Abstract
Antibiotic resistance is a growing health concern, and new avenues of antimicrobial drug design are being actively sought. One suggested pathway to be targeted for inhibitor design is that of iron scavenging through siderophores. Here we present a high throughput screen to the isochorismate-pyruvate lyase of Pseudomonas aeruginosa, an enzyme required for the production of the siderophore pyochelin. Compounds identified in the screen are high nanomolar to low micromolar inhibitors of the enzyme and produce growth inhibition in PAO1 P. aeruginosa in the millimolar range under iron-limiting conditions. The identified compounds were also tested for enzymatic inhibition of E. coli chorismate mutase, a protein of similar fold and similar chemistry, and of Y. enterocolitica salicylate synthase, a protein of differing fold but catalyzing the same lyase reaction. In both cases, subsets of the inhibitors from the screen were found to be inhibitory to enzymatic activity (mutase or synthase) in the micromolar range and capable of growth inhibition in their respective organisms (E. coli or Y. enterocolitica).
Keywords: siderophore, isochorismate pyruvate lyase, chorismate mutase, salicylate synthase
1. Introduction
Most pathogens require iron for survival because this metal serves as a required cofactor in many essential biological processes including DNA biosynthesis and cellular respiration [1]. While direct mechanisms for iron uptake (for example, uptake of heme) have evolved, indirect methods of iron acquisition, such as the use of high affinity iron chelators called siderophores, allow for iron scavenging regardless of the source [1]. Siderophore biosynthesis has been linked to virulence in many pathogenic bacteria [2–10]: without the ability to scavenge iron from the host, the bacteria are incapable of causing disease. Salicylate-capped siderophores are produced by several pathogenic bacteria, such as Yersinia spp. including the causative agent of plague, Mycobacterium tuberculosis, which is the causative agent of tuberculosis, and Pseudomonas aeruginosa, a nosocomial pathogen infecting susceptible hosts, including burn victims, cancer patients with hematological malignancies and those undergoing chemotherapy, patients with immune deficiencies such as AIDS patients, and Cystic Fibrosis (CF) patients [11]. Compounds that prevent the formation of salicylate or its incorporation into siderophores may prove to be a new class of antimicrobial drugs in this age of antibiotic drug resistance.
P. aeruginosa, Yersinia spp., and M. tuberculosis make chemically related siderophores that all use a salicylate cap (Figure 1A), the hydroxyl group of which is involved in iron chelation [12]. The salicylate 2 is formed in two steps from chorismate 4 with an isochorismate 1 intermediate (Figure 1B). The first step is the general acid-general base isomerization of chorismate to form isochorismate [13–15], whereas the second is the pericyclic migration of the enolpyruvyl tail [15, 16]. Because the cyclic transition state for this reaction includes a hydrogen, the result is the elimination of the tail and the formation of salicylate and pyruvate 3. For Yersinia spp. (siderophore: yersiniabactin) and M. tuberculosis (mycobactin), both reactions are catalyzed by a salicylate synthase, Irp9 and MbtI, respectively. The generation of the siderophore pyochelin by P. aeruginosa requires two enzymes, an isochorismate synthase (PchA) and an isochorismate-pyruvate lyase (PchB). Interestingly, PchA, Irp9 and MbtI are all structural homologues in the MST (menaquinone, siderophore and tryptophan biosynthesis) family, but as yet it is unknown why PchA cannot perform the pericyclic lyase reaction [17]. There are homologues of PchA that are isochorismate synthases, which also cannot perform the pericyclic lyase reaction. E. coli MenF [18, 19] is involved in menaquinone biosynthesis whereas EntC [20–23] from E. coli and VibC [24, 25] from Vibrio cholera are found in the biosynthetic pathways for dihydroxybenzoate capped siderophores. For MenF, EntC and VibC, the inability to perform the lyase reaction is biologically logical, since isochorismate is required for the formation of the biosynthetic products whereas salicylate is not [17].
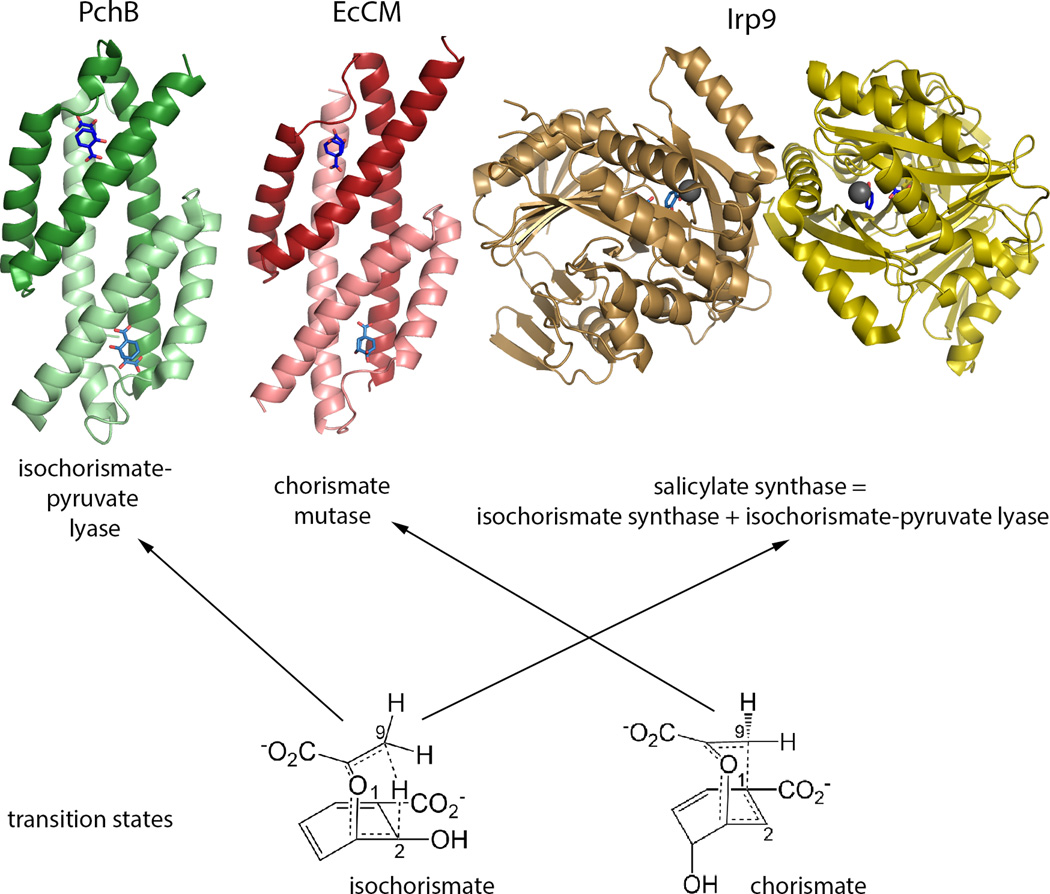
Figure 1. Salicylate-capped Siderophores and the Enzymes Targeted.
a. The salicylate-capped siderophores generated by P. aeruginosa (pyochelin), Yersinia spp. (yersiniabactin) and M. tuberculosis (mycobactin). b. The isochorismate-pyruvate lyase from Pseudomonas aeruginosa (PchB) is the specific enzyme of screen, and performs the pericyclic cleavage of isochorismate 1 to form salicylate 2 and pyruvate 3. Inhibitors were also tested against the salicylate synthase from Yersinia enterocolitica (Irp9), which is a chorismate-utilizing enzyme and a structural homologue of the isochorismate synthase PchA from P. aeruginosa. Irp9 and PchA catalyze the conversion of chorismate 4 to isochorismate 1. Irp9 subsequently catalyzes the same lyase reaction as PchB from an isochorismate intermediate, whereas PchA does not. The compounds were also tested for inhibition of E. coli chorismate mutase (EcCM), also a chorismate-utilizing enzyme, which performs a pericyclic reaction similar to that catalyzed by PchB, generating prephenate 5.
The current work was initiated as a high throughput screen for the isochorismate-pyruvate lyase from P. aeruginosa (PchB), which performs the second, pericyclic reaction in the generation of the pyochelin siderophore. PchB is easily generated, we have adopted an effective microbial production system for obtaining the substrate isochorismate [26], and the salicylate product is fluorescent, allowing for the development of an enzymatic assay suitable for automation. PchB is a structural homologue of E. coli chorismate mutase (EcCM) of the AroQ structural class (Figure 2) [27, 28]. EcCM is the N-terminal domain of the well-studied bifunctional P-protein [29], the terminal enzyme of the shikimate biosynthetic pathway that is the branch point for the production of aromatic amino acids [30]. Both PchB and EcCM perform pericyclic reactions with similar transition states, differing in the alignment of the enolpyruvyl tail over the ring and thus the atomic composition of the cyclic transition state (Figure 2) [17]. The shikimate pathway is also recognized as an attractive target for antimicrobial drug design, since mammals do not synthesize aromatic amino acids. Rational design of inhibitors has been conducted for the chorismate mutases [31–38], including the TSA (transition state analogue), an oxabicyclic acid generated by Bartlett [34, 35]. EcCM is refractory to screening, because it lacks a good spectrophotometric handle and is difficult to make in sufficient quantities.
Figure 2. Enzyme scaffolds and transition states.
The X-ray crystallographic structures for PchB (PBD code: 3REM) with salicylate and pyruvate bound, EcCM (1ECM) with Bartlett’s TSA inhibitor bound, and Irp9 (2FN1) with Mg2+, salicylate and pyruvate are depicted as cartoons. Each of these enzymes is a homodimer, with one monomer shaded darker than the other, and the active sites identified by the ligands shown as sticks. PchB and EcCM share the same fold – they are AroQ enzymes, whereas Irp9 is in the MST family of enzymes. The transition states for the reactions catalyzed, isochorismate-pyruvate lyase (left) and chorismate mutase (right), are shown below. The transition states are similar, differing only in the alignment of the pyruvylenol tail over the ring to make a cycle at the 1 (mutase) or 2 (lyase) carbon. PchB and Irp9 perform the same chemistry using different scaffolds, whereas PchB and EcCM perform related reactions in the same scaffold. It should be noted that PchB has adventitious mutase activity, albeit very low.
While pyochelin has been implicated in virulence [4, 10], any compounds that are effective inhibitors against PchB would need to be used in combination with inhibitors to the other iron scavenging pathways in P. aeruginosa, including that of the high affinity siderophore pyoverdin. Salicylate synthases (for example, Irp9 and MbtI mentioned above) perform the same pericyclic reaction as PchB (Figure 2), but are found in siderophore biosynthetic pathways necessary for virulence by organisms that are more limited in variety of iron uptake systems (most particularly, in Yersinia spp. [2, 3, 6] and M. tuberculosis [5, 7–9]). While salicylate synthases perform the same reaction, they do so using a different protein scaffold (Figure 2). Salicylate synthesis and activation of salicylate by adenylation for incorporation into siderophores have been the targets of inhibition studies to produce lead compounds for antimicrobial drug design. Rational design of inhibitors to enzymes such as isochorismate synthase, salicylate synthase and salicylate adenylase has been documented [39–59], as well as high throughput screening for inhibitors of salicylate synthase [60].
We hypothesized that compounds identified as inhibitors in an enzymatic screen may also be effective against an enzyme that performs the same chemistry albeit using a differing protein fold. By the same token, an inhibitor identified by screening methods may also be effective against a protein with a homologous protein scaffold and similar transition states, in this case, allowing for a way to identify inhibitors to an enzyme that is not suitable for screening. Therefore, we report a high-throughput screen designed to identify inhibitors of the isochorismate-pyruvate lyase from P. aeruginosa, measuring decreased salicylate production from isochorismate in the presence of an inhibitory compound. The compounds were further tested for inhibition of salicylate production from chorismate by the Y. enterocolitica salicylate synthase Irp9 which performs the same pericyclic lyase reaction but using a different protein fold. An orthogonal assay was used to measure inhibition of pyruvate production for both PchB and Irp9. Due to similarities in protein fold, substrate and transition state, identified compounds were also tested for inhibition of E. coli chorismate mutase activity, measuring chorismate disappearance. Finally, growth inhibition was measured for P. aeruginosa, E. coli and Y. enterocolitica for two of the most promising compounds.
2. Results
2.1 High Throughput Screen
Our assay measures the conversion of isochorismate to salicylate and pyruvate by the isochorismate-pyruvate lyase, PchB. The assay takes advantage of the fluorescent properties of salicylate, and is amenable to standard lab and high throughput conditions. Using the lab conditions and concentrations derived from steady state kinetics [27] as a starting point, a high-throughput assay was developed. Optimization of the assay conditions were tested by varying enzyme and substrate concentration and incubation times. The assay was insensitive to DMSO up to concentrations of 2%.
The assay was automated at the University of Kansas High Throughput Screening Laboratory and used to screen PchB versus the ChemDiv, ChemBridge, Microsource, Prestwick and the University of Kansas Chemical Methodologies and Library Development libraries. A pilot screen identified compound 6 as a positive control. Wells in each plate were reserved for no enzyme negative controls and 10 µM compound 6 positive controls to allow for the calculation of plate-to-plate statistics and to detect irregularities in the screen. Each compound was screened at a concentration of 10 µM. Substrate and enzyme dilutions were prepared fresh each day, with 30 plates run per day. Many compounds (258) were auto-fluorescent (showed > 100% inhibition) and were eliminated from further analysis (Supplemental Figure 1A). Compounds that showed inhibition effects greater than plate median plus three standard deviations but less than/equal to 100% (422 compounds) were deemed “actives” – a hit rate of 0.4%. Z’ factors on individual plates ranged from 0.5 to 0.9 and was 0.8 overall (Supplemental Figure 1B). The signal to baseline ranged from 10 to 30. Active compounds (231 of the 422) were confirmed by rescreening from the source plates in the high-throughput assay. Based on score (IC50 values from dose response data), availability and price, five compounds from three structural classes (9–13) were purchased for further evaluation (Figure 3). Compounds 6–8 were no longer commercially available and so were synthesized as described in the Methods.
Figure 3. Compounds selected from screen for further evaluation.
2.2 Inhibition of isochorismate-pyruvate lyase (PchB)
The eight selected hits from the high-throughput screen were more rigorously examined with the same conditions that were used to obtain the original kinetic constants [27]. Half maximal inhibitory concentration (IC50) values for the prospective inhibitors were calculated from dose response curves based on percent inhibition of PchB (Table 1). All compounds were effective with IC50 values ranging from 100 nM to 10 µM. KI values were determined for compounds 6, 9 and 10: 44 +/− 2 nM, 80 +/− 8 nM, and 350 +/− 40 nM, respectively, and all three were competitive inhibitors. An orthogonal assay was adopted to measure pyruvate production (as opposed to salicylate production) using an absorbance based assay. The calculated IC50 values for this assay were comparable to those from the original salicylate fluorescence assay.
Table 1.
IC50 values determined for the isochorismate-pyruvate lyase PchB, the salicylate synthase Irp9, and the chorismate mutase EcCM.
| Compound | IC50 (µM) | ||||
|---|---|---|---|---|---|
| PchB | Irp9 | EcCM | |||
| Salicylate Production Assay |
Pyruvate Production Assay |
Salicylate Production Assay |
Pyruvate Production Assay |
Chorismate Disappearance Assay |
|
| 6 | 0.10 ± 0.01 | 0.18 ± 0.09 | NI | NI | 30 ± 3 |
| 7 | 0.25 ± 0.01 | 0.22 ± 0.01 | NI | NI | 170 ± 20 |
| 8 | 0.24 ± 0.01 | 0.094 ± 0.001 | NI | NI | 200 ± 0 |
| 9 | 0.10 ± 0.10 | 0.07 ± 0.01 | 80 ± 20 | NI | 82 ± 2 |
| 10 | 0.87 ± 0.02 | 0.53 ± 0.09 | 25 ± 3 | 5 ± 2 | 32 ± 3 |
| 11 | 3.3 ± 0.2 | 1.8 ± 0.0 | 160 ± 10 | NI | 45 ± 3 |
| 12 | 10 ± 1 | 10 ± 1 | 60 ± 20 | 160 ± 10 | 120 ± 10 |
| 13 | 1.4 ± 0.0 | 1.4 ± 0.1 | 42 ± 3 | NI | 85 ± 2 |
NI= not inhibitory
2.3 Inhibition of salicylate synthase (Irp9)
Since Irp9 performs the same chemistry as PchB, albeit by an enzyme of a different structural class, the same assays (salicylate production and pyruvate production) were used to determine IC50 values for the eight selected hits from the high-throughput screen. The compounds of the first structural class (6–8) were not inhibitory for Irp9 in either assay (Table 1). The remaining compounds showed IC50 values ranging from 25 to 160 µM in the salicylate production assay, but these values were only recapitulated in the pyruvate production assay for compounds 10 and 12. A KI value was determined for compound 10: 0.8 +/− 0.2 µM, a competitive inhibitor.
2.4 Inhibition of chorismate mutase (EcCM)
EcCM and PchB are of the same structural class and both perform pericyclic reactions on similar substrates (chorismate and isochorismate). Therefore, the eight selected hits from the high-throughput screen were also tested for inhibition of EcCM using a chorismate disappearance absorbance assay. Compounds 6 through 13 gave IC50 values between 30 and 200 µM (Table 1). KI values were determined for compounds 6 and 10: 30 +/− 10 µM, and 40 +/− 20 µM, respectively, and both were competitive inhibitors.
2.5 Bacterial Growth Inhibition
To determine if the compounds were effective at inhibiting bacterial growth, half maximal effective concentration (EC50) values were determined for P. aeruginosa, Y. enterocolitica, and E. coli. The HTS and subsequent screens provided a minimal structure-activity relationship for the two compound classes represented by compounds 6 and 10. Therefore, these two compounds were chosen for EC50 determination. The EC50 values were determined in iron-rich (200 µM) or iron-poor (5 µM) media for both P. aeruginosa and Y. enterocolitica to determine if inhibition is related to iron uptake. The EC50 values were not determined in iron-free media, because the bacteria could not be reliably grown in the absence of iron even without potential inhibitors. For P. aeruginosa, EC50 values were determined for the wildtype PAO1 strain, for a pyoverdin-minus PAO1 strain, and for a pyochelin-minus PAO1 strain. The pyoverdin-minus strain is a ΔpvdA knockout strain, deficient in the ornithine hydroxylase enzyme that is the first committed step for production of the high-affinity pyoverdin siderophore. The pyochelin-minus strain is a ΔpchE knockout strain, deficient in the nonribosomal peptide synthetase required for salicylate incorporation. These data are normalized relative to bacteria grown in the absence of inhibitor and plotted as relative growth. The highest concentration of the compounds tested in these assays was 5 mM, limited by solubility and vehicle (DMSO) toxicity.
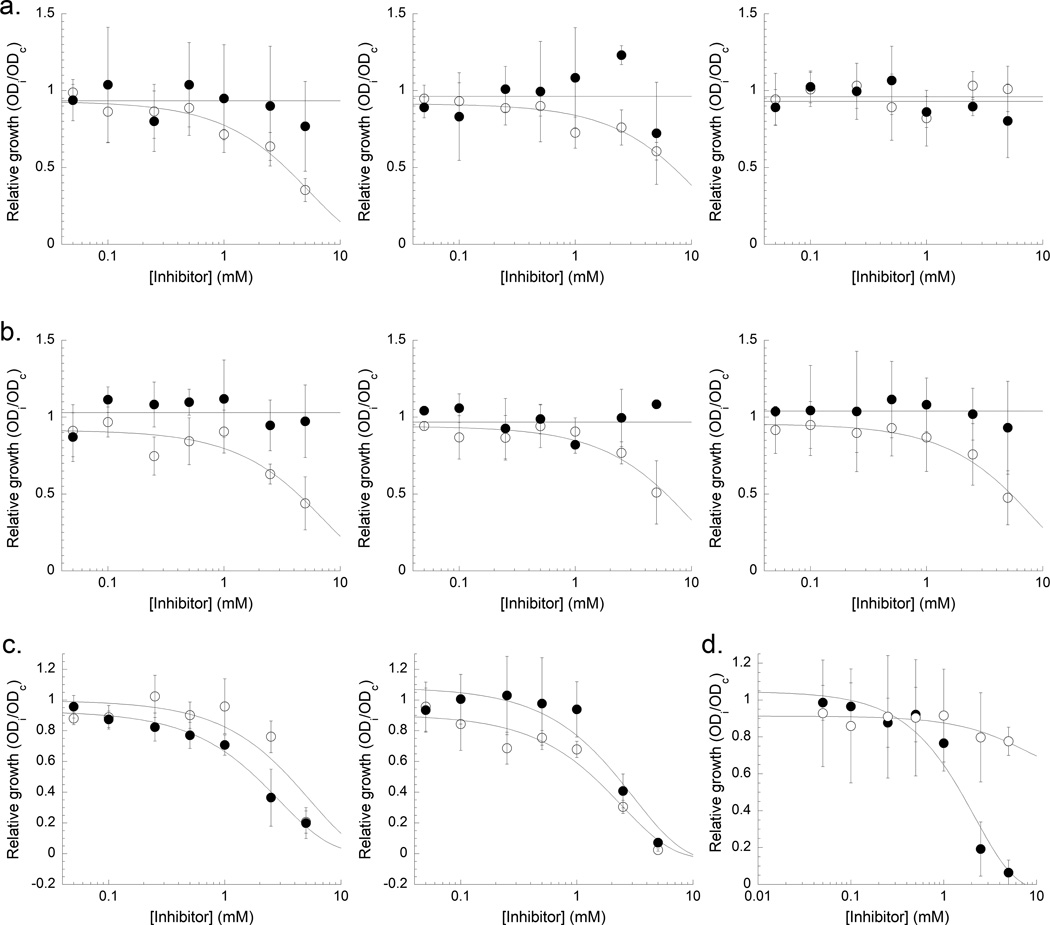
Compounds 6 and 10 are sub-micromolar inhibitors of the isochorismate-pyruvate lyase required for pyochelin production. These compounds did not cause growth inhibition in iron-rich media, regardless of P. aeruginosa, strain (Figure 4A and 4B, Table 2). This was anticipated, since the bacteria were not reliant on siderophore production for iron uptake when the media is replete with iron. Under iron-limiting conditions, if the compounds were specific for inhibition of PchB, we would expect the strongest inhibition in pyoverdin-minus strain, as this strain is more reliant on the pyochelin pathway for iron-uptake. The wildtype PAO1 strain could show some growth inhibition or this could be overcome by the functional and more efficient pyoverdin pathway. The pyochelin-deficient strain should show no inhibition in relative growth, since the pyochelin production pathway was genetically disrupted. Both the wildtype PAO1 strain and the pyoverdin-minus strain showed growth inhibition for both compounds at concentrations that were >5 mM. The pyochelin-minus strain was not inhibited by compound 6 but was inhibited at > 5mM by compound 10. These data suggest that compound 6 maybe more specific for the pyochelin pathway than compound 10. However, these data suggest that under iron-limiting conditions there may be a second target that is also inhibited by compound 10. P. aeruginosa has chorismate mutases that may be inhibited. Additionally, P. aeruginosa is highly antibiotic resistant due in part, to the presence of efflux pumps for the export of xenobiotics. Growth inhibition would likely be augmented in the presence of an efflux pump inhibitor, such as phenylalainine-arginine-β-naphthylamide [61], a future direction of this work.
Figure 4. Growth inhibition of Pseudomonas aeruginosa, Yersinia enterocolitica and E. coli growth.
a. P. aeurginosa growth inhibition by compound 6. b. P. aeurginosa growth inhibition by compound 10. For both A and B, left: WT PAO1 strain; middle: ΔpvdA PAO1, a pyoverdin minus strain; right: ΔpchE PAO1, a pyochelin minus strain. c. Y. enterocolitica growth inhibition by compound 6 (left) and compound 10 (right). For A–C: symbols: ∘ iron poor, ∙ iron rich. d. E. coli growth inhibition by compound 6 (∘) and compound 10 (∙) under iron rich conditions.
Table 2.
Half maximal value of bacterial growth inhibition
| EC50 (mM) | ||||||
|---|---|---|---|---|---|---|
| Compound | PAO1 p.a. |
ΔpvdA p.a. |
ΔpchE p.a. |
Y. enterocolitica |
E. coli | |
| low | 6 | > 5 | > 5 | NI | > 5 | |
| iron | 10 | > 5 | > 5 | > 5 | 2. 3 ± 0.1 | |
| high | 6 | NI | NI | > 5 | 3 ± 1 | > 5 |
| iron | 10 | NI | NI | NI | 3.1 ± 0.9 | 3.2 ± 0.2 |
NI= not inhibitory
Compound 6 was not an enzymatic inhibitor of the Y. enterocolitica salicylate synthase Irp9, but compound 10 was a micromolar inhibitor. If our hypothesis was correct and compound 10 was specific for yersiniabactin biosynthesis, then neither should inhibit Y. enterocolitica bacterial growth under iron rich conditions, and only compound 10 should inhibit under iron poor conditions. If the inhibitors are non-selective, then both may inhibit bacterial growth under both iron rich and iron poor conditions. If the inhibitors work against both the yersiniabactin biosynthesis and another enzymatic process, then compound 10 should be more effective under iron poor conditions. Based on the data in Figure 4C and Table 2, both compounds give EC50 concentrations in the millimolar range under both iron rich and iron poor conditions (indeed, the differing iron conditions were within error of each other), suggesting that compound 10 is not selective for siderophore biosynthesis and that both inhibitors are causing growth inhibition by some other mechanism.
Compounds 6 and 10 are low micromolar inhibitors of E. coli chorismate mutase. In growth inhibition assays against Seattle strain E. coli, compound 6 arguably showed no growth inhibition, while compound 10 gave an EC50 value of 3 mM (Figure 4D).
3. Discussion
Two inhibitors of PchB are reported in the literature: Bartlett’s TSA (Ki ~2 µM) and adamantane-1-phosphonate (Ki >5 mM) [62]. The work reported here identifies three compounds with Ki values in the nanomolar range: 6 (Ki 44 +/− 2 nM), 9 (Ki = 80 +/− 8 nM) and 10 (Ki = 350 +/− 40 nM). In addition, compounds 6 and 10 show growth inhibition of P. aeruginosa at millimolar concentrations under iron-limiting conditions. Therefore, these compounds represent the best known inhibitors of PchB, and may serve as initial lead compounds for the development of new antibtiotics.
To our knowledge, the best known inhibitor for E. coli chorismate mutase is Bartlett’s TSA. The literature reports inhibition of this and similar compounds as I50/Km1, with a value of 0.008 for the TSA [34, 35]. Similar calculations were performed for compounds 6 and 10 versus EcCM, and values of 0.05 and 0.053 were determined, better than all of the reported compounds save the TSA. These compounds therefore represent new scaffolds suitable for optimization as chemical probes and inhibitors of chorismate mutases. Furthermore, compound 10 also was effective at growth inhibition of E. coli in the millimolar range.
Rational design of inhibitors against isochorismate and salicylate synthases and similar chorismate-utilizing enzymes have primarily targeted varying the pyruvylenol tail of the substrate and have produced inhibitors with low micromolar Ki values [47, 51, 52, 63] Interestingly, in two cases, PchB was used to convert isochorismate generated by an isochorismate synthase into salicylate for detection but there is no mention as to whether the inhibitors were tested for inhibition of PchB alone [51, 52]. A high-throughput screen against the salicylate synthase from M. tuberculosis (MbtI) identified two structural classes of inhibitors, which showed IC50 values in the low micromolar range [60]. The first was a diarylsulfone reminiscent of compound 9; however, compounds of this scaffold have been identified as pan assay interference compounds [64] and are therefore unlikely for further development. The second was a benzimidazole-2-thione similar to compounds 10–12. We likewise determined a micromolar IC50 values for compound 10 with the Y. enterocolitica salicylate synthase Irp9, which shows growth inhibition of Y. enterocolitica in the low millimolar range. These fused heterocyclic ring systems may be an excellent starting point for inhibition in the MST enzyme class.
We hypothesized that an inhibitor identified through a high throughput screen may be effective against enzymes of similar scaffold or the same catalytic function. The screen against the isochorismate-pyruvate lyase identified competitive inhibitors that were found to likewise be competitive inhibitors against chorismate mutase (similar fold and active site structure) and salicylate synthase (same catalytic mechanism). While one may speculate that the ability to “target hop” may be only possible with competitive inhibitors, exploiting similarity of the active site and/or transition state, it is also possible to envision a scenario where an allosteric regulator binding site may also be similarly employed. The bacterial inhibition studies suggest that the compounds may be multi-selective. In other words, the compounds inhibit proteins of similar structure and/or function in vitro, and may do so in vivo as well. All of the bacterial cultures grown generate chemically-related siderophores with salicylate or dihydroxybenzoate caps utilizing enzymes that are structurally or functionally related the PchB and/or Irp9. These bacterial species also produce aromatic amino acids, thereby utilizing a chorismate mutase of the same structural family as EcCM. However, these studies do not rule out the possibility that these inhibitors are non-selective, inhibiting a target dissimilar and completely independent to those tested. Identification of the cellular target(s) is the next important step in this experimentation. .
4. Conclusions
The HTS screen presented here identified inhibitors of the isochorismate-pyruvate lyase from P. aeruginosa. Lead compounds were scrutinized in an orthogonal assay and for bacterial growth inhibition. The inhibitors were similarly tested against an enzyme that performs identical chemistry in a different protein scaffold (salicylate synthase) and against an enzyme with a homologous protein scaffold that performs similar chemistry (chorismate mutase). Inhibitors were identified that are better than those previously reported in the literature, and also cause growth inhibition of P. aeruginosa in a seemingly iron-dependent way. A subset of the identified inhibitors were also effective against enzymes of similar scaffold (EcCM) or identical chemistry (Irp9). The result is a series of compounds are among the most effective inhibitors to date for all three enzymes and may prove useful in the discovery of future antibiotics.
5. Methods
5.1 Preparation of substrates
Isochorismate was isolated from Klebsiella pneumoniae 62-1 harboring the entC plasmid pKS3-02 [26] with only minor changes, as described previously [27]. Chorismate was isolated from Klebisella pneumonia 62-1 [65] with only minor changes, as previously described [14].
5.2 Preparation of Overexpression Plasmids
The PchB overexpression plasmid (without a histidine tag) was generated as previously described [28]. The Irp9 overexpression plasmid was generated as previously described [66].
E. coli chorismate mutase (EcCM) is a domain of a larger pheA gene. The EcCM domain portion of the pheA gene was amplified from Escherichia coli K-12 substrain genomic MG1655 DNA (ATCC) by polymerase chain reaction by use of Master Mix (Eppendorf) with 1.5 mM magnesium acetate. The forward primer (5’-ATC GTC ATA TGA CAT CGG AAA ACC CGT TA-3’) includes an NdeI site (underlined) and the reverse primer (5’-TTA AGC TTT CAG AGA AAA GCG ATG CGT GCT G-3’) contains a stop codon and a HindIII site (underlined). The amplified 352 base pair fragment was digested with NdeI and HindIII and ligated into the pET28b plasmid (Novagen) digested with the same enzymes. The resultant plasmid encodes the chorismate mutase domain of pheA with an N-terminal histidine tag and no mutations.
5.3 Protein overexpression and purification
PchB without the histidine tag was overexpressed and purified as previously described [28]. Irp9 was overexpressed and purified as previously described [66].
BL21 Star (DE3) pLysS E. coli containing the EcCM expression plasmid were grown in LB medium containing 50 µg/mL kanamycin at 37 °C with shaking (250 rpm) until an OD600 of ~0.8 was reached. The culture temperature was reduced to 30 °C and the cells were harvested by centrifugation (6 000g, 10 min, 4 °C) after 4 h. The cell pellet was resuspended in 15 mL of 20 mM Tris-HCl pH 8.8, 500 mM NaCl, 5 mM imidazole (buffer A) per liter of culture. Cells were disrupted by use of a French pressure cell (35 000 psi), and cellular debris was removed by centrifugation (12 000g, 30 min, 4 °C). The supernatant was applied to a chelating Sepharose fast-flow column (Amersham Biosciences) charged with nickel chloride and pre-equilibrated in buffer A. EcCM protein eluted at 250 mM imidazole in a linear gradient of 5–500 mM imidazole in buffer A. The pooled fractions were applied to a Superdex 75 size-exclusion column (Amersham Biosciences) equilibrated with 25 mM sodium phosphate pH 8.5, 150 mM KCl, 10% glycerol. The fractions containing EcCM were pooled and concentrated by use of an Amicon stirred cell with a YM-10 membrane to 417 µM as determined by absorbance at 280 nm (ext. coeff 1,490 M−1 cm−1) and stored at −80 °C.
5.4 High-throughput screening
To identify inhibitors of PchB required for salicylate synthesis, high throughput screening was performed using an assay in which isochorismate is converted to salicylate by PchB. The fluorescence of salicylate was measured with an excitation wavelength of 300 nm and an emission wavelength of 400 nm. The assay was adapted to 384-well (Costar #3575) format and used to screen the compound libraries available at the University of Kansas High Throughput Screening Laboratory, Lawrence, Kansas. The compound collection was derived from both commercially available compound libraries (Prestwick (1120 compounds), Microsource Spectrum collection (2000 compounds), ChemDiv library (50,000 compounds), Chembridge library (43,736 compounds)) and the compounds (3360) synthesized at the University of Kansas Center for Chemical Methodologies and Library Development (CMLD). All enzyme and substrate stocks were prepared in reaction buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1 mM TCEP. The assay was initiated by adding PchB (12 nM final concentration) enzyme mixture to compounds (10 µM final concentrations) in a 384-well plate and incubated 5 mins at room temperature. The reaction was initiated by adding substrate (isochorismate, 3 µM final which is 1.5-times the Km) solution to the wells containing the compounds and enzyme mixture. The fluorescence intensity was measured at 0 and 20 mins, during which time the reaction was linear at 25 °C, using a Tecan Safire2 plate reader. Each plate had 16 wells of positive controls (DMSO with no compound) and 14 wells for minus protein negative controls. A compound identified from a previous ChemBridge library screen (ChemBridge #5325243) was included in two wells as a positive assay control (6 µM). In summary, a total number of 100,216 compounds were screened.
5.5 HTS data analysis
Relative Fluorescence Units (RFU) per minute was calculated by subtracting the fluorescence intensity from pre-reads (before the addition of isochorismate) from that of the end reads (20 mins after the addition of isochorismate). The percent inhibition was calculated from median RFU/min in all DMSO/compound containing wells. A hit rate of 0.4% was obtained with 422 compounds showing >50% inhibition in salicylate synthesis over the DMSO treated controls. An average Z’ value of 0.80 ± 0.05 was obtained across all plates screened.
5.6 Secondary screening at HTS facility
All 422 compounds were cherry picked and tested for activity in an 8-point dose-response curve with 2-fold serial dilutions starting at 30 µM. The primary screening assay was used to confirm activity of the cherry-picked compounds. A reconfirmation rate of 50% was obtained and 131 compounds were found to be dose-responsive, with IC50 values ranging from 410 nM to >30 µM.
5.7 Preparation of compounds for confirmatory assays and minimum inhibitory concentration
Compounds 9–13 were purchased from ChemDiv. Compounds 6, 7, and 8 were no longer available from ChemBridge and were prepared by a modified procedure that was originally reported by Miller and colleagues [67].
5.7.1 General procedure
Anthranilic acid (177 mg, 1.29 mmol) was added to a solution of Na2CO3 (5 mL, 0.63 M) at 60°C. The appropriate sulfonyl chloride (1.55 mmol, 1.2 equiv) was added in three portions over a period of 10 min. The reaction was heated to 75°C for 30 min and then raised to 85°C and quenched with a solution of HCl (1 mL, 6 M). The solution was cooled to room temperature and the resulting precipitate collected by vacuum filtration and was dried under vacuum overnight. All materials were obtained from commercial suppliers and used without further purification. 1H NMR and 13C NMR were recorded on a Bruker AV-500 with cryoprobe. Chemical shifts (δ) are reported in ppm downfield from TMS and are referenced to the residual DMSO solvent peak. High resolution mass spectroscopy (HRMS) was performed using a LCT Premier (Micromass Ltd., Manchester UK) time of flight mass spectrometer with an electrospray ion source. Melting points are uncorrected and were measured on a Thomas Hoover Capillary Melting Point Apparatus.
5.7.2 5-(N-(2-carboxyphenyl)sulfamoyl)-2-hydroxybenzoic acid (Compound 6)
1H NMR (500 MHz, DMSO) δ 10.99 (s, 1H), 8.14 (d, J = 2.5 Hz, 1H), 7.89 (dd, J = 7.9, 1.5 Hz, 1H), 7.84 (dd, J = 8.8, 2.5 Hz, 1H), 7.57 (m, 1H), 7.49 (dd, J = 8.3, 0.9 Hz, 1H), 7.14 (td, J = 8.0, 1.2 Hz, 1H), 7.08 (d, J = 8.8 Hz, 1H). 13C NMR (126 MHz, DMSO) δ 170.19, 169.76, 164.32, 139.62, 134.63, 133.47, 131.64, 130.06, 128.84, 123.78, 119.07, 118.66, 117.30, 113.97. HRMS: [M-H]− calcd 336.0178, found 336.0183. Melting point: 221–222°C (dec.)
5.7.3 2-(3-carboxyphenylsulfonamido)benzoic acid (Compound 7)
1H NMR (500 MHz, DMSO) δ 11.21 (s, 1H), 8.26 (t, J = 1.6 Hz, 1H), 8.15 (m, 1H), 8.02 (ddd, J = 7.9, 2.0, 1.1 Hz, 1H), 7.88 (dd, J = 7.9, 1.5 Hz, 1H), 7.69 (dd, J = 11.8, 4.0 Hz, 1H), 7.57 (m, 1H), 7.50 (dd, J = 8.3, 0.9 Hz, 1H), 7.15 (td, J = 7.9, 1.2 Hz, 1H). 13C NMR (126 MHz, DMSO) δ 169.68, 165.77, 139.33, 139.20, 134.58, 134.04, 132.03, 131.62, 130.91, 130.33, 127.31, 123.97, 119.24, 117.65. HRMS: [M-H]− calcd 320.0229 found 320.0229. Melting point: 234–236°C (dec.)
5.7.4 2-bromo-5-(N-(2-carboxyphenyl)sulfamoyl)benzoic acid (Compound 8)
1H NMR (500 MHz, DMSO) δ 11.16 (s, 1H), 8.09 (d, J = 2.4 Hz, 1H), 7.91 (t, J = 8.4 Hz, 1H), 7.90 (s, 1H), 7.80 (dd, J = 8.4, 2.4 Hz, 1H), 7.57 (ddd, J = 8.5, 7.4, 1.6 Hz, 1H), 7.47 (dd, J = 8.3, 0.8 Hz, 1H), 7.17 (td, J = 7.9, 1.1 Hz, 1H). 13C NMR (126 MHz, DMSO) δ 169.59, 165.85, 138.99, 138.25, 135.53, 134.61, 134.58, 131.68, 130.25, 128.73, 126.01, 124.10, 119.30, 117.84. HRMS: [M-H]−calcd 397.9334, found 397.9333. Melting point: 219–220°C (dec.)
5.8 Salicylate fluorescence assay
For the PchB assays, 50 nM enzyme was added to 50 mM potassium phosphate, pH 8 and the reaction was initiated by the addition of isochorismate. For the Irp9 assays, 100 nM enzyme was added to 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10% (v/v) glycerol and the reaction was initiated by the addition of chorismate. Initial velocities were determined at 25 °C by measuring the accumulation of salicylate by fluorescence with an excitation wavelength of 310 nm and an emission wavelength of 430 nm using a Cary Eclipse fluorescence spectrometer (Varian) with temperature controller. Initial velocities were measured for 120 seconds during which the reaction was linear. The substrate concentration was set at the Km of 2 µM for IC50 determination, and the inhibitor concentrations varied. For the Ki determination for compounds 9 and 10, the PchB concentration was 50 nM and the isochorismate concentrations ranged from 0–75 µM with inhibitor concentrations ranging from 0–100 nM for compound 9 and 0–1.5 µM for compound 10. The data was collected as for the IC50 determination measuring the fluorescence of salicylate at 430 nm upon excitation at 310 nm.
The Ki was determined for compound 6 using a TgK Scientific SF-61DX2 stopped-flow apparatus equipped with a photomuliplier detector and a mercury-xenon lamp at 25 °C. Equal volumes of enzyme with inhibitor were mixed with isochorismate (for PchB Ki determination) or chorismate (for Irp9 Ki determination) and initial velocity measurements were collected as an increase in salicylate fluorescence upon excitation at 310 nm using a cutoff filter at 360 nm. Initial velocities determined using the stopped-flow were measured for 30 seconds during which the reaction was linear. The final concentration of PchB was 50 nM, the isochorismate concentration ranged from 0–50 µM and the inhibitor concentration ranged from 0–100 nM in a reaction buffer of 50 mM potassium phosphate, pH 8, 1 mM TCEP. The final concentration of Irp9 was 100 nM, the chorismate concentration ranged from 0–40 µM and the inhibitor concentration ranged from 0–10 µM in a reaction buffer of 50 mM Tris, pH 7.5, 10 mM MgCl2, 10% glycerol.
5.9 Pyruvate production assay
This assay is based on the method described by Anthon and Barrett [68]. For the PchB pyruvate production assays, 50 nM enzyme was added to 50 mM potassium phosphate, pH 8 and the reaction was initiated by the addition of isochorismate. For Irp9, 200 nM enzyme was used and the reaction was initiated by the addition of chorismate. Initial velocities were determined at 25 °C by adding 70 µl of sample at 2 min time points to 70 µl 0.25 g/L 2,4-dinitrophenylhydrazine in 1 M HCl and incubated 10 min at 37 °C. The absorbance at 515 nm was read upon addition of 70 µl 1.5 M NaOH using a Cary 50 MPR microplate reader (Varian) and converted to pyruvate concentration by means of a standard curve. The substrate concentration was set above the Km of 10 µM for IC50 determination, and the inhibitor concentrations varied. All experiments were performed in triplicate.
5.10 Chorismate disappearance assay
For the EcCM assays, the disappearance of chorismate was determined using a TgK Scientific SF-61DX2 stopped-flow apparatus equipped with a photomuliplier detector and a mercury-xenon lamp operated at 37 °C. Equal volumes of enzyme with inhibitor were mixed with chorismate in reaction buffer consisting of 50 mM Tris-HCl, pH 8, 2.5 mM EDTA, 20 mM βME, 0.01 % BSA and initial velocity measurements were collected as a decrease in absorbance at 310 nm for 30 seconds during which the reaction was linear. Pre-injection enzyme, substrate, and inhibitor concentration were twice the final concentration in the cell (post-mixing). The post-mixing concentrations were 500 nM for the enzyme, substrate chorismate concentration was set at the Km of 600 µM, and the inhibitor concentration varied from 0–500 µM. All experiments were performed in triplicate. For the Ki determination of compounds 6 and 10, the final concentration of EcCM was 500 nM, the chorismate concentration ranged from 0–2 mM and the inhibitor concentration ranged from 0–50 µM.
5.11 Bacterial strains used for growth inhibition assays
Wild type strains include E. coli strain Seattle (ATCC, 25922), Yersinia enterocolitica (ATCC 9610) and P. aeruginosa PAO1 (provided by Dr. Jeffrey L. Urbauer, University of Georgia, Athens, GA). P. aeruginosa deletion mutant strains were acquired from a P. aeruginosa PAO1 transposon mutant library (University of Washington [69]) and include a P. aeruginosa PAO1 PvdA deletion mutant (PW5011) and a P. aeruginosa PAO1 PchE deletion mutant (PW8175).
5.12 Growth inhibition assasy
P. aeruginosa and Y. enterocoltica were grown overnight in LB media and diluted to an OD600 of 0.5 with PMH-D media [70]. Dose response curves were determined by adding 10 µl diluted cells with 90 µl of PMH-D media with 5 µM FeCl3 (iron-limited) or 200 µM FeCl3 (iron-rich) containing inhibitor concentrations of 0–5 mM. The plates were incubated at 37 °C with shaking (200 rpm) for 24 hrs at which time the well solution was centrifuged to pellet the cells and the media with residual inhibitor was removed. The cells were resuspended in 100 µl LB media and the absorbance at 600 nm was read using a Cary 50 MPR microplate reader (Varian). E. coli were grown overnight in LB media and diluted to an OD600 of 0.2 with LB media. Dose response curves were determined by adding 10 µl diluted cells with 90 µl of tryptic soy broth containing inhibitor concentrations of 0–5 mM. The plates were incubated at 37 °C with shaking (200 rpm) for 10 hrs and the absorbance was read at 600 nm. All experiments were performed in triplicate.
Supplementary Material
Acknowledgements
We are grateful to Frank Schoenen and Scott Hefty for helpful discussions. This publication was made possible by funds from Kansas Masonic Cancer Research Institute, from the NIH grants numbered R01 AI77725 and K02 AI093675 from the National Institute for Allergy and Infectious Disease (A.L.L), NIH grant number P20 RR016475 from the INBRE Program of the National Center for Research Resources, and by the Graduate Training Program in Dynamic Aspects of Chemical Biology NIH grant number T32 GM08545 (A. P. R.). The University of Kansas High Throughput Screening Laboratory is funded in part by NIH COBRE Grant 8 P30 GM103495 (Timmermann, PI) and the NCI Cancer Support Grant 5 P30 CA168524 (Jensen, PI).
Abbreviations
- EcCM
E. coli chorismate mutase
- EntC
isochorismate synthase from E. coli
- Irp9
salicylate synthase from Yersinia enterocolitica
- MbtI
salicylate synthase from Mycobacterium tuberculosis
- MenF
isochorismate synthase from E. coli
- MST
protein family containing proteins of menaquinone, siderophore or tryptophan biosynthesis
- PchA
isochorismate synthase from Pseudomonas aeruginosa
- PchB
isochorismate-pyruvate lyase from Pseudomonas aeruginosa
- TSA
transition state analogue
- VibC
isochorismate synthase from Vibrio cholera
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bartlett and colleagues reported inhibition of chorismate mutase/prephenate dehydrogenase by their transition state analogues as I50 /Km values [34, 35], where they defined I50 as “the concentration of inhibitor giving 50% inhibition when the substrate concentration equals Km” [34]. For the sake of direct comparison, we likewise make these calculations.
Supplementary data
Supplemental figure 1: assay readout, Z’ and signal-to-baseline plots. This material is available free of charge via the internet at http://pubs.acs.org.
Reference
- 1.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65(5):1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearden SW, Perry RD. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32(2):403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 4.Cox CD. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect Immun. 1982;36(1):17–23. doi: 10.1128/iai.36.1.17-23.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE., 3rd The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. U S A. 2000;97(3):1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fetherston JD, Bertolino VJ, Perry RD. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32(2):289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- 7.Golden CA, Kochan I, Spriggs DR. Role of mycobactin in the growth and virulence of tubercle bacilli. Infect Immun. 1974;9(1):34–40. doi: 10.1128/iai.9.1.34-40.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy PV, Puri RV, Chauhan P, Kar R, Rohilla A, Khera A, Tyagi AK. Disruption of mycobactin biosynthesis leads to attenuation of Mycobacterium tuberculosis for growth and virulence. J. Infect. Dis. 2013;208(8):1255–1265. doi: 10.1093/infdis/jit250. [DOI] [PubMed] [Google Scholar]
- 9.Snow GA. Isolation and structure of mycobactin T, a growth factor from Mycobacterium tuberculosis. Biochem. J. 1965;97(1):166–175. doi: 10.1042/bj0970166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokol PA. Surface expression of ferripyochelin-binding protein is required for virulence of Pseudomonas aeruginosa. Infect Immun. 1987;55(9):2021–2025. doi: 10.1128/iai.55.9.2021-2025.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salyers AA, Whitt DD. Bacterial Pathogenesis: A Molecular Approach. 2002 [Google Scholar]
- 12.Miller MC, Parkin S, Fetherston JD, Perry RD, Demoll E. Crystal structure of ferric-yersiniabactin, a virulence factor of Yersinia pestis. J. Inorg. Biochem. 2006;100(9):1495–1500. doi: 10.1016/j.jinorgbio.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 13.He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. Conservation of mechanism in three chorismate-utilizing enzymes. J. Am. Chem. Soc. 2004;126(8):2378–2385. doi: 10.1021/ja0389927. [DOI] [PubMed] [Google Scholar]
- 14.Meneely KM, Luo Q, Dhar P, Lamb AL. Lysine221 is the general base residue of the isochorismate synthase from Pseudomonas aeruginosa (PchA) in a reaction that is diffusion limited. Arch. Biochem. Biophys. 2013;538(1):49–56. doi: 10.1016/j.abb.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zwahlen J, Kolappan S, Zhou R, Kisker C, Tonge PJ. Structure and mechanism of MbtI, the salicylate synthase from Mycobacterium tuberculosis. Biochemistry. 2007;46(4):954–964. doi: 10.1021/bi060852x. [DOI] [PubMed] [Google Scholar]
- 16.DeClue MS, Baldridge KK, Kunzler DE, Kast P, Hilvert D. Isochorismate Pyruvate Lyase: A Pericyclic Reaction Mechanism? J. Am. Chem. Soc. 2005;127(43):15002–15003. doi: 10.1021/ja055871t. [DOI] [PubMed] [Google Scholar]
- 17.Lamb AL. Pericyclic reactions catalyzed by chorismate-utilizing enzymes. Biochemistry. 2011;50(35):7476–7483. doi: 10.1021/bi2009739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolappan S, Zwahlen J, Zhou R, Truglio JJ, Tonge PJ, Kisker C. Lysine 190 is the catalytic base in MenF, the menaquinone-specific isochorismate synthase from Escherichia coli: implications for an enzyme family. Biochemistry. 2007;46(4):946–953. doi: 10.1021/bi0608515. [DOI] [PubMed] [Google Scholar]
- 19.Parsons JF, Shi KM, Ladner JE. Structure of isochorismate synthase in complex with magnesium. Acta crystallographica. 2008;D64(Pt 5):607–610. doi: 10.1107/S0907444908005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Duncan K, Walsh CT. Nucleotide sequence of a cluster of Escherichia coli enterobactin biosynthesis genes: identification of entA and purification of its product 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. J. Bacteriol. 1989;171(2):791–798. doi: 10.1128/jb.171.2.791-798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Quinn N, Berchtold GA, Walsh CT. Overexpression, purification, and characterization of isochorismate synthase (EntC), the first enzyme involved in the biosynthesis of enterobactin from chorismate. Biochemistry. 1990;29(6):1417–1425. doi: 10.1021/bi00458a012. [DOI] [PubMed] [Google Scholar]
- 22.Rusnak F, Liu J, Quinn N, Berchtold GA, Walsh CT. Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and substrate specificity of isochorismatase. Biochemistry. 1990;29(6):1425–1435. doi: 10.1021/bi00458a013. [DOI] [PubMed] [Google Scholar]
- 23.Sridharan S, Howard N, Kerbarh O, Blaszczyk M, Abell C, Blundell TL. Crystal structure of Escherichia coli enterobactin-specific isochorismate synthase (EntC) bound to its reaction product isochorismate: implications for the enzyme mechanism and differential activity of chorismate-utilizing enzymes. J Mol Biol. 2010;397(1):290–300. doi: 10.1016/j.jmb.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 24.Keating TA, Marshall CG, Walsh CT. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry. 2000;39(50):15513–15521. doi: 10.1021/bi001651a. [DOI] [PubMed] [Google Scholar]
- 25.Wyckoff EE, Smith SL, Payne SM. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J. Bacteriol. 2001;183(5):1830–1834. doi: 10.1128/JB.183.5.1830-1834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt K, Leistner E. Microbial Production of (+)-trans-Isochorismic Acid. Biotech. & Bioeng. 1995;45:285–291. doi: 10.1002/bit.260450402. [DOI] [PubMed] [Google Scholar]
- 27.Luo Q, Olucha J, Lamb AL. Structure-function analyses of isochorismate-pyruvate lyase from Pseudomonas aeruginosa suggest differing catalytic mechanisms for the two pericyclic reactions of this bifunctional enzyme. Biochemistry. 2009;48:5239–5245. doi: 10.1021/bi900456e. [DOI] [PubMed] [Google Scholar]
- 28.Zaitseva J, Lu J, Olechoski KL, Lamb AL. Two crystal structures of the isochorismate pyruvate lyase from Pseudomonas aeruginosa. J Biol Chem. 2006;281(44):33441–33449. doi: 10.1074/jbc.M605470200. [DOI] [PubMed] [Google Scholar]
- 29.Lee AY, Karplus PA, Ganem B, Clardy J. Atomic structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J. Am. Chem. Soc. 1995;117(12):3627–3628. [Google Scholar]
- 30.Halsam E. Shikimic acid: metabolism and metabolites. New York: Wiley; 1993. [Google Scholar]
- 31.Agrawal H, Kumar A, Bal NC, Siddiqi MI, Arora A. Ligand based virtual screening and biological evaluation of inhibitors of chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv. Bioorg. Med. Chem. Lett. 2007;17(11):3053–3058. doi: 10.1016/j.bmcl.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 32.Andrews PR, Cain EN, Rizzardo E, Smith GD. Rearrangement of chorismate to prephenate. Use of chorismate mutase inhibitors to define the transition state structure. Biochemistry. 1977;16(22):4848–4852. doi: 10.1021/bi00641a015. [DOI] [PubMed] [Google Scholar]
- 33.Andrews PR, Smith GD, Young IG. Transition-state stabilization and enzymic catalysis. Kinetic and molecular orbital studies of the rearrangement of chorismate to prephenate. Biochemistry. 1973;12(18):3492–3498. doi: 10.1021/bi00742a022. [DOI] [PubMed] [Google Scholar]
- 34.Barlett PA, Johnson CR. An Inhibitor of Chorismate Mutase Resembling the Transition State Conformation. J. Am. Chem. Soc. 1985;107:7792–7793. [Google Scholar]
- 35.Barlett PA, Nakagawa Y, Johnson CR, Reich SH, Luis A. Chorismate Mutase Inhibitors: Synthesis and Evaluation of Some Potential Transition-State Analogues. J. Org. Chem. 1988;53:3195–3210. [Google Scholar]
- 36.Clarke T, Stewart J, Ganem B. Transition-state analogue inhibitors of chorismate mutase. Tetrahedron. 1990;46:731–748. [Google Scholar]
- 37.Husain A, Galopin CC, Zhang S, Pohnert G, Ganem B. S-(−)-dinitrobiphenic acid: a selective inhibitor of Escherichia coli chorismate mutase based on prephenate mimicry. J. Am. Chem. Soc. 1999;121:2647–2648. [Google Scholar]
- 38.Mandal A, Hilvert D. Charge optimization increases the potency and selectivity of a chorismate mutase inhibitor. J. Am. Chem. Soc. 2003;125(19):5598–5599. doi: 10.1021/ja029447t. [DOI] [PubMed] [Google Scholar]
- 39.Chi G, Manos-Turvey A, O'Connor PD, Johnston JM, Evans GL, Baker EN, Payne RJ, Lott JS, Bulloch EM. Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis. Biochemistry. 2012;51(24):4868–4879. doi: 10.1021/bi3002067. [DOI] [PubMed] [Google Scholar]
- 40.Cisar JS, Ferreras JA, Soni RK, Quadri LE, Tan DS. Exploiting ligand conformation in selective inhibition of non-ribosomal peptide synthetase amino acid adenylation with designed macrocyclic small molecules. J. Am. Chem. Soc. 2007;129(25):7752–7753. doi: 10.1021/ja0721521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duckworth BP, Wilson DJ, Nelson KM, Boshoff HI, Barry CE, 3rd, Aldrich CC. Development of a Selective Activity-Based Probe for Adenylating Enzymes: Profiling MbtA Involved in Siderophore Biosynthesis from Mycobacterium tuberculosis. ACS Chem. Biol. 2012;7(10):1653–1658. doi: 10.1021/cb300112x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreras JA, Ryu JS, Di Lello F, Tan DS, Quadri LE. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 2005;1(1):29–32. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- 43.Gupte A, Boshoff HI, Wilson DJ, Neres J, Labello NP, Somu RV, Xing C, Barry CE, Aldrich CC. Inhibition of siderophore biosynthesis by 2-triazole substituted analogues of 5'-O-[N-(salicyl)sulfamoyl]adenosine: antibacterial nucleosides effective against Mycobacterium tuberculosis. J. Med. Chem. 2008;51(23):7495–7507. doi: 10.1021/jm8008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerbarh O, Bulloch EM, Payne RJ, Sahr T, Rebeille F, Abell C. Mechanistic and inhibition studies of chorismate-utilizing enzymes. Biochem. Soc. Trans. 2005;33(Pt 4):763–766. doi: 10.1042/BST0330763. [DOI] [PubMed] [Google Scholar]
- 45.Kozlowski MC, Barlett PA. Synthesis of a Potential Transition-State Analogue Inhibitor of Isochorismate Synthase. J. Am. Chem. Soc. 1991;113:5897–5898. [Google Scholar]
- 46.Kozlowski MC, Tom NJ, Seto CT, Sefler AM, Barlett PA. Chorismate-Utilizing Enzymes Isochorismate Synthase, Anthranilate Synthase, and p-Aminobenzoate Synthase: Mechanistic Insight through Inhibitor Design. J. Am. Chem. Soc. 1995;117:2128–2140. [Google Scholar]
- 47.Manos-Turvey A, Bulloch EM, Rutledge PJ, Baker EN, Lott JS, Payne RJ. Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI) ChemMedChem. 2010;5(7):1067–1079. doi: 10.1002/cmdc.201000137. [DOI] [PubMed] [Google Scholar]
- 48.Miethke M, Bisseret P, Beckering CL, Vignard D, Eustache J, Marahiel MA. Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. Febs J. 2006;273(2):409–419. doi: 10.1111/j.1742-4658.2005.05077.x. [DOI] [PubMed] [Google Scholar]
- 49.Neres J, Labello NP, Somu RV, Boshoff HI, Wilson DJ, Vannada J, Chen L, Barry CE, 3rd, Bennett EM, Aldrich CC. Inhibition of siderophore biosynthesis in Mycobacterium tuberculosis with nucleoside bisubstrate analogues: structure-activity relationships of the nucleobase domain of 5′-O-[N-(salicyl)sulfamoyl]adenosine. J. Med. Chem. 2008;51(17):5349–5370. doi: 10.1021/jm800567v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neres J, Wilson DJ, Celia L, Beck BJ, Aldrich CC. Aryl acid adenylating enzymes involved in siderophore biosynthesis: fluorescence polarization assay, ligand specificity, and discovery of non-nucleoside inhibitors via high-throughput screening. Biochemistry. 2008;47(45):11735–11749. doi: 10.1021/bi801625b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Payne RJ, Bulloch EM, Kerbarh O, Abell C. Inhibition of chorismate-utilising enzymes by 2-amino-4-carboxypyridine and 4-carboxypyridone and 5-carboxypyridone analogues. Org. Biomol. Chem. 2010;8(15):3534–3542. doi: 10.1039/c004062b. [DOI] [PubMed] [Google Scholar]
- 52.Payne RJ, Bulloch EM, Toscano MM, Jones MA, Kerbarh O, Abell C. Synthesis and evaluation of 2,5-dihydrochorismate analogues as inhibitors of the chorismate-utilising enzymes. Org. Biomol. Chem. 2009;7(11):2421–2429. doi: 10.1039/b901694e. [DOI] [PubMed] [Google Scholar]
- 53.Payne RJ, Toscano MD, Bulloch EM, Abell AD, Abell C. Design and synthesis of aromatic inhibitors of anthranilate synthase. Org. Biomol. Chem. 2005;3(12):2271–2281. doi: 10.1039/b503802b. [DOI] [PubMed] [Google Scholar]
- 54.Qiao C, Gupte A, Boshoff HI, Wilson DJ, Bennett EM, Somu RV, Barry CE, 3rd, Aldrich CC. 5'-O-[(N-acyl)sulfamoyl]adenosines as antitubercular agents that inhibit MbtA: an adenylation enzyme required for siderophore biosynthesis of the mycobactins. J. Med. Chem. 2007;50(24):6080–6094. doi: 10.1021/jm070905o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sikora AL, Wilson DJ, Aldrich CC, Blanchard JS. Kinetic and inhibition studies of dihydroxybenzoate-AMP ligase from Escherichia coli. Biochemistry. 2010;49(17):3648–3657. doi: 10.1021/bi100350c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somu RV, Boshoff H, Qiao C, Bennett EM, Barry CE, 3rd, Aldrich CC. Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J. Med. Chem. 2006;49(1):31–34. doi: 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- 57.Stirrett KL, Ferreras JA, Jayaprakash V, Sinha BN, Ren T, Quadri LE. Small molecules with structural similarities to siderophores as novel antimicrobials against Mycobacterium tuberculosis and Yersinia pestis. Bioorg. Med. Chem. Lett. 2008;18(8):2662–2668. doi: 10.1016/j.bmcl.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vannada J, Bennett EM, Wilson DJ, Boshoff HI, Barry CE, 3rd, Aldrich CC. Design, synthesis, and biological evaluation of beta-ketosulfonamide adenylation inhibitors as potential antitubercular agents. Organic letters. 2006;8(21):4707–4710. doi: 10.1021/ol0617289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziebart KT, Dixon SM, Avila B, El-Badri MH, Guggenheim KG, Kurth MJ, Toney MD. Targeting multiple chorismate-utilizing enzymes with a single inhibitor: validation of a three-stage design. J. Med. Chem. 2010;53(9):3718–3729. doi: 10.1021/jm100158v. [DOI] [PubMed] [Google Scholar]
- 60.Vasan M, Neres J, Williams J, Wilson DJ, Teitelbaum AM, Remmel RP, Aldrich CC. Inhibitors of the salicylate synthase (MbtI) from Mycobacterium tuberculosis discovered by high-throughput screening. ChemMedChem. 2010;5(12):2079–2087. doi: 10.1002/cmdc.201000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunzler DE, Sasso S, Gamper M, Hilvert D, Kast P. Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from combinatorial mutagenesis and selection. J Biol Chem. 2005;280(38):32827–32834. doi: 10.1074/jbc.M506883200. [DOI] [PubMed] [Google Scholar]
- 63.Payne RJ, Kerbarh O, Miguel RN, Abell AD, Abell C. Inhibition studies on salicylate synthase. Org. Biomol. Chem. 2005;3(10):1825–1827. doi: 10.1039/b503800f. [DOI] [PubMed] [Google Scholar]
- 64.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of medicinal chemistry. 2010;53(7):2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 65.Rieger CE, Turnbull JL. Small scale biosynthesis and purification of gram quantities of chorismic acid. Preparative biochemistry & biotechnology. 1996;26(1):67–76. doi: 10.1080/10826069608000051. [DOI] [PubMed] [Google Scholar]
- 66.Meneely KM, Luo Q, Lamb AL. Redesign of MST enzymes to target lyase activity instead promotes mutase and dehydratase activities. Arch. Biochem. Biophys. 2013;539(1):70–80. doi: 10.1016/j.abb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Surman MD, Mulvihill MJ, Miller MJ. Novel 1,4-benzodiazepines from acylnitroso-derived hetero-Diels-Alder cycloadducts. Organic letters. 2002;4(1):139–141. doi: 10.1021/ol017036w. [DOI] [PubMed] [Google Scholar]
- 68.Anthon GE, Barrett DM. Modified method for the determination of pyruvic acid with dinitrophenylhydrazine in the assessment of onion pungency. J. Sci. Rood Agric. 2003;83:1210–1213. [Google Scholar]
- 69.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Staggs TM, Perry RD. Identification and cloning of a fur regulatory gene in Yersinia pestis. J. Bacteriol. 1991;173(2):417–425. doi: 10.1128/jb.173.2.417-425.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.