Abstract
Histone H2B ubiquitination is a dynamic modification that promotes methylation of histone H3K79 and H3K4. This crosstalk is important for the DNA damage response and has been implicated in cancer. Here, we show that in engineered yeast strains, ubiquitins tethered to every nucleosome promote H3K79 and H3K4 methylation from a proximal as well as a more distal site, but only if in a correct orientation. This plasticity indicates that the exact location of the attachment site, the native ubiquitin-lysine linkage and ubiquitination cycles are not critical for trans-histone crosstalk in vivo. The flexibility in crosstalk also indicates that other ubiquitination events may promote H3 methylation.
Keywords: chromatin, crosstalk, Dot1, histone ubiquitination, Set1
Introduction
The DNA in the nucleus is wrapped around octamers of histone proteins. Histones can be modified and these post-translational modifications (PTMs) regulate most processes that occur at the DNA. In turn, histone PTMs are regulated by signaling events in the cell, such as other histone PTMs. This crosstalk can occur between PTMs on the same histone, but also between histones.
An evolutionarily conserved trans-histone crosstalk is that between ubiquitin on the C-terminus of histone H2B and methylation of histone H3 on lysine 79 (H3K79) and lysine 4 (H3K4) (Fig 1A). In yeast, H2B is ubiquitinated on lysine 123 (H2BK123) by the Rad6/Bre1 complex, which enhances the overall activity of Dot1 (Fig 1B) and activates the Set1 complex (COMPASS) ([1] and references therein). Whereas H2Bub is a transient mark, H3K4 and especially H3K79 methylation are more stable and will remain after H2Bub has been removed [2,3]. In mammalian cells, ubiquitination of H2B by RNF20/40 promotes H3K79 methylation by DOT1L and H3K4 methylation by several Set1/MLL complexes [4,5]. As proliferation in MLL-rearranged leukemias depends on RNF20, DOT1L and MLL1, this trans-histone crosstalk may have a role in cancer [6–8].
Figure 1. Location of H2B ubiquitin and its role in H3K79 methylation.
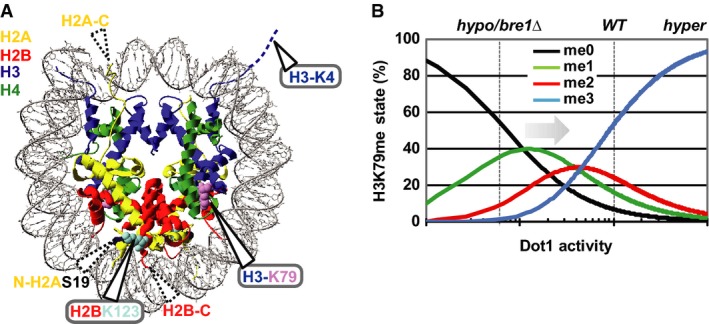
- Nucleosome core particle in which H2BK123, H3K79 and H3K4 are indicated as well as the sites of linear ubiquitin attachment described below. Most of the unstructured N- and C-terminal histone tails are absent from this structure; the histone ends fused to ubiquitin (see below) are indicated with dashed triangles.
- Dependence of H3K79me levels on Dot1 activity, modified from [19]. Dashed lines indicate approximate H3K79me levels in WT or bre1Δ cells. Arrow indicates the direction of the changes expected upon positive histone crosstalk.
In general, despite intensive studies, how monoubiquitin acts as a signal often remains unknown [9]. For the role of ubiquitinated H2B (H2Bub) in promoting Dot1 activity, two non-exclusive models have been suggested: a wedge model and a bridge model (reviewed in [10,11]). The wedge model, in which the ubiquitin moiety disrupts chromatin structure and enables Dot1 to access the nucleosome core surface, is supported by in vitro experiments that showed that ubiquitin on H2B disrupts chromatin compaction [12]. However, since the crosstalk can occur on mononucleosomes [13,14], the opening of the chromatin cannot be the only mechanism behind the crosstalk. Moreover, in vivo, H2Bub may actually lead to chromatin compaction [15–17]. In the bridge model, ubiquitin functions as a bridge to recruit or activate Dot1. However, Dot1 has a similar affinity for both ubiquitinated and unmodified nucleosomes [13] and the only putative ubiquitin-binding domain in Dot1 [18] is not required for the crosstalk [19].
Although the molecular mechanisms remain elusive, in vitro studies have revealed that there is some plasticity in this remarkable histone crosstalk. Ubiquitin confers crosstalk when linked to various residues in the vicinity of H2BK123, and the ubiquitin-like modifier Nedd8 can stimulate Dot1 as strongly as ubiquitin itself [14]. Whether similar rules apply to the crosstalk in vivo is an important next question that remains to be addressed. However, studying the function of one specific ubiquitination event on one particular target in vivo is challenging [20]. We took advantage of the possibility to engineer complex histone mutants in budding yeast, to learn which molecular features of ubiquitinated H2B are important for the trans-histone crosstalk to H3 methylation in vivo. Our experiments using ubiquitins tethered to the nucleosome reveal that the crosstalk from ubiquitination to methylation has a high degree of plasticity in vivo. Our study leads to new insights about the molecular determinants and provides support for a new model for the crosstalk toward H3K79 methylation.
Results
Ubiquitin tethered to the N-terminus of H2A confers crosstalk to H3K79 methylation
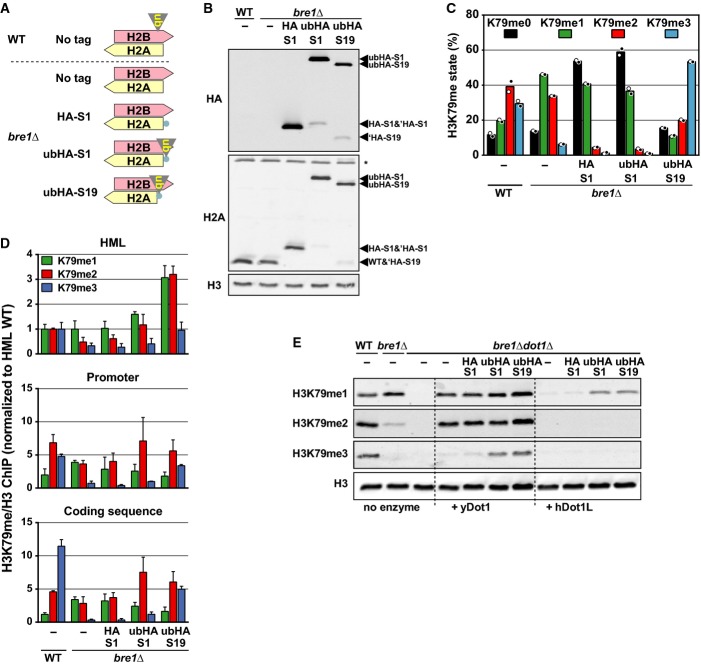
To specifically determine the features of H2Bub that promote histone methylation at H3K79 and H3K4 in vivo, we designed yeast strains in which the endogenous H2Bub pathway was eliminated by the deletion of BRE1 and in which a ubiquitin moiety was specifically re-introduced on the nucleosome. We used a plasmid shuffle assay to create strains in which an engineered H2A gene was the only source of H2A protein (Supplementary Fig S1A and B). Ubiquitin (ub) was tethered via a short HA tag to H2A at S1 or at S19, in close proximity to H2BK123 (Fig 1A). The terminal glycine of ub was removed to prevent ub-H2A cleavage (Supplementary Fig S1D). Strains expressing HA-S1-H2A, ubHA-S1-H2A or ubHA-S19-H2A were viable (Supplementary Fig S1B–D), suggesting that these fusions showed no gross defects in histone transport, assembly and chromatin packaging. The HA-S19-H2A-expressing bre1Δ strain was not viable and was eliminated from further analyses, see Supplementary Fig S1B. Immunoblot analysis confirmed that the H2A fusion proteins were expressed, that no wild-type H2A was present, and that the majority of the fusions were full-length (Fig 2B). ChIP-qPCR experiments confirmed that tagged H2A was incorporated into chromatin (Supplementary Fig S1E).
Figure 2. Ubiquitin on the H2A N-terminus promotes H3K79 methylation in vivo and in vitro.
- Cartoon of H2A/H2B in the used strains.
- Immunoblot analysis shows tagged H2A and the absence of endogenous wild-type H2A. Asterisk indicates non-specific signal. ‘HA indicates ubHA-H2A proteins that lost the ub moiety due to the activity of ubiquitin-specific proteases.
- MS analysis of H3K79 methylation levels in strains shown in (A) and (B) (mean and individual data points of two biological replicates). See also Supplementary Fig S1F.
- ChIP-qPCR data of the different methylation states at three loci. Values for each methyl antibody were normalized to H3, after which all values were normalized per methyl antibody to wild-type at the HML locus. Primer sets against HML, the promoter of the GAL1 gene and the coding sequence of SPA2 were used. Mean ± SD of three biological replicates is shown.
- Immunoblot analysis of the in vitro methyltransferase activity of purified 10× His-tagged yeast or human Dot1 protein toward chromatin templates isolated from yeast strains lacking Dot1 and Bre1 and expressing wild-type H2A or N-terminal fusions of H2A. See also Supplementary Fig S1H.
Source data are available online for this figure.
Multiple methylation by Dot1 occurs by a distributive mechanism, which leads to a characteristic shift in the relative abundance of the H3K79 methylation states when the activity of Dot1 increases or decreases (Fig 1B) [19,21]. The ubiquitin moiety at H2AS19, close to the native position of ubiquitination, caused a shift from H3K79me0/me1 to H3K79me2/me3, while Dot1 expression was not increased (Fig 2C and Supplementary Fig S1F). Unexpectedly, the more distal ubiquitin at S1 of H2A was also able to promote H3K79 methylation, albeit with a somewhat lower efficiency (Fig 2C). This did not require the formation of ubiquitin branches, since K0 mutants of ubiquitin could still promote H3K79 methylation (Supplementary Fig S1G).
H3K79 methylation is not distributed uniformly across the genome, but is very low in heterochromatin, intermediate in intergenic regions and high in coding sequences (see Fig 2D) [22]. In a bre1Δ strain, a shift to lower methylation states was observed at all loci (in line with the distributive mechanism, see Fig 1B). Tethered ubiquitins increased methylation at all tested loci. The H3K79 methylation in strains expressing histone-ubiquitin fusions was more evenly distributed than in wild-type cells with native H2Bub (Fig 2D), confirming that this crosstalk contributes to the establishment of the H3K79 methylation pattern. However, since the pattern was still partially maintained, additional mechanisms of regulation must exist, such as histone turnover [19,23] or the protection of nucleosomes by binding of the Sir complex [24].
Together, these findings show that a ubiquitin constitutively tethered to the N-terminus of H2A by a linear fusion confers trans-histone crosstalk in vivo, throughout the genome. To eliminate indirect effects of the ub-H2A on H3K79 methylation, we performed in vitro methyltransferase assays using chromatin templates isolated from the engineered yeast strains. Ubiquitin tethered to the H2A N-terminus promoted the activity of yeast Dot1 as well as human DOT1L in vitro, suggesting that it has a direct effect on Dot1 activity (Fig 2E and Supplementary Fig S1H).
Ubiquitin tethered to the C-terminus of H2A or H2B confers modest crosstalk to H3K79 methylation
To find the limits of the observed plasticity, and thereby determine which features are important for the crosstalk, we created C-terminal fusions between histone H2A or H2B and ubiquitin (Supplementary Fig S2A). The C-terminal glycine of ub was changed to valine to avoid conjugation of the fusion protein to ubiquitination targets. Yeast strains expressing an H2A-ub or H2B-ub fusion protein were viable in the absence of untagged versions of the respective histones (Supplementary Fig S2B). Expression of the fusion proteins and the absence of untagged histone protein were confirmed by immunoblot analysis (Fig 3B).
Figure 3. Ubiquitins tethered to the C-terminus of H2A and H2B can modestly promote H3K79 methylation in vivo.
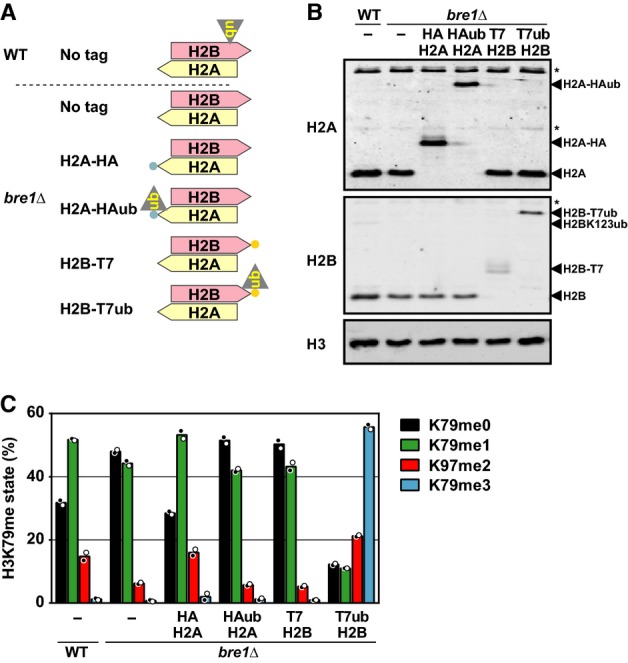
- Cartoon of H2A/H2B in the used strains.
- Immunoblot analysis of tagged and untagged histones. Asterisk indicates non-specific signal.
- MS analysis of H3K79 methylation levels in strains expressing H2A or H2B C-terminal ub fusions (mean and individual data points of two biological replicates). See also Supplementary Fig S2C.
Source data are available online for this figure.
In bre1Δ strains, lacking endogenous H2Bub, ubiquitins tethered to the C-terminus of H2A or H2B led to more H3K79me1/me2 and less H3K79me0, while Dot1 expression was not increased (Fig 3C and Supplementary Fig S2C). However, they bypassed the lack of endogenous H2Bub only partially; ubiquitin tethered to the H2A or H2B C-terminus was much less potent than ubiquitin tethered to the N-terminus of H2A (see Fig 2C).
Ubiquitin crosstalk to H3K4 methylation
Having established that ubiquitin tethered to the nucleosome can take over the role of native H2Bub in promoting Dot1 activity, we asked whether they could also be used to gain insight into the crosstalk from H2Bub to H3K4 methylation. Using the engineered yeast strains, we found that ub-H2A fully restored H3K4me1 and me2 and partially restored H3K4me3 in the absence of endogenous H2Bub (Fig 4A). H2A-ub did not promote H3K4 methylation and H2B-ub had a modest effect (Fig 4A). Taken together, the findings indicate that also the crosstalk to H3K4 methylation is plastic and that Dot1 and Set1 require overlapping but also distinct structural features of chromatin containing H2Bub (Supplementary Table S1).
Figure 4. Tethered ubiquitins can promote H3K4 methylation and DNA damage checkpoint activation.
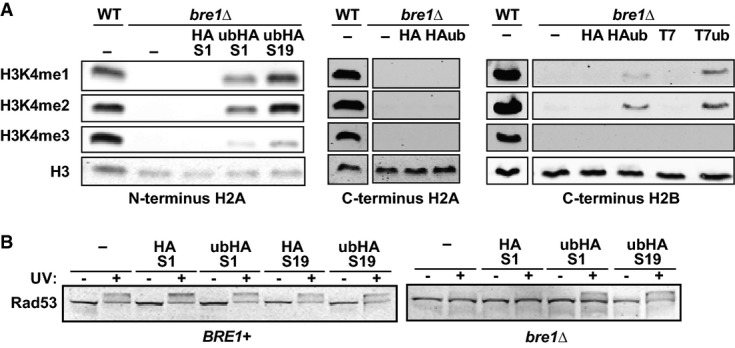
- Immunoblot analysis of H3K4 methylation in strains expressing various histone-ubiquitin fusions.
- Immunoblot analysis of activation of the central DNA damage checkpoint kinase Rad53 upon exposure of yeast cells arrested in G1/S to 100 J m−2 UV irradiation. Rad53 is activated by phosphorylation, which leads to a mobility shift.
Tethered ubiquitins substitute for native H2B ubiquitination in the DNA damage response
H2B ubiquitination and H3K79 methylation are required for a timely induction of the DNA damage response [25]. However, whether dynamic changes in H2Bub or histone methylation are required is unknown. Two general models have been proposed: (i) due to global H2Bub and H3K79me, checkpoint proteins are at the DNA and can be phosphorylated upon DNA damage and (ii) a local increase in H2Bub and H3K79me at the site of lesion recruits and/or activates checkpoint proteins [26–28]. We investigated this problem, using phosphorylation of the central kinase Rad53 as the readout of checkpoint activation after UV irradiation of G1-arrested cells. Whereas bre1Δ cells poorly phosphorylated Rad53 upon UV irradiation (Fig 4B), tethered ubiquitins restored Rad53 phosphorylation in bre1Δ cells (visible as an upward shift of the protein, Fig 4B and Supplementary Fig S3A). Since the constitutive ubiquitin moieties on each nucleosome are sufficient for checkpoint activation after UV, we conclude that a local accumulation of H2Bub specifically at the site of damage is not required. Instead, the data suggest a role of global chromatin modification in modulating the DNA damage checkpoint. In addition, since the ubiquitin tethers are static, our results demonstrate that cycles of ubiquitination and deubiquitination are not required for DNA damage checkpoint activation.
Discussion
The crosstalk between H2B ubiquitination and H3K4 and H3K79 methylation is evolutionary conserved from yeast to metazoans. Since many other chromatin proteins are also subject to ubiquitination, an important question is which molecular features of ubiquitinated H2B are important for the trans-histone crosstalk in vivo. We have shown that various tethered ubiquitins, expressed as linear histone-ubiquitin fusions, can substitute for native H2BK123ub in the crosstalk to H3K79 and H3K4 methylation. This reveals an unexpected plasticity in the crosstalk in vivo.
The extent of the plasticity in the crosstalk in vivo is remarkable. Previously, in vitro experiments already revealed some plasticity in the site of ubiquitin attachment [14,29]. However, the in vivo plasticity of the crosstalk observed here goes beyond what has so far been observed in vitro. This allows us to draw several conclusions about the features of H2Bub that are important for crosstalk to H3 methylation.
First, the precise location of ubiquitin attachment to the nucleosome core surface is not critical for nucleosome crosstalk since H3K79 and H3K4 methylation were enhanced by ubiquitin tethered to various positions on the nucleosome. This is in line with the finding that ubiquitination of H2BK34 by MSL2 stimulates H3K79 and H3K4 methylation in human cell lines and in vitro [30]. In contrast to the previous finding that a short linker between ubiquitin and H2B decreases the crosstalk considerably in vitro [14], ubiquitin tethered to H2AS1 was approximately as effective as ubiquitin on H2AS19. Note that the ubiquitin tethered to H2AS1 is attached to the flexible H2A N-terminus and thereby not in itself located on the nucleosome core surface.
Second, the specific branched structure of ubiquitin linked to a lysine side chain via an isopeptide bond is not a critical feature, since the genetically encoded ubiquitin fusions represent linear proteins and also lack the terminal GG motif. Therefore, the specificity must originate from other parts of ubiquitin.
Third, modification–demodification cycles of H2Bub are not required for crosstalk in vivo. Steady-state levels of H2Bub are the result of ongoing ubiquitination by Bre1/Rad6 and deubiquitination by Ubp8 and Ubp10 [3]. It has been proposed that turnover of histone modifications could be an important determinant of epigenetic regulation [31,32]. However, since our non-cleavable ubiquitins could promote H3K79 and H3K4 methylation, we conclude that H3 methylation can be promoted without H2Bub turnover.
Fourth, Bre1 activity toward non-nucleosomal substrates is not a requirement for the crosstalk, since ubiquitins tethered to the nucleosome confer crosstalk to H3 methylation in the absence of Bre1. It has been suggested that the Bre1/Rad6 ubiquitination machinery, rather than H2BK123ub, promotes maximal crosstalk, potentially by ubiquitinating Swd2 [33,34]. However, this mechanism is still under debate [35]. In our strains, the presence of ubiquitin on the nucleosome is uncoupled from Bre1 activity. Our finding that tethered ubiquitins can promote H3 methylation in the absence of Bre1 argues against a critical role of the ubiquitination machinery. However, since even the most efficient tethered ubiquitins we engineered did not fully rescue H3K4me3 and H3K79me3, it is still possible that the Bre1-dependent ubiquitination of a non-histone protein has an additional effect.
Fifth, the orientation of the ubiquitin seems to be a critical structural feature. The C-terminus of H2B and the N-terminus of H2A are at approximately the same distance from H2BK123 (Supplementary Fig S3B and C). Yet, ubiquitin tethered to H2AS1 was much more effective in histone crosstalk. One key structural difference is that the ubiquitin on the N-terminus of H2A is presented in the same orientation as the native ub on H2BK123, whereas ubiquitin on the C-terminus of H2B is in the opposite orientation.
We believe that our in vivo results, together with previous in vitro findings, are most parsimonious with a model in which ubiquitin can interact with the nucleosome surface and thereby increase the chance of a productive encounter of Dot1 with H3K79 on the nucleosome. This could be achieved by creating a physical road block or crash barrier that coaches Dot1 in the right direction (see Fig 5). A similar fold-back mechanism has previously been suggested for ubiquitin on PCNA [36]. Ubiquitin tethered to the N-terminus of H2A, both at S19 and S1, would be able to fold back just like the native H2Bub. The ubiquitins tethered to the C-termini, which are in the opposite orientation, would not be able to do this. The modest crosstalk that is still observed for these tethers may occur through another mechanism, possibly as proposed in the wedge or bridge model.
Figure 5. Schematic representation of a crash barrier model for the role of H2Bub in promoting histone methylation.
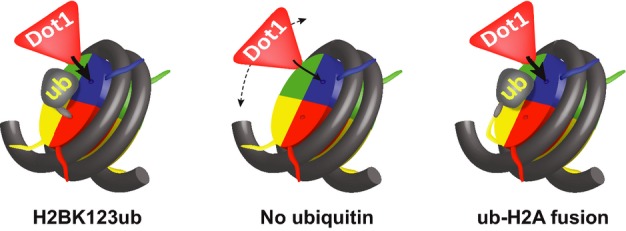
Ubiquitin, if in the correct orientation, can interact with the nucleosome and coach Dot1 toward H3K79, thereby increasing Dot1′s chances of a productive encounter. See text.
The finding that ubiquitin moieties at locations other than H2B can promote histone methylation suggests that nucleosome crosstalk could also be affected by other ubiquitination events in the cell. This is consistent with the crosstalk that has been observed between H2BK34ub and H3 methylation [30], but the site of ubiquitination does not have to be limited to H2B or even the nucleosome core. It will be interesting to determine whether the extensive chromatin ubiquitination during DNA damage response in mammalian cells [25], especially the ubiquitination at the N-terminus of H2A [37], influences the methylation of resident or newly assembled histones during the process of DNA repair. The tethered ubiquitins that we describe here are genetically encoded and amenable to genetic manipulation. Therefore, they will provide opportunities to further delineate the role of ubiquitin in the control of histone methylation and other aspects of chromatin structure and function.
Materials and Methods
Strains, plasmids and growth conditions
Yeast media and growth conditions, and strains and plasmids are described in the Supplementary Methods.
Immunoblotting and mass spectrometry
Histone methylation and expression were determined by immunoblotting and mass spectrometry as described previously [19]. See Supplementary Methods.
In vitro methyltransferase assays
Recombinant 10× His-Dot1 proteins were incubated with yeast nuclear chromatin templates and analyzed by immunoblot analysis as described previously [21]. See Supplementary Methods.
DNA damage checkpoint activation
Yeast cells were arrested in G1 for 2 h, exposed to 100 J/m2 UV irradiation and recovered for 30 min before protein extracts were made, as described previously [21]. See Supplementary Methods.
Acknowledgments
We thank T. Sixma, F. Mattiroli, I. Stulemeijer and C. McLean for critical reading of the manuscript, M. Terweij for the initial characterization of the RITE strains, Y. Bollen for advice on protein spacer length calculations, T. Richmond for advice and sharing unpublished observations, L. O'Neil for providing the H3K4me1 antibody and L. Janssen for the hUb(K0) plasmids. FvL and HV were supported the Dutch Cancer Society (KWF2009-4511) and the Netherlands Genomics Initiative.
Author contributions
HV and FvL designed the studies, analyzed the data and wrote the paper. HV and TvW performed the plasmid and strain constructions, yeast manipulations and methyltransferase assays. ELdG, AFMA and AJRH were responsible for mass spectrometry measurements. DO constructed conditional ub-tag cassettes under supervision of PSS.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
Supplementary Information
Review Process File
Source Data for Figure 2C
Source Data for Figure 3C
References
- 1.Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi Y, Chandrasekharan MB, Sun Z-W, Osley MA, Strahl BD, Jaspersen SL, et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu B, Zheng Y, Pham A-D, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Kim J-A, McGinty RK, Nguyen UTT, Muir TW, Allis CD, Roeder RG. The n-SET Domain of Set1 Regulates H2B Ubiquitylation-Dependent H3K4 Methylation. Mol Cell. 2013;49:1121–1133. doi: 10.1016/j.molcel.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang E, Kawaoka S, Yu M, Shi J, Ni T, Yang W, Zhu J, Roeder RG, Vakoc CR. Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110:3901–3906. doi: 10.1073/pnas.1301045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1 l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 10.Braun S, Madhani HD. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 2012;13:619–630. doi: 10.1038/embor.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandrasekharan MB, Huang F, Sun Z-W. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5:460–468. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 15.Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandrasekharan MB, Huang F, Sun Z-W. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming AB, Kao C-F, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Oh S, Jeong K, Kim H, Kwon CS, Lee D. A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem Biophys Res Commun. 2010;399:512–517. doi: 10.1016/j.bbrc.2010.07.100. [DOI] [PubMed] [Google Scholar]
- 19.De Vos D, Frederiks F, Terweij M, van Welsem T, Verzijlbergen KF, Iachina E, de Graaf EL, Altelaar AFM, Oudgenoeg G, Heck AJR, et al. Progressive methylation of ageing histones by Dot1 functions as a timer. EMBO Rep. 2011;12:956–962. doi: 10.1038/embor.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson A, Werner A, Rape M. The colossus of ubiquitylation: decrypting a cellular code. Mol Cell. 2013;49:591–600. doi: 10.1016/j.molcel.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat Struct Mol Biol. 2008;15:550–557. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- 22.Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9:e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Côté J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 27.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang S-Y, Eppink B, Chung YM, Shalev G, Shema E, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiloh Y, Shema E, Moyal L, Oren M. RNF20-RNF40: a ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 2011;585:2795–2802. doi: 10.1016/j.febslet.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Lee SY, Zhou B, Nguyen UTT, Muir TW, Tan S, Dou Y. ASH2L Regulates Ubiquitylation Signaling to MLL: trans-Regulation of H3 K4 Methylation in Higher Eukaryotes. Mol Cell. 2013;49:1108–1120. doi: 10.1016/j.molcel.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. The RING finger protein MSL2 in the MOF complex Is an E3 ubiquitin ligase for H2B K34 and Is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell. 2011;43:132–144. doi: 10.1016/j.molcel.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry KW, Wyce A, Lo W, Duggan LJ, Emre NCT, Kao C, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thornton JL, Westfield GH, Takahashi Y-H, Cook M, Gao X, Woodfin AR, Lee J-S, Morgan MA, Jackson J, Smith ER, et al. Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev. 2014;28:115–120. doi: 10.1101/gad.232215.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10:1365–1371. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- 35.Soares LM, Buratowski S. Histone crosstalk: H2Bub and H3K4 methylation. Mol Cell. 2013;49:1019–1020. doi: 10.1016/j.molcel.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freudenthal BD, Gakhar L, Ramaswamy S, Washington MT. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat Struct Mol Biol. 2010;17:479–484. doi: 10.1038/nsmb.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattiroli F, Vissers JHA, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File
Source Data for Figure 2C
Source Data for Figure 3C