Abstract
The early postnatal period is a critical window for intestinal and immune maturation. Intestinal development and microbiome diversity and composition differ between breast- (BF) and formula-fed (FF) infants. Mechanistic examination into host-microbe relationships in healthy infants has been hindered by ethical constraints surrounding tissue biopsies. Thus, a statistically rigorous analytical framework to simultaneously examine both host and microbial responses to dietary/environmental factors using exfoliated intestinal epithelial cells was developed. Differential expression of ~1,200 genes, including genes regulating intestinal proliferation, differentiation and barrier function, was observed between BF and FF term infants. Canonical correlation analysis uncovered a relationship between microbiome virulence genes and host immunity and defense genes. Lastly, exfoliated cells from preterm and term infants were compared. Pathways associated with immune cell function and inflammation were up-regulated in preterm, whereas cell growth-related genes were up-regulated in the term infants. Thus, coordinate measurement of the transcriptomes of exfoliated epithelial cells and microbiome allows inquiry into mutualistic host-microbe interactions in the infant, which can be used to prospectively study gut development or, retrospectively, to identify potential triggers of disease in banked samples.
Keywords: infant, intestine, nutrition, microbiome, gene expression, exfoliated cells
Introduction
Intestinal epithelial cells and the commensal microbiota are in close and intimate contact. Studies emanating from germ free and gnotobiotic animals have provided conclusive evidence of the critical role of the intestinal microbiota in regulating gut development and gene expression [1,2], mucosal and systemic immunity [3], the enteric nervous system [4], gut brain axis [5] and host metabolism [6,7]. Recent studies have dispelled the concept that amniotic fluid and meconium are sterile under normal conditions [8]. Meconium, which is formed primarily by ingestion of amniotic fluid by the fetus in utero, also contains exfoliated intestinal cells and mucus. The meconium microbiome is influenced by maternal factors, including clinical conditions [9] and probiotic use [10], and may impact child health outcomes [8, 9, 11]. Thus, host-microbe interactions and education of the neonatal immune system begin in the womb [8].
Immediately after delivery, the human infant acquires a much more complex microbiota, whose composition is influenced by an interplay between genetic and environmental factors [12], of which nutrition is a key component [13]. At the same time, the gastrointestinal tract undergoes rapid structural and functional adaptation, which differs between breast-fed (BF) and formula-fed (FF) infants [14,15]. Although human milk contains growth factors and bioactive proteins and lipids that may directly promote the growth of the gastrointestinal tract [16,17], we speculated that dissimilarities in the composition of the microbiota between breast- and formula-fed infants [17,18] could also be contributing to the enhanced gut development observed by mode of nutrition [14,15].
Our long-term goal is to determine the role of host-microbe interactions within the neonatal intestine on infant development and to define how these cross-functional communications are affected by diet. Among the components of human milk that shape the composition of the microbiota are the human milk oligosaccharides (HMO). The HMO are comprised of a mixture of up to 200 complex oligosaccharides that constitute the third most predominant component of human milk [19]. The HMO content and composition is influenced by the mothers’ genetics (FUT-2 secretor status and Lewis blood group) [20], preterm delivery [21] and, to a lesser degree, the stage of lactation, where sialic acid containing HMO decline, while fucosylated HMO increase or stay constant over the course of lactation [19]. The potential physiological roles of HMO for the developing infant is far reaching in that their multifunctional actions range from regulation of intestinal cell proliferation, functional differentiation and apoptosis [22, 23], gene expression [24], immune function [25–27], pathogen protection [28, 29], and prebiotic activities, including serving as substrates for fermentation [30, 31] and promoting growth of specific bifidobacteria [32], bacteroides [33] and Lactobacillus [34] species (reviewed in [35]).
We hypothesize that nutrition is a central regulator of host-microbe interactions in early life. As noted above, the composition of the microbiota of BF and FF infants differs in terms of overall diversity as well as composition [12, 13, 18]. Epidemiological studies have demonstrated that human milk protects against common infectious diseases in infancy (otitis media, respiratory syncytial virus, urinary tract infection), necrotizing enterocolitis (NEC) in preterm infants as well as immune-mediated disorders in later childhood, including allergy, asthma, atopic dermatitis, inflammatory bowel disease, Celiac Disease, Type 1 and Type 2 diabetes mellitus, and leukemia (ALL and AML) [36]. Recently, Walker [37] proposed that a diverse balanced microbiota is necessary for the development of an appropriate innate and adaptive immune response. This is further supported by studies associating dysbiosis in early life with immune-mediated childhood disorders [38–40] and obesity [41, 42]. Dysbioses can arise from common pediatric practices, including preterm delivery, formula feeding, cesarean section, and use of antibiotics [42, 43] (Figure 1). Interestingly, cesarean section [43] and antibiotic use [44] are independently associated with an increased susceptibility to immune-mediated disease, potentially through dysregulation of host immune homeostasis [44, 45]. It is important to note that all of these practices are amenable to changes in clinical protocols, and, as such, should be a priority for pediatric practice.
Figure 1.
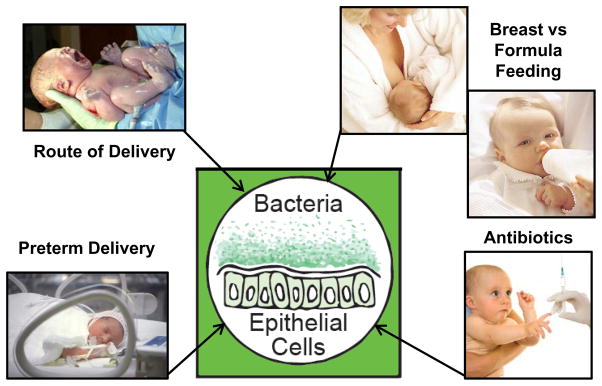
Common pediatric practices that impact gut microbiota and host microbe interactions.
Given the evidence that early life nutritional exposures program long-term health outcomes, potentially through host-microbe interactions, our research group set out to systematically integrate genomic data from both the infant (host mucosa) and gut microbiota in order to define host gene-diet interactions within the context of the structure and operations of gut microbial communities. Until recently, no investigators had comprehensively profiled intestinal gene expression during early postnatal development due to limited availability of intestinal tissue from healthy infants. Thus, the potential for exfoliated epithelial cells to provide a non-invasive readout of intestinal gene expression was investigated [46–48].
Use of Exfoliated Cells to Assess Host Gene Expression
Each day, ~1/3rd to 1/6th of normal adult epithelial cells are shed [49], which corresponds to ~10 billion (1010) cells per day. Exfoliation of intestinal epithelial cells from the villus tips in the small intestine and crypt surface in the colon is ab active biochemical process linked to intestinal epithelial homeostasis [50]. Exfoliation typically induces anoikis, rather than apoptosis, which is a form of programmed cell death induced by anchorage-dependent cells detaching from the surrounding extracellular matrix. Detachment also induces autophagy, which is a survival mechanism to loss of nutrients [51]. The exfoliated cells enter into a quiescent state and appear to maintain viability for differing lengths of time depending on the sources of cells. For example, quiescent exfoliated epithelial cells without signs of apoptosis were recovered in gastric fluid aspirates obtained from preterm infants [52]. Furthermore, exfoliated quiescent epithelial cells can be cultured, evidenced by the ability to use exfoliated cells to forms lumens in 3-dimensional epithelial cell culture [47, 48], suggesting that detachment–induced autophagy contributes to the viability of these cells.
This vast reservoir of host cells generated by exfoliation sparked interest from both basic and clinical translational investigators due to their potential utility to non-invasively assess cellular markers of gastrointestinal disease, predominantly colon cancer [53, 54]. Subsequently, exfoliated epithelial cells had been used as sentinels of in vivo exposure to nutritional regimens [55, 56] or as markers of disease states, including cancer in adults [57, 58] and children with inflammatory bowel disease [59]. More recently, the feasibility of identifying protein markers and amplifying genes by polymerase chain reaction was demonstrated in exfoliated gastric cells from preterm infants [52, 60]. Kaeffer and colleagues studied isolated exfoliated cells obtained from gastric aspirates [52, 60] and stool samples [52] obtained from preterm infants. The gastric exfoliated cells were confirmed to be of epithelial origin by cytokeratin 18 expression and mRNA for beta-actin, clock genes and SLC26-A7-1, an apical Cl−/HCO3−/sulfate exchanger present on parietal cells, was detected [52]. They subsequently reported that exfoliated epithelial cells isolated from gastric aspirates were quiescent and expressed membrane bound H+/K+ ATPase. Importantly, when the H+/K+ ATPase-positive cells were established in cell culture, Pouf5F1-Oct4 expression, a biomarker of progenitor status, was maintained [52].
Our goal was to investigate genome-wide markers by interrogating the transcriptome through gene microarray [46] or, more recently, RNAseq [48]. Because the number of intact cells that can be isolated from fecal material is low [53], the Chapkin laboratory developed a noninvasive mRNA-based method as a highly sensitive technique for detecting molecular markers of intestinal development and function [57, 58]. This methodology has the advantage of using host exfoliated cell mRNA directly isolated from feces, which contain sloughed small intestinal and colon cells. The method is capable of isolating and quantifying specific mRNAs under various intestinal conditions and has been tested in adult humans [58, 61]. A summary of the studies that have applied exfoliated epithelial cells in pediatric populations is shown in Table 1 and will be discussed below.
Table 1.
Studies in Pediatric Populations using Exfoliated Epithelial Cells to Measure the Impact of Diet, Gestational Age or Probiotic Administration on Gene Expression.
| Author | Population Studied | Tissue | How Obtained | Key Findings |
|---|---|---|---|---|
| Chapkin et al. 2010 [46] | BF (n=12) and FF (n=10) term | Intestine | Stool | 1,214 genes were differentially expressed between BF and FF. Major gene pathways were related to signal transduction, cytoskeletal remodeling, cell adhesion and immune response. |
| Kaeffer et al. 2007 [52] | Preterm (n=96); 24–36 wk GA | Stomach; Intestine | Gastric aspirates; Stool | Exfoliated cells isolated from gastric aspirates expressed housekeeping, cytokeratin, SLC-26-A7-I, period 2 and clock genes by PCR |
| Kaeffer et al. 2011 [60] | Preterm (n=24); 25–32 wk GA | Stomach | Gastric aspirates; Collected weekly after birth or every 3h over a 24h period | H+/K+-ATPase positive cells expressing markers of progenitor status were recovered from gastric fluid; aspirates in preterm infants. Exfoliation increased with postnatal/post-conceptual age and volume of enteral feeds and was greater in infants fed preterm formula than preterm human milk. |
| Knight et al. 2014 [48] | Term (n=6) and preterm (n=6) | Intestine | Stool | RNAseq analysis of host mRNA in stool. Pathways associated with immune cell function and inflammation were up-regulated in preterm infants; cell growth related genes were up-regulated in term infants. |
| Rautava et al. 2012 [10] | Term infants delivered by cesarean section (n=29). Mothers were given placebo (n=10), 109 CFU Bifidobacterium. lactis (n=10) or 109 CFU B. lactis + 109 CFU Lactobacillus rhamnosus GG (=9) for 14 d prior to cesarean | Intestine | Meconium | PCR of host mRNA in meconium. Reduced TLR7 mRNA expression was detected in intestinal cells from meconium of infants whose mothers received B. lactis, while maternal B. lactis +Lactobacillus GG decreased TLR6 mRNA expression in fetal intestinal cells from meconium. |
| Schwartz et al. 2012 [47] | BF (n=6) and FF (n=6) term | Intestine | Stool | Virulence genes in the bacterial metagenome were correlated with immunity and defense genes in host exfoliated epithelial cells. |
Abbreviations: BF, breastfed; FF, CFU, colony forming units; formula-fed; GA, gestational age; PCR, polymerase chain reaction; TLR, toll like receptor
Stool-derived Eukaryotic mRNA can be used to Non-invasively Assess the Impact of Nutrition on Intestinal Gene Expression in Term Infants
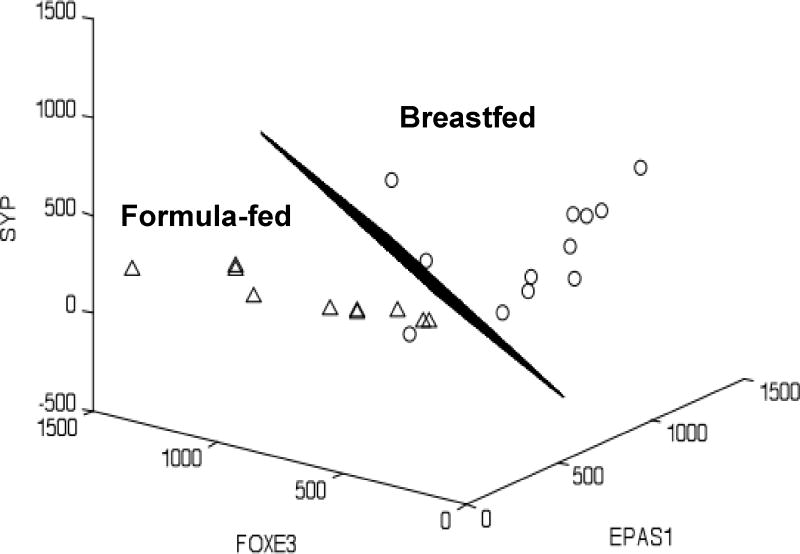
In a proof-of-principle study, stool samples containing exfoliated host cells and luminal bacteria were collected from 3-month-old exclusively BF or FF infants [46]. A total of 1,214 genes were significantly differentially-expressed between BF and FF infants, however, we focused our analyses on the 146 genes that were included in a list of 529 genes that we had a priori hypothesized could be differentially expressed based on prior knowledge. Analysis of gene networks reflected broad differences with respect to Signal Transduction (WNT, NOTCH, TGF-β), Cytoskeletal Remodeling; Cell Adhesion and Immune Response [46]. Linear Discriminant Analysis (LDA) was used to identify genes that best “classified” or discriminated BF from FF infants and the top up- and down-regulated genes and their fold-changes are shown Table 2. Although these human milk-regulated genes were identified in term infants, they could have particular importance to infants with impaired gut development and function, including preterm infants. These genes included glucocorticoid receptor (NRC31), as glucocorticoids are key contributors to gut maturation, Z01 (TJP1) a critical tight junction protein, and a number of genes encoding proteins involved in cell-cell interactions, including integrins, cadherins and syntaxin, and proteins involved in cell proliferation and apoptosis [46]. Interestingly, the most highly ranked gene for identifying BF versus FF infants was Endothelial PAS domain-containing protein 1 (EPAS-1; also known as Hypoxia-inducible factor-2α [HIF-2α]). It is well known that human milk protects preterm infants from the development of NEC, one of the most common causes of morbidity and mortality in very low birth weight infants [62, 63]. We postulate that induction of this gene may provide a mechanism whereby the intestines of premature infants fed human milk are better able to tolerate episodes of hypoxia and are thereby less likely to develop NEC. In addition to being the best single classifier, EPAS-1 was an even stronger predictor when combined with other genes in ten separate 2- or 3-gene combinations (an example is shown in Figure 2), thus providing potential biomarkers for nutritional modulation of gut development [46].
Table 2.
Early Nutrition Differentially Regulates Epithelial Gene Expression. Listed are the most highly up- and down-regulated genes in breastfed (BF) vs. formula-fed (FF) infants.
| Gene Symbol and Name | Function of Encoded Gene | Fold Change (BF/FF) |
|---|---|---|
| Up-regulated in BF Compared to FF | ||
| NR3C1; Glucocorticoid receptor | Transcription factor that binds to glucocorticoid response elements in the promoters of glucocorticoid responsive genes | 5.5 |
| BAD; BCL-2-associated agonist of apoptosis | Protein positively regulates cell apoptosis by forming heterodimers with BCL-xL and BCL-2, and reversing their death repressor activity | 4.0 |
| PCDH7; Protocadherin | Gene product is an integral membrane protein that is thought to function in cell-cell recognition and adhesion | 3.9 |
| EPAS1; Endothelial PAS domain protein 1 | Transcription factor involved in the induction of genes regulated by oxygen, which is induced as oxygen levels fall. | 3.3 |
| NR5A2; Nuclear receptor subfamily 5, group A, member 2 or liver receptor homolog-1 (LRH-1) | LRH-1 is important for maintaining pluripotence of stem cells during embryonic development | 2.8 |
| MYB; v-myb avian myeloblastosis viral oncogene homolog | Plays an important role in the control of proliferation and differentiation | 2.8 |
| Stx2; Syntaxin-2 | Gene product regulates epithelial-mesenchymal interactions and epithelial cell morphogenesis and activation | 2.5 |
| ITGB2; Integrin Beta-2 (CD18) | ICAM Receptor; gene product is known to participate in cell adhesion as well as cell-surface mediated signaling | 2.5 |
| TJP1; ZO-1 | The N-terminal may be involved in transducing a signal required for tight junction assembly, while the C-terminal may have specific properties of tight junctions | 2.2 |
| Down-regulated in BF Compared to FF | ||
| FOXE3 | Forkhead box protein E3; cell proliferation | 0.32 |
| EGFR | EGF Receptor; cell proliferation and migration | 0.33 |
| WNT7B | Wingless-type MMRV integration site family, 7B; cell fate and cell patterning | 0.58 |
| LYZL6 | Lysozyme-like 6; bacteriostatic factor | 0.70 |
| HIF3A | Hypoxia-inducible factor 3 alpha; negative regulator of hypoxia- inducible gene expression | 0.71 |
| DAPK1 | Death-associated protein kinase 1; induction of apoptosis | 0.73 |
| APC | Adenomatous polyposis coli; tumor suppressor | 0.77 |
| TDGF1 | Teratocarcinoma-derived growth factor 3; cell growth | 0.80 |
| SPARC | Secreted-protein, acidic, cysteine-rich (osteonectin); cell proliferation | 0.82 |
From: Chapkin et al. 2010 [40]
Figure 2.
Identification of three-feature combination gene sets for diet. A strong multivariate (three gene) discriminator of breast-fed (circles) versus formula-fed (triangles) subjects is shown. Note that there is a clear separation between the two groups, except for one outlier. SYP, synaptophysin; FOXE3, forkhead box protein E3; EPAS1, endothelial PAS domain-containing protein 1 (hypoxia-inducible factor 2α). From: Chapkin et al., 2010 [40]
Stool-derived Eukaryotic mRNA and Intestinal Microbiota DNA can be used to Non-mInvasively Evaluate Host-Microbe Interactions in the Intestine of Term Infants
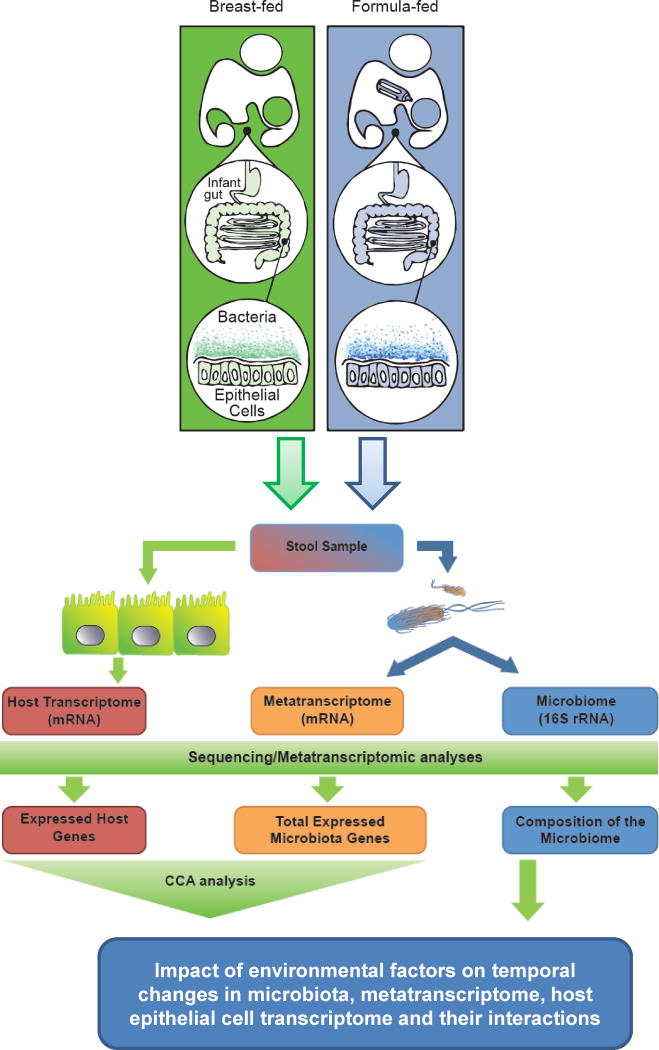
In an extension of this work, we created a novel methodology designed to assess the multivariate relationship between the microbiome metagenomic functional profile and the host transcriptome [47] as shown in Figure 3. By examining the multivariate structure underlying the bacterial metagenome or metatranscriptome and gut exfoliated cell transcriptome, our approach leverages richer and fuller information content compared to analyses focusing on single data sets (e.g., only host transcriptome data or only bacterial metatranscriptome data) and only single variables (e.g., gene by gene differential expression testing). We propose that this “integrative” strategy will help identify intestinal genes that are responsive to diet and influenced by factors known to cause dysbiosis in term and preterm infants (e.g. route of delivery, antibiotic use, formula feeding, parenteral nutrition).
Figure 3.
Experimental pipeline for simultaneous analysis of the host transcriptome and bacterial microbiome and metatranscriptome.
Using this approach, phyla-level differences in the microbiota of BF and FF infants were observed [47]. Microbiota functional characteristics were mapped to functional SEED categories. Because of the hierarchical structure of the SEED classification system, aggregating reads into coarser classifications provided for a more informed analysis. Virulence was the one SEED category that differed between the bacterial metagenome of BF and FF infants [47]. To examine the intrinsic relationship between host and microbiome, host transcriptome and metagenomic data were combined and integrated using the multivariate technique of canonical correlation analysis (CCA) [47]. We examined whether a relationship existed between microbiota virulence genes and sets of host immunity and defense genes (n=660), intestinal biology genes (n=660) or a random genes (n=459). A robust multivariate structure relating microbiota virulence genes and host immunity and defense genes was observed. Seven of the top eleven immunity and defense host genes that were related to the microbiota were down-regulated in BF versus FF infants, including ALOX5, a lipoxygenase involved in arachidonic acid and leukotriene synthesis, the cytokine IL1α, and binding proteins for natural killer cells (KLRF1), T-lymphocytes (AOC3) and LPS (BPILI) [47]. These findings indicate that the overall impact of breastfeeding was to reduce inflammatory genes in the gut potentially promoting tolerance to the luminal microbes.
Stool-Derived Eukaryotic mRNA can be used to Non-Invasively Assess the Impact of Preterm Delivery on Intestinal Gene Expression
We next applied our exfoliated cell transcriptomic approach to the preterm infant [48]. Preterm birth, which affected 15 million children in 2010, is a major determinant of neonatal morbidity and is the second leading cause of death in children under 5 years [64, 65]. Infants born at <32 weeks gestational age (GA) are faced with a unique set of challenges due to their developmental immaturity. In the U.S., the costs associated with preterm birth were more than $26.2 billion in 2005 [66]. Emerging evidence has clearly demonstrated direct and interactive links between diet, the intestinal microbiome and immune development [67,68]. Many common diseases afflicting preterm infants are associated with dysregulated immune function [69–73].
To determine whether sufficient exfoliated epithelial cells could be isolated from preterm infants and whether they would inform developmental differences, host transcript abundance in healthy full term (>38 weeks GA) and extremely preterm (24–30 weeks GA) infants were measured using RNA-Seq [48]. Approximately, 5500 genes were detected (FPKM>1) on average in both preterm and term samples. Several key observations were made. First, gene expression in preterm infants was more heterogeneous amongst themselves and compared to term infants. Second, exfoliated cells express genes associated with specific intestinal cell types including absorptive enterocytes (lactase and sucrose-isomaltase), Goblet cells (mucin-2), enteroendocrine cells (chromogranin A), and Paneth cells (lysozyme). Lastly, the transcriptional landscape is dramatically altered in the preterm versus term infant intestine [48].
Gene pathways that were over-expressed in preterm versus term or term versus preterm intestine were evaluated. Although none of the infants were clinically ill at the time the stool samples were collected, preterm over-expressed genes related to immune function. Several cytokines, including IL-1α and IL-33 were up-regulated in preterm versus term. In addition, several genes that regulate the expression of cytokines and other immune genes were expressed at 3- (NFKB1a) to 6-fold (CASP1) higher levels in preterm versus term infant exfoliated cells [48]. This is consistent with work by Nanthakumar and colleagues who showed that immortalized cells isolated from fetuses (H4 cells) or tissue explants from fetuses mount a more robust proinflammatory cytokine response (IL-8) after inflammatory stimulation with lipopolysaccharide or IL-1β than cells from adult tissue (Caco-2) or explants from older children [74]. The excessive inflammatory response of the immature intestine appeared to be in part due to a developmental under-expression of IkB [75] coupled with overexpression of the NFkB/MyD88 innate inflammatory genes (TLR2, TLR4, MyD88, TRAF-6, NFkB1 and IL-8) and reduced expression of negative regulator genes (SIGIRR, IRAK-M, A-20 and TOLLIP) in fetal intestine relative to other children [75]. Thus, it appears that immaturity of the intestinal innate immune response may contribute to excessive inflammation in the intestine in response to colonizing bacteria, which is a hallmark of NEC [76].
In contrast, in term infants, up-regulated immune genes were involved in balancing the immune system, e.g. promoting T-cell development (LCP2; 3.6-fold greater than preterm), while inhibiting macrophage activation (LENG9; 16-fold greater than preterm). The majority of genes were involved in cell turnover, by regulating proliferation and apoptosis. One of the most highly differentially-expressed genes was an anti-apoptotic factor (MTRNR2L6; 5-fold higher in term than preterm). Another interesting gene was SP3 (~2-fold higher in term than preterm), which is a transcription factor that can be regulated through short-chain fatty acid induced acetylation [77, 78], potentially supporting the role of products of microbial metabolism in regulating normal gut growth in term infants.
The underlying reason for differences in intestinal gene expression between preterm and term infants is multifactorial and may be a result of the developmental immaturity of the preterm gut, coupled with specific environmental exposures that are unique to the post-natal course of this population. Although well-controlled studies are needed to evaluate these exposures, this study highlights the potential of using the described noninvasive technology. We anticipate that this approach will allow investigators to elucidate how diet and bedside clinical management of this immature population influences intestinal development and immune ontogeny over time and has the potential for generating comprehensive, diagnostic gene sets for the noninvasive identification/prediction of different intestinal phenotypes in infants.
In addition to the host gene networks that are affected by the developing microbiota, the metabolic products from both microbes and host can give rise to signaling and inflammatory pathways that can affect numerous organs such as the lung, liver and brain in addition to the intestinal tract [73]. Undoubtedly, the comprehensive analysis of microbiota, intestinal transcriptome and metabolites will provide an overall picture of the milieu that is associated with many of the major morbidities seen in preterm infants.
Eukaryotic mRNA -derived for Meconium can be used to Non-invasively Assess the Impact of Maternal Probiotic treatment on Fetal Intestinal Immune Gene Expression In Utero
The long-held belief that the fetus existed in a sterile intrauterine environment and that introduction of bacteria into the amniotic fluid would lead to an adverse pregnancy outcome has been challenged by recent studies showing that the placenta, amniotic fluid and fetus contain a diversity bacteria and that these differ between term and preterm infants [79, 80]. Although dysbiosis may trigger preterm delivery [11, 81], other studies have described a relatively complex placenta microbiota comprised of nonpathogenic commensal microbiota from the Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes, and Fusobacteria phyla [9, 79, 80] that, surprisingly, clusters more similarly to the maternal oral than vaginal or gut microbiota [79, 82]. These results suggest that the placental and intrauterine bacteria do not ascend from the vagina, but may be delivered through the circulation [82]. Rautava [10] administered probiotics in order to investigate whether microbes in placenta or amniotic fluid affected fetal innate immune gene expression during late pregnancy and whether innate immune gene expression profiles in the placenta and the fetal gut may be modulated by dietary supplementation with specific probiotics. In a double-blind clinical trial, pregnant women were administered either placebo, Bifidobacterium lactis (B. lactis) or B. lactis and Lactobacillus rhamnosus GG (LGG) for 14 days before elective cesarean section at full term [10]. Bacterial DNA was detected in all placenta samples by PCR. Meconium samples were collected and host mRNA extracted from fetal exfoliated epithelial cells using the method of Chapkin and colleagues [46]. An association between the presence of microbial DNA in amniotic fluid and placenta and changes in toll like receptor (TLR)-related gene expression in the fetal intestine was observed. Additionally maternal probiotic supplementation significantly modulated the expression of TLR-related genes both in the placenta and in the fetal gut. Compared to mRNA expression in exfoliated cells of infants exposed to the placebo, TLR6 mRNA expression was down-regulated nearly 90% in exfoliated cells from infants whose mothers consumed B. lactis + LGG, whereas TLR7 was down-regulated 70% in infants of mothers administered B. lactis alone [10]. TLR are important mediators of innate immunity through their recognition of highly conserved microbial-associated molecular patterns. Specfically, TLR6 interacts with TLR2 to mediate cellular response to bacterial lipoproteins. TLR7 recognizes single-stranded RNA in endosomes, which is a common feature of viral genomes [83]. These findings support the hypothesis that microbial programming begins in fetal life through host-microbe interactions in utero [8], which can be manipulated by maternal probiotic intervention [10].
Summary
We have recently validated a novel molecular methodology that utilizes stool samples containing intact sloughed epithelial cells to noninvasively quantify intestinal gene expression profiles in the developing human neonate, which is “an important first step towards a more comprehensive understanding of the biological mechanisms underlying the parallel development of the host and microbiome in early life” [84]. This approach enables repeated assessment of the same infant overtime to assess temporal changes in gene expression [40]. Furthermore, we have expanded upon this methodology by combining host gene expression with the bacterial metagenome from the same infant [47]. This approach is statistically rigorous and is sensitive to dietary intake [46, 47] and the stage of gestation [48]. In conclusion, we propose that the investigation of host-microbiome interaction will fill an important gap in our understanding of the coordinated development of gut microbiota and the infant intestine. In the long-term, it is anticipated that nutritional strategies to improve the development of the microbiome and intestine will enhance the clinical care of high-risk infants.
Acknowledgments
Support: This work was supported by National Institute of Health grants R01 CA129444 (RSC), U01 CA162077 (RSC), R25 TCA090301 (RSC), P30 ES023512 (RSC), R01 HD61929 (SMD), Hatch project ILLU-971-346 through the Division of Nutritional Sciences Vision 20/20 program (SMD) and USDA–NIFA Grant Designing Foods for Health 2010-34402-20875 (RSC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chowdhury SR, King DE, Willing BP, Band MR, Beever JE, Lane AB, Loor JJ, Marini JC, Rund LA, Schook LB, Van Kessel AG, Gaskins HR. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics. 2007;8:215. doi: 10.1186/1471-2164-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El Aidy S, Merrifield CA, Derrien M, van Baarlen P, Hooiveld G, Levenez F, Doré J, Dekker J, Holmes E, Claus SP, Reijngoud DJ, Kleerebezem M. The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut. 2013;62:1306–14. doi: 10.1136/gutjnl-2011-301955. [DOI] [PubMed] [Google Scholar]
- 3.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Collins J, Borojevic R, Verdu EF, Huizinga JD, Ratcliffe EM. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol Motil. 2014;26:98–107. doi: 10.1111/nmo.12236. [DOI] [PubMed] [Google Scholar]
- 5.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18:666–73. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 6.Bäckhed F. Host responses to the human microbiome. Nutr Rev. 2012;70(Suppl 1):S14–7. doi: 10.1111/j.1753-4887.2012.00496.x. [DOI] [PubMed] [Google Scholar]
- 7.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rautava S, Luoto R, Salminen S, Isolauri E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol. 2012;9:565–76. doi: 10.1038/nrgastro.2012.144. [DOI] [PubMed] [Google Scholar]
- 9.Gosalbes MJ, Llop S, Vallès Y, Moya A, Ballester F, Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy. 2012;43:198–211. doi: 10.1111/cea.12063. [DOI] [PubMed] [Google Scholar]
- 10.Rautava S, Collado MC, Salminen S, Isolauri E. Probiotics modulate host-microbe interaction in the placenta and fetal gut: A randomized, double-blind, placebo-controlled trial. Neonatology. 2012;102:178–84. doi: 10.1159/000339182. [DOI] [PubMed] [Google Scholar]
- 11.Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, Murgas-Torrazza R, Sharma R, Hudak ML, Triplett EW, Neu J. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE. 2014;9(3):e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholtens PAMJ, Oozeer R, Martin R, Ben Amor K, Knol J. The early settlers: Intestinal microbiology in early life. Annu Rev Food Sci Technol. 2012;3:425–47. doi: 10.1146/annurev-food-022811-101120. [DOI] [PubMed] [Google Scholar]
- 13.Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: Role of human milk oligosaccharides. Adv Nutr. 2012;3:450S–5S. doi: 10.3945/an.112.001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catassi C, Bonucci A, Coppa GV, Carlucci A, Giorgi PL. Intestinal permeability changes during the first month: the effect of natural versus artificial feeding. J Pediatr Gastroenterol Nutr. 1995;21:383–6. doi: 10.1097/00005176-199511000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Cummins AG, Thompson FM. Postnatal changes in mucosal immune response: a physiological perspective of breast feeding and weaning. Immunol Cell Biol. 1997;75:419–29. doi: 10.1038/icb.1997.67. [DOI] [PubMed] [Google Scholar]
- 16.Donovan SM. Role of human milk components in gastrointestinal development: Current knowledge and future needs. J Pediatr. 2006;149 (Suppl 3):49–61. [Google Scholar]
- 17.Donovan SM, Odle J. Growth factors in milk as mediators of infant development. Annu Rev Nutr. 1994;14:147–67. doi: 10.1146/annurev.nu.14.070194.001051. [DOI] [PubMed] [Google Scholar]
- 18.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, Doré J Other Members of the INFABIO Team. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 19.Thurl S, Munzert M, Henker J, Boehm G, Müller-Werner B, Jelinek J, Stahl B. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br J Nutr. 2010;104:1261–71. doi: 10.1017/S0007114510002072. [DOI] [PubMed] [Google Scholar]
- 20.Totten SM, Zivkovic AM, Wu S, Ngyuen U, Freeman SL, Ruhaak LR, Darboe MK, German JB, Prentice AM, Lebrilla CB. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J Proteome Res. 2012;11:6124–33. doi: 10.1021/pr300769g. [DOI] [PubMed] [Google Scholar]
- 21.De Leoz ML, Gaerlan SC, Strum JS, Dimapasoc LM, Mirmiran M, Tancredi DJ, Smilowitz JT, Kalanetra KM, Mills DA, German JB, Lebrilla CB, Underwood MA. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J Proteome Res. 2012;11:4662–72. doi: 10.1021/pr3004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hester SN, Donovan SM. Individual and combined effects of nucleotides and human milk oligosaccharides on proliferation, apoptosis and necrosis in a human fetal intestinal cell line. Food Nutr Sci. 2012;3:1567–76. [Google Scholar]
- 23.Holscher HD, Davis SR, Tappenden KA. Human milk oligosaccharides influence maturation of human intestinal Caco-2Bbe and HT-29 cell lines. J Nutr. 2014;144:586–91. doi: 10.3945/jn.113.189704. [DOI] [PubMed] [Google Scholar]
- 24.Lane JA, O’Callaghan J, Carrington SD, Hickey RM. Transcriptional response of HT-29 intestinal epithelial cells to human and bovine milk oligosaccharides. Br J Nutr. 2013;110:2127–37. doi: 10.1017/S0007114513001591. [DOI] [PubMed] [Google Scholar]
- 25.Newburg DS. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J Anim Sci. 2009;87(Suppl 13):26–34. doi: 10.2527/jas.2008-1347. [DOI] [PubMed] [Google Scholar]
- 26.Comstock SS, Wang M, Hester SN, Li M, Donovan SM. Select human milk oligosaccharides directly modulate peripheral blood mononuclear cells isolated from 10-day-old pigs. Br J Nutr. 2014;111:819–828. doi: 10.1017/S0007114513003267. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Liu S, Leone S, Newburg DS. Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. 2014 Apr 2; doi: 10.1038/mi.2014.20. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Monaco MH, Wang M, Comstock SS, Kuhlenschmidt TB, Fahey GC, Jr, Miller MJ, Kuhlenschmidt MS, Donovan SM. Human milk oligosaccharides shorten rotavirus diarrhea and modulate piglet mucosal immunity and colonic microbiota. ISME J. 2014 Feb 13; doi: 10.1038/ismej.2014.10. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Bauer LL, Chen X, Wang M, Kuhlenschmidt TB, Kuhlenschmidt MS, Fahey GC, Jr, Donovan SM. Microbial composition and in vitro fermentation patterns of human milk oligosaccharides differ between formula-fed and sow-reared piglets. J Nutr. 2012;142:681–9. doi: 10.3945/jn.111.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vester Boler BM, Rossoni Serao MC, Faber TA, Bauer LL, Chow J, Murphy MR, Fahey GC., Jr In vitro fermentation characteristics of select nondigestible oligosaccharides by infant fecal inocula. J Agric Food Chem. 2013;61:2109–19. doi: 10.1021/jf305056f. [DOI] [PubMed] [Google Scholar]
- 32.Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012;3:415S–21S. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bidart GN, Rodríguez-Díaz J, Monedero V, Yebra MJ. A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto-N-biose and lacto-N-biose in Lactobacillus casei. Mol Microbiol. 2014 Jun 18; doi: 10.1111/mmi.12678. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 35.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–62. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.American Academy of Pediatrics, Section on Breastfeeding. Breastfeeding and the use of human milk. Pediatrics. 2012;129:e827–41. doi: 10.1542/peds.2011-3552. [DOI] [PubMed] [Google Scholar]
- 37.Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab. 2013;63 (Suppl 2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 38.Li M, Wang M, Donovan SM. Early development of the gut microbiome and immune-mediated childhood disorders. Semin Reprod Med. 2014;32:74–86. doi: 10.1055/s-0033-1361825. [DOI] [PubMed] [Google Scholar]
- 39.Martin R, Nauta AJ, Ben Amor K, Knippels MJ, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010;1:367–82. doi: 10.3920/BM2010.0027. [DOI] [PubMed] [Google Scholar]
- 40.Nauta AJ, Ben Amor K, Knol J, Garssen J, van der Beek EM. Relevance of pre- and postnatal nutrition to development and interplay between the microbiota and metabolic and immune systems. Am J Clin Nutr. 2013;98(Suppl):586S–93S. doi: 10.3945/ajcn.112.039644. [DOI] [PubMed] [Google Scholar]
- 41.Ajslev TA, Andersen CS, Gamborg M, Sørensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond) 2011;35:522–9. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- 42.Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012;129:950–60. doi: 10.1542/peds.2011-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neu J, Rushing J. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol. 2011;38:321–31. doi: 10.1016/j.clp.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host–microbiota mutualism. Nat Rev Microbiol. 2011;9:233–43. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 45.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Björkstén B, Engstrand L, Andersson AF. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–66. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 46.Chapkin RS, Zhao C, Ivanov I, Davidson LA, Goldsby JS, Lupton JR, Mathai RA, Monaco MH, Rai D, Russell WM, Donovan SM, Dougherty ER. Non-invasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells. Am J Physiol - Gastrointest Liver Physiol. 2010;298:G582–9. doi: 10.1152/ajpgi.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz S, Friedberg I, Ivanov I, Davidson LA, Goldsby JS, Dahl DB, Herman D, Wang M, Donovan SM, Chapkin RS. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in developmental and immune responses. Genome Biol. 2012;13:R32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knight JM, Davidson LA, Herman D, Martin CR, Goldsby JS, Ivanov IV, Donovan SM, Chapkin RS. Non-invasive analysis of intestinal development in preterm and full term infants using RNA-SEQ: A pilot study. Nature Sci Rep. 2014;4:5453. doi: 10.1038/srep05453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potten CS, Schofield R, Lajtha LG. A comparison of cell replacement in bone marrow, testis and three regions of epithelium. Biochim Biophys Acta. 1979;560:281–99. doi: 10.1016/0304-419x(79)90022-2. [DOI] [PubMed] [Google Scholar]
- 50.Kaeffer B. Survival of exfoliated epithelial cells: A delicate balance between anoikis and apoptosis. J Biomed Biotech. 2011;2011:Article ID 534139. doi: 10.1155/2011/534139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Molec Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaeffer B, des Robert C, Alexandre-Gouabau MC, Pagniez A, Legrand A, Amarger V, Küster A, Piloquet H, Champ M, le Huërou-Luron I, Rozé JC. Recovery of exfoliated cells from the gastrointestinal tract of premature infants: a new tool to perform “noninvasive biopsies?”. Pediatr Res. 2007;62:564–9. doi: 10.1203/PDR.0b013e318155a402. [DOI] [PubMed] [Google Scholar]
- 53.Albaugh GP, Iyengar V, Lohani A, Malayeri M, Bala S, Nair PP. Isolation of exfoliated colonic epithelial cells, a novel non-invasive approach to the study of cellular markers. Int J Cancer. 1992;52:347–50. doi: 10.1002/ijc.2910520303. [DOI] [PubMed] [Google Scholar]
- 54.Bandaletova T, Bailey N, Bingham SA, Loktionov A. Isolation of exfoliated colonocytes from human stools as a new technique for colonic cytology. APMIS. 2002;110:239–46. doi: 10.1034/j.1600-0463.2002.100306.x. [DOI] [PubMed] [Google Scholar]
- 55.Kamra A, Kessie G, Chen JH, Kalavapudi S, Shores R, McElroy I, Gireesh T, Sudhakaran PR, Dutta SK, Nair PP. Exfoliated colonic epithelial cells: surrogate targets for evaluation of bioactive food components in cancer prevention. J Nutr. 2005;135:2719–22. doi: 10.1093/jn/135.11.2719. [DOI] [PubMed] [Google Scholar]
- 56.Cho Y, Kim H, Turner ND, Mann JC, Wei J, Taddeo SS, Davidson LA, Wang N, Vannucci M, Carroll RJ, Chapkin RS, Lupton JR. A chemoprotective fish oil- and pectin-containing diet temporally alters gene expression profiles in exfoliated rat colonocytes throughout oncogenesis. J Nutr. 2011;141:1029–35. doi: 10.3945/jn.110.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson LA, Jiang YH, Lupton JR, Chapkin RS. Non-invasive detection of putative biomarkers for colon cancer using fecal mRNA. Cancer Epidemiol Biomarkers Prev. 1995;4:643–7. [PubMed] [Google Scholar]
- 58.Davidson LA, Lupton JR, Miskovsky E, Fields AP, Chapkin RS. Quantification of human intestinal gene expression profiles using exfoliated colonocytes: a pilot study. Biomarkers. 2003;8:51–61. doi: 10.1080/1354750021000042268. [DOI] [PubMed] [Google Scholar]
- 59.Holland N, Harmatz P, Golden D, Hubbard A, Wu YY, Bae J, Chen C, Huen K, Heyman MB. Cytogenetic damages in blood lymphocytes and exfoliated epithelial cells of children with inflammatory bowel disease. Pediatr Res. 2007;61:209–214. doi: 10.1203/pdr.0b013e31802d77c7. [DOI] [PubMed] [Google Scholar]
- 60.Kaeffer B, Legrand A, Moyon T, Frondas-Chauty A, Billard H, Guzman-Quevedo O, Darmaun D, Rozé JC. Non-invasive exploration of neonatal gastric epithelium by using exfoliated epithelial cells. PLoS One. 2011;6:e25562. doi: 10.1371/journal.pone.0025562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lupton JR, Davidson LA, Chapkin RS. Non-invasive detection of candidate molecular biomarkers in subjects with a history of insulin resistance and colorectal adenomas. Cancer Prev Res. 2009;2:590–7. doi: 10.1158/1940-6207.CAPR-08-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sisk PM, Lovelady CA, Dillard RG, Gruber KJ, O’Shea TM. Early human milk feeding is associated with a lower risk of necrotizing enterocolitis in very low birth weight infants. J Perinatol. 2007;27:428–33. doi: 10.1038/sj.jp.7211758. [DOI] [PubMed] [Google Scholar]
- 63.Meinzen-Derr J, Poindexter B, Wrage L, Morrow AL, Stoll B, Donovan EF. Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J Perinatol. 2009;29:57–62. doi: 10.1038/jp.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beck S, Wojdyla D, Say L, Betran AP, Merialdi M, Requejo JH, Rubens C, Menon R, Van Look PFA. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ. 2010;88:31–8. doi: 10.2471/BLT.08.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Garcia CV, Rohde S, Say L, Lawn JE. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;2379:2162–72. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- 66.Petrou S. The economic consequences of preterm birth during the first 10 years of life. BJOG. 2005;112 (Suppl 1):10–5. doi: 10.1111/j.1471-0528.2005.00577.x. [DOI] [PubMed] [Google Scholar]
- 67.Frank DN, Zhu W, Sartor RB, Li E. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol. 2011;19:427–34. doi: 10.1016/j.tim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poroyko V, Morowitz M, Bell T, Ulanov A, Wang M, Donovan S, Bao N, Gu S, Hong L, Alverdy JC, Bergelson J, Liu DC. Diet creates metabolic niches in the “immature gut” that shape microbial communities. Nutr Hosp. 2011;26:1283–95. doi: 10.1590/S0212-16112011000600015. [DOI] [PubMed] [Google Scholar]
- 69.Madan JC, Salari RC, Saxena D, Davidson L, O’Toole GA, Moore JH, Sogin ML, Foster JA, Edwards WH, Palumbo P, Hibberd PL. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012;97:F456–62. doi: 10.1136/fetalneonatal-2011-301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morowitz MJ, Poroyko V, Caplan M, Alverdy J, Liu DC. Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics. 2010;125:777–85. doi: 10.1542/peds.2009-3149. [DOI] [PubMed] [Google Scholar]
- 71.Stewart CJ, Marrs ECL, Magorrian S, Nelson A, Lanyon C, Perry JD, Embleton ND, Cummings SP, Berrington JE. The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Pædiatrica. 2012;101:1121–7. doi: 10.1111/j.1651-2227.2012.02801.x. [DOI] [PubMed] [Google Scholar]
- 72.Kuypers K, Ophelders D, Jellema RK, Kunzmann S, Gavilanes AW, Kramer BW. White matter injury following fetal inflammatory response syndrome induced by chorioamnionitis and fetal sepsis: Lessons from experimental ovine models. Early Human Develop. 2012;8:931–6. doi: 10.1016/j.earlhumdev.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 73.Leviton A, Dammann O, Engelke S, Allred E, Kuban KCK, O’Shea TM, Paneth N for the ELGAN study investigators. The clustering of disorders in infants born before the 28th week of gestation. Acta Pædiatrica. 2010;99:1795–1800. doi: 10.1111/j.1651-2227.2010.01973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nanthakumar NN, Fusunyan RD, Sanderson I, Walker WA. Inflammation in the developing human intestine: a possible pathophysiological contribution to necrotizing enterocolitis. Proc Natl Acad Sci USA. 2000;23:6043–8. doi: 10.1073/pnas.97.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Claud EC, Lu L, Anton PM, Savidge T, Walker WA, Cherayil BJ. Developmentally regulated IkappaB expression in intestinal epithelium and susceptibility to flagellin-induced inflammation. Proc Natl Acad Sci USA. 2004;101:7404–8. doi: 10.1073/pnas.0401710101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nanthakumar N, Meng D, Goldstein AM, Zhu W, Lu L, Uauy R, Llanos A, Claud EC, Walker WA. The mechanism of excessive intestinal inflammation in necrotizing enterocolitis: An immature innate immune response. PLoS One. 2011;6(3):e17776. doi: 10.1371/journal.pone.0017776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeissig S, Fromm A, Mankertz J, Weiske J, Zeitz M, Fromm M, Schulzke JD. Butyrate induces intestinal sodium absorption via Sp3-mediated transcriptional up-regulation of epithelial sodium channels. Gastroenterology. 2007;132:236–48. doi: 10.1053/j.gastro.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 78.Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192:275–83. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 79.Prince AL, Antony KM, Chu DM, Aagaard KM. The microbiome, parturition, and timing of birth: more questions than answers. J Reprod Immunol. 2014 Apr 18; doi: 10.1016/j.jri.2014.03.006. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ganu RS, Ma J, Aagaard KM. The role of microbial communities in parturition: is there evidence of association with preterm birth and perinatal morbidity and mortality? Am J Perinatol. 2013;30:613–24. doi: 10.1055/s-0032-1329693. [DOI] [PubMed] [Google Scholar]
- 81.Stout MJ, Conlon B, Landeau M, Lee I, Bower C, Zhao Q, Roehl KA, Nelson DM, Macones GA, Mysorekar IU. Identification ofintracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol. 2013;208:226, e1–e7. doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aagaard K, Ma J, Antony KM, Anders A, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 84.Pop M. We are what we eat: how the diet of infants affects their gut microbiome. Genome Biol. 2012;13:152. doi: 10.1186/gb-2012-13-4-152. [DOI] [PMC free article] [PubMed] [Google Scholar]