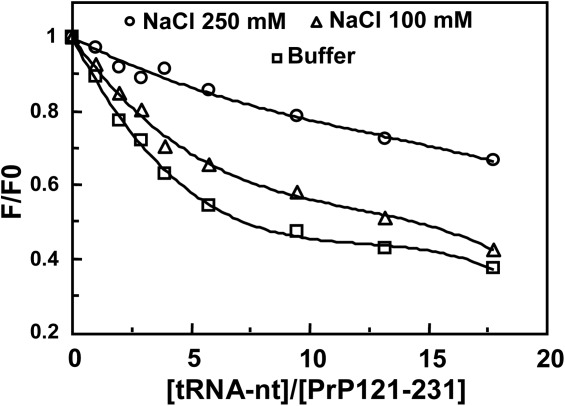
Nature of the binding interaction between moPrP 121–231 fragment and tRNA in 1 mM acetate buffer, pH 5 obtained from fluorescence titration of the single tryptophan moiety situated at 145 position of the prion protein helix 1. A 2 µM solution of the protein fragment was used. F0 is the fluorescence of the tryptophan in buffer, F is the same in the presence of tRNA. The titration results in the presence of 0, 0.1, and 0.25 M NaCl are shown. Excitation, 280 nm, emission at 350 nm; slit widths are 10 and 20 nm, respectively; temperature 20°C.
Exposure of the non-polar groups at pH 5
The dye, bis-ANS, in the presence of protein fragment at pH 7.2 showed an emission maximum at 520 nm which was identical to the same observed in buffer (not shown). The dye bis-ANS does not change its fluorescence property upon changing its surrounding pH. The emission spectra of the dye in the presence of PrP (121–231) fragment showed a gradual blue shift with the lowering of the pH. A plot of the ratio of the fluorescence intensities at 490 nm to that at 520 nm against pH of solutions ranging between 7.5 and 3 has been shown in Figure 9. The plot shows that a structural transition of the prion protein fragment occurred with the decrease in pH of the solution with the apparent pK of structural transition of 5. A similar pK value of 5 was obtained with the human prion protein 90–231 fragment during its structural change from neutral to acid pH using the same bis-ANS dye.28
Figure 9.
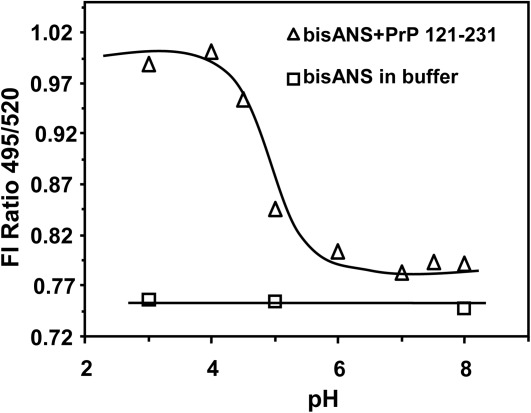
pH dependence of the fluorescence intensity of bis-ANS in the presence of moPrP (121–231) (triangle). In the experiment the protein (2 µM) was mixed with bis-ANS (8 µM) at different pH. After 30 min of incubation the individual spectrum was taken and the fluorescence intensity ratios at 495–520 nm at different pHs were plotted as a function of pH. The fluorescence intensities of bis-ANS remain between the pHs indicated (square).
Discussion
The results show that the secondary structure of the prion protein 121–231 fragment is not significantly altered when the pH of the solution is changed from neutral to pH 5 (Fig. 2). The thermal stability of the protein fragment against thermal denaturation also does not show any change with the variation in the pH as evidenced from Tm values of 66.5°C ± 0.4°C and 65.4°C ± 0.2°C at pH 7.2 and pH 5 buffers, respectively (Table1). Although nucleic acids do not influence the secondary structure of the protein fragment at neutral pH [Figs. 3(A) and 4(A)], there are perceptible changes in the secondary structure of the protein in the presence of nucleic acids at pH 5 [Figs. 3(B) and 4(B)]. The present results also show that nucleic acids, single or double stranded, destabilize the structure of the globular fragment 121–231 of the prion protein in acidic pH [Figs. 5(B) and 6(B); Table1]. The result obtained with the mouse prion protein fragment is expected to be valid for the huPrP 121–231 fragment because these two protein fragments have 95% sequence homology.23
Previous work by Marques et al. (2009) indicated that the prion protein stability enhanced coupled to DNA recognition. It is also indicated that the stability of a soluble, 1:1 complex formed between an 18 base-pair DNA fragment and the full-length recombinant prion protein. In fact DNA confers a gain in PrP stability against urea and guanidinium denaturation, which is enhanced at lower pHs and in moderate concentrations of NaCl.29 These findings support our current results.
Our previous results showed that nucleic acid by binding to the positively charged N-terminal segment of the full-length prion protein forms functional nucleoprotein complex. We envisaged that the polymerization of the prion protein through its C-terminal 121–231 segment would require direct interaction of nucleic acid with this fragment.9,24 Our present data show that nucleic acids can interact with the purified prion protein 121–231 fragment that is dominated by electrostatic interaction at pH 5 (Figs. 8 and 9). We believe that similar interaction of nucleic acid with this segment also occurs in the full-length protein. Support of this possibility comes from another protein-nucleic acid system viz. X-ray crystallographic data of the enzyme Bam HI show that nucleic acid can interact with the enzyme through different mechanisms inducing ordering of the N-terminal part and unwinding of the C-terminal segment of the molecule.30
Of note, nucleic acids also bind to the globular domain PrP (121–231), in addition to the N-terminal domain. However, we did not see any structural contribution from nucleic acids towards protein helical peaks in CD spectra [Supporting Information Fig. 1 (Fig. 1s)]. The CD spectra of the free PrP (121–231), free tRNA or gcDNA were compared with CD spectra of protein-nucleic acids complexes. As expected protein helical structure characterized by two different peaks at 222 and 210 nm were observed. There is no characteristic signal around 210 or 222 nm for the tRNA. Experiments have also performed with gcDNA. CD spectral data indicated that the structural elements contributed from tRNA or gcDNA is not interfering with the helical structural peaks of PrP (121–231) (Supporting Information Fig. 1s). As described in our previous findings that the secondary or tertiary structure of nucleic acids were affected by the interacting PrP protein.8 However, we did not see the direct helical structural contribution in CD spectra from nucleic acids towards PrP (121–231) helices. Therefore, our present observation of the unfolding of PrP globular domain by nucleic acid is not contributed from the structural changes of nucleic acid. Besides, our current data is an agreement with the recent finding to what has been observed to the binding of low molecular weight heparin to murine PrP at pH 5.5 in comparison to neutral pH. The interaction between recombinant PrP and heparin (low molecular weight) at pH 5.5 had a stoichiometry of 2:1 (Hep:rPrP 23–231), in contrast to a 1:1 binding ratio at pH 7.4.31,32
Previously we suggested that extremely hydrophilic helix 1 spanning the residues 144–154 (DWEDRYYRENN) in the PrP fragment is destabilized by nucleic acid and partial unfolding of the protein by low concentrations of nucleic acids lead to the oligomerization and polymerization of the PrP 121–231 protein fragment.9,24 The absence of ANS binding to the PrP 121–231 amyloid indicated the absence of hydrophobic surface/contacts in the amyloid where the dye can bind. This agrees with the observation that the helix 1 has a low capacity to form hydrophobic contacts and its regular secondary structure is stabilized by electrostatic interactions [Fig. 1(B)].33,34 NMR results reveal that although at neutral pH, the end residues of helix 1 adopt a 310-helix conformation, they show a less regular conformation at acidic pH most possibly due to the perturbation of the electrostatic interaction which stabilizes prion helix 1.33
It is possible that the protonation of His 140 at acidic pH would tend to alter helix 1 structure by the introduction of an unfavorable second positive charge close to Arg148.33 This is probably reflected in a very small change in the profile of secondary structure and exposure of the non-polar groups from the protein interior at pH 5 (Figs. 2 and 9). We suggest that phosphate groups of the nucleic acids through ion-induced destabilization of salt bridges Asp144-Arg148 and/or Asp147-Arg 151 of helix-1 would lead to the unfolding of the protein at pH5 [Figs. 5(B) and 6(B); Table1].35 It has been suggested that the helix 2 at acidic pH has reduced stability that can arise from the protonation of His187, as positively charged histidine side chains in the middle of α-helices have been shown to have a destabilizing effect because of unfavorable interaction with the helix macrodipole.36
From NMR and model building studies it has been suggested that the C-terminal residues of prion helix 2 might play a role in the transition to an aggregation competent state. This result and the observation that the secondary structure of PrPSc has a high percentage of β-sheet content imply that the whole protein molecule unfolds substantially in going from PrPC to an aggregation prone state.37 However, unfolding simulation of Syrian hamster PrP (109–219) at low pH showed that the prion protein core, consisting of helix 2 and helix 3, remained unchanged in the PrPC and aggregated PrPSc forms, whereas a conformational transition and instability was identified at the N-terminal part of the protein and within helix −1 and the following β-sheet (second β-sheet).38 Further, a structural model of PrPSc based on electron crystallography39,40 suggested that helix 2 and helix 3 have a very similar conformation in both PrPC and PrPSc. Therefore we consider that the conformation of the prion helix 2 is not altered in nucleic acid solution.
The electrostatic interaction between proteins and nucleic acids, in general, increases the stability of the protein41 and this interaction is not expected to destabilize the prion protein fragment 121–231. This is also evidenced from the results at neutral pH where nucleic acids do not have any effect on the thermal stability of the protein fragment [Figs. 5(A) and 6(A)]. In agreement of these present findings, previous data demonstrated that the structure of prion protein is influenced by pH (on NMR structure and stability) as well as prion protein structure is affected by pH-dependent interaction with membranes.33,42 Both these cases structural stability influenced at the globular domain of the prion protein.
The above explanation of mechanism of protein unfolding based only on the perturbation of the salt linkage in helix 1 and modification of the interaction of the protonated His 187 of helix 2 with the macor-dipole of the helix 2 by phosphate ion is not sufficient to explain the differences in the stabilities of the protein fragment in the presence of different nucleic acids. This is due to the observations that at the same nucleotide concentrations (phosphate to the protein ratio) Poly C decreases the Tm value of the protein fragment by ∼10°C whereas the Tm values of the protein in mDNA or tRNA solutions decrease by ∼20°C from the buffer value of Tm at pH 5 (Table1). This suggests that nucleic acids can influence the structural stability of the prion protein fragment which could depend on base composition, sequence or nature of the secondary structure of the nucleic acids.
The binding of the fluorophor bis-ANS shows that there is exposure of nonpolar groups from the interior of the PrP 121–231 fragment in acidic pH indicating a structural change of the protein (Fig. 8). Although this conformational change did not significantly alter either the secondary structure or the thermal stability of the protein fragment in acidic pH (Fig. 2; also see above), there was gross unfolding of the protein structure in the presence of nucleic acids at pH 5 as evidenced from a large decrease of ∼20°C in the Tm value (Table1). In contrast, we did not observe any difference in the Tm values at pH 7.2 in the presence of nucleic acids [Figs. 5(A) and 6(A)].
On the basis of these considerations, we suggest that destabilization of the PrP (121–231) globular fragment in the presence of nucleic acids at pH 5 can arise from favorable interaction(s) of the newly exposed nonpolar groups with nucleic acid bases through more favorable dispersion interaction compared to that of the nonpolar groups with water. This favorable interaction between the nonpolar groups of the protein fragment with the nucleic acid bases would stabilize the unfolded state of the protein reflected in the lowering of the Tm values in the presence of nucleic acids. The dispersion force between the protein non-polar groups and nucleic acid could depend upon base composition, sequence or nature of the secondary structure of the nucleic acids.43–47 At present we do not know which nonpolar groups will be exposed from the interior of the globular protein fragment at pH 5 that will make favorable contacts with the nucleic acid bases.
We have mentioned previously that the titration of the huPrP (90–231) fragment showed nearly 300-fold increase in the bound bis-ANS fluorescence at pH 5.8,16 Compared to that, there was only 30% increase in the bis-ANS fluorescence when (121–231) fragment was titrated. Our unpublished data shows that at identical nucleic acid concentrations (double or single stranded), the decrease in the Tm values of the 90–231 fragment are comparable to the decrease in the Tm values of huPrP (121–231) or moPrP (121–231) prion protein fragments in the same nucleic acid solutions. The consensus hydrophobic domains of the full-length human or mouse prion proteins span amino acid residues 112–122 (VAGAAAAGAVV). This amino acid sequence in the 90–231 prion protein fragment present in the unstructured region of the protein is expected to remain in contact with the external solvent at neutral pH. However the results of titration of 90–231 prion protein fragment with pH variation and in the presence of bis-ANS showed that between pH 9 and 6, the dye does not bind to the protein. This suggests that the above hydrophobic region of the protein is not accessible to the solvent at neutral pH [Figs. 1(A,B) and 9]. This hydrophobic region becomes accessible to the solvent to bind to the dye due to its exposure as a consequence in change in the protein conformation at pH 5.28 Because the decrease in the Tm values of the 90–231 and 121–231 prion protein fragments are nearly same in the presence of nucleic acids at pH 5 (see above), we suggest that the interaction of nucleic acid with the exposed nonpolar groups in the 121–231 region of the protein is relevant for the structural destabilization of both the protein fragment. By extension we consider that this explanation would be applicable to the unfolding of the full length prion protein which leads to amyloid formation in nucleic acid solution. We believe that exposed nonpolar 112–122 region of the full-length prion protein with nucleic acids at pH 5 can explain the formation of lager nucleoprotein complexes which are insensitive to the salt.
In conclusion, the present study shows the possibility that in the cell, nucleic acid can catalyze the conversion of the cellular prion protein to a β-sheet rich PrPSc form by inducing unfolding of the structured region of the prion protein and therefore can act as a cofactor in the prion diseases.
Materials and Methods
The recombinant mouse prion protein fragment moPrP (121–231) was expressed in E coli. and isolated by anion exchange and size exclusion chromatography.23 The isolated protein fragment has the desired disulfide bond between residues 178 and 213 and in the SDS-polyacrylamide gel it shows a single band with a molecular mass 13.3 kDa verified by electron mass spectrometry. An absorbance value of 1.55 at 280 nm in 1 cm cell for 1 mg of protein was used to calculate the protein concentration.
Single and double stranded nucleic acids were obtained from Sigma-Aldrich and were used without further purification. The concentrations of different nucleic acids were determined as follows. An OD (at 260 nm) of 1 corresponds to 50 µg ml−1 for double stranded DNA (mDNA and gcDNA) and 40 µg ml−1 for single-stranded DNA (Poly A, Poly C, and Poly U) and tRNA were used to calculate the respective concentrations. The ratio between OD values at 260 and 280 nm (OD260/OD280) were above 1.9 for the nucleic acids used.
Secondary structure of the protein fragment in the far UV region was studied by circular dichroism spectra in a JASCO 810 Spectropolarimeter fitted with a Peltier thermostat. The spectra were recorded at 20°C. The stability of the protein against thermal denaturation was monitored from the changes in the CD values of the protein α-helical peak at 222 nm by increasing the solution temperature by 1°C min−1 (Nandi et al., 2002).24 The preceding studies were carried out in either 0.1M sodium acetate buffer (pH 5) or 0.1M Tris-Cl buffer (pH 7.2) in a 0.1cm cuvette at 20°C. Protein at 20 µM concentration was used for the experiments. Five individual scans of the spectrum were averaged, and the spectra are reported after subtraction of the nucleic acid spectra. The effects of different nucleic acids on the thermal stability of the protein have been compared after normalization with respect to nucleotide concentrations. Fluorophor 4,4′-Dianilino-1,1′-binaphthyl-5,5′-disulfonic acid dipotassium salt, bis-ANS, was obtained from Molecular Probes, Eugene, Oregon and has been used to study structural change of the prion protein in buffer using a Hitachi 4500 Spectrofluorometer fitted with a circulating water thermostat.
The study on the effect of pH on the structural change of the protein fragment based on fluorescence properties of the dye bis-ANS fluorescence was carried out by exciting the solution at 460 nm. Solutions containing 2 µM of moPrP (121–231) fragment and 8 µM of bis-ANS at different pHs were incubated for 30 min after which the spectra were recorded by exciting the solutions at 360 nm and recording the spectra between 460 and 550 nm. For different pHs, the citrate-phosphate (pH 3 to 4), acetate (pH 5), phosphate (pH 6) and Tris-HCl for higher pHs buffer were used keeping the ionic strength as 0.1M. After 30-min incubation in the dark, fluorescence of bis-ANS was measured in a Hitachi 4500 Spectrofluorometer using low excitation slit (5 nm) to minimize the photo bleaching.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information Figure 1.
References
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Weissman JS. Deadly conformations—protein misfolding in prion disease. Cell. 1997;89:499–510. doi: 10.1016/s0092-8674(00)80232-9. [DOI] [PubMed] [Google Scholar]
- Wadsworth JD, Hill AF, Joiner S, Jackson GS, Clarke AR, Collinge J. Strain-specific prion-protein conformation determined by metal ions. Nat Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- Cordeiro Y, Machado F, Juliano L, Juliano MA, Brentani RR, Foguel D, Silva JL. DNA converts cellular prion protein into the beta-sheet conformation and inhibits prion peptide aggregation. J Biol Chem. 2001;276:49400–49409. doi: 10.1074/jbc.M106707200. [DOI] [PubMed] [Google Scholar]
- Nandi PK, Leclerc E. Polymerization of murine recombinant prion protein in nucleic acid solution. Arch Virol. 1999;144:1751–1763. doi: 10.1007/s007050050702. [DOI] [PubMed] [Google Scholar]
- Adler V, Zeiler B, Kryukov V, Kascsak R, Rubenstein R, Grossman A. Small, highly structured RNAs participate in the conversion of human recombinant PrP(Sen) to PrP(Res) in vitro. J Mol Biol. 2003;332:47–57. doi: 10.1016/s0022-2836(03)00919-7. [DOI] [PubMed] [Google Scholar]
- Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- Bera A, Nandi PK. Biological polyamines inhibit nucleic-acid-induced polymerisation of prion protein. Arch Virol. 2007;152:655–668. doi: 10.1007/s00705-006-0907-8. [DOI] [PubMed] [Google Scholar]
- Bera A, Roche AC, Nandi PK. Bending and unwinding of nucleic acid by prion protein. Biochemistry. 2007;46:1320–1328. doi: 10.1021/bi0620050. [DOI] [PubMed] [Google Scholar]
- Deleault NR, Kascsak R, Geoghegan JC, Supattapone S. Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry. 2010;49:3928–3934. doi: 10.1021/bi100370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, Supattapone S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci USA. 2012;109:8546–8551. doi: 10.1073/pnas.1204498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmes AG, Moore RA, Fischer ER, Priola SA. Recombinant prion protein refolded with lipid and RNA has the biochemical hallmarks of a prion but lacks in vivo infectivity. PloS One. 2013;8:e71081. doi: 10.1371/journal.pone.0071081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar JG, Kellings K, Serban A, Groth D, Cleaver JE, Prusiner SB, Riesner D. Search for a prion-specific nucleic acid. J Virol. 2005;79:10796–10806. doi: 10.1128/JVI.79.16.10796-10806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrington E, Gabus C, Leblanc P, Chnaidermann J, Grave L, Dormont D, Swietnicki W, Morillas M, Marck D, Nandi P, Darlix JL. PrPC has nucleic acid chaperoning properties similar to the nucleocapsid protein of HIV-1. C R Biol. 2002;325:17–23. doi: 10.1016/s1631-0691(02)01388-4. [DOI] [PubMed] [Google Scholar]
- Nandi PK, Nicole JC. Nucleic acid and prion protein interaction produces spherical amyloids which can function in vivo as coats of spongiform encephalopathy agent. J Mol Biol. 2004;344:827–837. doi: 10.1016/j.jmb.2004.09.080. [DOI] [PubMed] [Google Scholar]
- Nandi PK, Bera A, Sizaret PY. Osmolyte trimethylamine N-oxide converts recombinant alpha-helical prion protein to its soluble beta-structured form at high temperature. J Mol Biol. 2006;362:810–820. doi: 10.1016/j.jmb.2006.07.060. [DOI] [PubMed] [Google Scholar]
- Gomes MP, Vieira TC, Cordeiro Y, Silva JL. The role of RNA in mammalian prion protein conversion, Wiley interdisciplinary reviews. RNA. 2012;3:415–428. doi: 10.1002/wrna.118. [DOI] [PubMed] [Google Scholar]
- Cordeiro Y, Silva JL. The hypothesis of the catalytic action of nucleic acid on the conversion of prion protein. Prot Peptide Lett. 2005;12:251–255. doi: 10.2174/0929866053587138. [DOI] [PubMed] [Google Scholar]
- Liu ML, Wen JJ, Xu XF, Zhao DM. Neurotoxic effect of the complex of the ovine prion protein (OvPrP(C)) and RNA on the cultured rat cortical neurons. Neurochem Res. 2011;36:1863–1869. doi: 10.1007/s11064-011-0506-2. [DOI] [PubMed] [Google Scholar]
- Gomes MP, Millen TA, Ferreira PS, e Silva NL, Vieira TC, Almeida MS, Silva JL, Cordeiro Y. Prion protein complexed to N2a cellular RNAs through its N-terminal domain forms aggregates and is toxic to murine neuroblastoma cells. J Biol Chem. 2008;283:19616–19625. doi: 10.1074/jbc.M802102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima T, Matsugami A, Nishikawa F, Nishikawa S, Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornemann S, Glockshuber R. A scrapie-like unfolding intermediate of the prion protein domain PrP(121–231) induced by acidic pH. Proc Natl Acad Sci USA. 1998;95:6010–6014. doi: 10.1073/pnas.95.11.6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liemann S, Glockshuber R. Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry. 1999;38:3258–3267. doi: 10.1021/bi982714g. [DOI] [PubMed] [Google Scholar]
- Nandi PK, Leclerc E, Nicole JC, Takahashi M. DNA-induced partial unfolding of prion protein leads to its polymerisation to amyloid. J Mol Biol. 2002;322:153–161. doi: 10.1016/s0022-2836(02)00750-7. [DOI] [PubMed] [Google Scholar]
- Gabus C, Auxilien S, Pechoux C, Dormont D, Swietnicki W, Morillas M, Surewicz W, Nandi P, Darlix JL. The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J Mol Biol. 2001;307:1011–1021. doi: 10.1006/jmbi.2001.4544. [DOI] [PubMed] [Google Scholar]
- Carra JH, Privalov PL. Energetics of folding and DNA binding of the MAT alpha 2 homeodomain. Biochemistry. 1997;36:526–535. doi: 10.1021/bi962206b. [DOI] [PubMed] [Google Scholar]
- Nandi PK. Evidence of molten globule like state(s) of interferon gamma in acidic and sodium perchlorate solutions. Int J Biol Macromol. 1998;22:23–31. doi: 10.1016/s0141-8130(97)00082-2. [DOI] [PubMed] [Google Scholar]
- Swietnicki W, Petersen R, Gambetti P, Surewicz WK. pH-dependent stability and conformation of the recombinant human prion protein PrP(90–231) J Biol Chem. 1997;272:27517–27520. doi: 10.1074/jbc.272.44.27517. [DOI] [PubMed] [Google Scholar]
- Marques AF, Cordeiro Y, Silva JL, Lima LM. Enhanced prion protein stability coupled to DNA recognition and milieu acidification. Biophys Chem. 2009;141:135–139. doi: 10.1016/j.bpc.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- Vieira TC, Cordeiro Y, Caughey B, Silva JL. Heparin binding confers prion stability and impairs its aggregation. FASEB J. 2014;28:2667–2676. doi: 10.1096/fj.13-246777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira TC, Reynaldo DP, Gomes MP, Almeida MS, Cordeiro Y, Silva JL. Heparin binding by murine recombinant prion protein leads to transient aggregation and formation of RNA-resistant species. J Am Chem Soc. 2011;133:334–344. doi: 10.1021/ja106725p. [DOI] [PubMed] [Google Scholar]
- Calzolai L, Zahn R. Influence of pH on NMR structure and stability of the human prion protein globular domain. J Biol Chem. 2003;278:35592–35596. doi: 10.1074/jbc.M303005200. [DOI] [PubMed] [Google Scholar]
- Morrissey MP, Shakhnovich EI. Evidence for the role of PrP(C) helix 1 in the hydrophilic seeding of prion aggregates. Proc Natl Acad Sci USA. 1999;96:11293–11298. doi: 10.1073/pnas.96.20.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetri AC, Surewicz WK. Atypical effect of salts on the thermodynamic stability of human prion protein. J Biol Chem. 2003;278:22187–22192. doi: 10.1074/jbc.M302130200. [DOI] [PubMed] [Google Scholar]
- Armstrong KM, Baldwin RL. Charged histidine affects alpha-helix stability at all positions in the helix by interacting with the backbone charges. Proc Natl Acad Sci USA. 1993;90:11337–11340. doi: 10.1073/pnas.90.23.11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima RI, Thirumalai D. Exploring the propensities of helices in PrP(C) to form beta sheet using NMR structures and sequence alignments. Biophys J. 2002;83:1268–1280. doi: 10.1016/S0006-3495(02)73899-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso DO, DeArmond SJ, Cohen FE, Daggett V. Mapping the early steps in the pH-induced conformational conversion of the prion protein. Proc Natl Acad Sci USA. 2001;98:2985–2989. doi: 10.1073/pnas.061555898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Michelitsch MD, Guenebaut V, Supattapone S, Serban A, Cohen FE, Agard DA, Prusiner SB. Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci USA. 2002;99:3563–3568. doi: 10.1073/pnas.052703499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Zhang GF, Baldwin MA, Cohen FE, Prusiner SB. Separation of scrapie prion infectivity from PrP amyloid polymers. J Mol Biol. 1996;259:608–621. doi: 10.1006/jmbi.1996.0343. [DOI] [PubMed] [Google Scholar]
- Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Re F, Sesana S, Barbiroli A, Bonomi F, Cazzaniga E, Lonati E, Bulbarelli A, Masserini M. Prion protein structure is affected by pH-dependent interaction with membranes: a study in a model system. FEBS Lett. 2008;582:215–220. doi: 10.1016/j.febslet.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu Rev Biophys Biomol Struct. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Nilsson L. Interaction of human SRY protein with DNA: a molecular dynamics study. Proteins. 1998;31:417–433. doi: 10.1002/(sici)1097-0134(19980601)31:4<417::aid-prot8>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Thornton JM. Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J Mol Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan HP, Garcia MA, Jones S, Thornton JM. Identifying DNA-binding proteins using structural motifs and the electrostatic potential. Nucleic Acids Res. 2004;32:4732–4741. doi: 10.1093/nar/gkh803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Figure 1.


