Abstract
Motivation: Off-target interactions of a popular immunosuppressant Cyclosporine A (CSA) with several proteins besides its molecular target, cyclophilin A, are implicated in the activation of signaling pathways that lead to numerous side effects of this drug.
Results: Using structural human proteome and a novel algorithm for inverse ligand binding prediction, ILbind, we determined a comprehensive set of 100+ putative partners of CSA. We empirically show that predictive quality of ILbind is better compared with other available predictors for this compound. We linked the putative target proteins, which include many new partners of CSA, with cellular functions, canonical pathways and toxicities that are typical for patients who take this drug. We used complementary approaches (molecular docking, molecular dynamics, surface plasmon resonance binding analysis and enzymatic assays) to validate and characterize three novel CSA targets: calpain 2, caspase 3 and p38 MAP kinase 14. The three targets are involved in the apoptotic pathways, are interconnected and are implicated in nephrotoxicity.
Contact: lkurgan@ece.ualberta.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
First identified in 1972, Cyclosporine A (CSA) was purified from the fungus Tolypocladium inflatum (Agathos et al., 1987). This drug is primarily used an immunosuppressant to prevent rejection in solid organ transplants (Tedesco and Haragsim, 2012). The mechanism of action of CSA involves binding to its therapeutic target cyclophilin A and forming a complex with calcineurin (Tedesco and Haragsim, 2012). Cyclophilin A is a peptidylprolylisomerase located in the cytoplasm that is inhibited by binding of CSA (Takahashi et al., 1989). Peptidylprolylisomerases are involved in the isomerization of proline peptide bonds in oligopeptides and allow accelerated folding of a protein. Calcineurin is a calcium/calmodulin-dependent serine/threonine protein phosphatase that is specifically involved in the dephosphorylation of transcription factors involved in signaling events, including nuclear factor of activated T cells (NFAT). The binding of CSA to cyclophilin and calcineurin prevents the dephosphorylation of NFAT, thwarting transit to the nucleus. NFAT is critical for initiation of promoters that stimulate the production and activation of T cells; consequently, the immune response is shut down (Tedesco and Haragsim, 2012). CSA is also used to treat several disease states, including heart failure, psoriasis and rheumatoid arthritis. Unfortunately, use of CSA is associated with severe side effects, including nephrotoxicity, fibrosis, hepatotoxicity and cardiotoxicity, and their mechanistic details remain unclear (Rezzani, 2004; Tedesco and Haragsim, 2012). CSA was shown to interact with many proteins other than its molecular target, cyclophilin A. These off-target interactions could be involved in the activation of signaling pathways that lead to the undesirable symptoms, and their knowledge could be exploited in other disease states. As a few examples, CSA-induced nephrotoxicity involves renal tubular dysfunction that is thought to result from a blockade of the mitochondria permeability transition pore component cyclophilin D (Devalaraja-Narashimha et al., 2009). CSA also causes endoplasmic reticulum (ER) stress by inhibiting the ER-localized cyclophilin B (Lee et al., 2012) and indirectly activates other proteins, such as TGF-β that is involved in fibrosis (Wolf, 2006).
Motivated by the ubiquity of the protein–CSA interactions, we used inverse ligand binding predictions (Xie et al., 2011) to comprehensively determine putative partners of CSA. This prediction uses structures of a few protein–ligand (drug) complexes, in our case, the available CSA–cyclophilin complexes, to predict other targets of CSA on the structural human proteome scale. The predictions were generated with the ILbind method that was recently shown to accurately find distant targets (targets that have structurally different folds compared with the proteins that are known to interact with the given compound) for a diverse set of >30 small organic ligands (Hu et al., 2012). Similar inverse ligand binding-based approaches that used older predictors were used to find novel targets for other compounds, such as Comtan (Kinnings et al., 2009), Cholesteryl Ester Transfer Protein (CETP) inhibitors (Xie et al., 2009) and Raloxifene (Sui et al., 2012). We empirically show that predictive performance of ILbind is better when compared with other available predictors. We assessed selected top-ranked putative partners of CSA in the context of their involvement in the toxicity-related pathways, and we focused on several targets that are potentially associated with nephrotoxicity. We modeled their interactions with CSA using docking and then verified their binding and changes in the corresponding enzymatic activity using in vitro assays.
2 METHODS
The datasets, methods related to computational modeling, experimental analysis and evaluation protocols are described in the Supplementary Material. Briefly, we collected structures of protein–CSA complexes and used them to predict other protein targets of CSA in the structural human proteome using ILbind. The top-ranked targets were analyzed with Ingenuity platform, and their association with CSA was investigated based on manual scanning of relevant publications. Three targets that are involved in apoptotic pathways with links to nephrotoxicity were further analyzed using docking and molecular dynamics (MD) simulations, and were validated experimentally with surface plasmon resonance (SPR) and enzymatic assays.
3 RESULTS
3.1 Empirical evaluation of predictions
Predictions generated by ILbind were empirically compared against predictions from other relevant approaches: SMAP (Xie and Bourne, 2008) and FINDSITE (Brylinski and Skolnick, 2008). Native targets of CSA were collected from multiple sources, including Protein Data Bank (PDB) (Berman et al., 2000), BindingDB (Liu et al., 2007) and DrugBank (Knox et al., 2011). Details of the assessment are explained in the Supplementary Materials. First, we selected the best performing (based on the AUtpr100 value) output of SMAP and FINDSITE, which are raw score and identity score, respectively, to assess their predictions. Using ILbind, we identified 38% of the native targets of CSA in the top 0.3% of its predictions. The last column in Table 1 shows that ILbind found all native targets in the top 65% (66% when clustering proteins at 80%) of its predictions, compared with the top 72% (73%) and 92% (96%) of the predictions from SMAP and FINDSITE, respectively. Table 1 also reveals that ILbind obtains higher predictive performance (based on the area under the TPR curve, AUtpr, for 10, 20 and 100% of the top scoring predictions) compared with the other two predictors. The corresponding absolute improvements compared with the second best method are modest at ∼ 1% (relative improvements are 100% × (0.44 − 0.43)/ 0.43 = 2.3, 2.1 and 1.2% for AUtpr10, AUtpr20 and AUtpr100, respectively) and statistically significant, except for the AUtpr100 measure.
Table 1.
Benchmark results for ILbind, SMAP and FINDSITE for the prediction of CSA binding in the structural human proteome
| Dataset | Predictor | AUtpr10 | AUtpr20 | AUtpr100 | % proteome where all targets found |
|---|---|---|---|---|---|
| Clustered at 90% similarity | ILbind | 0.44 ± 0.09** | 0.48 ± 0.07** | 0.78 ± 0.04** | 65 ± 9** |
| SMAP | 0.43 ± 0.09* | 0.47 ± 0.08* | 0.77 ± 0.04** | 72 ± 9* | |
| FINDSITE | 0.40 ± 0.10* | 0.42 ± 0.10* | 0.66 ± 0.07* | 92 ± 6* | |
| Clustered at 80% similarity | ILbind | 0.39 ± 0.08** | 0.43 ± 0.07** | 0.76 ± 0.04** | 66 ± 5** |
| SMAP | 0.38 ± 0.08* | 0.41 ± 0.07* | 0.75 ± 0.03** | 73 ± 9* | |
| FINDSITE | 0.37 ± 0.08* | 0.39 ± 0.08* | 0.66 ± 0.07* | 96 ± 3* |
Note. We report mean ± SD and significance of differences between a given method and ILbind. *P-value < 0.05; **P-value ≥ 0.05 (see Supplementary Material for details).
We also performed a more detailed assessment of predictions for the top 100 predicted targets from each method. We expanded the annotation of the CSA targets collected from PDB, BindingDB and DrugBank (direct targets) with the targets found in articles from PUBMED (indirect targets). The indirect targets were identified by PUBMED search using names of the drug and a given putative target protein and assuming that the target interacts with CSA if there is at least one article that links them together using a meaningful association derived by reading the full article; the results for ILbind are given in Supplementary Table S1. Moreover, we included predictions performed using molecular docking with AUTODOCK 4 (Osterberg et al., 2002), motivated by popularity of this method (Sousa et al., 2006). Because docking is computationally expensive, we limited the scope to the top 100 targets predicted by ILbind, SMAP or FINDSITE. Docking for each of these targets was repeated 40 times; we used the four representative CSA conformation collected from the template targets (PDBid 3pmp_B, 2rmc_A, 1ikf_H and 2oju_A) to accommodate for flexibility of this drug, and we run each conformation 10 times to accommodate for the randomization related to the use of a genetic algorithm in AUTODOCK 4. The lowest docking energy over 40 runs was used to score predictions from docking. We used the top 100 targets with lowest energies from the docking-based results to perform assessment and then compared these results with the top 100 targets generated by ILbind, SMAP and FINDSITE; each set of the top 100 targets was clustered at 80% identity, and we assessed the results for the corresponding clusters. The TPRs of ILbind, AUTODOCK, SMAP and FINDSITE are 0.54 ± 0.07, 0.45 ± 0.05, 0.43 ± 0.04 and 0.35 ± 0.06, respectively. This means that 54% (32 of 59) of the clusters in the top 100 targets predicted by ILbind are known to bind CSA; this rate is significantly higher (based on the t-test) than the TPRs of the other four methods (P < 0.01).
3.2 Analysis of targets predicted with ILbind
We analyzed the putative CSA–protein interactions provided by ILbind and investigated whether these interactions could be responsible for the side effects experienced by patients taking CSA. We focused on the top 199 high-scoring targets generated by ILbind; these targets and their annotations are available in Supplementary Table S1. They were annotated as supported by direct evidence of binding to CSA (based on PDB, BindingDB and DrugBank) or indirect evidence (based on search in PUBMED); otherwise they were annotated as having no prior evidence of binding to CSA.
Supplementary Figure S5A shows the cumulative number of putative targets in each of these three types of annotations and their ILbind scores. The direct evidence exists for cyclophilins and ILbind generated high scores for the corresponding top 42 cyclophilins and cyclophilin-like folds. We found 48 targets with indirect evidence and another 109 targets that were annotated as not being supported by evidence. The ILbind scores for these targets were relatively high, >0.65, which was indicative of a high likelihood of interaction. Supplementary Figure S5B, which shows scores over the entire structural human proteome, reveals that below the 0.65 cutoff, the ILbind scores plateau and would be less useful to discriminate the off-targets from non-binding targets. Overall, the results show that our top putative targets include both confirmatory and new results.
After removing similar proteins among the 199 targets, e.g. we had multiple targets that included cyclophilin A domain, we performed IPA analysis with Ingenuity release November 1 2012 for 111 targets of the 144 unique targets; the remaining 33 targets were not recognized by Ingenuity. The results are summarized in Table 2. The top canonical pathways are IL-17 signaling, acute phase response signaling and dendritic cell maturation, all critical for immune system activation, corresponding with the CSA’s intended purpose. The top associated molecular and cellular function is cell death and survival with 64 of the considered 111 targets being involved. We also found a wide range of significantly associated toxicities, including hepatotoxicity, cardiotoxicity and nephrotoxicity, each supported by over a dozen of our putative CSA targets that are identified in Supplementary Table S1. Supplementary Figure S6 provides a detailed overview of the various types of heart, liver and kidney toxicities and their association with the putative targets that we identified. Each of the corresponding 14 toxicity types was connected with at least seven targets and is associated with targets that were categorized as having indirect and no prior evidence. In short, we found that our putative targets of CSA are associated with relevant pathways and cellular function and with several types of toxicities that are observed in patients who take this drug.
Table 2.
Top scoring pathways, molecular and cellular functions and toxicities generated by Ingenuity for the considered putative CSA targets
| Top pathways, functions or toxicities | P-values | Number of targets |
|---|---|---|
| Top canonical pathways | ||
| IL-17 Signaling | 8.51e-10 | 9 |
| Acute phase response signaling | 9.31e-10 | 12 |
| Dendritic cell maturation | 1.48e-09 | 12 |
| Top molecular and cellular functions | ||
| Cell death and survival | 3.91e-14 | 64 |
| Cellular function and maintenance | 7.76e-14 | 53 |
| Cell morphology | 5.04e-11 | 50 |
| Cellular movement | 1.55e-10 | 42 |
| Lipid metabolism | 4.47e-10 | 45 |
| Top toxicity functions | ||
| Hepatotoxicity | ||
| Liver necrosis/cell death | 5.12e-10 | 15 |
| Liver proliferation | 7.24e-08 | 12 |
| Liver inflammation | 2.10e-06 | 8 |
| Liver damage | 6.43e-06 | 9 |
| Liver hepatitis | 2.36e-05 | 8 |
| Cardiotoxicity | ||
| Cardiac necrosis/cell death | 3.02e-08 | 13 |
| Heart failure | 2.81e-05 | 9 |
| Cardiac hypertrophy | 8.03e-05 | 13 |
| Cardiac proliferation | 1.16e-04 | 5 |
| Cardiac damage | 6.90e-04 | 4 |
| Nephrotoxicity | ||
| Kidney failure | 3.65e-08 | 12 |
| Renal inflammation | 3.66e-08 | 13 |
| Renal nephritis | 3.66e-08 | 13 |
| Renal necrosis/Cell death | 3.06e-06 | 14 |
| Renal fibrosis | 2.38e-04 | 4 |
Note. P-values are calculated using the right-tailed Fisher exact test to evaluate whether the targets are involved in a given function, considering the number of molecules in the Ingenuity Knowledge Base that are included in the corresponding network. ‘Number of targets’ indicates the number of putative CSA targets that overlap with a given function.
3.3 Analysis of interactions of CSA with CAPN2, CASP3 and MAPK14
We selected three putative targets that were identified by ILbind, calpain 2 (CAPN2), caspase 3 (CASP3) and p38 mitogen-activated protein kinase 14 (MAPK14), to further characterize and experimentally validate their interactions with CSA. This selection was motivated by our focus on targets that participate in the apoptotic response seen with the CSA treatment (de Arriba et al., 2013; Eckstein et al., 2005; Sato et al., 2011) and because enzymatic assays to monitor their binding and activity were readily available. Our analysis shows that CAPN2, CASP3 and MAPK14 are involved in renal, cardiac and liver necrosis (Supplementary Fig. S6). These targets can be also linked to each other via the apoptotic pathway (Supplementary Fig. S7). CAPN2 was identified to be involved in the regulation of the apoptotic CASP3 by cleaving and reducing CASP3 activity and potentially playing a protective role (Bizat et al., 2003). As apoptosis progresses, CASP3 becomes involved in degrading an endogenous calpain inhibitor, calpastatin, leading to CAPN2-dependent plasma membrane disruption and necrosis (Neumar et al., 2003). Furthermore, direct linkage between CASP3 and MAPK14 was found in human and rat cells (Alvarado-Kristensson et al., 2004; McLaughlin et al., 2001). We hypothesized that binding of CSA to these novel targets may trigger apoptosis and account for the severity of some side effects of this compound.
3.3.1 Molecular docking
We performed MD simulations to model flexibility of the drug molecule and the three target proteins and docking with AUTODOCK 4 to provide putative molecular-level details of the protein–drug interactions (see Supplementary Materials for details). In total, 300 docking simulations (10 MD-generated conformations of CSA and 10 of each of the 3 targets) were performed. These docking results were ranked by their binding energies, and top 10 hits per target were used as a starting point for an all-atom solvated MD simulation to investigate stability and pose of CSA within the corresponding binding pockets of each target. We repeated docking using the molecular operating environment-induced fit algorithm (www.chemcomp/com) to validate AUTODOCK results. Although there are relatively minor variations in the docking results, the overall binding poses generated by the two methods are similar for the three targets (Supplementary Fig. S8). We focus on confirming that CSA binds these targets rather than on providing precise binding modes.
The binding poses of CSA for the three targets generated with AUTODOCK that correspond to the minimal docking energy among the 10 simulations are shown in Supplementary Figure S9. To compare, the binding poses for the four representative positive controls (targets in complex with CSA), which were also computed combining MD and docking, are in Supplementary Figure S10. In spite of a limited flexibility of the backbone of the CSA peptide, its fairly flexible side chains allowed it to adopt distinctive conformations (Supplementary Fig. S4) and to have different geometries in binding sites. The topologies of the binding sites for the positive controls ranged from a flat and open pocket where CSA circulates a hydrophobic protrusion (Supplementary Fig. S10D) to a compact and closed pocket where CSA is buried in a deep cleft (Supplementary Fig. S10A). Similar binding poses are observed for the three putative targets, where the interaction with MAPK14 (Supplementary Fig. S9C) is characterized by the lowest docking energy. The energies for CASP2, CAPN2 and MAPK14 are −40.5, −45.5 and −60.7 kcal/mol, respectively. To compare, the docking energies of the positive controls are at −42.1 kcal/mol for FAB fragment IGG1-kappa, −55.1 for Cyclophilin A, −60.4 for Cyclophilin C and −60.9 for the cyclophilin-like protein. The fact that the docking energies obtained for the putative targets are similar to the energies obtained for the known targets suggests that CSA may bind these putative targets.
3.3.2 SPR and enzymatic validation
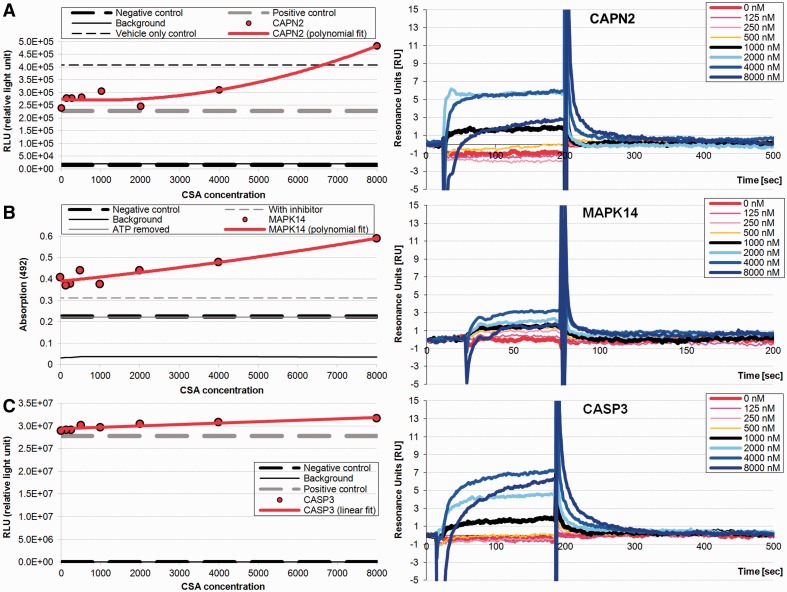
We performed SPR analysis for CAPN2, MAPK14 and CASP3 (Fig. 1), the positive control, cyclophilin A, and the negative control, NF-kappa-B (NFKB) (Supplementary Fig. S9). Our prior experience with SPR showed that higher coupling amounts generate more non-specific interactions for small molecules; thus, we monitored the total coupling and limited it to <2500 relative units (RU). We run a dilution series of CSA (8000, 4000, 2000, 1000, 500, 250, 125 and 0 nM) in triplicate. Clinical use of CSA is usually between 100 and 1600 nM (Hauser, 1998) with this range covering most clinical and physiological situations. Using BiaEvaluation, we computed kinetic association and dissociation rate constants and affinity KD (Supplementary Table S2) to describe drug–target complex formation and stability. We used specific enzymatic assays to monitor the effect of CSA on the enzymatic activity of CAPN2, MAPK14 and CASP3 (Fig. 1), further validating the CSA interactions and presenting a possible hypothesis regarding the effect of CSA on cellular signaling events.
Fig. 1.
Purified enzyme activity measurements (on the left) and real-time sensorgrams (on the right) for the interaction of CSA with calpain 2 (CAPN2; panel A), p38 (MAPK14; panel B) and caspase 3 (CASP3; panel C); see Supplementary Material for details. All measurements were done in triplicate, and the corresponding averages are shown. The sensorgrams are at 8000, 4000, 2000, 1000, 500, 250, 125 and 0 nM concentration of CSA using Biacore 3000
The positive control (ILbind score of 0.86) shows robust binding (Supplementary Fig. S11A) and strong kinetics. Our measurements, with the average KD of 30 nM, 0.253 uM−1s−1 association rate and 6.6 × 10−3 s−1 dissociation rate (Supplementary Table S2), are in agreement with previous work where KD was in the range of 7–40 nM (Kawai et al., 1998; Wear et al., 2005) and association and dissociation rates were 0.86 uM−1s−1 and 10 × 10−3 s−1, respectively (Wear and Walkinshaw, 2006). The negative control NFKB (ILbind score of 0.58) has no appreciable binding (Supplementary Fig. S11B), and thus, kinetics could not be calculated; to the best of our knowledge, NFKB has not been identified to interact with CSA. These results show that SPR discriminates between positive and negative targets. The 0.58 score is low, as the ILbind scores range between 0.32 and 0.86, with majority of them <0.65 (Supplementary Fig. S5B). NFKB is ranked 2312 among the 9652 considered proteins.
CAPN2 is a cytosolic member of calpain family that is activated by intracellular calcium signaling and is involved in homeostasis and apoptotic signaling (Smith and Schnellmann, 2012). SPR shows a substantial increase in binding with the addition of increasing concentrations of CSA (Fig. 1A). Using a luciferase-based activity assay that measures enzymatic cleavage of a specific substrate used by CAPN2, we found that CSA directly binds to CAPN2 and affects its enzymatic cleavage activity in a significant manner; a 40% increase in activity observed at the highest concentration of CSA (Fig. 1A). Calculation of activity constants and curve fitting shows that CSA affects activity of CAPN2 in a quadratic manner over a 2 log concentration of CSA (Fig. 1A).
MAPK14 is a central kinase that is involved in cellular signaling and apoptosis (Wada and Penninger, 2004). Similar to CAPN2, SPR indicates that CSA binds to MAPK14 at higher CSA concentrations (Fig. 1B). Using a MAPK14-specific enzymatic activity assay that measures direct phosphorylation of a MAPK14 substrate in the presence of different concentrations of CSA, we show a 50% increase in the MAPK14 enzymatic activity, which is approximated with a quadratic fit (Fig. 1B).
CASP3 is implicated in apoptosis and is one of the primary caspases that is responsible for cellular damage (Slee et al., 1999). We observe increasing binding of CSA to CASP3 with greater concentrations of CSA (Fig. 1C). Using a luciferase-based enzymatic activity assay, we determined that CSA had a relatively minor effect on CASP3 activity (Fig. 1C), i.e. ∼ 10% increase in activity with higher concentrations of CSA is found.
Interestingly, CAPN2, MAPK14 and CASP3 have similar ILbind scores (0.67, 0.65 and 0.68, respectively; Supplementary Table S1) and comparable SPR curves and RU values (Fig. 1). Although the RU values are lower relative to the values observed for cyclophilin A (Supplementary Fig. S11A), other studies that considered similar small molecules showed comparably low RU values to demonstrate binding (Du et al., 2006; Nordin et al., 2005). Moreover, these lower values also reflect lower coupling to the SPR sensor chip of these three proteins when compared with cyclophilin A. The corresponding kinetic analysis (Supplementary Table S2) reveals that CSA interacts with these three targets, although not as strongly as with the cyclophilin A. We hypothesize that the physiological significance of CSA binding to these off-target proteins should be relevant, as some of the toxic side effects may be caused by these binding events that we show affects the enzymatic properties of these proteins. A more detailed discussion, which links these targets to nephrotoxicity (Peyrou et al., 2007; Ramesh and Reeves, 2005) that is observed in patients who take CSA, is provided in Section 5 in the Supplementary Material.
4 DISCUSSION
We performed a large-scale structural human proteome-wide study to find protein off-targets of CSA using state-of-the-art computational method, ILbind. We empirically demonstrate that predictions generated by ILbind offer good predictive performance. We compiled a comprehensive list of 100+ putative targets of this drug and then linked these targets with an assortment of cellular functions, canonical pathways and toxicities, which are typical for patients who take CSA. We used a complementary arsenal of approaches, including ILbind, molecular dynamics, molecular docking, SPR and enzymatic assays to identify and characterize in detail three novel targets of CSA. We show that CSA likely binds to and may be an exogenous agonist of CAPN2 and MAPK14, and that it also activates CASP3 but to a lesser extent. A detailed discussion of the functional roles of these interactions is provided in the Supplementary Materials. Our study may also lead to new discoveries for a wide range of toxic responses to CSA for which we found previously unknown putative proteins targets. Moreover, our pipeline could be used to provide insights into side effects and mechanisms of action of other small molecule drugs that target proteins.
Funding: This work was supported by Natural Science Foundation of China [grant numbers 31050110432 L.K. and J.R., 31150110577 L.K. and J.R., 11301286 to K.W., 11101226 to G.H.]; Natural Sciences and Engineering Research Council of Canada [grant number 298328 to L.K.]; Canadian Institutes of Health Research [M.M. and J.G.]; and Dissertation Fellowship by University of Alberta [M.J.M.].
Conflict of interest: none declared.
Supplementary Material
REFERENCES
- Agathos SN, et al. The fungal production of cyclosporine. Ann. N. Y. Acad. Sci. 1987;506:657–662. doi: 10.1111/j.1749-6632.1987.tb23863.x. [DOI] [PubMed] [Google Scholar]
- Alvarado-Kristensson M, et al. p38-MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J. Exp. Med. 2004;199:449–458. doi: 10.1084/jem.20031771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizat N, et al. In vivo calpain/caspase cross-talk during 3-nitropropionic acid-induced striatal degeneration—Implication of a calpain-mediated cleavage of active caspase-3. J. Biol. Chem. 2003;278:43245–43253. doi: 10.1074/jbc.M305057200. [DOI] [PubMed] [Google Scholar]
- Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc. Natl Acad. Sci. USA. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Arriba G, et al. Cyclosporine A-induced apoptosis in renal tubular cells is related to oxidative damage and mitochondrial fission. Toxicol. Lett. 2013;218:30–38. doi: 10.1016/j.toxlet.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Devalaraja-Narashimha K, et al. Cyclophilin D gene ablation protects mice from ischemic renal injury. Am. J. Physiol. Renal. Physiol. 2009;297:F749–F759. doi: 10.1152/ajprenal.00239.2009. [DOI] [PubMed] [Google Scholar]
- Du L, et al. Binding investigation of human 5-lipoxygenase with its inhibitors by SPR technology correlating with molecular docking simulation. J. Biochem. 2006;139:715–723. doi: 10.1093/jb/mvj084. [DOI] [PubMed] [Google Scholar]
- Eckstein LA, et al. Cyclosporin a inhibits calcineurin/nuclear factor of activated T-cells signaling and induces apoptosis in retinoblastoma cells. Invest. Ophthalmol. Vis. Sci. 2005;46:782–790. doi: 10.1167/iovs.04-1022. [DOI] [PubMed] [Google Scholar]
- Hauser IA, et al. Therapeutic concentrations of cyclosporine A, but not FK506, increase P-glycoprotein expression in endothelial and renal tubule cells. Kidney Int. 1998;54:113–1149. doi: 10.1046/j.1523-1755.1998.00095.x. [DOI] [PubMed] [Google Scholar]
- Hu G, et al. Finding protein targets for small biologically relevant ligands across fold space using inverse ligand binding predictions. Structure. 2012;20:1815–1822. doi: 10.1016/j.str.2012.09.011. [DOI] [PubMed] [Google Scholar]
- Kawai R, et al. Physiologically based pharmacokinetics of cyclosporine A: extension to tissue distribution kinetics in rats and scale-up to human. J. Pharmacol. Exp. Ther. 1998;287:457–468. [PubMed] [Google Scholar]
- Kinnings SL, et al. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PloS Comp. Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, et al. Cyclosporine A suppresses immunoglobulin G biosynthesis via inhibition of cyclophilin B in murine hybridomas and B cells. Int. Immunopharmacol. 2012;12:42–49. doi: 10.1016/j.intimp.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Liu T, et al. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin B, et al. p38 Activation is required upstream of potassium current enhancement and caspase cleavage in thiol oxidant-induced neuronal apoptosis. J. Neurosc. 2001;21:3303–3311. doi: 10.1523/JNEUROSCI.21-10-03303.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumar RW, et al. Cross-talk between calpain and caspase proteolytic systems during neuronal apoptosis. J. Biol. Chem. 2003;278:14162–14167. doi: 10.1074/jbc.M212255200. [DOI] [PubMed] [Google Scholar]
- Nordin H, et al. Kinetic studies of small molecule interactions with protein kinases using biosensor technology. Anal. Biochem. 2005;340:359–368. doi: 10.1016/j.ab.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Osterberg F, et al. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- Peyrou M, et al. Calpain inhibition but not reticulum endo-plasmic stress preconditioning protects rat kidneys from p-aminophenol toxicity. Toxicol. Sci. 2007;99:338–345. doi: 10.1093/toxsci/kfm105. [DOI] [PubMed] [Google Scholar]
- Ramesh G, Reeves WB. p38 MAP kinase inhibition ameliorates cisplatin nephrotoxicity in mice. Am. J. Physiol. Renal. Physiol. 2005;289:F166–F174. doi: 10.1152/ajprenal.00401.2004. [DOI] [PubMed] [Google Scholar]
- Rezzani R. Cyclosporine A and adverse effects on organs: histochemical studies. Prog. Histochem. Cytochem. 2004;39:85–128. doi: 10.1016/j.proghi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Sato M, et al. Cyclosporine A induces apoptosis of human lung adenocarcinoma cells via caspase-dependent pathway. Anticancer Res. 2011;31:2129–2134. [PubMed] [Google Scholar]
- Slee EA, et al. Serial killers: ordering caspase activation events in apoptosis. Cell Death Differ. 1999;6:1067–1074. doi: 10.1038/sj.cdd.4400601. [DOI] [PubMed] [Google Scholar]
- Smith MA, Schnellmann RG. Calpains, mitochondria, and apoptosis. Cardiovasc. Res. 2012;96:32–37. doi: 10.1093/cvr/cvs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SF, et al. Protein-ligand docking: Current status and future challenges. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- Sui SJH, et al. Raloxifene attenuates Pseudomonas aeruginosa pyocyanin production and virulence. Int. J. Antimicrob. Agents. 2012;40:246–251. doi: 10.1016/j.ijantimicag.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, et al. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- Tedesco D, Haragsim L. Cyclosporine: a review. J. Transplant. 2012;2012:230386. doi: 10.1155/2012/230386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–2849. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- Wear MA, et al. A surface plasmon resonance-based assay for small molecule inhibitors of human cyclophilin A. Anal. Biochem. 2005;345:214–226. doi: 10.1016/j.ab.2005.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear MA, Walkinshaw MD. Thermodynamics of the cyclophilin-A/cyclosporin-A interaction: a direct comparison of parameters determined by surface plasmon resonance using Biacore T100 and isothermal titration calorimetry. Anal. Biochem. 2006;359:285–287. doi: 10.1016/j.ab.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Wolf G. Renal injury due to renin-angiotensin-aldosterone system activationof the transforming growth factor-beta pathway. Kidney Int. 2006;70:1914–1919. doi: 10.1038/sj.ki.5001846. [DOI] [PubMed] [Google Scholar]
- Xie L, Bourne PE. Detecting evolutionary relationships across existing fold space, using sequence order-independent profile-profile alignments. Proc. Natl Acad. Sci. USA. 2008;105:5441–5446. doi: 10.1073/pnas.0704422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, et al. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PloS Comp. Biol. 2009;5:e100038. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, et al. Structure-based systems biology for analyzing off-target binding. Curr. Opin. Struct. Biol. 2011;21:189–199. doi: 10.1016/j.sbi.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.