Abstract
Cryptococcus neoformans is a facultative intracellular pathogen, which can replicate in the acidic environment inside phagolysosomes. Deletion of the enzyme inositol-phosphosphingolipid-phospholipase-C (Isc1) makes C. neoformans hypersensitive to acidic pH likely by inhibiting the function of the proton pump, Plasma Membrane ATPase (Pma1). In this work, we examined the role of Isc1 on Pma1 transport and oligomerization. Our studies showed that Isc1 deletion did not affect Pma1 synthesis or transport, but significantly inhibited Pma1 oligomerization. Interestingly, Pma1 oligomerization could be restored by supplementing the medium with phytoceramide. These results offer insight into the mechanism of intracellular survival of C. neoformans.
Keywords: Cryptococcus neoformans, plasma membrane, inositiol phosphosphingolipid phospholipase C (Isc1), plasma membrane ATPase (Pma1), sphingolipid
1. Introduction
Cryptococcus is an environmental pathogen and the cause of the disease cryptococcosis. Cryptococcus neoformans is the major species causing the disease, although other strains such as Cryptococcus gattii can also cause severe infections. C. neoformans is a facultative intracellular pathogen capable of survival and replication both in the extracellular spaces (with neutral and alkaline pH) and inside the phagolysosomes (with acidic pH).1-3 Sphingolipids play an important role in regulating C. neoformans survival and pathogenicity at these different environments.4, 5 The glucosylceramide synthase 1 (GCS1) gene regulates the synthesis of the lipid glucosylceramide, which confers the fungus the ability to grow at neutral and alkaline pH,4 while the inositol phosphosphingolipid phospholipase C 1 (ISC1) gene plays a significant role in C. neoformans growth in acidic conditions.5, 6 The enzyme Isc1 degrades inositol phosphorylceramide (IPC) and other complex sphingolipids to inositol phosphate and phytoceramide in yeast.5, 7 The absence of Isc1 in C. neoformans leads to longer lag time in growth in acidic pH and decreased survival inside the macrophages as shown in the cryptococcal mutant strain Δisc1.5
Two lines of evidence suggest that the hypersensitivity of C. neoformans Δisc1 strain to acidic pH could be due to inhibition of the function of the proton pump plasma membrane-ATPase1 (Pma1), which is critical for maintaining the intracellular pH within the physiological range.8-10 First, the Δisc1 strain is hypersensitive to the Pma1 inhibiting drug ebselen under acidic conditions;5 secondly, this strain has significantly lower levels of C26-phytoceramide compared to the wild type.6 C26 bound ceramides and fatty acids have been shown to be important in stabilizing newly synthesized Pma1,11-13 and a reduction in their level suggests an alteration in Pma1 function.
Pma1 is a homooligomer synthesized in the endoplasmic reticulum and transferred to the Golgi apparatus before reaching the plasma membrane.14 At the plasma membrane Pma1 is localized in lipid microdomains (i.e. lipid rafts).11, 12, 15, 16 Pma1 functionality is necessary for C. neoformans survival and it has been suggested as a potential antifungal target.5, 10 Previous studies in the model yeast Saccharomyces cerevisiae have shown that efficient function of Pma1 is greatly dependent on sphingolipids. The sphingoid-base of sphingolipids has been shown to play a role in oligomerization and surface stability of Pma1 in S. cerevisiae,17, 18 while lipid chain length plays a role in lipid raft association of the protein.12, 19 However, some conflicting reports exist regarding the surface stability of Pma1 in S. cerevisiae;15, 18 and although it has been shown that sphingolipids synthesis up to IPC is important for the surface delivery of Pma1,11 the role of IPC degradation (regulated by the enzyme Isc1) in Pma1 transport is not clear suggesting the need for further mechanistic studies on the role of lipids in Pma1 function.
In this study, we investigated the role of Isc1 deletion in Pma1 transport and oligomerization in C. neoformans. Our findings show that Isc1 deletion and the consecutive lack of degradation of complex sphingolipids leads to defects in Pma1 oligomerization, which explains the sensitivity to acidic environment that was previously shown.5 These results can have important implications for the mechanisms of intracellular growth and pathogenesis of C. neoformans.
2. Materials and methods
2.1. Strains and growth media
C. neoformans var. grubii serotype A strain H99 (obtained from the Duke University Medical Center, Durham, USA) henceforth referred to as wildtype (WT), C. neoformans Δisc1 mutant and the reconstituted Δisc1REC strain were used in this study. All strains were maintained on yeast extract, peptone, dextrose (YPD) agar plates and grown in yeast nitrogen base (YNB) medium supplemented with amino acids and containing 20 g/L glucose and 25 mM HEPES at 30 °C. The pH of this medium was adjusted to 4.0 or 7.0 using HCl or NaOH, respectively. Phytoceramide supplementation studies were performed using YNB as medium and adding C6-phytoceramide (dissolved in methanol) at desired concentrations.
2.2. Tagging yeast strains with hemagglutinin (HA)
Previous attempts to study Pma1 using antibody was not successful due to lack of strong antibody recognizing C. neoformans Pma1. Therefore, we tagged the PMA1 gene with HA epitope using the following strategy. First, the 5'UTR-PMA1-HA fragment was obtained by PCR using genomic DNA from C. neoformans wild-type H99 as a template and primers PMTAG (5‘-CAT GAG CTC CAC TTT CTT CGG TCG TGC TGC CAC TCT TGT-3’) and PMTAGHA (5‘-CAA GGA TCC CTA AGC GTA GTC TGG GAC GTC GTA TGG GTA CGC CGC GGG CCT GGA GTG GGC ACG GGT-3’). This generated the fragment of 5' UTR-PMA1-HA containing Sac1 and BamH1 restriction site (underlined). This PCR fragment was inserted into pCR2.1-TOPO plasmid and cloned in DH5α. This construct was first sequenced and the 5'UTR-PMA1-HA fragment was released from pCR2.1-TOPTO-5'UTR-PMA1-HA by digesting with Sac1 and BamH1. Secondly, the nourseothricin acetyltransferase gene (NAT1) under the control of H99 actin promoter constructed in pCR2.1-TOPO-NAT120 was digested with Sac1 and BamH1 and the 5'UTR-PMA1-HA fragment was ligated into the plasmid pCR2.1-TOPO-NAT1 generating a pCR2.1-TOPO-5'UTR-PMA1-HA construct. This construct was amplified in DH5α and the extracted plasmid was biolistically delivered into H99 (WT) and Δisc1 mutant. Stable nourseothricin-resistant transformants were selected and southern hybridization with 5'UTR probe was performed to identify a double-crossover event at the PMA1 locus (supplementary Figure 1).
2.3. Confocal microscopy
Confocal imaging was performed as described previously.21 Briefly, cells were grown at pH 4.0 for the specified amount of time and fixed using 2.5 mL of 5x fixation reagent (46 mL of 0.5 M potassium phosphate and 54 mL of formaldehyde) per 10 mL of culture. After 2 hours of shaking, the cells were centrifuged at 1700 g for 5 minutes, resuspended in 1x fixation reagent and left in a shaker incubator at room temperature overnight. The day after, cells were centrifuged at 1700 g for 5 minutes and resuspended in SHA buffer (1 M sorbitol, 100 mM HEPES, 50 mM sodium azide in water, pH=7.5). After washing twice with 1 mL of SHA buffer, cells were counted and 103 cells were resuspended in 0.5 mL of WT buffer (100 mM HEPES, 0.3 M NaCl, 2 mM sodium azide, 10 g bovine serum albumin, 0.2 mL of Tween20 in 200 mL of water) with 8 µg/mL of anti-giantin polyclonal or anti-HA monoclonal (Covance, Princeton, NJ) antibodies, and incubated with shaking at room temperature overnight. The day after, cells were washed with 1 mL of WT buffer for four times and then resuspended in WT buffer with Alexa Fluor® 488-(green) or 633-(red) conjugated anti-rabbit or anti-mouse IgG secondary antibodies (Life Technologies, Carlsbad, CA). Cells were incubated with shaking at room temperature for one hour, washed with 1 mL of WT buffer four times and resuspended in 50 µL of WT buffer. Twenty µL of this suspension were added to a glass slide coated with poly-L-lysine. Five microliters of ProLong® Gold Antifade mounting solution (Life Technologies) were added to the slides and confocal imaging was performed using a LSM 510 META laser-scanning microscope (Zeiss, Jena, Germany). DAPI staining of the nucleus was performed following the above-mentioned procedure for fixing and using a DAPI-containing mounting solution (Life Technologies).
2.4. Extraction of detergent resistant membranes
Detergent resistant membranes (DRM) were extracted following the method of Siafakas et al.22 with minor modifications. Briefly, cells were grown on YPD plates for 72 hours at 30 °C. Five plates of cells were scraped into two centrifuge tubes each containing 20 mL of 0.9% (wt/vol) saline. The cells were vortex mixed and then pelleted by centrifugation for 15 min at 2000 g. The pellet was washed once with saline and once with morpholineethanesulfonic acid (MES)-buffered saline (25 mM MES, 150 mM NaCl, 2 mM EDTA, pH=6.5). The pellet was snap-frozen in liquid nitrogen and resuspended in 3 mL of resuspension buffer (MES-buffered saline, 0.1% (vol/vol) Triton X-100, and a HaltTM protease inhibitor cocktail, Sigma Aldrich, St. Louis, MO). Aliquots of 1 ml were transferred to conical tubes containing 1 mL of glass beads and were homogenized in a MiniBeadbeater-16 cell disrupter (Biospec, Bartlesville, OK) at 4°C for three cycles of 1 min, alternating with a 1-min cooling period on ice. The cell lysate was transferred to centrifuge tubes and centrifuged at 3500 g for 10 min at 4 °C. The supernatant was collected and retained, the pellet was further broken by probe sonication (5 cycles of 10 seconds on, 10 seconds off) and centrifuged at 3500 g for 15 min at 4 °C. This supernatant was combined with the first supernatant, added to a 1 mL thick wall polycarbonate centrifuge tube (Beckman-Coulter, Danvers, MA) and centrifuged at 135000 g for 1 h at 4 °C in an ultracentrifuge (Beckman-Coulter) using a TLA120 rotor. The pellet was resuspended in 200 µL of ice-cold 1% (vol/vol) Triton X-100 in MES-buffered saline, dispersed by a few seconds of probe sonication and held on ice for 1 hour. Then, 113 µL of this solution were added to the bottom of another centrifuge tube and mixed with 170 µL of 68% sucrose in resuspension buffer forming an opaque solution with a final sucrose concentration of 40%. On top of this solution, 1.07 mL of 30% sucrose in resuspension buffer were carefully added forming a distinct clear solution at the top. The tube was centrifuged at 201000 g for 18 h at 4°C. Eleven fractions of 110 µL were collected from the top to the bottom, injected into dialysis bags and dialyzed in water at 4 °C for 20 hours. The contents of the dialysis bags were collected and used for lipid extraction. The top fractions showed increased amount of sterols, glucosylceramide and phospholipids were used for western blotting.
2.5. Lipid and protein extraction
Lipid extraction was performed according to Folch's method.23 Briefly, 700 µL of methanol and 1.4 mL of chloroform were added to each 100 µL of dialyzed fractions and vortex mixed for 30 seconds. Then, 1.1 mL of water were added on top, vortex mixed for another minute and kept at 4 °C for phase separation to occur. The lower phase was aspirated and dried in a centrifugal evaporator (SavantTM SPD 2010 Speed Vac, ThermoScientific, Pittsburgh, PA). The dried lipid was dissolved in 20 µL of chloroform and loaded on thin layer chromatography (TLC) plates. A mobile phase of chloroform/methanol/water (65/25/4 by volume) was used for lipid separation. Lipids were visualized using iodine vapor.
Total protein extraction was performed from an overnight C. neoformans culture grown in YNB. Cells were washed twice and resuspended in PBS. Two hundred µL of lysis buffer (1 mL water, 9 mL 1M Tris pH 8.0, 1.5 mL glycerol and 1% protease cocktail inhibitor) were added to 100 µL of cells in screw-cap tubes. The solution was vortex mixed and disrupted by adding 1 mg of glass beads and disrupting in bead beater for four times (40 seconds each with 1 minute interval on ice). The tube was then centrifuged for 10 minutes at 2500 g at 4 °C. The supernatant was transferred to another tube and protein concentration was determined using a Bradford protein assay kit (Bio-rad, Hercules, CA)
2.6. Western blotting
Western blotting was performed with the top fractions of sucrose density gradient (DRM fractions) or with the total protein extracts of the cells. In both cases, the total amount of protein was estimated and the amount of solution equal to 20 µg of protein was added to a polyacrylamide gel electrophoresis (PAGE) gel. A gel concentration of 10% (with or without sodium dodecyl sulfate) was used for studies with total protein extract with phytoceramide addition, whereas a discrete native gel (6% at the top, 20% at the bottom) was used for DRM fractions to allow for the visualization of the HA band and the marker at the same time. The gel was run for 16 hours at 60 V. The gel was then transferred overnight to a nitrocellulose membrane (Bio-rad). The day after, the membrane was blocked for one hour using 5% non-fat dry milk and rinsed with PBST (1x PBS with 0.05% Tween 20) buffer. The membrane was then incubated with 1000 times diluted anti-HA monoclonal antibody overnight at 4 °C. The day after, the membrane was washed with PBST for five times (five minute each) and then incubated with 10000 times diluted horse-radish peroxidase (HRP)-conjugated IgG antibody (Santacruz Biotechnology, Santacruz, CA) for two hours. The membrane was washed for five times using PBST buffer and the protein band was visualized by addition of the chemiluminescent substrate SuperSignal (ThermoScientific) and visualized using a film developer (SRX 101A, Konica Minolta, Long Island City, NY).
3. Results
3.1. Pma1 is transferred to the plasma membrane even in the absence of Isc1
To understand the role of Isc1 deletion in the growth defect of C. neoformans in acidic pH, the transport of Pma1 to the plasma membrane was studied in the WT and the Δisc1 strain using confocal microscopy. In order to facilitate the imaging of Pma1 protein, the protein was tagged with human influenza hemagglutinin (HA) epitope both strains and antibody against HA was used for imaging and detection of the protein. Western blot analysis confirmed that the HA epitope was successfully added and could be detected in both WT and Δisc1 strains regardless of the pH (supplementary Figure 2).
The transport of Pma1 to the plasma membrane was examined using antibodies against the Golgi specific protein, Giantin24 and Pma1-HA at acidic pH (Figure 1, a and b). In both strains, Pma1 was strongly associated with the plasma membrane as confirmed by a ring-like fluorescent signal from the cell membrane. Such membrane association suggests that the enzyme Isc1 is not essential for the transport of Pma1 to the cell membrane. A similar trend was observed for both strains at neutral pH as the protein could still be observed around the cell membrane; however, at this pH protein localization was not specific to the cell membrane and fluorescence signal could be also observed inside the cell (Figure 1, c and d). Given that the main function of Pma1 is to maintain physiological pH inside the cell, it is likely that at pH= 7 protein level is regulated at the membrane by a slower protein transport or rerouting it to vacuoles for degradation, thus resulting in excess intracellular signal. Although the secretory pathway of Pma1 does not include the nucleus, it has been shown that defects in the intracellular trafficking of this protein in yeasts can lead to stacked membranes that are closely associated with the nucleus, especially at acidic pH.25 This possibility was tested by staining the nucleus with DAPI fluorescent probe and overlaying the fluorescence signal with that of Pma1-HA. Similar to the Giantin-staining, Pma1 was primarily associated with the plasma membrane with little overlap with the nucleus (supplementary Figure 3). These results suggest that Pma1 was efficiently transferred to the cell membrane and its transfer was not affected by the deletion of Isc1.
Figure 1.
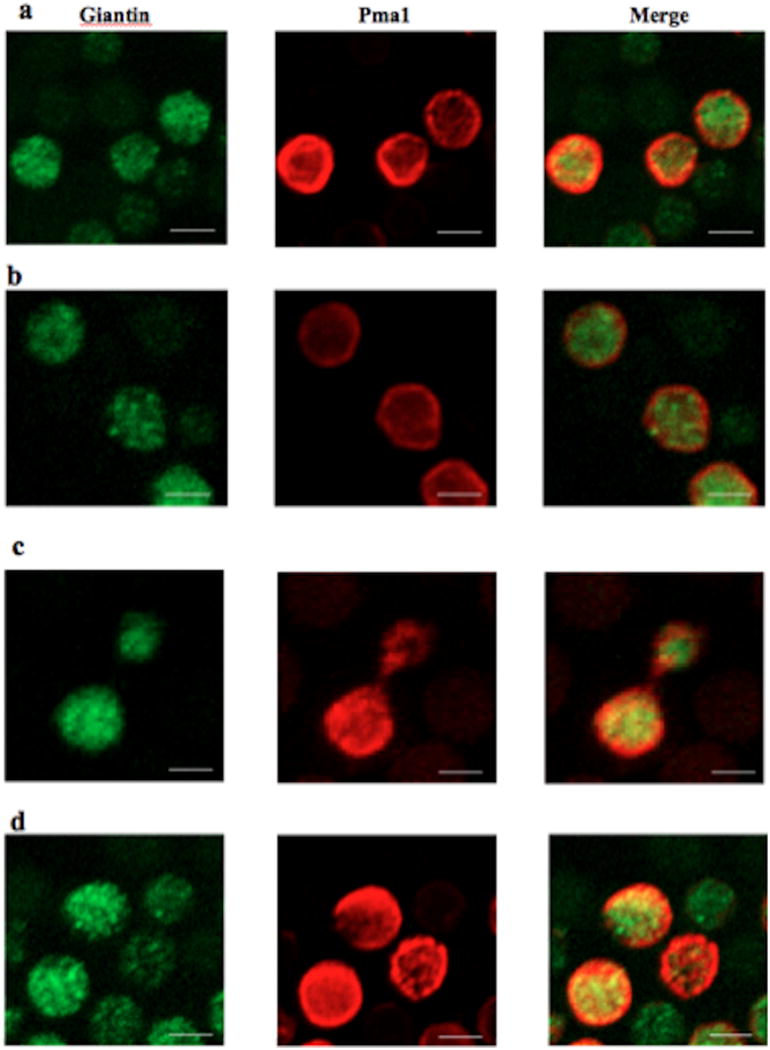
Confocal imaging showing the localization of Pma1 (red) with regard to Giantin, a marker of the Golgi apparatus (green). (A) ISC1-PMA1::HA (WT), pH=4; (B) Δisc1-PMA1::HA, pH=4; (C) ISC1-PMA1::HA (WT), pH=7; (D) Δisc1-PMA1::HA, pH=7. All images were acquired after 2 hours of culture growth followed by fixing and staining. Scale bar= 10 µm.
3.2. Pma1 is produced in higher levels in Δisc1 compared to the WT
Since no difference in the localization of Pma1 was found between the strains, the Pma1 level was examined to understand whether a smaller level of Pma1 has contributed to the pH sensitivity of the Δisc1 strain. Western blot analyses were performed from SDS-PAGE gels at different pH values and time points to examine potential changes in protein level with acidity or time. Interestingly, the mutant strain showed higher levels of Pma1 compared to the WT (Figure 2). This trend was consistent at both acidic and neutral pH and was observed regardless of culture time, although larger protein levels were observed at 16 hours compared to 4 hours. No significant changes were observed in the levels of Giantin or β-actin, indicating that the Golgi apparatus was not affected and also confirming that the differences observed in the levels of Pma1 are not caused by differences in protein loading in the gel.
Figure 2.
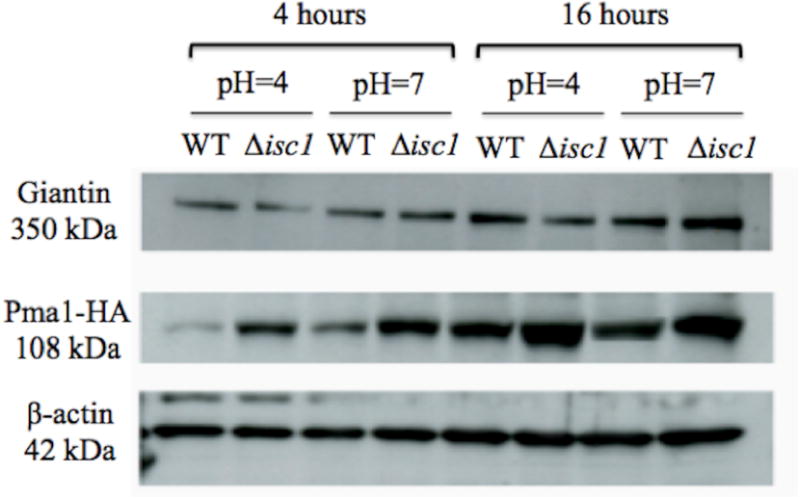
Western blotting of total protein extracts of C. neoformans ISC1-PMA1::HA (WT) and Δiscl-PMA1::HA (Δisc1) using anti-HA monoclonal antibody. The Δisc1 strain showed increased production of Pma1-HA at both acidic and neutral pH after 4 and 16 hours of growth. Giantin and β–actin were used as loading control.
3.3. Pma1 oligomerization is affected by Isc1 deletion
Western blot experiment performed on the SDS-PAGE demonstrated that changes in Pma1 level were not responsible for the pH sensitivity of Δisc1. However, it was likely that the deletion of Isc1 affects protein oligomerization and its functionality at the plasma membrane. Pma1 is a raft-associated protein and is shown to be concentrated in detergent-resistant membrane fractions (DRMs) in other fungi.22, 26 Thus, to study the protein in the plasma membrane, DRMs were separated from C. neoformans following a previously published procedure22 (Figure 3, top panel). Siafakas et al.22 reported that DRM fractions of C. neoformans are mainly composed of ergosterol (Erg), glucosylceramide (GlcCer) and phospholipids such as phosphatidyl choline (PC) and phosphatidyl ethanolamine (PE). A similar lipid profile was observed in the top fractions of the sucrose density gradient in the current study, this similar profile along with the fact that DRMs are known to float on a sucrose density gradient suggested that the top fractions of the gradient are primarily detergent resistant.27
Figure 3.
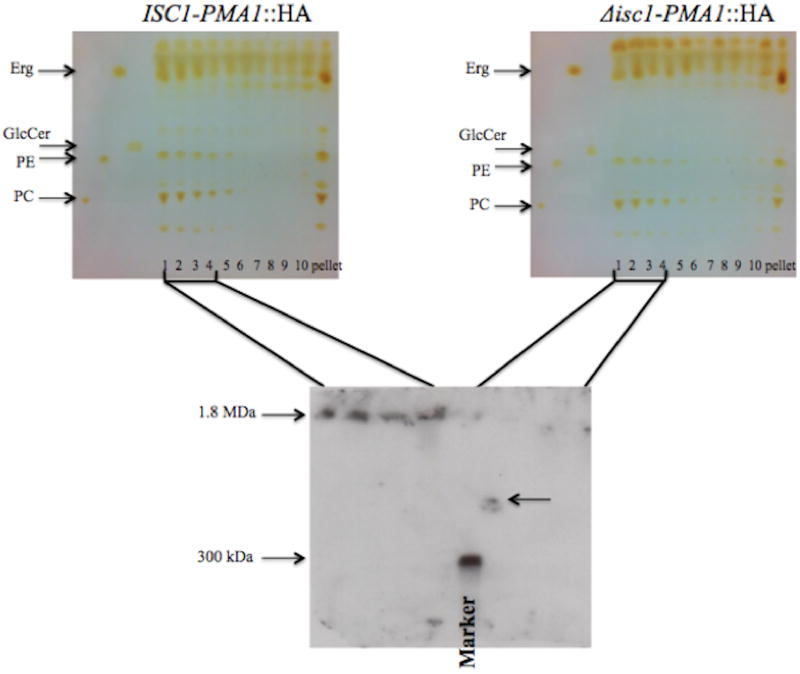
Top panel: Lipid profile of fractions (numbered from top to bottom) obtained by sucrose density gradient ultracentrifugation after cell lysis and exposure to ice cold Triton X-100 to separate the detergent resistant membrane (DRM) fractions. ISC1-PMA1::HA (left) and Δisc1-PMA1::HA (right) show similar lipid profiles in the top (detergent resistant) fractions and their lipid profile is similar to those reported by Siafakas et al.22 PC: phosphatidyl choline, PE: phosphatidyl ethanolamine, Erg: ergosterol, GlcCer: glucosylceramide. Bottom panel: Western blotting of the native-PAGE of the top four fractions of the ISC1-PMA1::HA (left) and Δisc1-PMA1::HA (right). The Δisc1 strain showed substantially lower levels of oligomeric Pma1, a band with a molecular weight lower than the native form in the first fraction of Δisc1 (right to left arrow) confirmed inefficient oligomerization. Marker band= 300 KDa.
The top four fractions of the sucrose density gradient were examined for the native form of Pma1, by performing a Western blot on a native-PAGE from these fractions (Figure 3, bottom panel). The large molecular weight of Pma1 oligomers (1.8 mega Daltons17) caused it to remain at the top of the gel, while staining with the anti-HA antibody ensured that the observed band was indeed Pma1. Interestingly, this experiment revealed a marked difference in the level of Pma1 oligomers between the WT and Δisc1. Observation of a band with a lower molecular weight in the first fraction of Δisc1 revealed that the protein was not in its functional oligomeric form in the mutant and either due to a deficiency in oligomerization or degradation to non-functional forms. This observation suggests that the main difference between the strains is not in the levels of Pma1, but its oligomerization efficiency.
3.4. Pma1 oligomerization can be restored by introducing phytoceramide
The enzyme Isc1 degrades complex sphingolipids into phosphorylinositol and phytoceramide, particularly C26-phytoceramide. It is previously shown that the absence of this enzyme leads to a lack of C26-phytoceramide in C. neoformans in acidic conditions.6 Since the presence of long-chain lipids and fatty acids has been reported to be important in oligomerization of Pma1,11, 12 we examined the effect of reintroduction of phytoceramides into the culture medium on the functional levels of Pma1 at acidic pH. Since long chain lipids cannot be readily metabolized by C. neoformans, this experiment was performed using C6-phytoceramide. Interestingly, supplementation with phytoceramide not only increased the level of monomeric Pma1 (Figure 4-a), but also restored the oligomeric form of Pma1 in a dose-dependent manner (Figure 4-b) showing the importance of phytoceramide in the oligomerization of Pma1.
Figure 4.
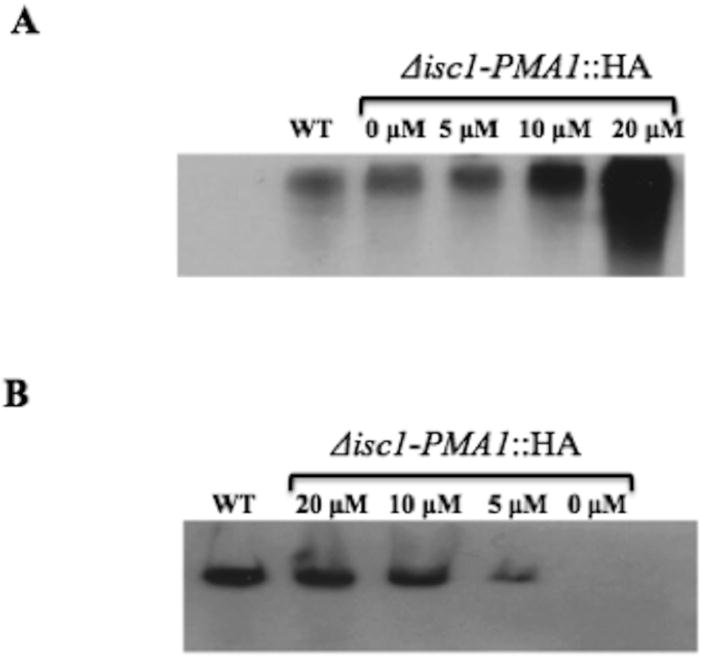
Addition of C6-phytoceramide increased the synthesis and oligomerization of Pma1 in the Δisc1 strain in a concentration-dependent manner. (A) SDS-PAGE of total protein extracts of the C. neoformans ISC1-PMA1::HA (WT) and Δisc1-PMA1::HA after the addition of 5 to 20 µM of phytoceramide using anti-HA monoclonal antibody. (B) Native-PAGE of total protein extracts of the ISC1-PMA1::HA (WT) and Δisc1-PMA1::HA after the addition of 5 to 20 µM of phytoceramide using anti-HA monoclonal antibody. Both gels were performed 2 hours after the addition of phytoceramide and in acidic environment (pH=4), values show the concentration of phytoceramide in the medium.
4. Discussion
The results of this study demonstrate that the enzyme Isc1, which hydrolyzes complex sphingolipids to phytoceramide regulates the function of Pma1 in C. neoformans through affecting Pma1 oligomerization, but without inhibiting protein synthesis or transport. Interestingly, the effect of the lack of enzyme on protein oligomerization could be restored by phytoceramide supplementation suggesting that this lipid plays an important role in Pma1 oligomerization.
Previous studies using S. cerevisiae as model yeast have established the role of sphingolipids in the transport of Pma1.11, 12, 15, 18 Using a S. cerevisiae mutant, deficient in sphingolipids synthesis it was shown that defects in sphingolipids synthesis leads to rerouting of Pma1 to the vacuole.15, 18 Also, using a series of inhibitory compounds, Gaigg et al.11 showed that sphingolipids synthesis up to IPC is required for Pma1 transport to the plasma membrane. Our membrane transport studies revealed that Pma1 was efficiently routed to the plasma membrane in both WT and the Δisc1 strain (Figure 1). Isc1 degrades IPC and other complex sphingolipids, but does not affect sphingolipid synthesis upstream of IPC. Thus, our results reaffirm the findings of Gaigg et al.11 and show that lack of degradation of complex sphingolipids does not affect Pma1 transport. Since the transport of Pma1 was unaffected in the Δisc1 strain, we examined the amount of Pma1 in the WT and the mutant to understand whether there is a difference in the protein level. SDS-PAGE analysis revealed that the mutant in fact contained a larger amount of Pma1 monomers compared to the WT. This phenomenon was observed at both 4 and 16 hours ruling out the possibility of impaired Pma1 synthesis (Figure 2). The mRNA level of the Pma1 gene was higher in the Δisc1 strain compared to the WT at acidic pH, but lower at neutral pH (data not shown). Thus, the increased Pma1 levels observed in the mutant is likely due to higher monomer stability.
The functionality of Pma1 requires efficient oligomerization, delivery to the plasma membrane and association with lipid rafts (tested by the ability of the protein to become detergent resistant). Previous reports suggest that Pma1 becomes associated with lipid rafts either in the ER17 or in the Golgi apparatus,15 before being routed to the plasma membrane. Protein functionality at the plasma membrane was examined by monitoring the amount of oligomeric protein in the lipid rafts using native PAGE analysis. Detergent resistant membrane fractions were separated by exposing cell lysates to ice-cold Triton X-100 and performing sucrose density gradient centrifugation. Top fractions showed an abundance of sterols and PE, PC and glucosylceramide as reported for C. neoformans lipid rafts.22 Analysis of these fractions for the native form of Pma1 revealed a significant reduction in the amount of Pma1 oligomers in the Δisc1 strain. This result suggests that the inability of C. neoformans to degrade complex sphingolipids to phytoceramide inhibits Pma1 oligomerization. This observation explains the growth defect of the Δisc1 strain at acidic pH, which was observed in our previous studies.5, 6
The presence of an intense band with a molecular weight significantly lower than 1.8 MDa confirmed that Pma1 was delivered to the detergent resistant fractions without proper oligomerization. It should be mentioned that the protein band observed in the detergent resistant fractions of the mutant strain corresponds to a multimeric protein (likely a pentamer). The conditions used for the native-PAGE experiment did not allow the observation of monomeric Pma1, due to the significant difference in the molecular weights of the monomeric (108 KDa vs. 1.8 MDa). It is likely that protein in its true monomeric form is not efficient in raft-association, a reduced association of the monomeric form of Pma1 with the detergent resistant membrane fractions of S. cerevisiae at 37 °C has been shown before.17
We have previously shown that the Δisc1 strain contains significantly lower amounts of C26-phytoceramide compared to the WT.6 The reduction of these long chain lipids is the most likely reason for defective oligomerization of Pma1. The importance of sphingolipids in Pma1 oligomerization in S. cerevisiae has been shown using a sphingolipid deficient mutant.17, 18 Since the Δisc1 strain can synthesize the sphingoid base, it is more likely that a specific interaction between phytoceramide and Pma1 is involved in oligomerization. It has also been suggested that long chain sphingolipids can play an indirect role in protein oligomerization by increasing raft formation, which might result in increased local concentration of Pma1 monomers leading to their oligomerization.17 In the current study, no significant differences were observed in the lipid profile of raft fractions between the WT and Δisc1 (Figure 3). Since the lipid composition of DRMs were similar according to TLC analysis, a substantial change in the amount of rafts seems unlikely, which further supports the hypothesis that a specific sphingolipid (i.e. phytoceramide) might be important in Pma1 oligomerization. However, the potential role of the absence of Isc1 on other membrane proteins such Gas1p, Hsp30, Nce2p (shown to be raft-associated by Bagnat et al.28) cannot be excluded; this is especially true for Hsp30, which has been shown to be closely associated with Pma1.29
The importance of phytoceramide in oligomerization of Pma1 was further corroborated by the significant and concentration-dependent improvement in oligomerization when the medium was supplemented with C6-phytoceramide (Figure 4). Although not directly observed due to the conditions of the native-PAGE experiments, it is possible that the addition of C6-phytoceramide also resulted in an increase in the monomeric form of Pma1 since the increase in band intensity of the native-PAGE upon addition of phytoceramide did not correlate with the increase in the band intensity of the SDS-PAGE especially at higher concentrations. Previously, Lee et al.17 showed that supplementation with phytosphingosine (PSH) increases the oligomerization of Pma1 in sphingolipid-deficient S. cerevisiae concluding that the level of sphingolipids in the cell is important for oligomerization. However, the Δisc1 strain is not defective in the sphingoid-base and only lacks the enzyme for the degradation of complex sphingolipids to phytoceramide. Thus, our results suggest that not only the sphingolipid levels; but, specifically the level of phytoceramide is important for Pma1 oligomerization. Further studies are needed to elucidate the interactions between Pma1 monomers and phytoceramide. In conclusion, we showed that deletion of the enzyme Isc1 inhibits the oligomerization of Pma1 resulting in a growth defect at acidic pH in the pathogenic yeast C. neoformans. These results can have important implications in the interactions of C. neoformans with the host especially in acidic media such as inside the phagolysosomes.
Supplementary Material
Highlights.
The role of Isc1 on Pma1 transport and stability in C. neoformans was studied.
Isc1 deletion does not inhibit Pma1 synthesis or its transport to the plasma membrane.
Pma1 oligomerization is inhibited in the absence of Isc1.
Addition of phytoceramide restores Pma1 oligomerization.
Acknowledgments
This work was supported by NIH grants AI56168, AI71142, AI87541 and AI100631 to MDP. Maurizio Del Poeta is Burroughs Wellcome Investigator in Infectious Diseases. We thank Navtej Kaur for help with the technical aspects of some of the experimental work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infection and Immunity. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levitz SM. Cryptococcus neoformans: intracellular or extracellular? Trends in Microbiology. 2001;9:417–418. doi: 10.1016/s0966-842x(01)02137-0. [DOI] [PubMed] [Google Scholar]
- 3.Tucker SC, Casadevall A. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proceedings of the National Academy of Sciences. 2002;99:3165–3170. doi: 10.1073/pnas.052702799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Hennig M, Luberto C, Del Poeta M. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. The Journal of Clinical Investigation. 2006;116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shea JM, Kechichian TB, Luberto C, Del Poeta M. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infection and Immunity. 2006;74:5977–5988. doi: 10.1128/IAI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry J, Guillotte A, Luberto C, Del Poeta M. Characterization of inositol phospho-sphingolipid-phospholipase C 1 (Isc1) in Cryptococcus neoformans reveals unique biochemical features. FEBS Letters. 2011;585:635–640. doi: 10.1016/j.febslet.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- 8.Holyoak C, Stratford M, McMullin Z, Cole M, Crimmins K, Brown A, Coote P. Activity of the plasma membrane H (+)-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Applied and Environmental Microbiology. 1996;62:3158–3164. doi: 10.1128/aem.62.9.3158-3164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano R, Kielland-Brandt MC, Fink GR. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- 10.Soteropoulos P, Vaz T, Santangelo R, Paderu P, Huang DY, Tamás MJ, Perlin DS. Molecular characterization of the plasma membrane H+-ATPase, an antifungal target in Cryptococcus neoformans. Antimicrobial Agents and Chemotherapy. 2000;44:2349–2355. doi: 10.1128/aac.44.9.2349-2355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaigg B, Timischl B, Corbino L, Schneiter R. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. Journal of Biological Chemistry. 2005;280:22515–22522. doi: 10.1074/jbc.M413472200. [DOI] [PubMed] [Google Scholar]
- 12.Gaigg B, Toulmay A, Schneiter R. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. Journal of Biological Chemistry. 2006;281:34135–34145. doi: 10.1074/jbc.M603791200. [DOI] [PubMed] [Google Scholar]
- 13.Toulmay A, Schneiter R. Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast. Biochimie. 2007;89:249–254. doi: 10.1016/j.biochi.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira T, Mason AB, Slayman CW. The yeast Pma1 proton pump: a model for understanding the biogenesis of plasma membrane proteins. Journal of Biological Chemistry. 2001;276:29613–29616. doi: 10.1074/jbc.R100022200. [DOI] [PubMed] [Google Scholar]
- 15.Bagnat M, Chang A, Simons K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Molecular Biology of The Cell. 2001;12:4129–4138. doi: 10.1091/mbc.12.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong X, Chang A. A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface. Proceedings of the National Academy of Sciences. 2001;98:9104–9109. doi: 10.1073/pnas.161282998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MC, Hamamoto S, Schekman R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. Journal of Biological Chemistry. 2002;277:22395–22401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Chang A. Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proceedings of the National Academy of Sciences. 2002;99:12853–12858. doi: 10.1073/pnas.202115499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenkolb M, Zenzmaier C, Leitner E, Schneiter R. A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Molecular Biology of The Cell. 2002;13:4414–4428. doi: 10.1091/mbc.E02-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heung LJ, Luberto C, Plowden A, Hannun YA, Del Poeta M. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. Journal of Biological Chemistry. 2004;279:21144–21153. doi: 10.1074/jbc.M312995200. [DOI] [PubMed] [Google Scholar]
- 21.Rhome R, Singh A, Kechichian T, Drago M, Morace G, Luberto C, Del Poeta M. Surface localization of glucosylceramide during Cryptococcus neoformans infection allows targeting as a potential antifungal. PLoS One. 2011;6:e15572. doi: 10.1371/journal.pone.0015572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryotic Cell. 2006;5:488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folch J, Lees M, Sloane-Stanley G. A simple method for the isolation and purification of total lipids from animal tissues. Journal of Biological Chemistry. 1957;226:497–509. [PubMed] [Google Scholar]
- 24.Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Molecular Biology of The Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Kerchove d'Exaerde A, Supply P, Dufour JP, Bogaerts P, Thinès D, Goffeau A, Boutry M. Functional complementation of a null mutation of the yeast Saccharomyces cerevisiae plasma membrane H-ATPase by a plant H-ATPase gene. Journal of Biological Chemistry. 1995;270:23828–23837. doi: 10.1074/jbc.270.40.23828. [DOI] [PubMed] [Google Scholar]
- 26.Tagliari L, Toledo MS, Lacerda TG, Suzuki E, Straus AH, Takahashi HK. Membrane microdomain components of Histoplasma capsulatum yeast forms, and their role in alveolar macrophage infectivity. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2012;1818:458–466. doi: 10.1016/j.bbamem.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Brown DA. Isolation and use of rafts. Current Protocols in Immunology. 2002:11.10.11–11.10.23. doi: 10.1002/0471142735.im1110s51. [DOI] [PubMed] [Google Scholar]
- 28.Bagnat M, Keranen S, Shevchenko A, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proceedings of The National Academy of Sciences. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braley R, Piper PW. The C-terminus of yeast plasma membrane H+-ATPase is essential for the regulation of this enzyme by heat shock protein Hsp30, but not for stress activation. FEBS Letters. 1997;418:123–126. doi: 10.1016/s0014-5793(97)01359-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


