1. Introduction
1.1. Kinetoplastid Parasites as Pathogenic Parasites: Overview, Life Cycle, Relevance of Life Stages to Health
Over the past decade, neglected tropical diseases (NTDs) have seen a surge in research interest in the area of drug discovery. Protozoan pathogens that cause three of these NTDs, Chagas disease, human African trypanosomiasis (HAT), and leishmaniasis, have been the focus of increasing numbers of reported drug discovery focused publications. This has been fueled by the elucidation of the pathogen genomes, and the ability to map targets between parasite and human enzymes, for which a large amount of target-based drug discovery has been performed, both inside and outside the pharmaceutical industry. It has been aided by the parasites that cause these diseases being cultivatable and amenable to reverse genetic manipulation and by the existence of mouse infection model systems. Their analysis has resulted in important fundamental discoveries, but there remain substantial gaps in knowledge, especially with respect to characteristics that can be the foundation for needed therapeutic interventions. In humans, kinases have been a significant focus of the drug discovery efforts, representing nearly a third of the “druggable genome”,1 and many of these enzymes have been validated with respect to a wide range of therapeutic indications.
With this background in mind, this review will focus on tools and approaches for understanding the essentiality of kinase targets in Trypanosoma brucei that causes HAT, T. cruzi that causes Chagas disease, and Leishmania spp. that cause the various types of leishmaniasis, and it will describe recent efforts to translate the understanding of these targets into new therapeutic approaches. While kinases phosphorylate a wide variety of molecules (e.g., lipids, carbohydrates, amino acids, nucleotides, etc.), this review is primarily focused on protein kinases because of their great diversity both in humans and their pathogens. In addition, these kinases play critical biological roles and have been shown to be valid drug targets. We do include in the Chemistry section, however, examples of small molecule discovery efforts that focused on nonprotein kinases.
These three parasites belong to the order Kinetoplastidae and cause extensive human suffering and death worldwide, as well as significant economic damage due to diseases that they cause in livestock (Table 1).2 Although they are insect-borne, this review will center upon the human host specific life stages of the parasite because of the relevance to drug discovery. The full life cycles for these three pathogens are reviewed elsewhere in this issue.3 The diseases due to these parasites are quite widespread, but their transmission by insect vectors largely limits their range to tropical and subtropical regions and primarily to poor populations and travelers to the regions with infected vectors. However, transmission also occurs by blood transfusion and ingestion of contaminated foods and infrequently by direct transfer between animals.
Table 1. Summary of Epidemiological Characteristics of Chagas Disease, Human African Trypanosomiasis, and Leishmaniasisa.
| disease | causative agent | geographic distribution | vector(s) reservoir | clinical outcome | incidence | prevalence |
|---|---|---|---|---|---|---|
| Chagas disease | T. cruzi | Latin America | triatominae (especially Triatoma, Rhodnius and Panstrongylus) | acute phase; chronic phase: indeterminate, cardiac and digestive infections | ∼ 50 000 new cases of Chagas disease per year | ∼8 M T. cruzi infections |
| human African trypanosomiasis | T. b. rhodesiense T. b. gambiense | Africa | glossinidae G. morsitanssspp. G. fuscipessspp. G. palpalissspp. | sleeping sickness | ∼10 000 new cases per year | 50 000–70 000 cases |
| leishmaniasis, visceral | L. donovani L. infantum | Asia and East Africa (L..don.) Mediterranean region, Latin America (L.inf.) | Phlebotomusspp. Phlebotomusspp. Lutzomyiaspp. | visceral leishmaniasis, post kala-azar dermal leishmaniasis | ∼500 000 new cases visceral leishmaniasis per year | |
| leishmaniasis, cutaneous and mucocutaneous | L. major L. tropica L. aethiopica L. braziliensis L. mexicana | worldwide | Phlebotomusspp. Lutzomyiaspp. | cutaneous leishmaniasis, diffuse cutaneous leishmaniasis, mucocutaneous leishmaniasis | ∼1.5 to 2 M new cutaneous leishmaniasis cases per year | NA |
Taken from the WHO report of Research Priorities for Chagas disease, human African trypanosomiasis, and leishmaniasis.2
These pathogens are related but cause distinct diseases with unique characteristics that are critical for drug development. Their genomes have striking similarity with extensive conservation of gene content and synteny.4 However, the different parasites are transmitted by different insects and cause different diseases which is reflected in distinct genomic differences. T. brucei, but not the other two, has many hundreds of variant surface glycoprotein (VSG) genes and characteristic telomeric sites from which they are expressed one at a time which results in antigenic variation and hence immune evasion. T. cruzi has multiple genomic loci with numerous tandemly repeated surface protein genes which may also function in immune evasion although this has been studied less extensively compared to T. brucei.4,5Leishmania have smaller, more numerous chromosomes, and their differential gene content compared to the two trypanosomes may reflect their different means of avoiding the host immune responses. It is especially important for drug discovery strategies to consider the different biological compartments within which each of these parasites resides in the mammalian host and the damaging effects that they exert.
1.1.1. Trypanosoma brucei
The extracellular metacyclic trypomastigote T. brucei parasites are transmitted by the bite of an infected tsetse fly and initially proliferate in the bloodstream and lymphatic system of their mammalian host. They evade immune elimination by periodically switching the composition of their VSG coat (i.e., antigenic variation), and thus development of anti-HAT vaccines is not considered feasible.6 The mammalian stage parasites rely on glycolysis, which primarily occurs in the peroxisome-like glycosomes.7 The parasites’ reliance on glycolysis, a lack of an oxidative phosphorylation system while retaining the single mitochondrion with its hundreds of proteins, and the unusual organellar location of glycolysis has led to its becoming a focus for drug targeting.8 Other features that diverge from the host, such as trypanothione-based protection from oxidative stress,9 mRNA trans-splicing,10 and purine auxotrophy,11 have also been the targets of drug discovery efforts.
T. brucei subspecies have important differences. T. brucei rhodesiense causes East African HAT and has an animal reservoir while T. brucei gambiense causes West African HAT and has a more protracted disease course and more limited documentation of animal reservoirs.12 These species also have different drug susceptibilities. For example, on the basis of invitro results, eflornithine is less effective for T. brucei rhodesiense.13T. evansi and T. vivax are T. brucei variants that lack the ability to infect tsetse flies but are transmitted directly by biting flies or by sexual transmission between animals. There are infrequent cases of human infections by these species due to direct transmission apparently by biting flies.14 The bases for these differences in infectivity are unknown. However, the differences between human versus animal-infective T. brucei involves the differential susceptibility to human serum HDL which is lethal to subspecies that infect animals but not humans.15 Human infective subspecies contain a gene that encodes the serum resistance protein that is related to VSGs.15,16
The HAT infection proceeds through two stages: The first, (or hemolymphatic) stage of the infection occurs in the blood and lymph, and in this stage the patient exhibits flu-like symptoms. After a period of time the parasites move from the bloodstream into the central nervous system, at which time the progression of infection leads to the characteristic symptoms of sleeping sickness: disruption of the sleep cycle, coma, and eventually death. The localization of the pathogen in the CNS during this second stage of the disease therefore requires brain-penetrant drugs to treat the infection.
1.1.2. Trypanosoma cruzi
Trypanosoma cruzi are transmitted as infective trypomastigotes in the feces that are deposited by infected triatomine bugs when they feed, and are introduced by contact with a break in the skin or contact with a mucosal surface. However, they can be also transmitted by ingestion of contaminated food or beverage, congenitally, or by transfusion of infected blood. The trypomastigotes infect many different cell types and differentiate in the host cell cytoplasm into amastigotes that proliferate in this intracellular compartment. The amastigotes differentiate into nondividing trypomastigotes that are released upon rupture of the infected cell, and the trypomastigotes move to infect other cells. This process is repeated throughout this acute phase of the disease. The disease subsequently enters a chronic phase where few parasites, if any, are detectable in the bloodstream by microscopy and other methods such as PCR, and the infection that leads to critical pathogenesis is mainly localized in gut and heart muscle. The chronic phase can last for decades and, over the course of time, is accompanied by cardiac and enteric pathogenesis.17 Cardiomyopathy entails ventricular hypertrophy that can progress to debilitating arrhythmia, severe cardiac pathology, and associated debility and sudden death. The pathogenesis appears to have an immunopathological component.18T. cruzi strains have modest diversity that has been cataloged on the basis of isoenzyme (zymodeme) and mitochondrial DNA or kDNA (schizodeme) variations, and show some complex association with disease characteristics.19
1.1.3. Leishmania sp.
Leishmania are transmitted as metacyclic promastigotes to the mammalian host by the bite of an infected sand fly and infect the host macrophages that engulf them. The promastigotes differentiate into amastigotes within the phagolysosome of the macrophages where they proliferate. The phagolysosome is normally a hostile environment that contains degradative enzymes and becomes acidified upon infection with pathogens.20 However, Leishmania have evolved mechanisms that enable them to escape destruction, though these mechanisms are poorly understood.20 The infected macrophages lyse and release the proliferated amastigotes, which infect circulating and tissue macrophages with the localization of the parasites and pathology dependent on the Leishmania species and perhaps host factors.
Leishmania species are much more diverse than the T. brucei and T. cruzi subspecies and they cause a broad spectrum of diseases that generally correlate with the species. Indeed, L. braziliensis has a different number of chromosomes from the other species. The diseases range from self-resolving cutaneous disease that is associated with L. amazonensis, L. major, L. mexicana, and L. tropica to lethal visceral disease that is associated with L. chagasi/infantum, and L. donovani or the disseminating and disfiguring mucocutaneous disease that is associated with L. braziliensis and L. guyanensis. Host genetic differences undoubtedly contribute to the differences in pathogenesis.21Leishmania also infect wild and domestic animals, including dogs, which appear to be significant reservoirs of infection, and there appear to be many people with asymptomatic infections, potentially making them reservoirs for infection of others. The diversity of leishmaniasis is also reflected in its widespread geographical and socioeconomic ranges. Poor populations in East Africa and India are at risk of lethal visceral disease as are more affluent populations in the Mediterranean region. Cutaneous disease also affects rural populations who are more likely to come into contact with infected sandflies.
Leishmaniasis may, in its earlier stages, be silent or have signs such as fever, swollen lymph nodes, or redness at the site of bite which develops into an ulcerating lesion in the case of cutaneous leishmaniasis. A swollen spleen and/or liver often results in the case of visceral leishmaniasis. The pathogenesis for each Leishmania species appears to have a substantial immunological component which likely includes genetic characteristics of both the host and the parasite and whether or not the parasite harbors an RNA virus that interacts with the host immune system.22 The cutaneous lesions typically heal, albeit slowly, and leave a scar, but the parasites persist. Leishmania infections are thus chronic, possibly lifelong, and drug treatment may not be curative. It is an opportunistic pathogen, held in check by a fully functional immune system, but is proliferative and pathogenic in immunocompromised infected hosts. Visceral leishmaniasis is treated because it is potentially fatal. However, treatment can result in post kala-azar dermal leishmaniasis (PKDL), a long-lasting rash-like condition that affects skin over much of the body.
1.1.4. Neglected Tropical Diseases
Neglected tropical diseases (NTDs) are those diseases that primarily and disproportionately affect the poorest regions in the world. Since the highly expensive drug discovery process is, by and large, performed by the for-profit biopharmaceutical industry, and since any drugs for NTDs are unlikely to be profitable, there is only limited effort by industry. As a result, NTDs are treated by drugs that are suboptimal in safety, efficacy, cost, and convenience.
Kinetoplastid diseases are perhaps central examples of NTDs. For example, since there are only about 10 000 cases of HAT per year, this represents a DALY of disability-adjusted life years (DALYs) of 1.6 million per year,2 and thus is comparable to prostate cancer, which is currently a significant focus of the pharmaceutical industry. However, since these infections occur foremost among the very poor in sub-Saharan Africa, there is no commercial market for developing new anti-HAT drugs. Despite this, the World Health Organization has targeted HAT for elimination using a combination of vector control and drug treatment. Importantly, this disease has been on the brink of eradication many times only to reoccur episodically. As an additional confounding factor, NTDs are often complicated by coinfection with HIV. There is a substantial population at risk of infection with one of these three diseases which have a combined incidence estimated at ∼2 million new cases a year.
1.2. Need for New Drugs for Kinetoplastid Diseases
Most drugs for these diseases were developed in the last century, and they are unsuitable because of insufficient efficacy, toxicity, prohibitive cost, and/or increasing drug resistance.
1.2.1. Current Drugs for Human African Trypanosomiasis
Two drugs, pentamidine and suramin, are approved for this acute first stage of the infection and are efficacious, though there are toxicity concerns and resistance is developing.23 The parasites subsequently invade the central nervous system, resulting in second stage disease which leads to several pathogenic consequences, with severe symptoms of lethargy, sleep disturbances, coma, and eventually death if not treated. There are two drugs for the second stage: one, the organoarsenical drug melarsoprol, is highly toxic and leads to mortality in approximately 5% of patients taking the drug. The other drug, eflornithine, is less toxic, though it is only efficacious for the gambiense subspecies and requires large amounts of drug which must be delivered by infusion over 14 days. There is a new combination therapy now used in lieu of monotherapy in gambiense infections: an eflornithine–nifurtimox combination therapy (NECT), which reduces the eflornithine dose to 7 days of twice-daily infusions.24 In addition, clinical trials continue on the benzoxoborole SCYX-715825 and the nitroaromatic compound fexinidazole.26 The WHO has set aspirational goals for new drugs for HAT, seeking orally bioavailable treatments with lower toxicity, and efficacy in the blood and CNS infections of both of the infective subspecies.27
1.2.2. Current Drugs for Chagas Disease
Recently, a comprehensive overview of Chagas treatments was presented that highlights the grim status of chemotherapeutics for this disease.28 Currently, Chagas disease is treated using nifurtimox or benzidazole, both requiring 1–2 months of therapy. This long treatment regimen leads to low treatment compliance, and both drugs exert toxicity that becomes apparent over time. Importantly, these drugs have only been definitively shown to adequately treat the acute infection (with a modest success rate); there has been no drug demonstrated to adequately treat the chronic stage of infection.29
Recent work to repurpose existing antifungal agents as Chagas disease treatments30 has met with mixed results in the clinic.31 Because of the inability to adequately monitor infection rates in the chronic disease, clinical trials for drugs against Chagas disease are exceedingly challenging, made further so by the low-resource regions in which the clinical trials must be performed.
1.2.3. Current Drugs for Leishmaniasis
With typical cure rates from 80% to 100%, the frontline treatment for leishmaniasis consists of pentavalent antimonial compounds: sodium stibogluconate and meglumine antimonate. As an alternative, ambisome, a liposomal formulation of amphotericin B, is very efficacious for visceral leishmaniasis, but this formulation is expensive and often out of economic reach for most of those who need treatment. As a result, ambisome is often utilized as a second line treatment, for patients for whom pentavalent antimonial drugs fail. Indeed, antimony resistance in endemic areas is increasing.32 Use of paromomycin (mostly for cutaneous leishmaniasis),33 and the repurposed anticancer drug miltefosine34 have also shown utility.
1.3. Protein Kinases as Druggable Targets
A wide range of approaches have been pursued in the search for new drugs for kinetoplastid diseases, including target-based35 or cell-based36 high-throughput small molecule screens and identification of putative essential targets by detailed cell biology. Following identification of chemical matter in this way, further optimization is required in order to ensure high potency, cellular selectivity (low host toxicity), and the ability of new drugs to meet the target-product profiles for HAT,37 leishmaniasis,38 and Chagas disease.39 In humans, the primary focus of drug discovery efforts has historically been upon essential targets within the so-called “druggable genome”;1 kinases represent the largest group of druggable targets in the human genome (22%). Therefore, not surprisingly, a significant amount of drug discovery focus has been placed on modulation of kinase signaling, and this historical knowledge provides a compelling case for drug discovery efforts in NTDs to focus in these areas.
Trypanosomatid genomes code for a large number of protein kinases. Searches for protein kinase active site motifs in predicted proteins show that there are approximately 176, 190, and 199 protein kinases encoded in the T. brucei, T. cruzi, and Leishmania major genomes, respectively.40 This predicted kinome is a relatively large proportion of all predicted protein coding genes, at 2%. Therefore, the kinase gene family also represents a rich family of potential biological targets for pursuit for antikinetoplastid agents.
2. Biology
2.1. Overview of Kinases in Trypanosomatid Parasites
The trypanosomatid kinomes contain nearly all the protein kinase groups that occur in humans, despite being smaller.40 It completely lacks members of the receptor-linked tyrosine and tyrosine kinase-like kinases although tyrosine phosphorylation occurs, perhaps via dual-specificity protein kinases. The CMGC and STE groups and NEK family kinases are expanded relative to humans. In addition, some kinases cannot be definitively assigned to any kinase groups, suggesting that kinases have diverged substantially, possibly for Trypanosomatid-specific functions. Most protein kinases are highly conserved among T. brucei, T. cruzi, and Leishmania sp., suggesting that efforts that identify essential kinases or effective kinase inhibitors in one species would likely aid similar studies in the other trypanosomatid species.
2.2. Strategies for Exploring the Kinetoplastid Kinome
Protein kinase functions, and their potential as drug targets, can be explored by genetic or chemical inhibition approaches, summarized in Table 2. Importantly, studies for target validation must be performed in the disease stage of the parasite. The genetic approaches examine the consequences of manipulating the genes of interest by targeted gene knockout, or alteration of expression levels or gene protein product function. These approaches need to contend with the fact that kinetoplastids are diploids and that paralogous genes may provide functional redundancy. Importantly, since cells with an essential kinase knocked-down generally cannot be propagated (cells die), a conditional knockdown approach is typically necessary to study the phenotypic impacts of the loss of an essential kinase. Chemical inhibition of specific kinases can also be used to study kinases and can provide potent and acute inhibition if specificity can be achieved.
Table 2. Strategies: Advantages and Disadvantages.
| approach | advantages | disadvantages |
|---|---|---|
| Genetic Approaches | ||
| gene knockout | completely removes the gene of interest | cannot be used if the gene is necessary for cell survival. |
| plasmid shuffle | completely removes the gene of interest | requires several steps of genetic manipulation; can only analyze a population over time, phenotypic analysis difficult |
| RNAi | requires minimal genetic manipulation. | incomplete knockdown of the gene of interest; off-target effects could occur; only available in T. brucei |
| conditional null (transcriptional repression) | potent repression of the gene of interest | requires several steps of genetic manipulation; only available in T. brucei |
| degradation domain | relatively fast loss of protein; can be used in systems lacking RNAi machinery or transcriptional regulatory tools | incomplete knockdown could occur, and varies from protein to protein; protein of interest must tolerate tags |
| Chemical Approaches | ||
| specific inhibitors | reflect enzymatic inhibition versus protein loss; immediate potent inhibition | specific inhibitors’ creation generally involves a tedious chemical optimization project |
| ATP analogue sensitive alleles | reflect enzymatic inhibition versus protein loss; immediate potent inhibition | cells expressing ATP sensitive analogues must be created |
2.2.1. Genetic Approaches
Most genetic approaches study the loss-of-function of a protein by eliminating the protein or reducing its cellular level. In some cases conditional expression of functionally mutated genes can be achieved. These approaches study the protein of interest at the DNA, RNA, or protein level.
2.2.1.1. DNA Level
Elimination of the DNA sequence that encodes the gene of interest is a reliable way to ensure complete elimination of the encoded protein’s function. This is typically done by replacing the target gene with a selectable marker that confers drug resistance. Examples of the use of this approach in T. brucei and Leishmania can be found in Table 3. For this, DNA constructs with a selectable marker are created with sequences that are 5′ and 3′ to the kinase coding region for targeting specific recombination. Double recombination, i.e., at both the 5′ and 3′ ends of the gene, leads to deletion of the coding sequence of the targeted gene. To create such gene knockout cell lines, these DNA constructs are electroporated into an appropriate cell line, and the transfected cells are selected by growth in media containing the antibiotic that corresponds to the selectable marker and checked for the proper genetic change, typically by PCR. This approach is repeated to eliminate the second allele, i.e., generate cells that are null for the gene. If stable cell lines are obtained this is considered definitive proof that the gene under study is not essential for cell viability. The inability to obtain null cell lines is evidence that the gene may be essential. For example, evidence of Leishmania mexicana CDC2-related kinase 1 and 3 (LmCRK1 and LmCRK3) essentiality was indicated by the inability to knockout both alleles in this diploid.41 Since the inability to create a null cell line can also result for technical reasons, more thorough approaches need to be used to further assess essentiality.
Table 3. Essential Kinases in the Disease Stage of Trypanosomatids.
| systematic ID | name | species | description | evidence of essentiality |
|---|---|---|---|---|
| Protein Kinases | ||||
| Tb927.10.1070 | CRK1 | T. brucei Leishmania sp. | cyclin dependent kinase involved in progression out of G1 | RNAi (T. brucei), cannot produce null (Leishmania)41a |
| Tb927.10.4990 | CRK3 | T. brucei Leishmania sp. | cyclin dependent kinase involved in progression out of G2/M | RNAi (T. brucei),48b,55 cannot produce null (Leishmania)41b |
| Tb927.2.4510 | CRK9 | T. brucei | cyclin dependent kinase involved in transcriptional control | RNAi56 |
| Tb11.01.4130 | CRK12 | T. brucei | cyclin dependent kinase | conditional null, RNAi47,48 |
| Tb11.01.0330 | AUK1 | T. brucei | aurora kinase involved in progression through G2 | RNAi60 |
| Tb927.7.6310 | PLK | T. brucei | polo-like kinase involved in progression through mitosis | RNAi57 |
| Tb927.7.5770 | PK53 | T. brucei | nuclear DBF-2-related (NDR) kinase | RNAi61 |
| Tb927.10.4940 | PK50 | T. brucei | nuclear DBF-2-related (NDR) kinase | RNAi61 |
| Tb927.10.13780 | GSK3 | T. brucei | glycogen synthase kinase 3 | RNAi62 |
| Tb11.01.4230 | CLK1 | T. brucei | CDC2-like kinase | RNAi46a |
| Tb927.10.5140 | ERK8 | T. brucei | RNAi48b | |
| Tb927.5.800 | CK1.2 | T. brucei | casein kinase 1 (CK1) | RNAi, cannot produce null63 |
| Tb927.1.1930 | TOR4 | T. brucei | target of rapamycin 4 | RNAi64 |
| LmjF36.6470 | MPK1 | Leishmania sp. | mitogen-activated protein (MAP) kinase | null65 |
| LmjF19.1440 | MPK4 | Leishmania sp. | mitogen-activated protein (MAP) kinase | cannot produce null66 |
| Carbohydrate | ||||
| Tb927.3.3270 | PFK | T. brucei | phosophofructokinase (PFK) | RNAi67 |
| Tb927.10.14140 | PyK1 | T. brucei | pyruvate kinase (PyK) | RNAi67 |
| Tb927.1.720 | PGKA, PGKB, PGKC | T. brucei | phosphoglycerate kinase (PGK) | RNAi68 |
| Tb927.1.710 | ||||
| Tb927.1.700 | ||||
| Tb927.10.2010 | HK1, HK2 | T. brucei | hexokinase | RNAi67,69 |
| Tb927.10.2020 | ||||
A more thorough approach to determine essentiality at the DNA level is using an approach based on the “plasmid shuffling” approach established in yeast.42 In L. major, this approach was used to show that 5,10-methylene tetrahydrofolate dehydrogenase (DHCH) was an essential gene.43 With this approach, both chromosomal copies of the target gene were knocked out while an episomal copy of DHCH was present in the background to support viability. A negative selectable marker present on the episomal vector was included to drive loss of this episomal vector when desired, and a positive selectable marker (GFP) was present to verify the presence or absence of the episomal vector. To test for essentiality, cells were grown in media to drive the loss of the episome, and flow cytometry was used to sort cells that either retained or lost the episome. It was found that cloned cell lines could only grow out when the episome was present, suggesting that expression of DHCH is essential for cell growth. This approach can be used in cases where cell viability can be supported with an episome. However, this approach requires a substantial amount of genetic manipulation and cell sorting to achieve a conclusive result.
2.2.1.2. RNA Level
RNAi approaches can be used to specifically degrade the mRNA for a target kinase. This approach can only be used in systems with robust RNAi machinery. As a consequence, RNAi approaches have been used routinely in T. brucei but have not been successfully used in T. cruzi or Leishmania sp.44 In T. brucei, RNAi is performed by inserting a transgene that conditionally expresses the dsRNA that is specific to a fragment of the mRNA of the target gene upon the addition of tetracycline. Libraries of cells that contain RNAi transgenes that target mRNAs from random regions of the genome can also be used in conjunction with high-throughput sequencing approaches to screen RNAi knockdown effects on a genome-wide level.45 RNAi knockdown in T. brucei employs a single straightforward transfection but has the disadvantages that the knockdown can be incomplete, which leads to nondefinitive results, and may affect off-target mRNAs. This approach has been widely used to identify likely essential kinases in T. brucei in a gene-by-gene approach (see Table 2) or by higher-throughput RNAi screens.45,46
Transcriptional regulation of a gene expression can also be used to eliminate or reduce expression of a gene of interest. This approach has been used in T. brucei in which tetracycline (tet)-regulatory approaches have been established. For this, a tet-regulatable copy of the gene is inserted at an exogenous locus in a strain that expresses a copy of the tet-repressor protein that is necessary for the conditional regulation. When this additional gene copy is expressed in the presence of tet, the two endogenous alleles can be knocked out as outlined above. Expression of the gene of interest can then repressed by growing cells in media lacking tet. This approach was used to show that CDC2-related kinase 12 (CRK12) was essential in T. brucei(47) as was observed upon RNAi knockdown.48 A disadvantage to this approach is that it requires several steps of genetic manipulation and has only been successfully used in T. brucei.
2.2.1.3. Protein Level
Expression of a protein of interest can be specifically down-regulated by knocking in a copy of the gene coding the kinase with a destabilizing domain (DD) tag.49 DD tags are protein domains that are properly folded only in the presence of a compound. When unfolded, the DD and fused protein will be specifically targeted for proteasomal degradation. When other endogenous copies of these genes are knocked out, expression of this protein is then reliant on the presence of a compound. This approach has successfully been used in trypanosomatids and Plasmodium sp., including the Plasmodium falciparum protein kinase PfCDPK5.50 One limitation of this approach is that all proteins may not be able to be successfully targeted this way because the toleration of tags by proteins and their targeting to the proteasome is unpredictable. Another limitation is that the subcellular location of a protein may impede its destruction by the cellular protein degradation machinery.
2.2.2. Chemical Inhibition Approaches To Identify Essential Kinases
Kinases can be specifically inhibited using compounds with high selectivity. When this is possible, treatment with a potent inhibitor can lead to almost immediate inhibition of a specific target. Such an approach can also reveal the effects of acute inhibition of enzymatic activity versus elimination of protein.51
Inhibitors that are specific to a kinase of interest can be produced utilizing medicinal chemistry approaches. With these specific inhibitors cellular phenotypes can be examined and essentiality can be determined. The challenge with the use of inhibitors to examine phenotypic consequences is obtaining specific inhibition of the kinase of interest, since it is unlikely that absolute kinase selectivity will be achieved, which may allow confounding phenotype(s). Nonetheless, inhibitors that have been developed and may enable this approach are described later in this article.
2.2.3. ATP Analogue Sensitive Alleles
The conservation of the ATP-binding site allows for a general strategy for the inhibition of specific protein kinases. Within this binding site nearly all protein kinases contain a large hydrophobic “gatekeeper” residue.52 When this gatekeeper residue is mutated to a small residue (glycine or alanine), a unique pocket is formed that will allow for the binding of promiscuous inhibitors with bulky groups allowing specific inhibition of this mutant allele. Cell lines can be constructed that only contain this inhibitor-sensitive allele, allowing for potent and specific inhibition. This approach has revealed important insights on the differences between traditional genetic approaches to knockdown/knockout kinases of interest versus chemical inhibition.51 For example, genetic studies have suggested that depletion of mammalian cyclin dependent kinase 2 (CDK2) shows almost no effect whereas inhibition with chemical inhibition, such as this, shows dramatic effects that cannot be attributed to off-target effects.53 This approach has been used in T. brucei to study the function of polo-like kinase (PLK), but could be applied to almost any protein kinase of interest. This approach also allows the study of essential protein kinases in systems with only a limited number of genetic approaches, such as T. cruzi or Leishmania species.
3. Characterization of Specific Kinases and Signaling Cascades
Specific kinases have generally been studied on a gene-by-gene basis with the genetic methods outlined in section 2.2.1 These kinases are discussed in the sections below and in Table 3. Recently, though, high-throughput screens have been performed to elucidate the roles and essentiality, of many, or all, kinases simultaneously.45,46b,47,48b These high-throughput findings have been thoroughly compiled by others recently46b and will therefore not be reviewed here.
3.1. Cell Cycle Protein Kinases
Many of the well-studied essential protein kinases in T. brucei are homologues of well-studied cell cycle regulators in other systems (reviewed elsewhere).54 Cell cycle kinases shown to date to be essential are tabulated in Table 3.
3.1.1. Cyclin Dependent Kinases
The trypanosomatid genomes code for 11 putative members of the cyclin dependent kinase family54a referred to as CDC2-Related Kinases (CRK). A majority of these 11 CRKs are essential in bloodstream form (BF) T. brucei, suggesting that this family may be an especially attractive target for chemical inhibitors. There is evidence that CRK3 is essential in both T. brucei(48b,55) and Leishmania.41b In T. brucei, CRK3 knockdown by RNA causes an increase in G2/M cells, suggesting that this kinase is essential for exit out of this phase of the cell cycle.55 In T. brucei, CRK1, CRK2, and CRK11 also appear to be essential, and their knockdown results in an accumulation of cells in G1 phase (unpublished results).55 CRK9 is essential in T. brucei, appears to play a role in transcriptional regulation, and may lead to minor defects in the cell cycle upon its depletion.56 Finally, CRK12 has been shown to be essential in BF T. brucei, but does not appear to have cell-cycle defects upon down-regulation.47,48
3.1.2. Polo-like Kinase (PLK)
Polo-like kinases are involved in progression through mitosis in other organisms. The T. brucei genome encodes one PLK and RNAi knockdown of TbPLK in BF T. brucei resulted in a strong growth defect. PLK has a basal body localization during early cell cycle stages and is redistributed to the flagellar activation zone upon later stages where PLK may promote initiation of cytokinesis,57,58 which is distinct from PLKs in other systems as being regulators of transition out of G2 phase.59
3.1.3. Dbf2-Related (NDR) Kinases
TbPK50 and TbPK53 are the only homologues in T. brucei of nuclear Dbf2-related (NDR) kinases. This family of kinases has been shown to be involved in cell cycle regulation and development in several organisms.70 Down-regulation of expression of each of these kinases by RNAi leads to a strong growth defect followed by cell death.61 Specific inhibition of these kinases leads to an accumulation of cells with two nuclei and two kinetoplast DNA bodies, which suggest that these kinases have roles in progression through cytokinesis.
3.1.4. Aurora Kinases
Aurora kinases have generally been found to be involved in chromosome segregation and cytokinesis.71 The T. brucei genome codes for three Aurora kinases and RNAi studies have found that at least one of these aurora kinaes, TbAUK1, is essential for cell growth. Knockdown of TbAUK1 or overexpression of a catalytically inactive mutant of TbAUK1 leads to pleotropic cell cycle defects including the typically conserved chromosome segregation and cytokinesis functions.60,72
3.2. Mitogen-Activated Protein (MAP) Kinases
Genetic analyses of mitogen-activated protein (MAP) kinases have shown that they have various roles in parasite biology, including parasite viability, virulence, environmental sensing, and flagellar biogenesis. An extensive analysis of MAPKs has been performed in Leishmania sp. and has been thoroughly reviewed elsewhere.73 There were 15 MAP kinases identified in Leishmania sp., and two have been found to be essential in the disease stages of the parasite in Leishmania mexicana. MPK1 has been shown to be essential for infection,65 and MPK4 has been shown to be essential in promastigote and amastigote stages.74 Interestingly, though, the orthologue of MPK4 was not found to be essential in the bloodform stage of T. brucei.66
3.3. Carbohydrate Kinases
Glycolysis is the highly conserved process of breaking down glucose to provide energy in the form of ATP. In kinetoplastids the components of the glycolytic pathway for the conversion of glucose to 3-phosphoglycerate are found in specialized peroxisome-related organelles called “glycosomes” with other components found in the cytosol.75 In the bloodstream form of T. brucei, import and utilization of glucose from the host is the sole source of ATP, due to the limited function of the mitochondria in this stage of the parasite. This makes glycolysis an especially appealing target for targeted therapeutics. The four kinases of the glycolysis pathway have been found to be essential in the bloodstream form of T. brucei: hexokinase (HXK) (two nearly identical enzymes, TbHK1 and TbHK, have been found to be independently essential),67,69 phosophofructokinase (PFK),67 pyruvate kinase (PYK),67 and phosphoglycerate kinase (PGK).68 Glycolysis kinases shown to be essential are tabulated in Table 3.
4. Identification of Kinase Targets Mediated by Small Molecules
Druggable kinases have been identified using several mass spectrometry-based approaches in combination with compounds. These approaches use cell lysates as a source of T. brucei kinase protein and specifically enrich for kinases using different matrices containing immobilized kinase inhibitors or other kinase-binding compounds.
4.1. Identification of Covalent Kinase Inhibitors
Hypothemycin, a natural polyketide that covalently inhibits a subset of protein kinases, was found to kill T. brucei and was used as probe to identify which protein kinases bind hypothemycin.46a Hypothemycin has been shown to covalently inhibit protein kinases through a cysteine residue preceding the conserved catalytic DXG motif found in all protein kinases (CDXG kinases).76 Mass spectrometry analysis, in combination with a hypothemycin-based probe, showed that 10 of 21 CDXG kinases in T. brucei covalently bind hypothemycin, and RNAi analysis showed that only two of these protein kinases were essential for growth. One of these essential CDXG kinases, CDC2-like kinase 1 (CLK1), was not known to be essential prior to this work. In the end, this approach was able to identify novel essential protein kinases that can readily bind an inhibitor in its active site, further suggesting that this kinase is druggable. This approach could theoretically be expanded to any other covalent kinase inhibitors in T. brucei to identify highly druggable targets.
4.2. Drug Elution from an ATP-Sepharose Matrix
Putative T. brucei kinase targets of compounds that kill T. brucei that were originally developed against human EGFR/VEGFR were identified using an ATP-sepharose enrichment, elution, and mass spectrometry approach.77 For this, T. brucei ATP-binding proteins were enriched with an ATP-sepharose matrix, and proteins were eluted with one of three different kinase inhibitors (lapatinib, canertinib, and AEE788). This study found that many protein kinases were eluted with these compounds and five were eluted with all three compounds. This specific elution suggests that the protein kinases may be targets of these compounds and that these protein kinases have accessible active sites.
4.3. Drug Elution from a Kinase Inhibitor Affinity Matrix
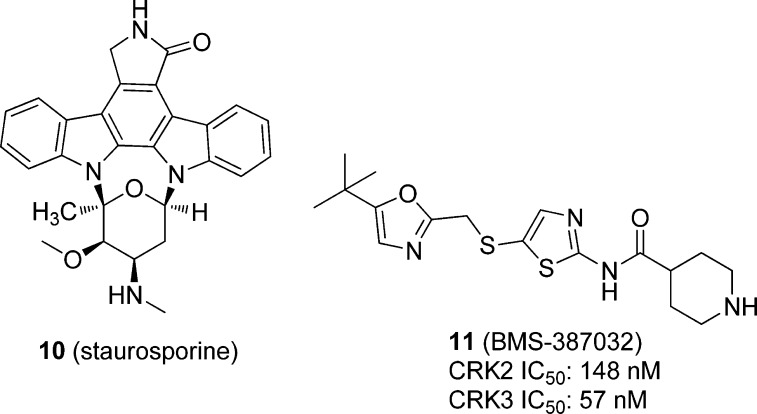
Another approach to determine potential targets of kinase inhibitors with a mass spectrometry approach was originally developed by Cellzome in efforts to identify the targets of ABL kinase inhibitors in humans.78 With this approach, protein kinases from lysate are enriched on a matrix containing several immobilized pan-kinase inhibitors. This is done with a lystate incubated with and without inhibitors. Protein kinases that bind an inhibitor of interest will bind the matrix less efficiently and will be found in less abundance when eluted from the matrix and measured with quantitative mass spectrometry (iTRAQ). This approach has been used to identify putative targets in T. brucei against staurosporine, BMS-387032, and several hit compounds against essential T. brucei protein kinases.79
5. Chemistry
5.1. Medicinal Chemistry of Protein Kinases
As the largest “druggable” family of targets in the human genome,1 it is perhaps not surprising that protein kinases have been of intense focus of drug discovery efforts over the past decade. A recent report notes that 90% of these programs have been for oncology indications, yet it appears that chronic indications are beginning to gather increasing levels of interest.80 The types of challenges inherent in discovery of inhibitors of kinases are manifold. First (and perhaps foremost), owing to the relatively high similarity one would expect from a family of enzymes that catalyze the same (phosphoryl transfer) reaction, achieving selectivity between kinases is a daunting task. This has been borne out experimentally.81 Others have suggested that complete selectivity may not be fully warranted, instead opting to look for some levels of nonselectivity.82 However, it has become clear in recent years that true kinase selectivity profiles often only become evident late in development. The classic example of this was in the discovery of imatinib (Gleevec); while it was originally touted as a PGDFR inhibitor, later studies uncovered its effectiveness was due to inhibition of other kinases (namely, cKIT and BCR-ABL).83
Thus, while much effort has focused on achieving kinase selectivity as ascertained by biochemical selectivity assays, there is movement toward increased focus on cellular assays to more faithfully evaluate the biological outcome that one wishes to effect. Indeed, cellular assays, while perhaps noisier and more challenging to deconvolute results, better inform the medicinal chemist with respect to the inhibitor effects in the presence of physiological concentrations of ATP (biochemical assays are most often performed at lower ATP concentrations, which makes the inhibitors appear more potent than they would be in the cellular environment). Cell assays can provide information regarding potential compensatory mechanisms, not to mention the ability of the inhibitor to permeate the cell membrane.
A prototypical kinase inhibitor is generally an aromatic heterocycle that presents the classic H-bond donor/acceptor motif, which enables binding to the hinge region of the targeted kinase. Also, increased lipophilicity often results from optimization of compounds that extend into lipophilic pockets near the binding site. As a result, such structural features can adversely impact upon compound solubility and absorption properties. Strategies to ameliorate this issue have emerged, such as installation of planarity-breaking functionality,84 or attachment of hydrophilic functionality that does not directly engage with the target, but extends toward solvent.85
In addition, an enormous amount of kinase-targeting medicinal chemistry has been enabled by structural biology studies that can help elucidate the binding modalities of compounds and assist in improving potency and selectivity profiles by informing key contacts between inhibitor and target. This has resulted in the identification of various classes of kinase inhibitors that bind to active kinase (type I, or DFG-in inhibitors), or that stabilize the inactive form of the kinase (type II, or DFG-out).86 This structural biology information on human targets can surely enrich and enable redirection of previous medicinal chemistry efforts onto kinases of other species (such as trypanosomatids).
5.2. Kinase Medicinal Chemistry Challenges
The medicinal chemistry challenges described above also apply to the discovery of kinase-targeting drugs for trypanosomatid parasites. For example, instead of working to ensure kinase selectivity within a single species, one must focus on achieving inhibitor selectivity against host and pathogen kinases. For tool compounds, which are molecules developed for the primary purpose of perturbing specific targets or pathways, a highly selective inhibitor is needed.. From a drug discovery perspective, however, enzyme selectivity within the parasitic kinome is unnecessary.
The physicochemical properties for drugs that target trypanosomatid parasites are also crucial beyond simply ensuring oral bioavailability. Indeed, the types of properties that make for good oral drugs are still applicable. However, additional consideration must be given to biodistribution. For example, compounds that would be effective agents for treating stage II HAT infections would need to be brain penetrant. Historical kinase-targeting drugs have higher molecular weight compared to other approved drugs;87 this is due to the extended structural motifs built to generate kinase selectivity by engaging pockets remote from the ATP binding site. Recent reports suggest that CNS-acting drugs need to be within a limited molecular weight, and lipophilicity/polarity range.88 Thus, there may be a trade-off between discovery of CNS-penetrant antitrypanosomal kinase inhibitor drugs and exquisite host kinase selectivity.
Another compound behavior driven by physicochemical properties (and heretofore not yet well-understood) is the ability of compounds to permeate into parasite cells and, for intracellular parasites, permeate into both the infected host cells and the parasite. Parasite cell permeation is therefore a consideration that should attract future attention.
While human kinase drug discovery programs have, to this point, been primarily focused on cancer chemotherapies (which is considered to be an time-bound indication compared to long-term, chronic diseases, such as diabetes), the therapeutic regimen often extends several months. As a result, tolerance for side effects that may emerge due to inhibition of other kinase targets is somewhat low (though not as low as would be required for diseases requiring life-long treatment). However, in the case of antiparasitic drug discovery, when considering the desired kinase selectivity profile of a new inhibitor one must realize that such a treatment is likely to last only a week or so (for example, see the target-product profile for HAT).37 Therefore, a kinase that is an undesirable off-target for longer-term treatments may not be an important off-target for short-term acute therapies such as an anti-infective indication. Clearly, inhibitor activity at antitargets such as the hERG ion channel (which is correlated to sudden and fatal cardiac arrhythmia),89 or those that are involved in any other sort of serious and acute toxicity, must be carefully monitored. This is a balance that must be made by antiparasitic drug hunters.
5.3. Kinase-Targeted Medicinal Chemistry in Protozoans Where the Biological Target Is Known
5.3.1. Trypanosoma brucei
5.3.1.1. Glycogen Synthase Kinase-3 Short (GSK-3)
Glycogen synthase kinase-3 (GSK-3), a protein kinase essential in the cell signaling pathway in mammals,90 has been targeted for therapeutics discovery for diseases such as Alzheimer’s disease and diabetes mellitus.91 The T. brucei homologue contains differences that should conceivably allow for inhibition selectivity over the mammalian GSK, a prediction that is borne out experimentally.62 Though two forms of GSK3β are expressed (long and short), RNAi assay data suggest the short form of TbGSK3β is essential for T. brucei parasite growth and viability in the bloodstream form.45
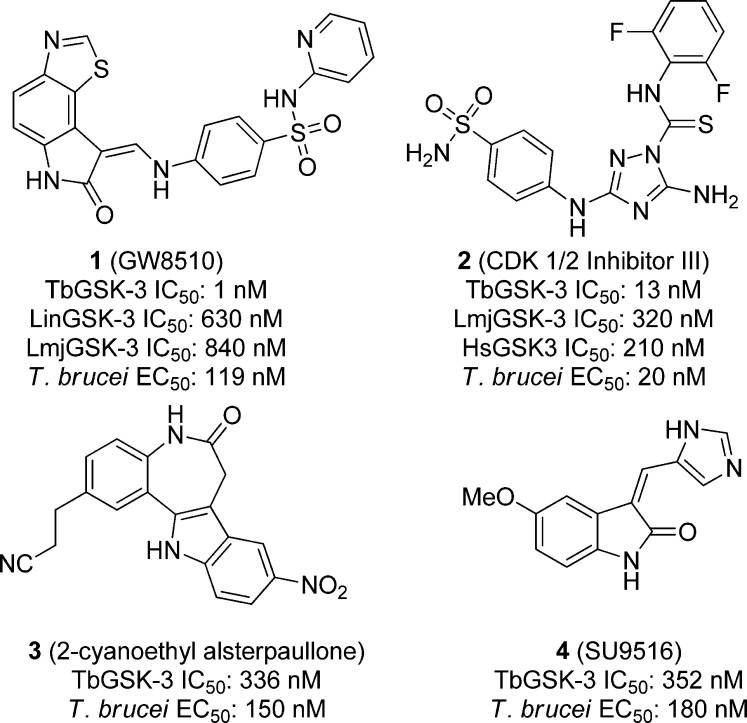
Excellent correlation was observed between the BSF cell activity and enzyme inhibition by compounds from a small molecule inhibitor library containing 48 known protein kinase inhibitors that were screened against T. brucei GSK3 short and T. brucei BSF parasites. GW8510 (1), shown in Figure 1, inhibited the T. brucei kinase and parasite growth with an IC50 of 1 nM against the parasite kinase, which translated to an EC50 of 119 nM against the BSF parasite. CDK 1/2 inhibitor III (2), 2-cyanoethyl alsterpaullone (3), and SU9516 (4, Figure 1) also showed submicromolar inhibition of both the kinase and cellular growth. Compound 2 was the most potent toward the whole cell parasite with an EC50 of 20 nM, and had an IC50 of 13 nM against TbGSK. A set of 255 known human GSK-3β inhibitors were also screened against the parasitic enzyme and the BF T. brucei, and again, excellent correlation between biochemical and cellular potency was observed, reconfirming that GSK-3 is a meaningful drug target for T. brucei(92) In a subsequent report (which used a different assay than above), compound 2 was 2-fold selective for the parasite enzyme over the host homologue: TbGSK IC50 of 0.12 μM versus HsGSK3β 0.21 μM, while compound 1 showed no selectivity over the human enzyme: TbGSK and HsGSK3β IC50s of 0.02 μM.93
Figure 1.
Protein kinase inhibitors screened against TbGSK and BSF T. brucei
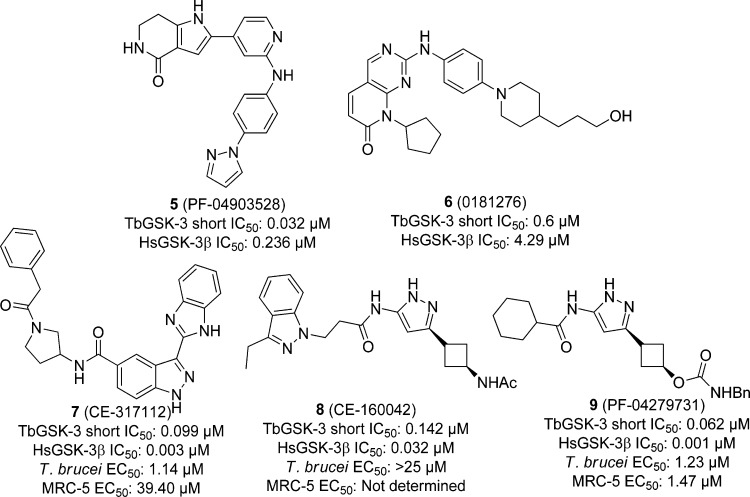
With an eye toward repurposing existing human GSK3β inhibitors, Pfizer screened 16 540 of these compounds in a HTS against TbGSK-3, counter-screened against the human homologue HsGSK3β. In the end, 362 had potency below 1 μM IC50, and 35 had potencies below 100 nM. Two of these compounds, PF-04903528 (5) and 0181276 (6) (Figure 2), showed 7-fold selectivity for the parasitic enzyme. There were 17 compounds with parasite kinase potencies of less than 100 nM, including 5 and 6, tested then against T. brucei cells and counter-screened against the human fetal lung fibroblast MRC-5 cell line. Despite the inverted biochemical selectivity between host and pathogen kinases (HsGSK3β IC50 = 3 nM vs TbGSK3 IC50 = 99 nM), compound CE-317112 (7), shown in Figure 2, displayed a 35-fold cellular selectivity margin. Although, generally speaking, a direct correlation between TbGSK3 and T. brucei cellular activity was observed, there were some exceptions. For example, CE-160042 (8) inhibited the TbGSK3 with an IC50 of 0.142 μM but showed no activity in the whole parasite assay (>25 μM). This is likely to be partially due to lack of cellular permeation (a hypothesis supported in part by lack of permeability in Caco-2 cells). From the set of screening hits, 13 compounds were tested against a panel of 40 human protein kinases at 10 μM to determine selectivity. Compound 8 showed high specificity by inhibiting only HsGSK3β, however, it was inactive against T. brucei cells. Compound PF-04279731 (9) showed moderate selectivity by inhibiting only two other human kinases besides HsGSK3β, though the compound was nonselective between T. brucei and MRC-5 cells. The authors conclude that, taken together, the desirable potency of 5 and selectivity of 6 in regards to the parasite kinase, the GSK-3β selectivity of 8, and the desirable cytotoxicity data of 7 all contribute to excellent starting points for future TbGSK inhibitors.94
Figure 2.
Human GSK inhibitors screened against TbGSK-3s.
Urbaniak et al. used a kinase-inhibitor matrix (kinobeads) that can measure a compound’s binding interaction with a wide variety of kinases simultaneously, to profile the known mammalian kinase inhibitors staurosporine (10) and BMS-387032 (11, Figure 3), along with four parasite kinase inhibitors identified by the University of Dundee’s Drug Discovery Unit (structures not disclosed). Staurosporine inhibited one-third of both the 44 human and trypanosome kinases tested with submicromolar potencies, 10 of which were under 100 nM. Compound 11, a known human CDK inhibitor, was found to inhibit most of the trypanosome CDKs, including CRK2 and CRK3, and two other kinases, CMGC and CAMK, with submicromolar potencies. Compound DDD85893 was found to inhibit T. brucei cells in vitro with good selectivity over human MRC5 cells. The kinobead profiling showed nanomolar inhibition of TbGSK3, as well as three other trypanosome CMGC kinases at micromolar potency. When the compound was profiled against human MRC5 cell lysates, GSK3α, GSK3β, and CDK9 were inhibited with nanomolar potency. The authors concluded that compound DDD85893 shows activity against TbGSK3 and other TbCMGC kinases; however, it shows no selectivity over the homologous human kinases.79
Figure 3.
Known mammalian kinase inhibitors profiled against the T. brucei kinome using kinobead technology.95
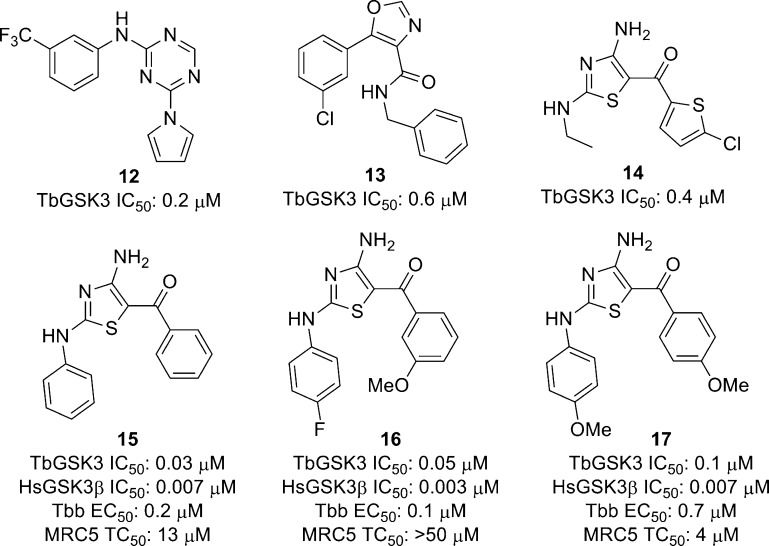
Recently, Woodland et al. reported the assessment of five series of compounds as TbGSK3 inhibitors based on a luminescence-based biochemical assay of 4110 compounds from the kinase set at the University of Dundee Drug Discovery Unit.96 While some TbGSK3 inhibitors were found to be nonspecific cellular toxins, the 2-amino-1,3,5-triazine 12 showed submicromolar activity. Out of over 100 oxazole-4-carboxamides tested, 11 showed activity against TbGSK3, including compound 13, shown in Figure 4, which had submicromolar inhibition. However, this series was not pursued due to generally high IC50 values and flat structure–activity relationships. On the other hand, the singleton 2,4-diaminothiazole, 14 in Figure 4, had an IC50 value of 0.4 μM and showed good physicochemical properties. However, this compound was nonselective over HsGSK3 (5 nM). Nonetheless, 21 analogues of the 2,4-diaminothiazole were designed on the basis of molecular modeling and tested against the parasite and mammalian GSK, and against parasite cells. Compounds 15–17 showed submicromolar inhibition of T. brucei cells with selectivity over the human MRC5 cells. While a range of potencies against TbGSK3 was observed, none were selective over HsGSK3β.96
Figure 4.
Inhibitors of TbGSK3.
In summary, the attractiveness of TbGSK3 remains open for discussion at present. While a handful of leads have been discovered with potencies in the nanomolar range, there is typically limited selectivity over the human homologue, and host cell toxicity effects are variable.
5.3.1.2. Phosophofructokinase (PFK)
The carbohydrate kinase phosphofructokinase (PFK) has been determined to be an attractive antitrypanosomal drug target,67,97 and because T. brucei PFK and human PFK have exceptionally different structures,98 there is an increased likelihood for selective drug design.
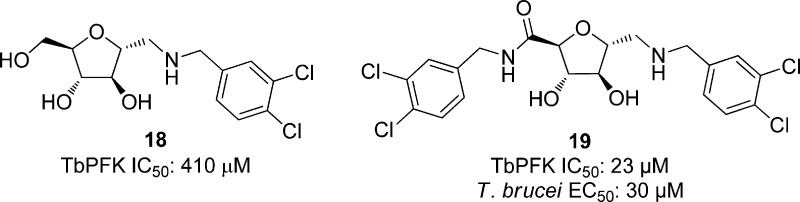
On the basis of previous work that showed 2,5-anhydro-d-mannitol-based compounds led to weak inhibition of T. brucei PFK,99 55 related analogues substituted with arylamino groups were synthesized and screened against TbPFK. Of these, 13 compounds showed over 50% inhibition at concentrations of 5 μM. Compound 18, with an IC50 of 410 μM, showed 6-fold potency over all other compounds screened. From compound 18, a series of compounds was synthesized and tested against TbPFK at 1 μM concentrations. All amides showed improved potency over 18, and compound 19, shown in Figure 5, gave the best potency against TbPFK with an IC50 of 23 μM. The eight most potent compounds against TbPFK were also screened against T. brucei cells. Increasing potency against TbPFK led to a similar trend of potency against the parasitic cells. While all compounds showed some growth inhibition, the best compound, 19, showed an EC50 of 30 μM. The correlation between biochemical and cellular activity was suggested by the authors to be evidence that the PFK is indeed the target of inhibition.100
Figure 5.
2,5-Anhydro-d-mannitol analogues screened against TbPFK.
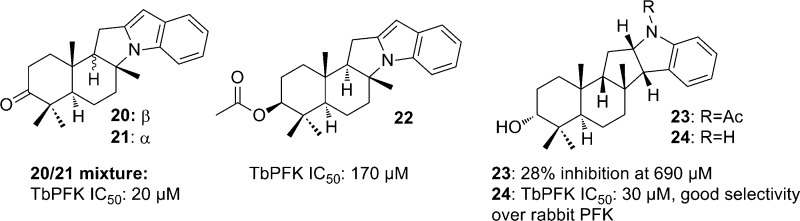
Ngantchou et al. tested alkaloids from the West African rainforest tree Polyalthia suaveolens against TbPFK (along with T. brucei GAPDH and aldolase enzymes). Compounds 20 (polysin) and 21 (greenwayodendrin-3-one) (Figure 6) were obtained from Polyalthia suaveolens bark and screened along with 3-O-acetyl greenwayodendrin, (22), N-acetyl polyveoline (23), and polyveoline (24). Compounds 20–22 and 24 showed activity against TbPFK and showed selectivity over rabbit muscle PFK. A mixture of compounds 20 and 21 was the most potent parasite enzyme inhibitor with an IC50 of 20 μM, only inhibiting rabbit muscle PFK 18% at 170 μM. Compound 24 had an IC50 of 30 μM and showed no inhibition of the rabbit PFK, while compound 22 was less potent toward the TbPFK with an IC50 of 170 μM. Kinetic studies showed that a mixture of compounds 20 and 21 competitively inhibits TbPFK. The Ki value of the mixture was measured to be 10 μM. In comparison to the natural substrate fructose-6-phosphate, the Ki/Km ratio was equal to 0.05, suggesting that the 20 and 21 mixture is 20 times more effective than fructose-6-phosphate. A small amount of purified 20 was isolated, and the Ki was calculated to be 9 μM, suggesting that compound 20 may be responsible for the activity and compound 21 may be acting synergistically.101
Figure 6.
Alkaloids screened against T. brucei PFK.
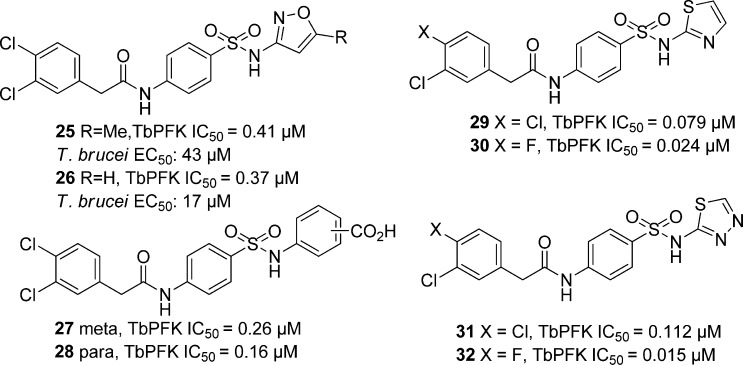
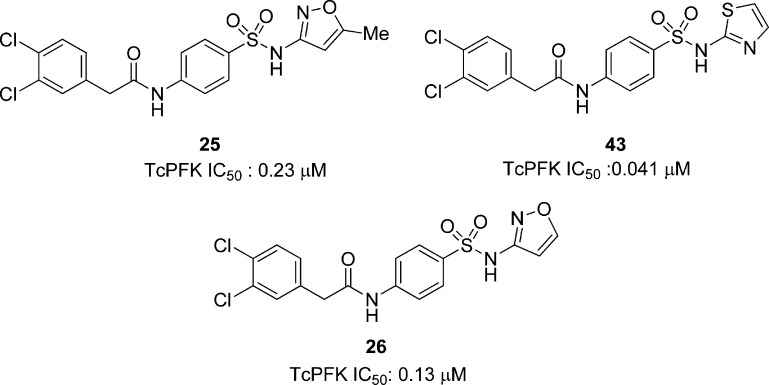
Recently, Brimacombe et al. screened 330 683 compounds against T. brucei PFK.102 The para-amidosulfonamide scaffold, typified by 25 (Figure 7), was identified as a hit series. A derivative of the antibiotic sulfamethoxazole, 25 was a 0.41 μM inhibitor of TbPFK. SAR studies showed that the para arrangement on the phenyl group core was necessary for activity, and replacement of this ring with a saturated ring, an alkyl chain, or a heteroaryl core all reduced activity. In addition, the sulfonamide and amide functionalities were found to be crucial for enzyme inhibition. Further exploration of the amide substituent revealed that the original 3,4-dichlorophenylacetamide functionality in 25 was optimal. Next, the isoxazole was probed. The 5-des-methyl analogue, shown as 26 in Figure 7, gave equivalent potency. Carboxylic acid functionality (27 and 28) further increased potency with IC50 values of 0.26 and 0.16 μM, respectively. The corresponding ester showed a sharp decrease in potency along with pyrimidine, phenol, and aniline replacements of the phenyl group. On the eastern end of the molecule, a thiazole-for-phenyl replacement resulted in improved activity (29, with TbPFK IC50 of 79 nM), while fluoro substitution on the benzyl group led to even greater potency (30, IC50 = 24 nM). The thiadiazole analogues 31 and 32 also gave good potency with IC50s of 112 and 15 nM for TbPFK, respectively. Compound 32 was shown to be selective over rabbit PFK. Upon screening select compounds against T. brucei BSF, 25 and 26 showed modest parasite dose-dependent toxicity with EC50 values equal to 34 and 17 μM, respectively, while 32, the most potent PFK inhibitor, did not show any significant parasite growth inhibition, for reasons that remain uncertain. Compounds 25, 26, and 32, however, showed no toxicity against MRC-5 human lung cell line and good ADME properties.
Figure 7.
Amidosulfonamide derivatives tested against TbPFK.
5.3.1.3. Phosphoglycerate Kinase (PGK)
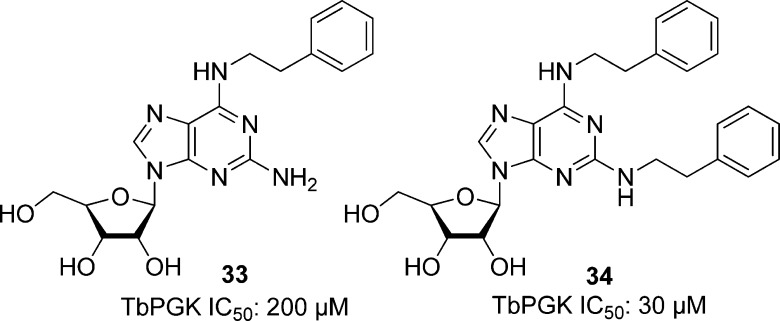
Targeting another carbohydrate kinase involved in glycolysis, Bernstein et al. targeted T. brucei phosphoglycerate kinase (TbPGK). From PGK crystallography and sequencing data103,103b−103d the researchers observed that the majority of the previously reported computational designs for inhibitors contain an adenosine component binding to the active site. In addition, previous SAR studies by the authors showed that the N6,2′-disubstituted adenosine analogues inhibited TbGAPDH.104,104b Therefore, several monosubstituted N6 and N2 adenosine derivatives were chosen to screen against TbPGK. Of these, 2-amino-N6-substituted analogues showed better activity against the parasite kinase compared with the N6 compounds that lacked the C2 amino group, although activity was still weak (Figure 8). A library of phenethyl analogues was created on the basis of the most potent compound, 2-amino-N6-(2″-phenylethyl)adenosine (33 in Figure 8), which had an IC50 of 200 μM. Attaching an additional phenethyl group to the adenine ring (69) resulted in increased potency (IC50 = 30 μM). At 100 μM, compound 34 was selective over rabbit muscle PGK. Compound 34 was also tested against BSF T. brucei brucei and T. brucei rhodesiense. Screens against both subspecies gave an EC50 of 20 μM, and 40 μM against murine fibroblasts, representing a 2-fold selectivity.105
Figure 8.
Adenosine derivatives tested against TbPGK and T. brucei.
5.3.1.4. Hexokinase
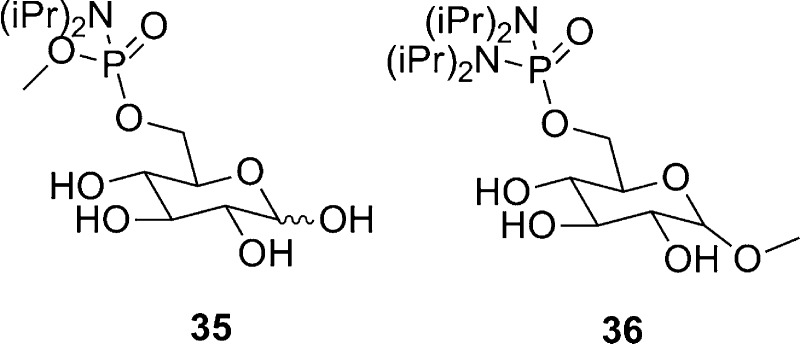
As a third example of a carbohydrate kinase targeted for inhibitor discovery, the T. brucei hexokinase is only 37% similar to the human homologue, suggesting the possibility of selective inhibitor design.8 Phosphorylation of glucose to glucose-6-phosphate is catalyzed by hexokinase, and several studies have shown that analogues of glucose, including glucosamine106 and 2-C-hydroxymethyl glucose107 derivatives, inhibit the reaction. Since glucose-6-phosphate has affinity toward the active site of T. brucei hexokinase, Willson et al. tested several glucose-6-phosphate analogues against T. brucei hexokinase. Compounds 35 and 36, shown in Figure 9, showed weak inhibition against T. brucei hexokinase, with 75% inhibition at 3 mM for 35 and 60% inhibition at 0.2 mM for 36.108
Figure 9.
Glucose-6-phosphate derivatives tested against T. brucei hexokinase.
5.3.2. Trypanosoma cruzi
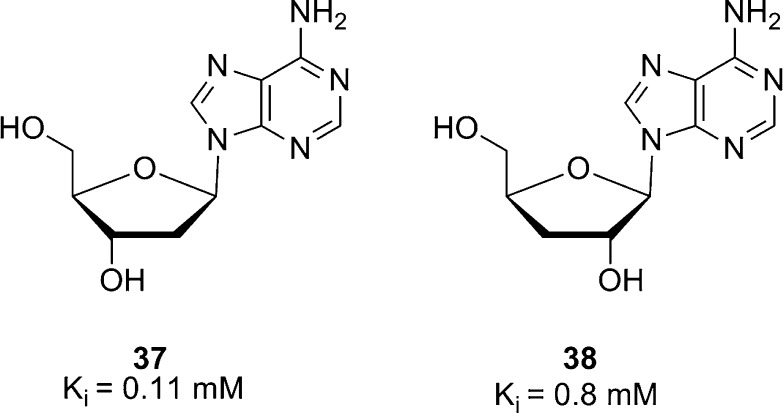
Protein kinase activity in T. cruzi has been studied since the late 1970s. It was found that T. cruzi’s protein kinase activity was independent of cyclic nuleotides and stimulated up to 4-fold by different nucleosides.109 Inosine stimulated protein kinase activity at low concentration, and adenosine showed maximal stimulation at 0.1 mM.109 Deoxyadenosines inhibited protein kinase activity in T. cruzi and T. gambiense; 2′ deoxyadenosine (37, Figure 10) inhibited protein kinase activity by 30% and 3′ deoxyadenosine (38) by 75%. Both deoxyadenosides are competitive inhibitors of ATP (Ki = 0.11 mM and 0.8 mM, respectively).109
Figure 10.
General protein kinase inhibitors in T. cruzi.
5.3.2.1. Arginine Kinase
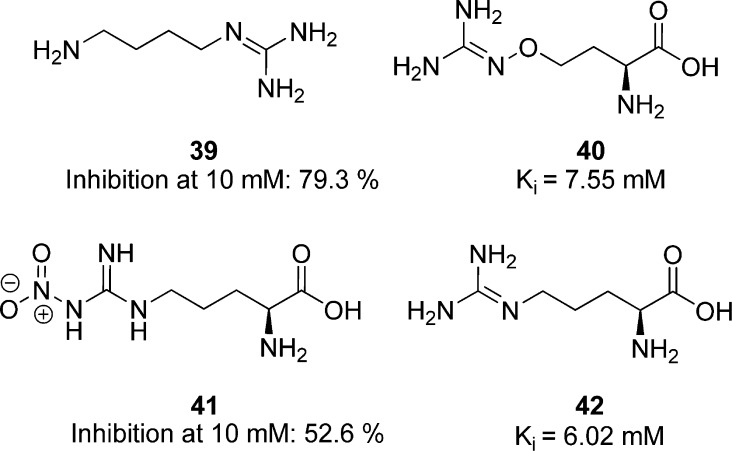
Arginine kinase belongs to the family of guanidine kinases. The guanidine kinases catalyze N-phosphorylated guanidino compounds by the reversible transfer of an ATP phosphoryl group to a guanidino acceptor in the enzyme. Phosphoarginine plays an important role as an energy reserve due to the high-energy phosphate transfer when a renewal of ATP is needed.110 A correlation between enzyme activity, nutrient availability, and cell density suggests that arginine kinases function as a regulator of energy reserves under starvation stress conditions.111T. cruzi arginine kinase is inhibited at 10 mM by the arginine analogues, agmatine (39) to 79.3%, canavanine (40) to 54.6%, nitroargine (41) to 52.6%, and homoarginine (42) to 38.2% (Figure 11). Additionally, canavanine and homoarginine inhibited the cell growth of epimastigotes of T. cruzi by 79.7% and 55.8% at a 10 mM drug concentration, and their arginine kinase Ki values were calculated to be 7.55 and 6.02 mM, respectively. These results suggest inhibition of cell growth mediated by the inhibition of the parasite’s arginine kinase, though the extraordinarily low potency of these inhibitors leaves room for additional study to confirm this.111
Figure 11.
Inhibitors of arginine kinase in T. cruzi.
5.3.2.2. Phosphofructokinase
Phosphofructokinase (PFK) has recently been identified to be a potential carbohydrate kinase drug target for T. brucei infections (see section 5.3.1.2). This enzyme is also present in T. cruzi with a 77% overall sequence identity and over 90% sequence identity in the enzyme active site when compared to the T. brucei PFK.102 The p-amidosulfonamide scaffold was identified as an inhibitor series for TbPFK that shows activity against the T. cruzi homologue. For example, compound 25 (Figure 12) inhibits TcPFK with an IC50 of 0.23 μM. As with T. brucei, the dichlorobenzyl motif in 25 was explored, and this SAR study confirmed the importance of the 3,4-dihalophenyl substitution. Additionally, removal, extension, or substitution of the benzylic methylene group resulted in a decrease in potency.
Figure 12.
Inhibitors of T. cruzi PFK.
Further exploration of the SAR was undertaken at the isoxazole position, giving compound 43 as the most potent analogue against TcPFK with an IC50 of 0.041 μM. Moreover, compound 26 (Figure 12) showed a modest potency of 0.13 μM against the enzyme, displaying no toxicity against MRC-5 human lung cell line, providing a promising selectivity profile.102 It should be noted that these three compounds are also submicromolar inhibitors of the T. brucei homologue (section 5.3.1.2, Figure 7). Taken together with promising ADME properties, these compounds represent a good start for exploring the role of TcPFK in T. cruzi infections.
5.3.3. Leishmania
5.3.3.1. Glycogen Synthase Kinase-3 Short (GSK-3)
Leishmania contains two GSK-3 orthologues, similar to those of T. brucei,112 and TbGSK3 and LmjGSK-3 short have 65% amino acid sequence identity.93 Therefore, compounds that inhibit TbGSK3 may also inhibit leishmanial GSK3.
The 11 kinase inhibitors tested against T. brucei GSK by Ojo et al. (see section 5.3.1.1) were also screened against L. major and L. infantum GSK-3 short. Compound 1, shown in Figure 1, inhibited LinfGSK-3 and LmjGSK-3 with IC50 values of 0.63 and 0.84 μM, respectively, but was more potent toward the human GSK. Compound 2 was the most potent against LmjGSK-3 with an IC50 of 320 nM, however, though it was a more potent inhibitor of HsGSK3 (0.21 μM).93
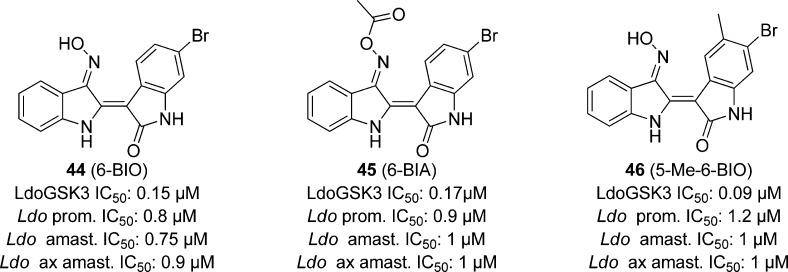
There were 16 indirubins, known inhibitors of mammalian cyclin-dependent kinases and GSK-3,113 tested against L. donovani promastigote growth, and 6-BIO (44), 6-BIA (45), and 5-Me-6-BIO (46, Figure 13) were found to be most potent with an IC50 of 0.8, 0.9, and 1.20 μM, respectively. Compounds 44–46 also inhibited L. donovani intracellular and axenic amastigotes with IC50s ranging from 0.75 to 1 μM. Derivatives of 44 and 45 showed that N1-methylation diminished parasitic growth inhibition. When the most potent compounds were tested against macrophage growth at 10 μM, no inhibition was observed, indicating that the effect observed was due to inhibition of parasitic targets rather than nonspecific host cell toxicity (though concentrations in excess of 25 μM did show cellular toxicity). The compounds were then screened against L. donovani GSK-3s and CRK3, since 6-bromo indirubins are known inhibitors of both kinase homologues found in mammals.113 Compound 44 showed an IC50 of 0.02 μM for LdoCRK3 and 0.15 μM for LdoGSK-3s, showing that CRK3 is 7-fold more potently inhibited by this analogue than GSK. Compound 45 was slightly more active against the leishmanial GSK-3s with an IC50 of 0.17 μM as opposed to 0.25 μM for CRK3. Only 46 showed significant selectivity of the LdoGSK-3 as the primary target with an IC50 of 0.09 μM, as compared with 0.65 μM for LdoCRK3.112
Figure 13.
Indirubins screened against L. donovani.
5.3.3.2. Pyruvate Kinase (PyK)
The carbohydrate kinase pyruvate kinase (PyK) and PFK are similar in trypanosomatidae, with over 70% sequence similarity;100 therefore, many PFK inhibitors are likely to also inhibit PyK. In addition, RNAi knockdown has shown PyK to be a target for trypanosomatids.8
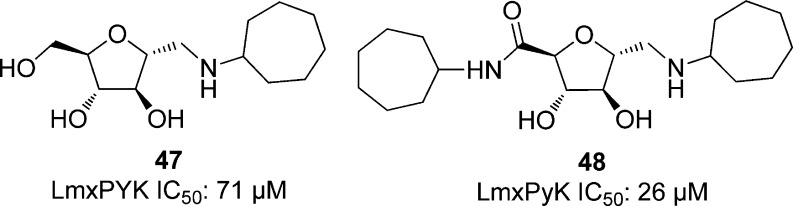
The 55 analogues of 2,5-anhydro-d-mannitol with alkylamino groups that were tested by Nowicki et al. against T. brucei PFK (see section 5.3.1.2, Figure 5) were also screened against L. mexicana PyK. Of these, 16 compounds showed over 50% inhibition of LmPyK. The most potent compounds had a hydrophobic group at the 1-position. Compound 47, shown in Figure 14, had an IC50 of 71 μM, over 10-fold more potent than any other compound tested. From compound 47, a series of compounds was prepared and tested for inhibition against LmPyK at concentrations of 1 mM. Compound 48 was the most potent against LmPyK with an IC50 of 26 μM.100
Figure 14.
2,5-Anhydro-d-mannitol derivatives tested against L. mexicanan PyK.
Several saccharin derivatives were identified as inhibitors from a 292 740 compound screen against L. mexicana PyK in a quantitative high-throughput screening, and NCGC00186526 (49), shown in Figure 15A, had an IC50 of 10 μM; however, the oxo linkage was hydrolyzed to the phenol under the assay conditions. The sulfur analogues, such as NCGC00188411 (50), proved to be more stable to hydrolysis. One related compound, DBS (51), was identified as a 2.9 μM enzyme inhibitor, with modest selectivity over the human PyK [IC50 of 8 and 16.3 μM for human tissue HsRPyK (from erythrocyte) and HsM2PyK (from embryonic or tumor cells), respectively]. This was confirmed crystallographically to be a covalent inhibitor of LmxPyK and the human homologue (Figure 15B), following reaction with the ε-amine of Lys355. This crystal structure was compared with the human PyK, and differences between the trypanosomatid and human binding pocket were noted in three amino acid side chains (R1, R2, and R3) around the Lys335 residue, shown in Figure 15B. This analysis suggests that the saccharin-binding pocket may allow for other selective PyK inhibitors to be developed.114
Figure 15.
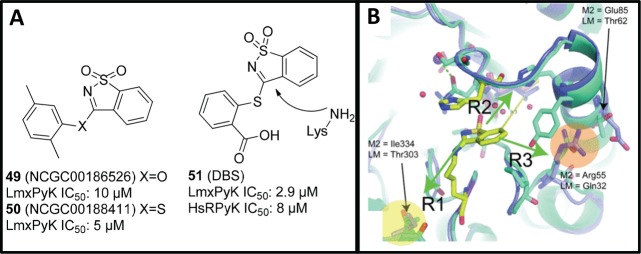
(A) Saccharin derivatives screened against L. mexicana PyK. (B) Overlaid X-ray structures of LmPyK and HsM2PyK with covalently bound inhibitor. Reprinted with permission from ref (114). Copyright 2012 The Biochemical Society.
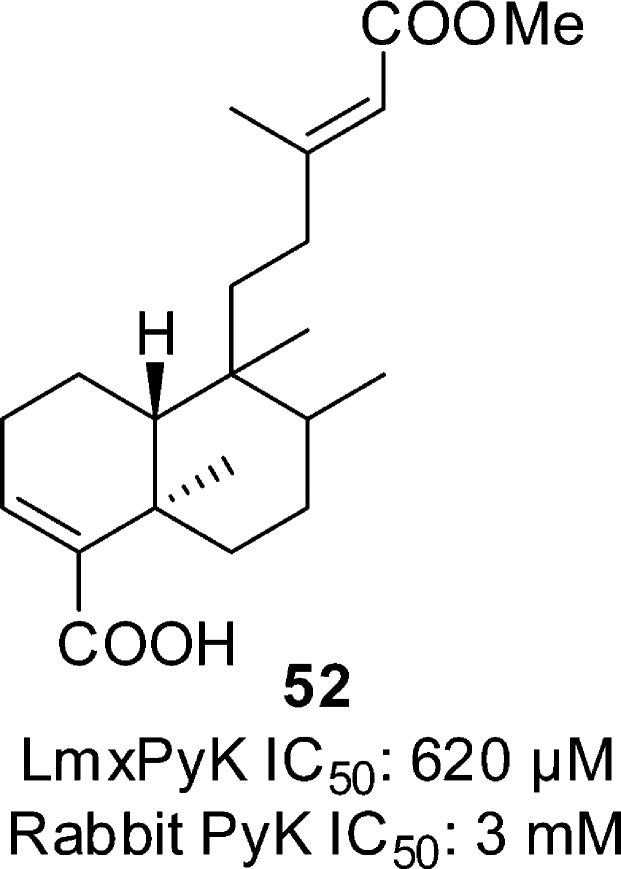
Nyasse et al. tested compound 52 (Figure 16), taken from the bark of the African plant Entada abyssinica against L. mexicana PyK. This compound showed weak inhibition (IC50 of 620 μM) and selectivity over the rabbit muscle PyK (IC50 > 3 mM).115
Figure 16.
Compound from the bark of the Entada abyssinica plant tested against LmPyK.
5.3.3.3. Casein Kinase 1 (CK1)
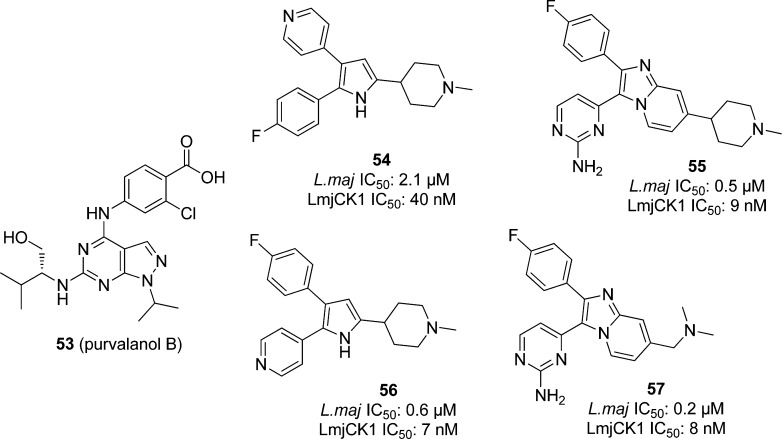
Casein kinase 1 (CK1), a multifunctional Ser/Thr protein kinase, may play a role in modulating parasite–host interactions.63 Trisubstituted pyrroles and imidazopyridines have been found to be potent inhibitors of parasite cGMP-dependent protein kinase (PKG) in protozoan parasites.116 Two CK1-like kinases found in Leishmania(117) are targeted by the cyclin-dependent kinase (CDK) inhibitor purvalanol B (53), shown in Figure 17, in L. mexicana, though no IC50 for this compound was reported for LmxCK1.118
Figure 17.
CK inhibitors purvalanol B, and pyrrole and imidazopyridine derivatives screened against Leishmania.
Allocco et al. screened several pyrrole and imidazopyridine PKG protein kinase inhibitors against LmjCK1 and L. major promastigotes, typified by compounds 54–57 in Figure 17. In whole cell assays, compounds 55–57 showed inhibition of L. major growth with submicromolar potency. Compound 55 had an IC50 of 0.5 μM, 3 gave 0.6 μM, and 4 showed 0.2 μM potency. Compound 54 was less active, with an IC50 of 2.1 μM. All compounds were tested against native LmjCK1 and bacterially expressed LmjCK1; inhibition of the kinases correlated with inhibition of the parasite. Compounds 55–57 were potent enzyme inhibitors, with IC50 ranging from 6 to 9 nM, while 53 was less active with an IC50 of 42 nM, thus suggesting that LmjCK1 is indeed a primary target of these inhibitors in L. major cells. Binding assays confirmed this hypothesis, showing that LmjCK1 isoform 2 is the primary high-affinity binding protein. All compounds were also tested against HeLa cells and showed no significant toxicity at >100 μM concentrations.119
5.3.3.4. Phospholipid-Dependent Protein Kinase C (PKC)
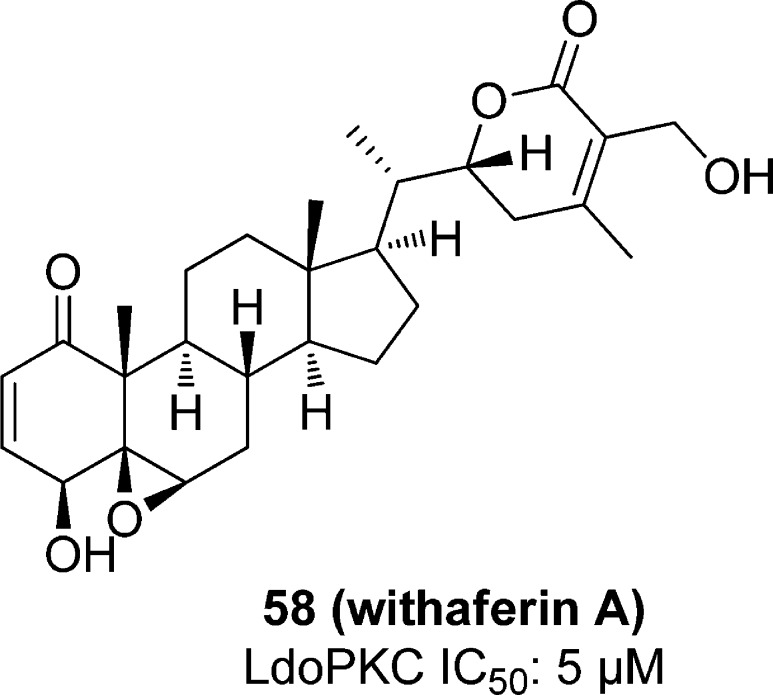
Withaferin A (58), an antitumor agent120 (Figure 18), inhibits L. donovani PKC in vitro and kills parasite cells by causing apoptosis. L. donovani cell lysate was treated with 58, and at 5 μM concentration, the phosphorylation of the LdoPKC substrate peptide HCV was inhibited by 50%. The compound also inhibited rat brain PKC 95% at 15 μM concentration. At that concentration, only 2% of L. donovani promastigotes survived after 7 h.121
Figure 18.
Compounds shown to inhibit L. donovani PKC.
5.3.3.5. Cyclin Dependent Kinase (CRK3)
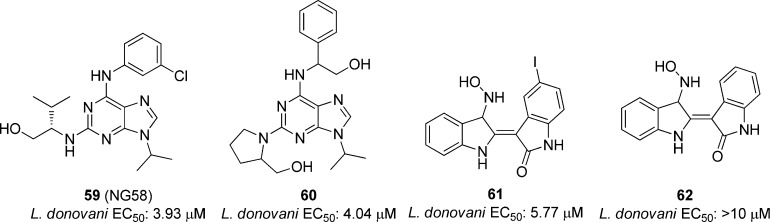
Several research laboratories have tested the parasite cyclin dependent kinase homologue CRK3 as a potential Leishmania target. Grant et al. screened 634 compounds, including analogues of known mammalian CDK inhibitors and plant natural products in a CRK3 histone H1 kinase assay at 100 μM concentrations. The 27 most potent compounds were tested against L. donovani amastigotes and L. mexicana promastigotes. Compounds NG58 (59) and 60 in Figure 19 showed modest inhibition toward the L. donovani amastigotes while 61 and 62 showed weak inhibition of the L. mexicana promastigotes (EC50 values not provided). Compounds that inhibited L. donovani amastigote infection were screened against the parasitic CRK3 and CDK1/cyclin B to determine if CRK3 is the main target. Of the 12 compounds tested, nine were more potent toward CDK1/cyclin B, and three showed comparable potency in regards to the two kinases. The authors concluded that, of the compounds that inhibited the parasite, none showed selectivity toward CRK3, suggesting that this was not the primary target causing growth inhibition.35h
Figure 19.
Compounds screened against L. donovani amastigotes and L. mexicana promastigotes.
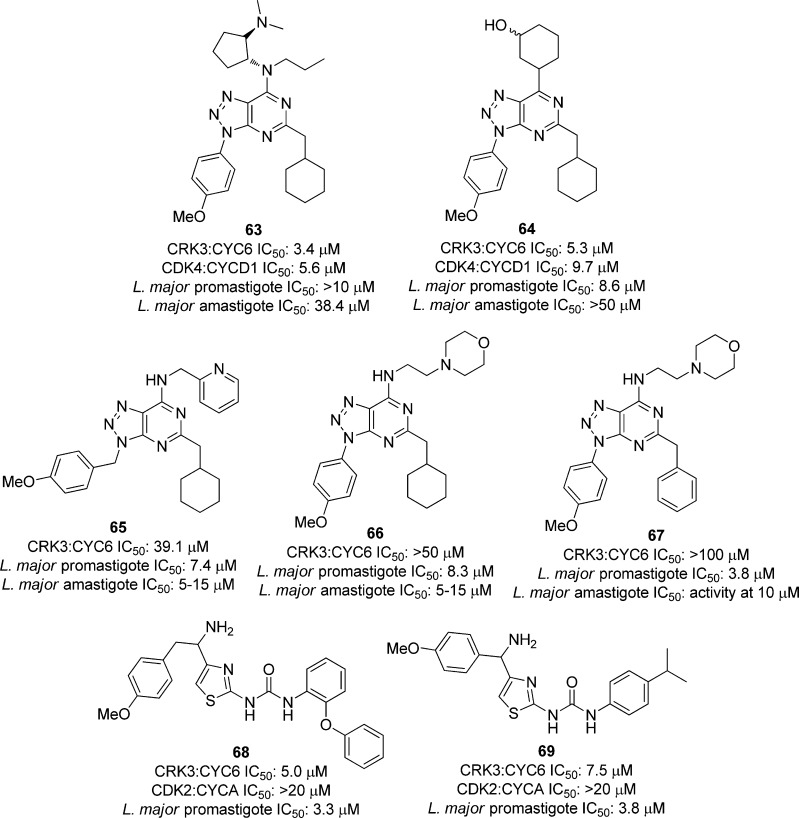
In a high-throughput screen, 25 000 compounds were tested against Leishmania CRK3:CYC6 and human CDK2:CycA. Of these, 43 compounds were selective toward the Leishmania kinase over the human CDK. Of the 16 most potent compounds (with IC50 values ranging from 2.6–11 μM), 12 were azapurine derivatives (Figure 20). To determine selectivity, the azapurine compounds were screened against 10 mammalian protein kinases, and all 12 inhibited Cdk4:CycD1 with IC50 < 30 μM. Eight of the most potent compounds against the parasite kinase were tested against L. major promastigotes and amastigotes, and compounds 63 and 64, shown in Figure 20, showed modest inhibition (IC50 values of 8.6 and 38.4 μM). Additional azapurine derivatives were synthesized and screened as well. Several compounds showed modest inhibition of the L. major cells, such as 65–67 in Figure 20; however, no correlation between CRK3:CYC6 and whole cell inhibition was observed. To further pursue these results, a second screen with a kinase-targeted library was carried out against Leishmania CRK3:CYC6 with 528 compounds at 20 μM concentrations. There were 13 compounds selective toward the parasite kinase, and of these, all were thiazole derivatives. There were 11 of these compounds tested against L. major promastigote cells, and 10 showed activity (IC50 values ranging between 3 and 27.5 μM), with compounds 68 and 69 in Figure 20 being the most active. Interestingly, two compounds that were inactive against CRK3:CYC6 were also tested against the whole parasite cells, and these showed inhibition, further suggesting that the link between the Leishmania CRK3:CYC6 potency and the parasite inhibition may be tenuous.122
Figure 20.
Compounds screened against Leishmania CRK3:CYC6, human CDK2:CycA, and L. major promastigote and amastigote cells.
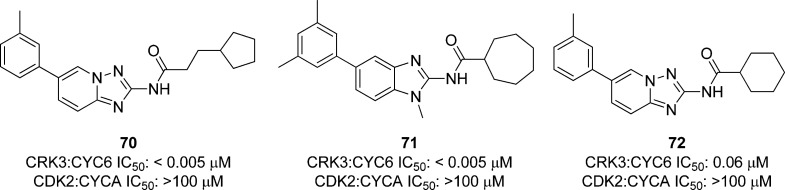
Cleghorn et al. screened 3383 compounds against Leishmania CRK3-CYC6 at 30 μM concentrations, leading to several series that were chosen to optimize selectivity of CRK3.35g From the primary screen results, several benzimidazole and triazole derivatives were synthesized and screened against leishmanial CRK3-CYC6 and human CDK2-CYCA. Several compounds gave excellent inhibition of the parasite kinase in the nanomolar range and showed no activity against the human homologue, such as 70 and 71 in Figure 21. The compounds showing activity and selectivity toward the Leishmania kinase were also screened against L. major promastigotes; however, only compound 72 showed moderate activity (2–10 μM). In addition, the physicochemical properties of the compounds were measured, and many compounds had properties suitable for membrane penetration. Since the researchers thought that cellular penetration was not the reason for lack of parasite activity, it was concluded that the results may have been because the compounds were effluxed from the cell. Alternative explanations included the possibility that CRK3 binds to a cyclin other than CYC6 in the parasitic cells, or that the CRK3 is not essential due to bypass mechanisms.35g
Figure 21.
Compounds screened against Leishmania CRK3:CYC6 and human CDK2:CycA.
Although Grant et al. and Mottram et al. have done gene-knockout experiments41a,123 showing that L. mexicana CRK3 is essential for the promastigote growth, each study concluded that no correlation between leishmanial CRK3 and parasite inhibition has been found, suggesting that CRK3 may not be a valid target for Leishmania species.
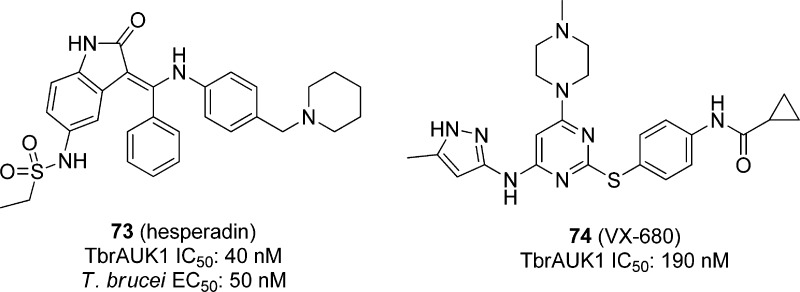
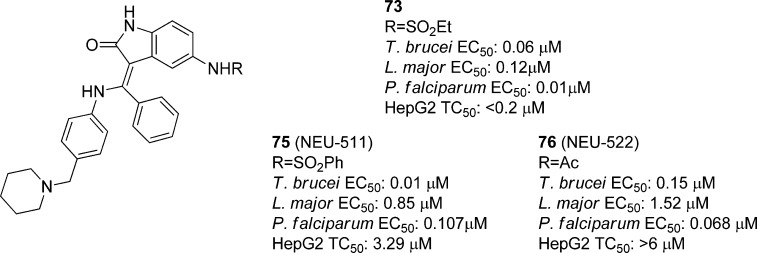
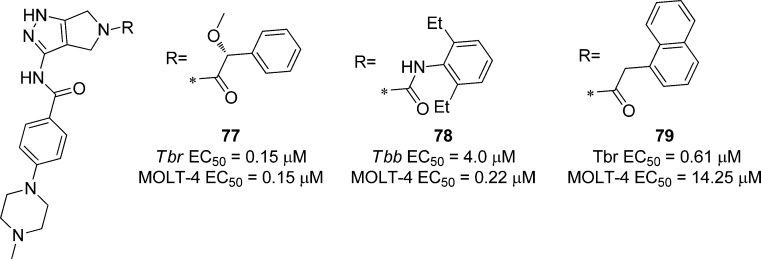
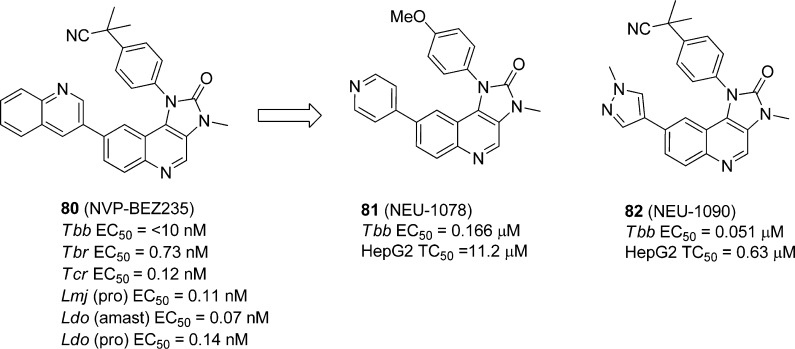
5.4. Cell-Based Optimization Strategies via Inhibitor Repurposing
An alternative method for identifying new kinase inhibitor chemotypes could be to identify classes of compounds that are known to be potent inhibitors of human homologues of essential kinases (or families of kinases) in parasites. Direct testing of these known inhibitor classes can reduce the requirement for large, random screening campaigns, and provide an entry into chemotypes that have been already shown to be drug-like, and able to be advanced into clinical trials (often referred to as “privileged” structures”).124 While human inhibitor compounds may already possess the desired potency, toxicity, and physicochemical properties for new parasitic agents (and thus be directly repurposed), it is more likely that these compounds’ properties will necessitate further optimization to become effective antiparasitic agents. This approach is often referred to as “piggy-back”125 or “target repurposing”126 drug discovery.
5.4.1. Aurora Kinase
Aurora kinases play an important role in cell division events such as mitotic spindle assembly, chromosomal separation, and cytokinesis. These kinases have been pursued as targets that have led to various inhibitors that are now in clinical trials for cancer. T. brucei expresses three aurora kinases, and TbAUK1 has been identified as an aurora kinase paralogue that inhibits nuclear division, cytokinesis, and growth in parasites.60 Moreover TbAUK1 is necessary for infection in mice and has been inhibited by the human aurora kinase inhibitors hesperadin (73)127 and VX-680 (74, Figure 22).128
Figure 22.
Human aurora kinase inhibitors.
An SAR exploration of the antiparasitic activity of the hesperadin chemotype was performed, resulting in compounds such as NEU-511 (75) and NEU-522 (76) (Figure 23).129 Compound 73 itself showed significant toxicity to HepG2 cells; however, modifying the ethyl sulfonamide provided a compound (75) that had an EC50 against T. brucei cells of 0.01 μM and 330-fold selectivity over HepG2 cells. Another analogue (76) was a 0.15 μM inhibitor of trypanosome growth, with no activity against host cells. Other analogues showed good potency against L. major, though most were inactive against T. cruzi.
Figure 23.
SAR exploration of the hesperadin chemotype.
Danusertib 77 (Figure 24), a phase II clinical candidate drug against solid tumors, and its predecessor analogue PHA-680632 78, were used to assess the SAR as its medicinal chemistry and structural biology profiles are well-known. Compounds 77 and 78 inhibited T. brucei growth (EC50 0.6 and 4.0 μM, respectively).130
Figure 24.
Repurposed human aurora inhibitors 1 and 2, and a new analogue with improved T. brucei selectivity.
Driven by docking experiments using a homology model of TbAUK1, analogues of 77 were designed that maintained the tetrahedral geometry of the carbon center adjacent to the carbonyl group in the headgroup. Analogues were tested against T. brucei rhodesiense, and none showed improved potency; however, analogue NEU-327 79 (Figure 8) showed increased selectivity over the MOLT-4 leukemia cell line. The analogues synthesized were docked into the homology model of TbAUK1, and the docking scores correlated with cellular EC50 values, suggesting that growth inhibition may indeed be mediated by TbAUK1 inhibition. However, confirmatory biochemical IC50 determinations were precluded by difficulties in expression of catalytically active TbAUK1.130
5.4.2. Phosphoinosotide 3-Kinases (PI3Ks) and Mammalian Target of Rapamycin (mTOR)
As examples of lipid kinases, PI3Ks control growth and metabolism, and in humans, inhibitors of these enzymes have drawn interest as a target for anticancer and anti-inflammatory agents, and inhibitors of PI3K and mTOR (selective, or cross-reactive) have been shown to be promising agents for cancer therapy.131
In the trypanosomatid parasites, there are at least 12 proteins belonging to the family of PI3K lipid kinases, some which are unique to the parasites. Some examples of these PI3Ks have been shown to be involved with trypanosomatid virulence and Golgi complex segregation,132and the downstream TOR complexes are critical for trypanosomal cell growth.64,133 Hypothesizing that these parasite kinases would be susceptible to inhibitors of their human homologues, various established PI3K and mTOR inhibitors were selected for testing against Leishmania and Trypanosoma species. From these inhibitors was identified NVP-BEZ235 (80) (Figure 25), an advanced drug candidate against solid tumors, which showed high potency against all three parasites.131 These results translated into observable cell growth phenotypes in T. brucei and L. major that were consistent with inhibition of the anticipated targets. In addition, in vivo efficacy was observed in a mouse model of T. brucei bloodstream infection (7 day life extension following 4 days of 10 mg/kg treatment), though efficacy was not observed in T. cruzi or Leishmania animal models.131
Figure 25.
NVP-BEZ235 and derivatives are potent trypanosomatid growth inhibitors. Tbb = T. brucei brucei; Tbr = T. brucei rhodesiense.
Subsequent analogue design and synthesis has been undertaken in order to improve the physicochemical properties (solubility, predicted CNS penetration) and to reduce these compounds’ potency against human PI3Ks and mTOR. Though these analogues showed reduced potency against T. brucei cells, compounds NEU-1078 (81) and NEU-1090 (82) were identified to have improved cellular and biochemical selectivity profiles and are predicted to be CNS-penetrant.88,134 Phosphoproteomics experiments indicated that these inhibitors, as well as 80, are indeed inhibiting the PI3K pathways, suggesting that inhibition of lipid phosphorylation may indeed be a fruitful approach for parasite growth inhibitor discovery.134a It is worth noting that another lipid kinase, phosphatidylinositol 4-kinase, has been identified to be required for Golgi maintenance in T. brucei,135 and the fact that the homologous enzyme in Plasmodium falciparum has been targeted with small molecules in Plasmodium falciparum(136) confirms druggability and bodes well for the development of inhibitors of the T. brucei enzymes.
5.4.3. Tyrosine Kinase Inhibitors
Though trypanosomatid parasites do not express tyrosine kinases,40 Tyr phosphorylation has been observed in the parasite. This suggests that the Tyr phosphorylation must therefore be achieved by dual specificity-enzymes that act on Ser/Thr and Tyr residues. The nonspecific tyrosine kinase inhibitor tyrphostin inhibited the key cellular process of transferrin uptake, which translated to inhibition of cell growth.137 This phenotype was replicated by lapatinib (83, Figure 26), a human epidermal growth factor receptor (EGFR) inhibitor and orally acting therapeutic for breast cancer, and this compound was a 1.54 μM inhibitor of T. brucei growth.137 It also cured the bloodstream form infection in 3 out of 4 mice,138 and four putative binding proteins were subsequently identified.77 In addition, eight analogues related to lapatinib killed T. brucei with EC50 in the low micromolar range, which prompted an SAR exploration of this chemotype.137 Three cycles of analogue design exploring each region of the molecule led to NEU-369 (84), and NEU-617 (85). Both compounds have an improved selectivity profile over HepG2 cells, and 85 is 37-fold more potent than lapatinib. Importantly, this analogue showed excellent plasma exposure following oral dosing in mice, though brain drug levels were low. Dosing infected animals at 40 mg/kg reduced parasite load below detectable levels, though toxicity was observed following extended dosing. However, IP administration of 10 mg/kg/day twice per day led to doubling of the survival rate compared to controls.137 Interestingly, while endocytosis of transferrin was inhibited in the trypanosome by 83, there was no effect on endocytosis of transferrin by 85. Instead, this compound blocked the duplication of kinetoplast and arrested cytokinesis without the disruption of the nucleus division on the parasite. This is clearly suggestive that 85 is inhibiting parasite growth via a mechanism different from lapatinib. A major limitation of compound 85 that is preventing good CNS exposure is the high cLogP value (7.1), and further optimization of physicochemical properties is thus needed. Importantly, cross-screening of these analogues against T. cruzi, L. major, and Plasmodium falciparum shows a range of parasite-specific SAR (unpublished results).
Figure 26.
Lapatinib and derivatives that show potent and selective T. brucei activity.
6. Perspective
Research performed over the last 15 years has made great headway in elucidating the importance of kinases in cellular signaling processes in trypanosomatid parasites, enabled by the advances in cellular and molecular biology approaches that uncover the functions and essentiality of a specific kinase or families of kinases. The resultant expanded understanding of this gene family strengthens the conviction that kinases are important targets for new antiparasitic therapeutics, as in other species. The specific similarities of parasite kinases to their human homologues, which have been repeatedly shown to be “druggable”, as well as key structural, functional, and cellular context differences make kinases attractive candidate targets for antiparasitic agents.
Trypanosomatid kinase medicinal chemistry is in its infancy compared to that of mammalian kinases, but the application of modern medicinal chemistry optimization approaches has quickly led to the identification of small (drug-like) molecules that inhibit kinase function and in some cases have effective antiparasitic activity. However, issues of selectivity over host kinases are neither resolved nor predictable. The properties that are shared by human and parasite kinases that suggest that effective parasite kinase inhibitors can be discovered on the basis of prior knowledge also raise concern about potential cross-species selectivity.
That said, it is currently unclear what characteristics of antiparasitic therapies need to be avoided because of potential effects on host kinase targets. Much of the existing understanding of the need for selectivity is based on off-target kinase effects and the expected dosage regimen for serious and/or chronic indications, such as cancer, diabetes, and inflammation. The considerations of off-target effects are likely to differ for parasitic diseases, especially in the acute or life threatening phase, where dosing regimens may be of shorter duration, resulting in less concern with long-term toxicity. In addition, the uptake, metabolism and retention of inhibitors by the parasite may either enhance or reduce efficacy during an infection. A better, research-based understanding of required selectivity over host kinases is therefore needed, and this may be best explored by focusing on short-term inhibition of the current set of “untouchable” kinase antitargets (i.e, a kinase antitarget that is linked to acute toxicity, or genotoxicity).
Parasite kinase inhibitor discovery has employed two complementary approaches: target-based, which focuses on selective inhibition of specific essential parasite kinases, and phenotype-based, which focuses on inhibition of cellular growth or other observable phenotypes that result from treatment with an investigational agent. On one hand, understanding mechanism-of-action may be useful, although a recent review of drug approvals indicated that the majority of approved drugs were discovered without initially knowing the target of action. In some cases the mechanism of action was elucidated during or after the drug development process was complete. This is more the case in infectious diseases, where targets and pathways are not as well-understood as in the mammalian system.139 In the NTD drug discovery space, this is illustrated by SCYX-7158, a new compound currently in phase II clinical trials for human African trypanosomiasis for which the mechanism of inhibition of parasite proliferation is not yet known.25
Combined target and cell-based approaches during drug optimization are useful for avoiding the apparent loss of potency that is typical when a compound’s activity is compared between biochemical and cellular assays. This can reflect characteristics of cellular permeability, competition with the inhibitor by cellular molecules such as ATP, and/or compensation by targets or pathways. Thus, driving medicinal chemistry optimization in a cellular assay factors in cellular permeability and provides a physiologically relevant system for testing the inhibitor. However, when testing inhibitors only in a cell assay we cannot say with certainty whether differences in potency among related compounds is due to differences in activity at a specific target, introduction of inhibition of other kinases, and/or cellular permeability. These considerations can complicate, but certainly not prevent, medicinal chemistry optimization. In a perfect situation, identification of trypanosome growth inhibitors would be followed by detailed mechanism of action studies, in order to learn what kinase inhibition profiles lead to successful antiparasitic drugs and to develop potent and selective probe molecules that can enable further research into parasite kinase function. However, the resource poor environment of neglected tropical diseases may make this combination of experiments a luxury, although it may be the most cost-effective in the long run.
While this review primarily discusses protein kinase inhibition as antitrypanosomatid approach, other nonprotein kinases (e.g., lipid kinases, carbohydrate kinases) warrant further work. As mentioned above, translation of understanding of kinase function in these parasites is still in its infancy compared to what is known about kinase function in humans. We expect, in time, that a broader and deeper expansion of small molecule discovery efforts into kinases involved in lipid metabolism or glycolysis will emerge.
In summary, great strides have been made toward understanding the utility of kinase inhibitors as antiparasitic agents, and progress appears to be accelerating, enabled and enhanced by new technologies and by redirecting existing technologies originally developed for use against mammalian kinases. Time will tell whether compounds of clinical utility can be found that target trypanosomatid kinases and which have the needed selectivity and physicochemical properties to be efficacious.
Acknowledgments
We acknowledge support from the National Institutes of Health (R01AI082577, to M.P.P., and R01AI078962, to K.S.), the National Science Foundation (Graduate Fellowship to L.E.S.), and the Tres Cantos Open Lab Foundation (to M.P.P.), and institutional support from Seattle Biomedical Research Institute and Northeastern University.
Biographies

Christopher Merritt received his B.S. degree in Molecular Genetics from The Ohio State University where he performed his thesis on C. elegans early embryonic development under the supervision of Dr. Russell Hill. He received his Ph.D. in Molecular Biology and Genetics from the Johns Hopkins School of Medicine. During his Ph.D. training, he prepared his thesis on C. elegans germ line development and post-transcriptional regulation under the supervision of Dr. Geraldine Seydoux. He is currently performing his postdoctoral work in the lab of Kenneth Stuart at the Seattle Biomedical Research Institute where he is studying T. brucei protein kinases.

Lisseth Silva received her B.S. degree from Roger Williams University as a double major in chemistry and biology. She first worked under Dr. Avelina Espinosa in her microbiology laboratory studying cell biology and behavior of Entamoeba histolytica and its relatives. Later on, she took an interest in organic chemistry and worked under Dr. Lauren Rossi where she worked on the synthesis of cucumerin A, an 8-C-glucosyl flavone. Additionally, she investigated the binding affinity of non- and C-glycosylated compounds with bovine serum albumin, a crucial protein for regulating the colloidal osmotic pressure of blood. She is currently pursuing her Ph.D. at Northeastern University and working under Dr. Michael Pollastri in his laboratory for neglected disease drug discovery.

Angela L. Tanner received her B.S. in chemistry at the University of Massachusetts, Boston, where she did her thesis in fluorination using organocatalysis under Dr. Wei Zhang. Angela is currently pursuing her Ph.D. at Northeastern University in Dr. Michael Pollastri’s lab working on design and synthesis of potential Trypanosoma brucei inhibitors.

Kenneth Stuart is Founder, President Emeritus, and Professor of the Seattle Biomedical Research Institute and Affiliate Professor of Global Health at the University of Washington. He received a B.A. in Biology from Northeastern University, M.A. in Biology from Wesleyan University, and Ph.D. in Zoology from the University of Iowa. He was a postdoctoral researcher in Biochemistry at the National Institute for Medical Research (London) and at SUNY Stony Brook. His research is on fundamental cell and molecular processes and drug discovery in kinetoplastid pathogens and employs genetic, genomic, proteomic, and other approaches. He made major contributions to the elucidation of RNA editing including its mechanism and machinery, kinetoplastid genomes including their sequencing and annotation, characterization of the mitochondrial proteome and its multiprotein complexes, and the functions and potential value as drug targets of critical kinetoplastid proteins including protein kinases, aminoacyl tRNA synthetases, and others.

Michael P. Pollastri is Associate Professor of Chemistry & Chemical Biology at Northeastern University. Following nearly 10 years in the pharmaceutical industry (Pfizer, Inc.), Dr. Pollastri transitioned into an academic career, with his research focused on hit-to-lead and lead optimization medicinal chemistry directed towards neglected tropical diseases. His research programs are primarily focused on repurposing kinase inhibitor compounds classes that have been shown to be effective against human enzymes by reoptimizing for parasite selectivity and appropriate physicochemical properties. He earned his A.B. in Chemistry at the College of the Holy Cross, his M.S. in Organic Chemistry from Duke University, and his Ph.D. from Brown University.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Hopkins A. L.; Groom C. R. Nat. Rev. Drug. Discovery 2002, 1, 727. [DOI] [PubMed] [Google Scholar]
- W.H.O. Tech. Rep. Ser. 2012, 1. [Google Scholar]
- Njoroge M.; Njuguna N.; Mutai P.; Ongarora D.; Smith P.; Chibale K. Chem. Rev. 2014, DOI: 10.1021/cr500098f. [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M.; Myler P. J.; Blandin G.; Berriman M.; Crabtree J.; Aggarwal G.; Caler E.; Renauld H.; Worthey E. A.; Hertz-Fowler C.; Ghedin E.; Peacock C.; Bartholomeu D. C.; Haas B. J.; Tran A. N.; Wortman J. R.; Alsmark U. C.; Angiuoli S.; Anupama A.; Badger J.; Bringaud F.; Cadag E.; Carlton J. M.; Cerqueira G. C.; Creasy T.; Delcher A. L.; Djikeng A.; Embley T. M.; Hauser C.; Ivens A. C.; Kummerfeld S. K.; Pereira-Leal J. B.; Nilsson D.; Peterson J.; Salzberg S. L.; Shallom J.; Silva J. C.; Sundaram J.; Westenberger S.; White O.; Melville S. E.; Donelson J. E.; Andersson B.; Stuart K. D.; Hall N. Science 2005, 309, 404.16020724 [Google Scholar]
- a El-Sayed N. M.; Myler P. J.; Bartholomeu D. C.; Nilsson D.; Aggarwal G.; Tran A. N.; Ghedin E.; Worthey E. A.; Delcher A. L.; Blandin G.; Westenberger S. J.; Caler E.; Cerqueira G. C.; Branche C.; Haas B.; Anupama A.; Arner E.; Aslund L.; Attipoe P.; Bontempi E.; Bringaud F.; Burton P.; Cadag E.; Campbell D. A.; Carrington M.; Crabtree J.; Darban H.; da Silveira J. F.; de Jong P.; Edwards K.; Englund P. T.; Fazelina G.; Feldblyum T.; Ferella M.; Frasch A. C.; Gull K.; Horn D.; Hou L.; Huang Y.; Kindlund E.; Klingbeil M.; Kluge S.; Koo H.; Lacerda D.; Levin M. J.; Lorenzi H.; Louie T.; Machado C. R.; McCulloch R.; McKenna A.; Mizuno Y.; Mottram J. C.; Nelson S.; Ochaya S.; Osoegawa K.; Pai G.; Parsons M.; Pentony M.; Pettersson U.; Pop M.; Ramirez J. L.; Rinta J.; Robertson L.; Salzberg S. L.; Sanchez D. O.; Seyler A.; Sharma R.; Shetty J.; Simpson A. J.; Sisk E.; Tammi M. T.; Tarleton R.; Teixeira S.; Van Aken S.; Vogt C.; Ward P. N.; Wickstead B.; Wortman J.; White O.; Fraser C. M.; Stuart K. D.; Andersson B. Science 2005, 309, 409.16020725 [Google Scholar]; b Berriman M.; Ghedin E.; Hertz-Fowler C.; Blandin G.; Renauld H.; Bartholomeu D. C.; Lennard N. J.; Caler E.; Hamlin N. E.; Haas B.; Bohme U.; Hannick L.; Aslett M. A.; Shallom J.; Marcello L.; Hou L.; Wickstead B.; Alsmark U. C.; Arrowsmith C.; Atkin R. J.; Barron A. J.; Bringaud F.; Brooks K.; Carrington M.; Cherevach I.; Chillingworth T. J.; Churcher C.; Clark L. N.; Corton C. H.; Cronin A.; Davies R. M.; Doggett J.; Djikeng A.; Feldblyum T.; Field M. C.; Fraser A.; Goodhead I.; Hance Z.; Harper D.; Harris B. R.; Hauser H.; Hostetler J.; Ivens A.; Jagels K.; Johnson D.; Johnson J.; Jones K.; Kerhornou A. X.; Koo H.; Larke N.; Landfear S.; Larkin C.; Leech V.; Line A.; Lord A.; Macleod A.; Mooney P. J.; Moule S.; Martin D. M.; Morgan G. W.; Mungall K.; Norbertczak H.; Ormond D.; Pai G.; Peacock C. S.; Peterson J.; Quail M. A.; Rabbinowitsch E.; Rajandream M. A.; Reitter C.; Salzberg S. L.; Sanders M.; Schobel S.; Sharp S.; Simmonds M.; Simpson A. J.; Tallon L.; Turner C. M.; Tait A.; Tivey A. R.; Van Aken S.; Walker D.; Wanless D.; Wang S.; White B.; White O.; Whitehead S.; Woodward J.; Wortman J.; Adams M. D.; Embley T. M.; Gull K.; Ullu E.; Barry J. D.; Fairlamb A. H.; Opperdoes F.; Barrell B. G.; Donelson J. E.; Hall N.; Fraser C. M. Science 2005, 309, 416.16020726 [Google Scholar]
- Barry D.; Carrington M. In The Trypanosomiases; Maudlin I., Holmes P. H., Miles M. A., Eds.; Commonwealth Agricultural Bureau International: Oxfordshire, U.K., 2004; p 25. [Google Scholar]
- Opperdoes F. In Structures and Organelles in Pathogenic Protists; de Souza W., Ed.; Springer: Berlin, 2010; Vol. 17, p 285. [Google Scholar]
- Verlinde C. L.; Hannaert V.; Blonski C.; Willson M.; Perie J. J.; Fothergill-Gilmore L. A.; Opperdoes F. R.; Gelb M. H.; Hol W. G.; Michels P. A. Drug Resist. Updates 2001, 4, 50. [DOI] [PubMed] [Google Scholar]
- Pal C.; Bandyopadhyay U. Antioxid. Redox Signaling 2012, 17, 555. [DOI] [PubMed] [Google Scholar]
- Gunzl A. Eukaryotic Cell 2010, 9, 1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning H. P.; Bridges D. J.; Burchmore R. J. FEMS Microbiol. Rev. 2005, 29, 987. [DOI] [PubMed] [Google Scholar]
- a Simo G.; Asonganyi T.; Nkinin S. W.; Njiokou F.; Herder S. Vet. Parasitol. 2006, 139, 57. [DOI] [PubMed] [Google Scholar]; b Funk S.; Nishiura H.; Heesterbeek H.; Edmunds W. J.; Checchi F. PLoS Comput. Biol. 2013, 9, e1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Hamill L. C.; Kaare M. T.; Welburn S. C.; Picozzi K. Parasites Vectors 2013, 6, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Njiokou F.; Laveissiere C.; Simo G.; Nkinin S.; Grebaut P.; Cuny G.; Herder S. Infect., Genet. Evol. 2006, 6, 147. [DOI] [PubMed] [Google Scholar]
- Burri C.; Brun R. Parasitol. Res. 2003, 90Supp 1S49. [DOI] [PubMed] [Google Scholar]
- Desquesnes M.; Holzmuller P.; Lai D. H.; Dargantes A.; Lun Z. R.; Jittaplapong S. BioMed Res. Int. 2013, 2013, 194176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzureau P.; Uzureau S.; Lecordier L.; Fontaine F.; Tebabi P.; Homble F.; Grelard A.; Zhendre V.; Nolan D. P.; Lins L.; Crowet J. M.; Pays A.; Felu C.; Poelvoorde P.; Vanhollebeke B.; Moestrup S. K.; Lyngso J.; Pedersen J. S.; Mottram J. C.; Dufourc E. J.; Perez-Morga D.; Pays E. Nature 2013, 501, 430. [DOI] [PubMed] [Google Scholar]
- Capewell P.; Clucas C.; DeJesus E.; Kieft R.; Hajduk S.; Veitch N.; Steketee P. C.; Cooper A.; Weir W.; MacLeod A. PLoS Pathog. 2013, 9, e1003686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassi A.; Rassi A.; Marin-Neto J. A. Lancet 2010, 375, 1388. [DOI] [PubMed] [Google Scholar]
- a Brener Z.; Gazzinelli R. T. Int. Arch. Allergy Immunol. 1997, 114, 103. [DOI] [PubMed] [Google Scholar]; b Andrade Z. A. Mem. Inst. Oswaldo Cruz 1999, 94Suppl 171. [DOI] [PubMed] [Google Scholar]
- Manoel-Caetano Fda S.; Silva A. E. Cad. Saude. Publica 2007, 23, 2263. [DOI] [PubMed] [Google Scholar]
- Kaye P.; Scott P. Nat. Rev. Microbiol. 2011, 9, 604. [DOI] [PubMed] [Google Scholar]
- Colmenares M.; Kar S.; Goldsmith-Pestana K.; McMahon-Pratt D. Trans. R. Soc. Trop. Med. Hyg. 2002, 96Suppl 1S3. [DOI] [PubMed] [Google Scholar]
- Hartley M. A.; Ronet C.; Zangger H.; Beverley S. M.; Fasel N. Front. Cell Infect. Microbiol. 2012, 2, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delespaux V.; de Koning H. P. Drug Resist. Updates 2007, 10, 30. [DOI] [PubMed] [Google Scholar]
- Priotto G.; Kasparian S.; Mutombo W.; Ngouama D.; Ghorashian S.; Arnold U.; Ghabri S.; Baudin E.; Buard V.; Kazadi-Kyanza S.; Ilunga M.; Mutangala W.; Pohlig G.; Schmid C.; Karunakara U.; Torreele E.; Kande V. Lancet 2009, 374, 56. [DOI] [PubMed] [Google Scholar]
- Jacobs R. T.; Nare B.; Wring S. A.; Orr M. D.; Chen D.; Sligar J. M.; Jenks M. X.; Noe R. A.; Bowling T. S.; Mercer L. T.; Rewerts C.; Gaukel E.; Owens J.; Parham R.; Randolph R.; Beaudet B.; Bacchi C. J.; Yarlett N.; Plattner J. J.; Freund Y.; Ding C.; Akama T.; Zhang Y. K.; Brun R.; Kaiser M.; Scandale I.; Don R. PLoS Neglected Trop. Dis. 2011, 5, e1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M.; Bray M. A.; Cal M.; Bourdin Trunz B.; Torreele E.; Brun R. Antimicrob. Agents Chemother. 2011, 55, 5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwaka S.; Hudson A. Nat. Rev. Drug. Discovery 2006, 5, 941. [DOI] [PubMed] [Google Scholar]
- Ribeiro I.; Sevcsik A.-M.; Alves F.; Diap G.; Don R.; Harhay M. O.; Chang S.; Pecoul B. PLoS Neglected Trop. Dis. 2009, 3, e484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina J. A.; Docampo R. Trends Parasitol. 2003, 19, 495. [DOI] [PubMed] [Google Scholar]
- Buckner F. S. Adv. Exp. Med. Biol. 2008, 625, 61. [DOI] [PubMed] [Google Scholar]
- Drugs for Neglected Diseases Initiative. http://www.dndi.org/media-centre/press-releases/354-media-centre/press-releases/1700-e1224.html (accessed 25 March 2014).
- Maltezou H. C. J. Biomed. Biotechnol. 2010, 2010, 617521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-On J.; Halevy S.; Grunwald M. H.; Weinrauch L. J. Am. Acad. Dermatol. 1992, 27, 227. [DOI] [PubMed] [Google Scholar]
- a Rahman M.; Ahmed B. N.; Faiz M. A.; Chowdhury M. Z.; Islam Q. T.; Sayeedur R.; Rahman M. R.; Hossain M.; Bangali A. M.; Ahmad Z.; Islam M. N.; Mascie-Taylor C. G.; Berman J.; Arana B. Am. J. Trop. Med. Hyg. 2011, 85, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bhattacharya S. K.; Sinha P. K.; Sundar S.; Thakur C. P.; Jha T. K.; Pandey K.; Das V. R.; Kumar N.; Lal C.; Verma N.; Singh V. P.; Ranjan A.; Verma R. B.; Anders G.; Sindermann H.; Ganguly N. K. J. Infect. Dis. 2007, 196, 591. [DOI] [PubMed] [Google Scholar]
- a Sharlow E. R.; Lyda T. A.; Dodson H. C.; Mustata G.; Morris M. T.; Leimgruber S. S.; Lee K.-H.; Kashiwada Y.; Close D.; Lazo J. S.; Morris J. C. PLoS Neglected Trop. Dis. 2010, 4, e659. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Smithson D. C.; Shelat A. A.; Baldwin J.; Phillips M. A.; Guy R. K. Assay Drug Dev. Technol. 2010, 8, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Brak K.; Doyle P. S.; McKerrow J. H.; Ellman J. A. J. Am. Chem. Soc. 2008, 130, 6404. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ferreira R. S.; Simeonov A.; Jadhav A.; Eidam O.; Mott B. T.; Keiser M. J.; McKerrow J. H.; Maloney D. J.; Irwin J. J.; Shoichet B. K. J. Med. Chem. 2010, 53, 4891. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Gunatilleke S. S.; Calvet C. M.; Johnston J. B.; Chen C.-K.; Erenburg G.; Gut J.; Engel J. C.; Ang K. K. H.; Mulvaney J.; Chen S.; Arkin M. R.; McKerrow J. H.; Podust L. M. PLoS Neglected Trop. Dis. 2012, 6, e1736. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Ben Khalaf N.; De Muylder G.; Ratnam J.; Ang K. K.-H.; Arkin M.; McKerrow J.; Chenik M. J. Biomol. Screening 2011, 16, 545. [DOI] [PubMed] [Google Scholar]; g Cleghorn L. A. T.; Woodland A.; Collie I. T.; Torrie L. S.; Norcross N.; Luksch T.; Mpamhanga C.; Walker R. G.; Mottram J. C.; Brenk R.; Frearson J. A.; Gilbert I. H.; Wyatt P. G. ChemMedChem 2011, 6, 2214. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Grant K. M.; Dunion M. H.; Yardley V.; Skaltsounis A.-L.; Marko D.; Eisenbrand G.; Croft S. L.; Meijer L.; Mottram J. C. Antimicrob. Agents Chemother. 2004, 48, 3033. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Morgan R. E.; Ahn S.; Nzimiro S.; Fotie J.; Phelps M. A.; Cotrill J.; Yakovich A. J.; Sackett D. L.; Dalton J. T.; Werbovetz K. A. Chem. Biol. Drug Des. 2008, 72, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ali B. R. S.; Pal A.; Croft S. L.; Taylor R. J. K.; Field M. C. Mol. Biochem. Parasitol. 1999, 104, 67. [DOI] [PubMed] [Google Scholar]; b Ferrins L.; Rahmani R.; Sykes M. L.; Jones A. J.; Avery V. M.; Teston E.; Almohaywi B.; Yin J. X.; Smith J.; Hyland C.; White K. L.; Ryan E.; Campbell M.; Charman S. A.; Kaiser M.; Baell J. B. Eur. J. Med. Chem. 2013, 66, 450. [DOI] [PubMed] [Google Scholar]; c Julianti T.; Hata Y.; Zimmermann S.; Kaiser M.; Hamburger M.; Adams M. Fitoterapia 2011, 82, 955. [DOI] [PubMed] [Google Scholar]; d Slusarczyk S.; Zimmermann S.; Kaiser M.; Matkowski A.; Hamburger M.; Adams M. Planta Med. 2011, 77, 1594. [DOI] [PubMed] [Google Scholar]; e Sykes M. L.; Baell J. B.; Kaiser M.; Chatelain E.; Moawad S. R.; Ganame D.; Ioset J.-R.; Avery V. M. PLoS Neglected Trop. Dis. 2012, 6, e1896. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Tatipaka H. B.; Gillespie J. R.; Chatterjee A. K.; Norcross N. R.; Hulverson M. A.; Ranade R. M.; Nagendar P.; Creason S. A.; McQueen J.; Duster N. A.; Nagle A.; Supek F.; Molteni V.; Wenzler T.; Brun R.; Glynne R.; Buckner F. S.; Gelb M. H. J. Med. Chem. 2014, 57, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Vodnala S. K.; Lundbaeck T.; Sjoeberg B.; Svensson R.; Rottenberg M. E.; Hammarstroem L. G. J. Antimicrob. Agents Chemother. 2013, 57, 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Dandapani S.; Germain A. R.; Jewett I.; Le Quement S.; Marie J.-C.; Muncipinto G.; Duvall J. R.; Carmody L. C.; Perez J. R.; Engel J. C.; Gut J.; Kellar D.; Lage de Siqueira-Neto J.; McKerrow J. H.; Kaiser M.; Rodriguez A.; Palmer M. A.; Foley M.; Schreiber S. L.; Munoz B. ACS Med. Chem. Lett. 2014, 5, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Germain A. R.; Carmody L. C.; Dockendorff C.; Galan-Rodriguez C.; Rodriguez A.; Johnston S.; Bittker J. A.; MacPherson L.; Dandapani S.; Palmer M.; Schreiber S. L.; Munoz B. Bioorg. Med. Chem. Lett. 2011, 21, 7197. [DOI] [PubMed] [Google Scholar]; j Keenan M.; Alexander P. W.; Chaplin J. H.; Abbott M. J.; Diao H.; Wang Z.; Best W. M.; Perez C. J.; Cornwall S. M. J.; Keatley S. K.; Thompson R. C. A.; Charman S. A.; White K. L.; Ryan E.; Chen G.; Ioset J.-R.; von Geldern T. W.; Chatelain E. Future Med. Chem. 2013, 5, 1733. [DOI] [PubMed] [Google Scholar]; k Engel J. C.; Ang K. K. H.; Chen S.; Arkin M. R.; McKerrow J. H.; Doyle P. S. Antimicrob. Agents Chemother. 2010, 54, 3326. [DOI] [PMC free article] [PubMed] [Google Scholar]; l Bell A. S.; Mills J. E.; Williams G. P.; Brannigan J. A.; Wilkinson A. J.; Parkinson T.; Leatherbarrow R. J.; Tate E. W.; Holder A. A.; Smith D. F. PLoS Neglected Trop. Dis. 2012, 6, e1625. [DOI] [PMC free article] [PubMed] [Google Scholar]; m De M. G.; Ang K. K. H.; Chen S.; Arkin M. R.; Engel J. C.; McKerrow J. H. PLoS Neglected Trop. Dis. 2011, 5, e1253. [DOI] [PMC free article] [PubMed] [Google Scholar]; n Osorio Y.; Travi B. L.; Renslo A. R.; Peniche A. G.; Melby P. C. PLoS Neglected Trop. Dis. 2011, 5, e962. [DOI] [PMC free article] [PubMed] [Google Scholar]; o Sharlow E. R.; Close D.; Shun T.; Leimgruber S.; Reed R.; Mustata G.; Wipf P.; Johnson J.; O’Neil M.; Grogl M.; Magill A. J.; Lazo J. S. PLoS Neglected Trop. Dis. 2009, 3, e540. [DOI] [PMC free article] [PubMed] [Google Scholar]; p Zhu X.; Pandharkar T.; Werbovetz K. Antimicrob. Agents Chemother. 2012, 56, 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugs for Neglected Disease Initiative. http://www.dndi.org/diseases-projects/diseases/hat/target-product-profile.html (accessed 25 March 2014).
- Drugs for Neglected Disease Initiative. http://www.dndi.org/diseases-projects/diseases/vl/tpp.html (accessed 25 March 2014).
- Drugs for Neglected Disease Initiative. http://www.dndi.org/diseases-projects/diseases/chagas/target-product-profile.html (accessed 25 March 2014).
- Parsons M.; Worthey E. A.; Ward P. N.; Mottram J. C. BMC Genomics 2005, 6, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Mottram J. C.; McCready B. P.; Brown K. G.; Grant K. M. Mol. Microbiol. 1996, 22, 573. [DOI] [PubMed] [Google Scholar]; b Hassan P.; Fergusson D.; Grant K. M.; Mottram J. C. Mol. Biochem. Parasitol. 2001, 113, 189. [DOI] [PubMed] [Google Scholar]
- Boeke J. D.; Trueheart J.; Natsoulis G.; Fink G. R. Methods Enzymol. 1987, 154, 164. [DOI] [PubMed] [Google Scholar]
- Murta S. M.; Vickers T. J.; Scott D. A.; Beverley S. M. Mol. Microbiol. 2009, 71, 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Baum J.; Papenfuss A. T.; Mair G. R.; Janse C. J.; Vlachou D.; Waters A. P.; Cowman A. F.; Crabb B. S.; de Koning-Ward T. F. Nucleic Acids Res. 2009, 37, 3788. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lye L. F.; Owens K.; Shi H.; Murta S. M.; Vieira A. C.; Turco S. J.; Tschudi C.; Ullu E.; Beverley S. M. PLoS Pathog. 2010, 6, e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ullu E.; Tschudi C.; Chakraborty T. Cell Microbiol. 2004, 6, 509. [DOI] [PubMed] [Google Scholar]
- Alsford S.; Turner D. J.; Obado S. O.; Sanchez-Flores A.; Glover L.; Berriman M.; Hertz-Fowler C.; Horn D. Genome Res. 2011, 21, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Nishino M.; Choy J. W.; Gushwa N. N.; Oses-Prieto J. A.; Koupparis K.; Burlingame A. L.; Renslo A. R.; McKerrow J. H.; Taunton J. eLife 2013, 2, e00712. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jones N. G.; Thomas E. B.; Brown E.; Dickens N. J.; Hammarton T. C.; Mottram J. C. PLoS Pathog. 2014, 10, e1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C.; Stuart K. Mol. Biochem. Parasitol. 2013, 190, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Monnerat S.; Almeida Costa C. I.; Forkert A. C.; Benz C.; Hamilton A.; Tetley L.; Burchmore R.; Novo C.; Mottram J. C.; Hammarton T. C. PLoS One 2013, 8, e67327. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mackey Z. B.; Koupparis K.; Nishino M.; McKerrow J. H. Chem. Biol. Drug. Des. 2011, 78, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L. A.; Chen L. C.; Maynard-Smith L. A.; Ooi A. G.; Wandless T. J. Cell 2006, 126, 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Armstrong C. M.; Goldberg D. E. Nat. Methods 2007, 4, 1007. [DOI] [PubMed] [Google Scholar]; b Dvorin J. D.; Martyn D. C.; Patel S. D.; Grimley J. S.; Collins C. R.; Hopp C. S.; Bright A. T.; Westenberger S.; Winzeler E.; Blackman M. J.; Baker D. A.; Wandless T. J.; Duraisingh M. T. Science 2010, 328, 910. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ma Y. F.; Weiss L. M.; Huang H. Int. J. Parasitol. 2012, 42, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Madeira da Silva L.; Owens K. L.; Murta S. M.; Beverley S. M. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight Z. A.; Shokat K. M. Cell 2007, 128, 425. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Bishop A.; Witucki L.; Kraybill B.; Shimizu E.; Tsien J.; Ubersax J.; Blethrow J.; Morgan D. O.; Shokat K. M. Chem. Biol. 1999, 6, 671. [DOI] [PubMed] [Google Scholar]
- Horiuchi D.; Huskey N. E.; Kusdra L.; Wohlbold L.; Merrick K. A.; Zhang C.; Creasman K. J.; Shokat K. M.; Fisher R. P.; Goga A. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, E1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Hammarton T. C. Mol. Biochem. Parasitol. 2007, 153, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Li Z. Eukaryotic Cell 2012, 11, 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X.; Wang C. C. J. Biol. Chem. 2004, 279, 20519. [DOI] [PubMed] [Google Scholar]
- Badjatia N.; Ambrosio D. L.; Lee J. H.; Gunzl A. Mol. Cell. Biol. 2013, 33, 1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graffenried C. L.; Ho H. H.; Warren G. J. Cell Biol. 2008, 181, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Hammarton T. C.; Kramer S.; Tetley L.; Boshart M.; Mottram J. C. Mol. Microbiol. 2007, 65, 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kumar P.; Wang C. C. Eukaryot. Cell 2006, 5, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V.; Glover D. M. Nat. Rev. Mol. Cell Biol. 2009, 10, 265. [DOI] [PubMed] [Google Scholar]
- Li Z.; Wang C. C. Eukaryotic Cell 2006, 5, 1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Benz C.; Grimaldi R.; Stockdale C.; Wyatt P.; Frearson J.; Hammarton T. C. J. Biol. Chem. 2010, 285, 15356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojo K. K.; Gillespie J. R.; Riechers A. J.; Napuli A. J.; Verlinde C. L.; Buckner F. S.; Gelb M. H.; Domostoj M. M.; Wells S. J.; Scheer A.; Wells T. N.; Van Voorhis W. C. Antimicrob. Agents Chemother. 2008, 52, 3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak M. D. Mol. Biochem. Parasitol. 2009, 166, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A.; Saldivia M.; Diaz R.; Bart J. M.; Vidal I.; Calvo E.; Hall M. N.; Navarro M. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese M. EMBO J. 1998, 17, 2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller I. B.; Domenicali-Pfister D.; Roditi I.; Vassella E. Mol. Biol. Cell 2002, 13, 3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M. A.; Haanstra J. R.; Hannaert V.; Van Roy J.; Opperdoes F. R.; Bakker B. M.; Michels P. A. J. Biol. Chem. 2005, 280, 28306. [DOI] [PubMed] [Google Scholar]
- Subramaniam C.; Veazey P.; Redmond S.; Hayes-Sinclair J.; Chambers E.; Carrington M.; Gull K.; Matthews K.; Horn D.; Field M. C. Eukaryotic Cell 2006, 5, 1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J. W.; Fowler M. L.; Morris M. T.; Morris J. C. Mol. Biochem. Parasitol. 2008, 158, 202. [DOI] [PubMed] [Google Scholar]
- Hergovich A.; Stegert M. R.; Schmitz D.; Hemmings B. A. Nat. Rev. Mol. Cell. Biol. 2006, 7, 253. [DOI] [PubMed] [Google Scholar]
- Carmena M.; Earnshaw W. C. Nat. Rev. Mol. Cell. Biol. 2003, 4, 842. [DOI] [PubMed] [Google Scholar]
- Tu X.; Kumar P.; Li Z.; Wang C. C. J. Biol. Chem. 2006, 281, 9677. [DOI] [PubMed] [Google Scholar]
- a Rotureau B.; Morales M. A.; Bastin P.; Spath G. F. Cell Microbiol. 2009, 11, 710. [DOI] [PubMed] [Google Scholar]; b Wiese M. Int. J. Parasitol. 2007, 37, 1053. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Melzer I. M.; Kruse M.; Sander-Juelch C.; Wiese M. Kinetoplastid Biol. Dis. 2005, 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperdoes F. R.; Borst P. FEBS Lett. 1977, 80, 360. [DOI] [PubMed] [Google Scholar]
- Schirmer A.; Kennedy J.; Murli S.; Reid R.; Santi D. V. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar S.; Kufareva I.; Behera R.; Thomas S. M.; Ogata Y.; Pollastri M.; Abagyan R.; Mensa-Wilmot K. PLoS One 2013, 8, e56150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M.; Eberhard D.; Abraham Y.; Bastuck S.; Boesche M.; Hobson S.; Mathieson T.; Perrin J.; Raida M.; Rau C.; Reader V.; Sweetman G.; Bauer A.; Bouwmeester T.; Hopf C.; Kruse U.; Neubauer G.; Ramsden N.; Rick J.; Kuster B.; Drewes G. Nat. Biotechnol. 2007, 25, 1035. [DOI] [PubMed] [Google Scholar]
- Urbaniak M. D.; Mathieson T.; Bantscheff M.; Eberhard D.; Grimaldi R.; Miranda-Saavedra D.; Wyatt P.; Ferguson M. A.; Frearson J.; Drewes G. ACS Chem. Biol. 2012, 7, 1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewry D. H.; Bamborough P.; Schneider K.; Smith G. K. In Kinase Drug Discovery; Ward R. A., Goldberg F., Eds.; The Royal Society of Chemistry: Cambridge, U.K., 2012; p 1. [Google Scholar]
- Karaman M. W.; Herrgard S.; Treiber D. K.; Gallant P.; Atteridge C. E.; Campbell B. T.; Chan K. W.; Ciceri P.; Davis M. I.; Edeen P. T.; Faraoni R.; Floyd M.; Hunt J. P.; Lockhart D. J.; Milanov Z. V.; Morrison M. J.; Pallares G.; Patel H. K.; Pritchard S.; Wodicka L. M.; Zarrinkar P. P. Nat. Biotechnol. 2008, 26, 127. [DOI] [PubMed] [Google Scholar]
- Morphy R. J. Med. Chem. 2010, 53, 1413. [DOI] [PubMed] [Google Scholar]
- Lorusso P. M.; Eder J. P. Expert Opin. Invest. Drugs 2008, 17, 1013. [DOI] [PubMed] [Google Scholar]
- Jones C. D.; Andrews D. M.; Barker A. J.; Blades K.; Byth K. F.; Finlay M. R.; Geh C.; Green C. P.; Johannsen M.; Walker M.; Weir H. M. Bioorg. Med. Chem. Lett. 2008, 18, 6486. [DOI] [PubMed] [Google Scholar]
- Swarbrick M. E. In Kinase Drug Discovery; Ward R. A., Goldberg F., Eds.; The Royal Society of Chemistry: Cambridge, U.K., 2012; p 79. [Google Scholar]
- a Mol C. D.; Fabbro D.; Hosfield D. J. Curr. Opin. Drug Discovery Dev. 2004, 7, 639. [PubMed] [Google Scholar]; b Liu Y.; Gray N. S. Nat. Chem. Biol. 2006, 2, 358. [DOI] [PubMed] [Google Scholar]
- Gill A. L.; Verdonk M.; Boyle R. G.; Taylor R. Curr. Top. Med. Chem. 2007, 7, 1408. [DOI] [PubMed] [Google Scholar]
- Wager T. T.; Chandrasekaran R. Y.; Hou X.; Troutman M. D.; Verhoest P. R.; Villalobos A.; Will Y. ACS Chem. Neurosci. 2010, 1, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M. C.; Tristani-Firouzi M. Nature 2006, 440, 463. [DOI] [PubMed] [Google Scholar]
- Plyte S. E.; Hughes K.; Nikolakaki E.; Pulverer B. J.; Woodgett J. R. Biochim. Biophys. Acta 1992, 1114, 147. [DOI] [PubMed] [Google Scholar]
- Meijer L.; Flajolet M.; Greengard P. Trends Pharmacol. Sci. 2004, 25, 471. [DOI] [PubMed] [Google Scholar]
- Ojo K. K.; Gillespie J. R.; Riechers A. J.; Napuli A. J.; Verlinde C. L. M. J.; Buckner F. S.; Gelb M. H.; Domostoj M. M.; Wells S. J.; Scheer A.; Wells T. N. C.; Van Voorhis W. C. Antimicrob. Agents Chemother. 2008, 52, 3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayode K. O.; Arakaki T. L.; Napuli A. J.; Inampudi K. K.; Keyloun K. R.; Zhang L.; Hol W. G. J.; Verlinde C. L. M. J.; Merritt E. A.; Van Voorhis W. C. Mol. Biochem. Parasitol. 2011, 176, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oduor R. O.; Ojo K. K.; Williams G. P.; Bertelli F.; Mills J.; Maes L.; Pryde D. C.; Parkinson T.; Van Voorhis W. C.; Holler T. P. PLoS Neglected Trop. Dis. 2011, 5, e1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak M. D.; Mathieson T.; Bantscheff M.; Eberhard D.; Grimaldi R.; Miranda-Saavedra D.; Wyatt P.; Ferguson M. A. J.; Frearson J.; Drewes G. ACS Chem. Biol. 2012, 7, 1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland A.; Grimaldi R.; Luksch T.; Cleghorn L. A. T.; Ojo K. K.; Van Voorhis W. C.; Brenk R.; Frearson J. A.; Gilbert I. H.; Wyatt P. G. ChemMedChem 2013, 8, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Fothergill-Gilmore L. A.; Michels P. A. M. Prog. Biophys. Mol. Biol. 1993, 59, 105. [DOI] [PubMed] [Google Scholar]; b Michels P. A. M.; Chevalier N.; Opperdoes F. R.; Rider M. H.; Rigden D. J. Eur. J. Biochem. 1997, 250, 698. [DOI] [PubMed] [Google Scholar]; c Vanschaftingen E.; Opperdoes F. R.; Hers H. G. Eur. J. Biochem. 1985, 153, 403. [DOI] [PubMed] [Google Scholar]
- Martinez-Oyanedel J.; McNae I. W.; Nowicki M. W.; Keillor J. W.; Michels P. A.; Fothergill-Gilmore L. A.; Walkinshaw M. D. J. Mol. Biol. 2007, 366, 1185. [DOI] [PubMed] [Google Scholar]
- Claustre S.; Denier C.; Lakhdar-Ghazal F.; Lougare A.; Lopez C.; Chevalier N.; Michels P. A.; Perie J.; Willson M. Biochemistry 2002, 41, 10183. [DOI] [PubMed] [Google Scholar]
- Nowicki M. W.; Tulloch L. B.; Worralll L.; McNae I. W.; Hannaert V.; Michels P. A.; Fothergill-Gilmore L. A.; Walkinshaw M. D.; Turner N. J. Bioorg. Med. Chem. 2008, 16, 5050. [DOI] [PubMed] [Google Scholar]
- Ngantchou I.; Nyasse B.; Denier C.; Blonski C.; Hannaert V.; Schneider B. Bioorg. Med. Chem. Lett. 2010, 20, 3495. [DOI] [PubMed] [Google Scholar]
- Brimacombe K. R.; Walsh M. J.; Liu L.; Vasquez-Valdivieso M. G.; Morgan H. P.; McNae I.; Fothergill-Gilmore L. A.; Michels P. A. M.; Auld D. S.; Simeonov A.; Walkinshaw M. D.; Shen M.; Boxer M. B. ACS Med. Chem. Lett. 2014, 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Blake C. C.; Evans P. R. J. Mol. Biol. 1974, 84, 585. [DOI] [PubMed] [Google Scholar]; b Bernstein B. E.; Michels P. A.; Hol W. G. Nature 1997, 385, 275. [DOI] [PubMed] [Google Scholar]; c Bernstein B. E.; Williams D. M.; Bressi J. C.; Kuhn P.; Gelb M. H.; Blackburn G. M.; Hol W. G. J. J. Mol. Biol. 1998, 279, 1137. [DOI] [PubMed] [Google Scholar]; d Bernstein B. E.; Hol W. G. J. Biochemistry 1998, 37, 4429. [DOI] [PubMed] [Google Scholar]
- a Aronov A. M.; Verlinde C. L. M. J.; Hol W. G. J.; Gelb M. H. J. Med. Chem. 1998, 41, 4790. [DOI] [PubMed] [Google Scholar]; b Aronov A. M.; Suresh S.; Buckner F. S.; Van Voorhis W. C.; Verlinde C. L. M. J.; Opperdoes F. R.; Hol W. G. J.; Gelb M. H. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressi J. C.; Choe J.; Hough M. T.; Buckner F. S.; Van Voorhis W. C.; Verlinde C. L. M. J.; Hol W. G. J.; Gelb M. H. J. Med. Chem. 2000, 43, 4135. [DOI] [PubMed] [Google Scholar]
- Otieno S.; Bhargava A. K.; Barnard E. A.; Ramel A. H. Biochemistry 1975, 14, 2403. [DOI] [PubMed] [Google Scholar]
- Sols A.; De La Fuente G.; Villarpalasi C.; Asensio C. Biochim. Biophys. Acta 1958, 30, 92. [DOI] [PubMed] [Google Scholar]
- Willson M.; Alric I.; Perie J.; Sanejouand Y. H. J. Enzyme Inhib. 1997, 12, 101. [DOI] [PubMed] [Google Scholar]
- Walter R. D.; Ebert F. Hoppe-Seyler’s Z. Physiol. Chem. 1977, 358, 23. [DOI] [PubMed] [Google Scholar]
- Alonso G. D.; Pereira C. A.; Remedi M. S.; Paveto M. C.; Cochella L.; Ivaldi M. S.; Gerez de Burgos N. M.; Torres H. N.; Flawia M. M. FEBS Lett. 2001, 498, 22. [DOI] [PubMed] [Google Scholar]
- Pereira C. A.; Alonso G. D.; Ivaldi S.; Bouvier L. A.; Torres H. N.; Flawia M. M. J. Eukaryotic Microbiol. 2003, 50, 132. [DOI] [PubMed] [Google Scholar]
- Xingi E.; Smirlis D.; Myrianthopoulos V.; Magiatis P.; Grant K. M.; Meijer L.; Mikros E.; Skaltsounis A. L.; Soteriadou K. Int. J. Parasitol. 2009, 39, 1289. [DOI] [PubMed] [Google Scholar]
- a Polychronopoulos P.; Magiatis P.; Skaltsounis A. L.; Myrianthopoulos V.; Mikros E.; Tarricone A.; Musacchio A.; Roe S. M.; Pearl L.; Leost M.; Greengard P.; Meijer L. J. Med. Chem. 2004, 47, 935. [DOI] [PubMed] [Google Scholar]; b Meijer L.; Skaltsounis A. L.; Magiatis P.; Polychronopoulos P.; Knockaert M.; Leost M.; Ryan X. Z. P.; Vonica C. A.; Brivanlou A.; Dajani R.; Crovace C.; Tarricone C.; Musacchio A.; Roe S. M.; Pearl L.; Greengard P. Chem. Biol. 2003, 10, 1255. [DOI] [PubMed] [Google Scholar]
- Morgan H. P.; Walsh M. J.; Blackburn E. A.; Wear M. A.; Boxer M. B.; Shen M.; Veith H.; McNae I. W.; Nowicki M. W.; Michels P. A. M.; Auld D. S.; Fothergill-Gilmore L. A.; Walkinshaw M. D. Biochem. J. 2012, 448, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyasse B.; Ngantchou I.; Tchana E. M.; Sonke B.; Denier C.; Fontaine C. Pharmazie 2004, 59, 873. [PubMed] [Google Scholar]
- Donald R. G.; Allocco J.; Singh S. B.; Nare B.; Salowe S. P.; Wiltsie J.; Liberator P. A. Eukaryotic Cell 2002, 1, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Vieira L. L.; Sacerdoti-Sierra N.; Jaffe C. L. Int. J. Parasitol. 2002, 32, 1085. [DOI] [PubMed] [Google Scholar]; b Sacerdoti-Sierra N.; Jaffe C. L. J. Biol. Chem. 1997, 272, 30760. [DOI] [PubMed] [Google Scholar]
- Knockaert M.; Gray N.; Damiens E.; Chang Y. T.; Grellier P.; Grant K.; Fergusson D.; Mottram J.; Soete M.; Dubremetz J. F.; Le Roch K.; Doerig C.; Schultz P.; Meijer L. Chem. Biol. 2000, 7, 411. [DOI] [PubMed] [Google Scholar]
- Allocco J. J.; Donald R.; Zhong T.; Lee A.; Tang Y. S.; Hendrickson R. C.; Liberator P.; Nare B. Int. J. Parasitol. 2006, 36, 1249. [DOI] [PubMed] [Google Scholar]
- Mohan R.; Hammers H. J.; Bargagna-Mohan P.; Zhan X. H.; Herbstritt C. J.; Ruiz A.; Zhang L.; Hanson A. D.; Conner B. P.; Rougas J.; Pribluda V. S. Angiogenesis 2004, 7, 115. [DOI] [PubMed] [Google Scholar]
- Sen N.; Banerjee B.; Das B. B.; Ganguly A.; Sen T.; Pramanik S.; Mukhopadhyay S.; Majumder H. K. Cell Death Differ. 2007, 14, 358. [DOI] [PubMed] [Google Scholar]
- Walker R. G.; Thomson G.; Malone K.; Nowicki M. W.; Brown E.; Blake D. G.; Turner N. J.; Walkinshaw M. D.; Grant K. M.; Mottram J. C. PLoS Neglected Trop. Dis. 2011, 5, e1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant K. M.; Hassan P.; Anderson J. S.; Mottram J. C. J. Biol. Chem. 1998, 273, 10153. [DOI] [PubMed] [Google Scholar]
- DeSimone R. W.; Currie K. S.; Mitchell S. A.; Darrow J. W.; Pippin D. A. Comb. Chem. High Throughput Screening 2004, 7, 473. [DOI] [PubMed] [Google Scholar]
- a Nallan L.; Bauer K. D.; Bendale P.; Rivas K.; Yokoyama K.; Horney C. P.; Pendyala P. R.; Floyd D.; Lombardo L. J.; Williams D. K.; Hamilton A.; Sebti S.; Windsor W. T.; Weber P. C.; Buckner F. S.; Chakrabarti D.; Gelb M. H.; Van Voorhis W. C. J. Med. Chem. 2005, 48, 3704. [DOI] [PubMed] [Google Scholar]; b Gelb M. H. Curr. Opin. Chem. Biol. 2007, 11, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri M. P.; Campbell R. K. Future Med. Chem. 2011, 3, 1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetton N.; Rothberg K. G.; Hubbard J. G.; Wise J.; Li Y.; Ball H. L.; Ruben L. Mol. Microbiol. 2009, 72, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Umeyama T.; Wang C. C. PLoS Pathog. 2009, 5, e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G.; Roncal N. E.; Lee P. J.; Leed S. E.; Erath J.; Rodriguez A.; Sciotti R. J.; Pollastri M. P. MedChemComm 2014, 5, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiana S. O.; Pandarinath V.; Wang Z.; Kapoor R.; Ondrechen M. J.; Ruben L.; Pollastri M. P. Eur. J. Med. Chem. 2013, 62, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Gonzalez R.; Kuhlmann F. M.; Galan-Rodriguez C.; Madeira da Silva L.; Saldivia M.; Karver C. E.; Rodriguez A.; Beverley S. M.; Navarro M.; Pollastri M. P. PLoS Neglected Trop. Dis. 2011, 5, e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. S.; Gabernet-Castello C.; Voak A.; Goulding D.; Natesan S. K.; Field M. C. J. Biol. Chem. 2006, 281, 27600. [DOI] [PubMed] [Google Scholar]
- a Barquilla A.; Crespo J. L.; Navarro M. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 14579. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Barquilla A.; Navarro M. Cell Cycle 2009, 8, 697. [DOI] [PubMed] [Google Scholar]; c Barquilla A.; Navarro M. Autophagy 2009, 5, 256. [DOI] [PubMed] [Google Scholar]; d de Jesus T. C.; Tonelli R. R.; Nardelli S. C.; da Silva Augusto L.; Motta M. C.; Girard-Dias W.; Miranda K.; Ulrich P.; Jimenez V.; Barquilla A.; Navarro M.; Docampo R.; Schenkman S. J. Biol. Chem. 2010, 285, 24131. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Saldivia M.; Barquilla A.; Bart J. M.; Diaz-Gonzalez R.; Hall M. N.; Navarro M. Biochem. Soc. Trans. 2013, 41, 934. [DOI] [PubMed] [Google Scholar]
- a Seixas J. D.; Luengo-Arratta S. A.; Diaz R.; Saldivia M.; Rojas-Barros D. I.; Manzano P.; Gonzalez S.; Berlanga M.; Smith T. K.; Navarro M.; Pollastri M. P. J. Med. Chem. 2014, 57, 4834. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wager T. T.; Hou X.; Verhoest P. R.; Villalobos A. ACS Chem. Neurosci. 2010, 1, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers M. J.; Albanesi J. P.; Phillips M. A. Eukaryotic Cell 2007, 6, 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara C. W.; Lee M. C.; Lim C. S.; Lim S. H.; Roland J.; Nagle A.; Simon O.; Yeung B. K.; Chatterjee A. K.; McCormack S. L.; Manary M. J.; Zeeman A. M.; Dechering K. J.; Kumar T. R.; Henrich P. P.; Gagaring K.; Ibanez M.; Kato N.; Kuhen K. L.; Fischli C.; Rottmann M.; Plouffe D. M.; Bursulaya B.; Meister S.; Rameh L.; Trappe J.; Haasen D.; Timmerman M.; Sauerwein R. W.; Suwanarusk R.; Russell B.; Renia L.; Nosten F.; Tully D. C.; Kocken C. H.; Glynne R. J.; Bodenreider C.; Fidock D. A.; Diagana T. T.; Winzeler E. A. Nature 2013, 504, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G.; Karver C. E.; Behera R.; Guyett P. J.; Sullenberger C.; Edwards P.; Roncal N. E.; Mensa-Wilmot K.; Pollastri M. P. J. Med. Chem. 2013, 56, 3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera R.; Thomas S. M.; Mensa-Wilmot K. Antimicrob. Agents Chemother. 2014, 58, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Swinney D. C. Clin. Pharmacol. Ther. 2013, 93, 299. [DOI] [PubMed] [Google Scholar]; b Swinney D. C.; Anthony J. Nat. Rev. Drug. Discovery 2011, 10, 507. [DOI] [PubMed] [Google Scholar]