Abstract
Phosphorylation is often used to promote protein ubiquitylation, yet we rarely understand quantitatively how ligase activation and ubiquitin (UB) chain assembly are integrated with phospho-regulation. Here we employ quantitative proteomics and live-cell imaging to dissect individual steps in the PINK1 kinase-PARKIN UB ligase mitochondrial control pathway disrupted in Parkinson’s Disease. PINK1 plays a dual role by phosphorylating PARKIN on its UB-like domain and poly-UB chains on mitochondria. PARKIN activation by PINK1 produces canonical and non-canonical UB chains on mitochondria, and PARKIN-dependent chain assembly is required for accumulation of poly-phospho-UB (poly-p-UB) on mitochondria. In vitro, PINK1 directly activates PARKIN’s ability to assemble canonical and non-canonical UB chains, and promotes association of PARKIN with both p-UB and poly-p-UB. Our data reveal a feed-forward mechanism that explains how PINK1 phosphorylation of both PARKIN and poly-UB chains synthesized by PARKIN drives a program of PARKIN recruitment and mitochondrial ubiquitylation in response to mitochondrial damage.
Introduction
Protein phosphorylation and ubiquitin (UB) systems function hand-in-hand to control signaling networks (Hunter, 2007). Two common paradigms are: 1) phosphorylation of substrates to promote their ubiquitylation, and 2) phosphorylation of UB ligases to regulate their activity. A major challenge for the field is to understand such systems in a quantitative manner. We rarely know the timing and stoichiometry of phosphorylation of substrates or E3s. Moreover, once the ubiquitylation process is initiated, we rarely understand the dynamics of site selection by E3s on their targets or the kinetics and UB linkage types that are assembled on substrates in vivo (Behrends and Harper, 2011; Kulathu and Komander, 2012). Here, we describe an experimental framework for quantitative analysis of the PINK1-PARKIN-UB pathway, a kinase-driven UB ligase signaling system critical for mitochondrial quality control.
The PARKIN (PARK2) E3 UB ligase is mutated in Parkinson’s Disease (PD), and controls mitochondrial autophagy (reviewed in Youle and Narendra, 2011). PARKIN acts as an effector while the mitochondrial protein kinase PINK1 functions as a sensor of mitochondrial damage. PINK1 is imported into mitochondria via the translocon of the outer membrane (TOM) complex and in healthy mitochondria is rapidly degraded (Jin et al., 2010; Lazarou et al., 2012). However, in damaged mitochondria, PINK1 accumulates on the mitochondrial outer membrane (MOM) where it promotes PARKIN recruitment from the cytoplasm and activation of PARKIN’s E3 activity (Narendra et al., 2008; Youle and Narendra, 2011; Kondapalli et al., 2012; Shiba-Fukushima et al., 2012). Once activated on the MOM, PARKIN promotes dynamic and site-selective ubiquitylation of numerous MOM proteins (Bingol et al., 2014; Sarraf et al., 2013). Despite significant advances, our understanding of how PINK1 activates PARKIN to promote substrate ubiquitylation remains incomplete.
PARKIN is a RING-IBR-RING UB ligases and functions via a so-called RING-HECT hybrid mechanism wherein a catalytic Cys residue in the RING2 domain receives UB from an E2 and transfers it to substrate (Wenzel et al., 2011; Lazarou et al., 2013; Zheng and Hunter, 2013). In the absence of activation, PARKIN exists in an auto-inhibited state (Chaugule et al., 2011; Riley et al., 2013; Trempe et al., 2013; Wauer and Komander, 2013). Two major structural elements contribute to auto-inhibition. First, a domain unique to PARKIN sterically precludes UB transfer from RING1-bound E2~UB (where “~” refers to a thioester intermediate) to the catalytic cysteine in RING2 (C431). Second, a “repressor” domain blocks E2~UB binding to RING1 (Riley et al., 2013; Trempe et al., 2013; Wauer and Komander, 2013). Available data imply that PARKIN undergoes substantial structural re-organization to reach the activated state.
A primary site of PARKIN phosphorylation by PINK1 is serine-65 (S65) (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012; Shiba-Fukushima et al., 2014), which is located in the N-terminal UBL domain, and mutation of this residue to alanine has been reported to delay PARKIN recruitment to mitochondria and reduce ubiquitylation of MOM proteins, although the extent of the phenotype observed is variable in different studies and an absolute requirement for S65 phosphorylation activation has been questioned (Kane et al., 2014; Sarraf et al., 2013; Shiba-Fukushima et al., 2012; Koyano et al., 2014; Shiba-Fukushima et al., 2014). In addition, phosphorylation of PARKIN by PINK1 can stimulate ubiquitin transfer by PARKIN in vitro and in the context of a cell-free depolarized mitochondria assay (Kondapalli et al., 2012; Lazarou et al., 2013). Nevertheless, the precise role of PARKIN phosphorylation and the extent to which phosphorylation of S65 is essential for PARKIN activation remains poorly defined. Recently, a role for direct PINK1 activation by PARKIN phosphorylation has been brought into question by the finding that PINK1 can also phosphorylate the equivalent residue (S65) in UB, and that monomeric p-UB can activate PARKIN’s E3 activity in vitro independent of direct PARKIN phosphorylation by PINK1 (Kane et al., 2014; Kazlauskaite et al., 2014b; Koyano et al., 2014), findings that we independently made in this study. While p-UB can activate PARKIN in vitro and overexpression of UBS65A delays PARKIN recruitment to mitochondria (Kane et al., 2014; Koyano et al., 2014), the precise role of p-UB in PARKIN activation and its relationship to potentially direct activation of PARKIN by PINK1 is murky.
Once activated on mitochondria, PARKIN ubiquitylates a host of MOM proteins but the UB linkage types produced in vivo and the kinetics of this process are poorly understood. UB can undergo chain extension on 7 lysines and the N-terminal methionine, with different fates for different linkage types (Behrends and Harper, 2011; Kulathu and Komander, 2012). As a RING-HECT hybrid, the chain linkage types made by PARKIN are thought to be dictated by PARKIN itself. However, prior studies both in vitro and in vivo have painted a bewildering picture of PARKIN chain assembly, with K27, K48, and K63 linked chains being independently reported on various PARKIN targets or on total mitochondrial proteins, most often using overexpressed UB mutants (in vivo) or fragments of PARKIN not activated by PINK1 (in vitro) that are prone to artifacts (Matsuda, 2006; Rankin et al., 2014; Riley et al., 2013; Chan et al., 2011; Geisler et al., 2010; Sims et al., 2012; van Wijk et al., 2012; Lazarou et al., 2013; Lim et al., 2013; Birsa et al., 2014). Thus, it is unclear the extent to which PARKIN is able to build particular types of UB chains on its substrates in vivo with endogenous UB, the extent to which the activation state of PARKIN determines its chain assembly activity, and whether particular PD patient-derived mutants alter chain linkage specificity.
Here, we use absolute quantification (AQUA)-based proteomics (Kirkpatrick et al., 2005; Phu et al., 2011), live-cell imaging, and in-depth in vitro biochemical studies to deconvolute the PINK1-PARKIN-UB pathway in vivo and in vitro, and to dissect the individual roles of PARKIN and UB phosphorylation. We find that mitochondrial depolarization leads to assembly of multiple UB chain linkage types (K6, K11, K48, and K63) on mitochondria in a manner that requires phosphorylation of PARKIN’s UBL on S65 and PARKIN’s catalytic activity. However, PARKIN phosphorylation on S65 in vivo does not require either PARKIN’s catalytic activity or the ability to stably associate with mitochondria. In vitro, phosphorylation of PARKIN by PINK1 dramatically activates its UB ligase activity and synthesis of K6, K11, K48, and K63 chain linkage types, providing to our knowledge the first example of a RING-HECT hybrid E3 that can intrinsically produce four specific linkage types (Kulathu and Komander, 2012; Ye and Rape, 2009). Moreover, we find that phosphorylation of UB conjugated to mitochondrial proteins requires new UB chain synthesis by phosphorylated PARKIN, indicating a step-wise role for PINK1 in PARKIN activation. Remarkably, PARKIN associates avidly with poly-UB chains only when both PARKIN and poly-UB are phosphorylated on S65, and we demonstrate that PINK1 has the capacity to phosphorylate preformed UB chains. Taken together with the translocation kinetics of PARKIN mutants, these data provide the first direct quantitative evidence of a multi-step feed-forward mechanism involving 1) phosphorylation of PARKIN by PINK1, 2) initial chain synthesis by activated PARKIN on mitochondria, 3) phosphorylation of poly-UB chains on mitochondria by PINK1, and 4) binding of phosphorylated PARKIN to phosphorylated poly-UB chains on mitochondria to retain PARKIN and thus enhance UB chain synthesis and substrate ubiquitylation by PARKIN. This model defines a dual role for PINK1 in activation of the PARKIN-UB system for mitochondrial quality control.
RESULTS AND DISCUSSION
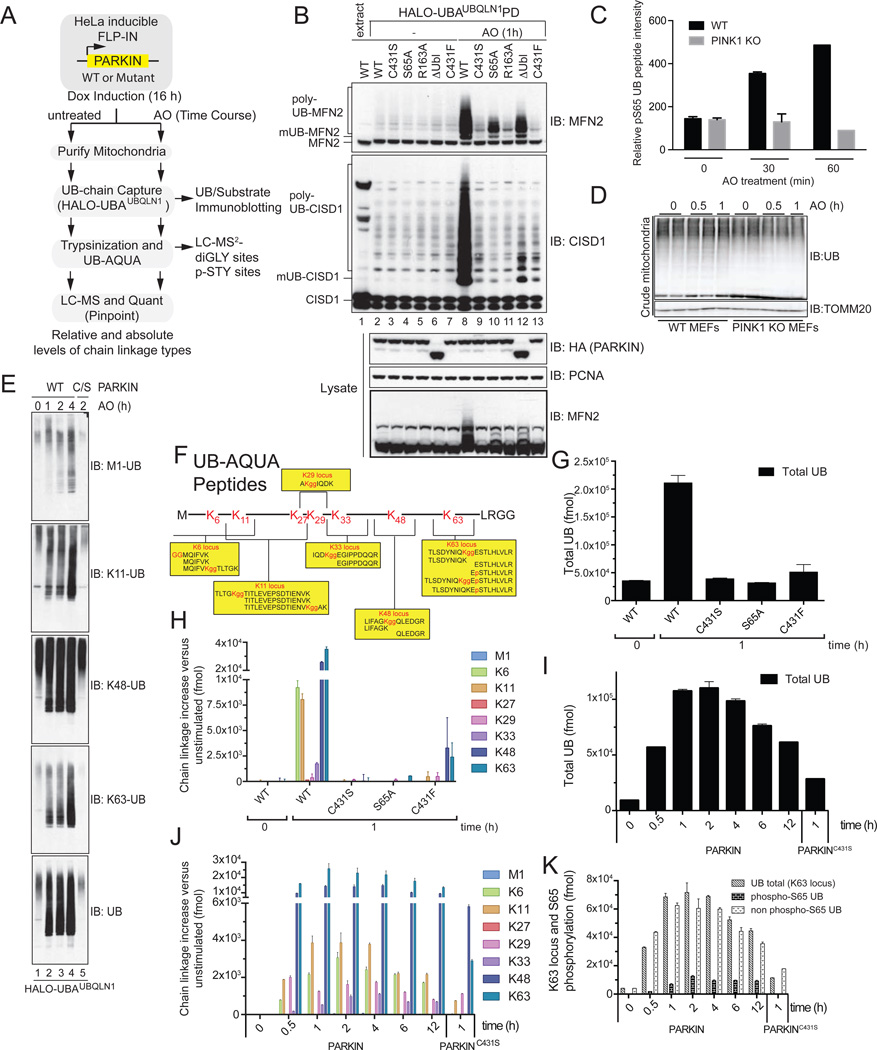
A System for Enrichment of Mitochondrial PINK1-PARKIN Pathway Targets Reveals Depolarization and PINK1-dependent poly-UB Phosphorylation at S65
We initially developed methodology that would allow mitochondrial UB chain linkage types to be identified and quantified. Because PARKIN overexpression affects its functional properties, we integrated a single Doxycycline (Dox) inducible HA-PARKIN construct (or mutants thereof) into PARKIN-negative HeLa Flp-In T-REx cells, thereby allowing for controlled expression (Figure 1A). This HA-tagged PARKIN protein displayed an activity profile indistinguishable from untagged PARKIN when examined for depolarization-dependent MFN2 ubiquitylation (Figure S1A). We then combined this system with sequential mitochondria purification and capture of all UB chain linkage types using 4 tandem immobilized UBA domains from UBQLN1 (Halo-UBAUBQLN1) (Emmerich et al., 2013), greatly increasing the sensitivity of UB chain detection on known PARKIN substrates in response to depolarization with AntimycinA and OligomycinA (AO) (Figure 1A,B). PARKIN C431F and C431S mutants lacking the catalytic Cys were inactive against both CISD1 and MFN2 while PARKINS65A was inactive against CISD1 and displayed residual activity towards MFN2 (Figure 1B). Using LC-MS/MS of these purified fractions, we identified 111 diGLY sites (including 62 previously identified sites) in 36 known MOM PARKIN targets (Sarraf et al., 2013) in response to depolarization with WT PARKIN, but no diGLY-containing peptides from these proteins were detected in cells expressing PARKINC431S (Table S1). Thus, Halo-UBAUBQLN1 provides a means by which to enrich for poly-ubiquitylated MOM proteins in response to depolarization.
Figure 1. Analysis of PARKIN and PINK1-dependent UB chain linkage formation on mitochondria in response to depolarization.
(A) Schematic UB-AQUA proteomics workflow.
(B) Ubiquitylated proteins present on purified mitochondria from HeLa Flp-In T-REx cells after depolarization were captured using immobilized Halo-UBAUBQLN1 prior to immunoblotting with α-MFN2 and α-CISD1.
(C,D) PINK1-dependent phosphorylation of S65 in UB. (C) Mitochondria from WT or PINK1−/− MEFs were subjected to 10-plex TMT at the indicated times post depolarization with AO and the S65 UB tryptic peptide quantified. (D) Total UB levels on mitochondria from Panel C.
(E) Mitochondria from cells subjected to the indicated treatments was used for poly-UB capture using Halo-UBAUBQLN1 and proteins immunoblotted with linkage specific antibodies.
(F) AQUA peptides used to quantify diGLY and phosphopeptides [after (Phu et al., 2011)].
(G–J) UB-AQUA proteomics of total UB (G) and individual UB chain linkage types (H) associated with mitochondria in response to depolarization with AO for 1 h in the presence of the indicated PARKIN protein (G,H) and over time (I,J). Error bars represent triplicate measurements (+/− SEM).
(K) UB-AQUA for total UB (K63 locus) and p-S65 UB accumulation on mitochondria in response to depolarization in the presence of WT or C431S PARKIN, and error bars represent triplicate measurements (+/− SEM).
Given that it has been proposed that PINK1 phosphorylates target proteins to promote PARKIN-dependent ubiquitylation (reviewed in Youle and Narendra, 2011), we searched for phosphopeptides in mitochondrial proteins isolated using immobilized Halo-UBAUBQLN1. While phosphopeptides derived from known PARKIN targets were not identified, we identified a UB peptide containing phospho-S65 (p-S65) (Table S1), indicating that depolarization leads to phosphorylation of UB chains conjugated to MOM proteins. Moreover, a 10-plex tandem mass tagging quantitative proteomic experiment revealed an increase of ~3-fold in p-S65 UB at 30 min and ~5-fold at 1 h post depolarization in PINK1+/+ MEFs but this increase was not observed in PINK1−/− MEFs (Figure 1C,D, S1B–F), indicating that PINK1 is required for phosphorylation of UB. Phosphorylation of S65 in UB by PINK1 was also reported while this paper was in preparation (Kane et al., 2014; Kazlauskaite et al., 2014b; Koyano et al., 2014).
The PINK1-PARKIN Pathway Promotes Canonical and Non-canonical UB Chain Synthesis on Damaged Mitochondria
We next examined the effects of PARKIN activation on the relative abundance of total and distinct UB chains using available chain linkage specific antibodies (Figure 1E), and while changes in the abundance of K11 and K63 chains were discernable at 4 h post depolarization, the non-quantitative nature of the approach and the lack of antibodies for all UB chain linkage types led us to apply an unbiased quantitative proteomic approach referred to as UB-AQUA (Kirkpatrick et al., 2005; Phu et al., 2011) to the problem. In this approach, MS1 peak intensities for experimental (light) tryptic peptides are compared with those of a set of 16 quantified reference (heavy) peptides representing UB itself as well as all 8 diGLY linkage types (Figure 1F, Figure S1G). In addition, we included AQUA peptides for quantification of p-S65 in UB with and without simultaneous ubiquitylation of K63 (K63gg) (Figure 1F, Figure S1G). HeLa Flp-In T-REx cells expressing WT HA-PARKIN or three mutants (C431F, C431S, or S65A) were treated with AO for 1 h, mitochondria isolated, and UB chains captured using Halo-UBAUBQLN1 prior to UB-AQUA proteomics (Figure 1A,G,H). The total abundance of UB on mitochondria was increased ~6-fold upon depolarization (Figure 1G). In contrast, little increase was observed with catalytically inactive PARKIN or with the non-phosphorylatable S65A mutant (Figure 1G), as expected if the increased UB signals observed primarily reflect the activity of the PINK1-PARKIN pathway. The increased UB content was primarily composed of canonical (K48 and K63) and non-canonical (K6 and K11) UB chains, with little evidence of M1, K27 and K29 chains, and required both the catalytic residue C431, and the S65 phosphosite (Figure 1H, S1H). We identified the same chain linkage types and a similar magnitude of UB chain synthesis in cells expressing untagged PARKIN depolarized with carbonyl cyanide m-chlorophenyl hydrazine (CCCP) for 1h and with more highly purified mitochondria after depolarization with AO (Figure S1J). Mitochondrial ubiquitylation was maximal by 1 h post-depolarization, and gradually decreased over 4–12 h (Figure 1I), and the chain linkage types were largely unchanged over time (Figure 1J, S1I). Thus, PARKIN promotes the synthesis of canonical and non-canonical poly-UB chains on depolarized mitochondria in a manner that depends upon its catalytic Cys residue and S65 within its UBL domain.
PARKIN Activity is Required for PINK1-dependent Poly-UB Phosphorylation in Response to Mitochondrial Depolarization
We next examined the stoichiometry of poly-p-S65-UB (poly-p-UB) on purified mitochondria in response to AO using AQUA. p-UB was essentially undetectable in association with mitochondria in the absence of depolarization, but accumulated to ~10% and ~18% of the total UB level at 1 and 2 h post-depolarization, and was maintained at ~20% of the total mitochondrial UB abundance over the 12h time course (Figure 1K). The doubly-modified p-S65/K63gg peptide represented ~5% of the total K63 locus found over the same time course (Figure S1L), indicating that p-S65 and K63gg can exist on the same molecule but are not enriched in the population. Accumulation of poly-p-UB on mitochondria in response to AO required active PARKIN (Figure 1K), despite the presence of significant pre-existing K48 and K63-linked chains under these conditions (Figure 1K, S1I), as discussed further below.
We also determined the abundance of p-UB in total cellular UB, obtained after collapsing all UB chains to monomers with USP2. Because the major tryptic S65-containing peptide identified by LC-MS/MS is a partial trypsinization product (residues 55–72) likely reflecting an inhibitory effect of phosphate at S65 (+2 position) on cleavage, we used both the partial and fully (residues 64–72) trypsinized AQUA peptides for analysis (Figure 1F, S1G). The amount of p-S65 UB we detected is ~2 orders of magnitude higher than that reported previously using only the residue 64–72 phosphopeptide as reference with endogenous PINK1 (Koyano et al., 2014). The total amount of p-S65 almost doubled (to 2.5%) 1h post AO and this increase was essentially eliminated in PINK1−/− HeLa Flp-In T-REx cells made by gene editing (Figure S1M,N).
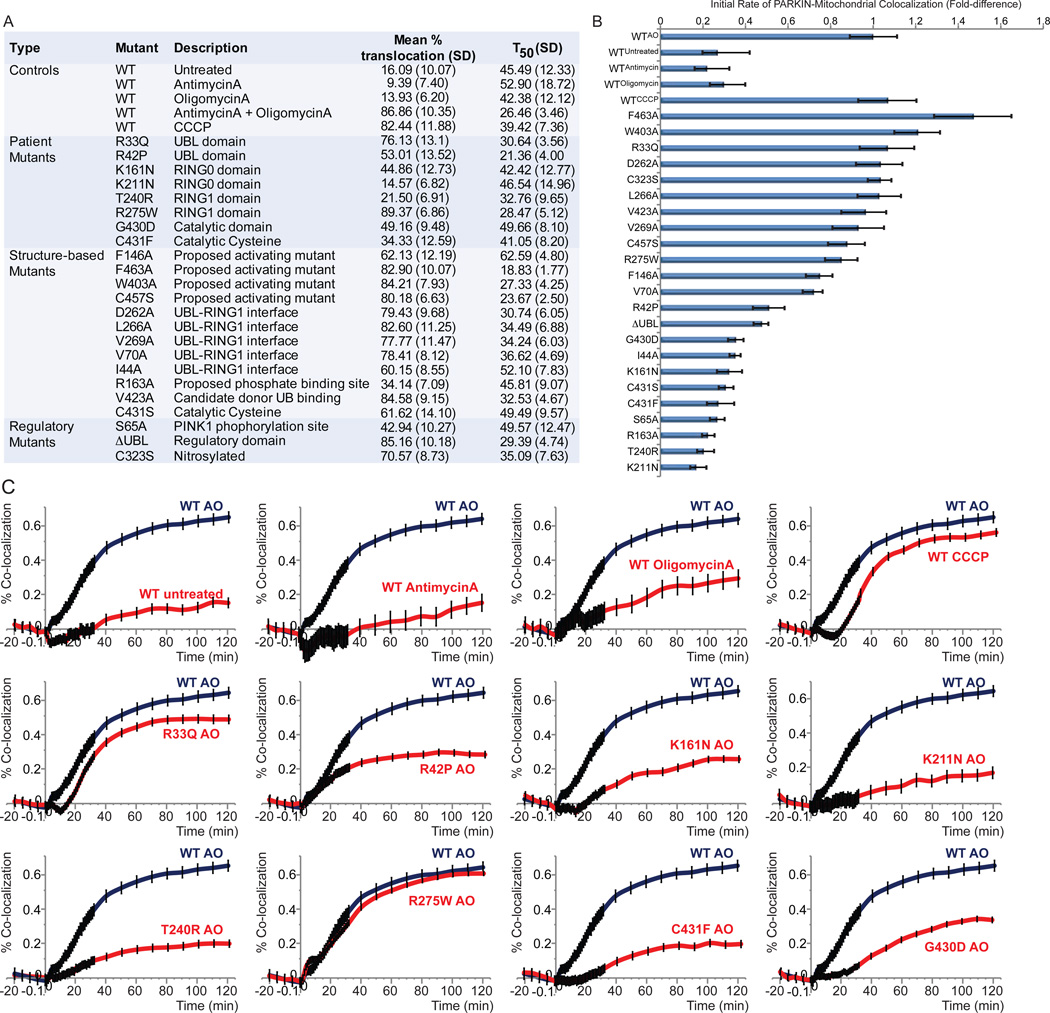
UB Chain Assembly on Depolarized Mitochondria by PD Patient and Structure-based Mutants Correlates with Rates of PARKIN Translocation
Patient-derived PARKIN mutants display diverse translocation and activity-related phenotypes (reviewed in Youle and Narendra, 2011). However, it is unclear the extent to which these mutants affect UB chain synthesis or linkage specificity and how any defects in assembly of UB chains is linked with defects in translocation. To address this question, we used a newly developed live-cell based PARKIN translocation assay (see Supplemental Experimental Procedures and Figure S2A–C) to examine the translocation kinetics of a large panel of PD patient and structure-based mutants (Figure 2A), and profiled a subset of mutants for alterations in chain synthesis properties using UB-AQUA. WT GFP-PARKIN could be seen accumulating on mitochondria as early as ~15 min post-depolarization with AO (Figure 2A–C and Movie S1), and displayed a half-time (T50) to maximal localization of ~26 min, consistent with previous studies using CCCP as depolarizing agent (Narendra et al., 2010; Trempe et al., 2013). As expected (Lazarou et al., 2013; Trempe et al., 2013), catalytically defective GFP-PARKINC431S displays greatly reduced translocation rates, approaching that seen with WT PARKIN in the absence of depolarization (Figure 2A,B and Movie S2). Similar defects were seen with several other PARKIN mutants, including S65A, ΔUBL, K161N, K211N, T240R, and G430D (Figure 2A–C, S2B). Two proposed activating mutations (F463A and W403A) (Trempe et al., 2013) slightly accelerated translocation, as previously observed (Figure 2B).
Figure 2. Systematic analysis of depolarization-dependent mitochondrial translocation of patient and structure-based PARKIN mutants in HeLa cells.
(A) Mutants analyzed and the rate constants for translocation, mean % translocation, and half-time (T50) for translocation (described in Supplemental Methods). Greater than 300 HeLa Flp-In T-REx WT or mutant GFP-PARKIN cells were imaged for 20 min prior to either no treatment or depolarization with AO, AntimycinA or OligomycinA, and cells imaged every minute for 30 min and every 10 min up to 2 h.
(B) Relative initial rates of translocation are derived from the approximated slope of averaged co-localization traces for 0–30 min post depolarization (see Figure S2).
(C) Plot for percent coincidence of GFP-PARKIN and mitochondria in response to depolarization with AO (>300 cells). Error bars represent (+/− SEM).
See also Figure S2.
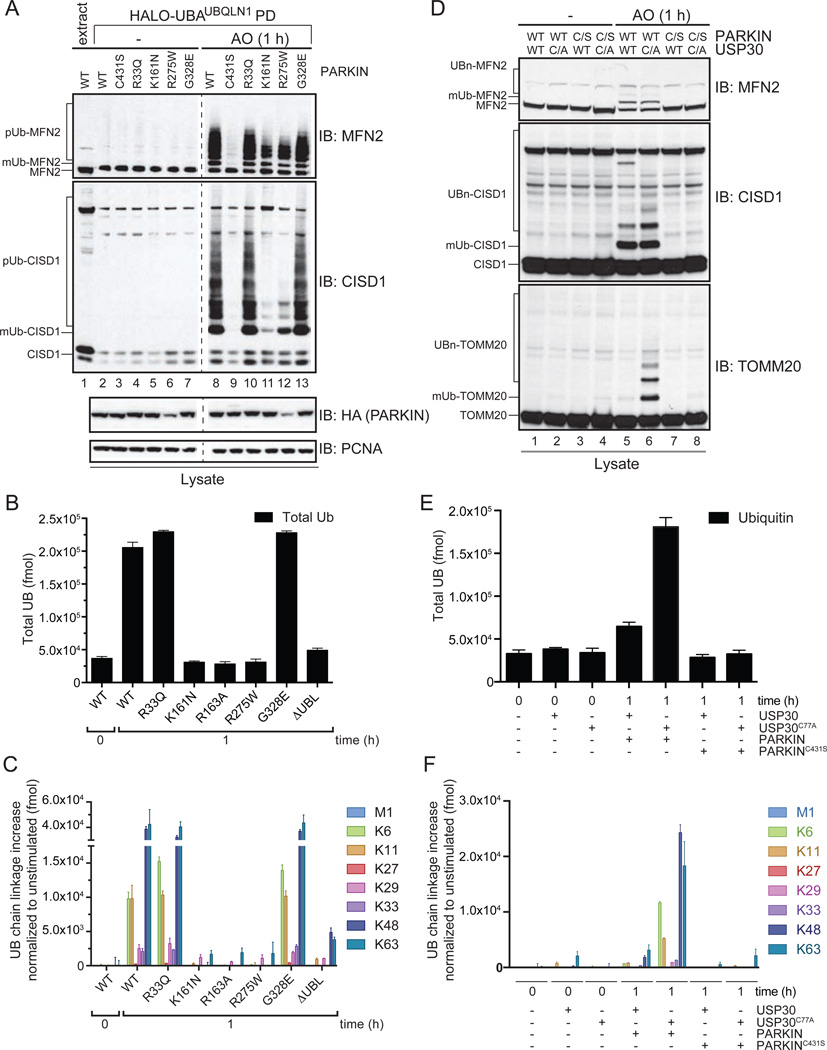
We next selected 6 mutants for mitochondrial UB chain linkage analysis in vivo. R33Q and G328E mutants displayed near WT UB chain synthesis activity and linkage specificity by AQUA, as well as when examined for modification of CISD1 and MFN2 substrates by western blotting (Figure 3A–C). In contrast, the translocation-defective proteins R163A, ΔUBL, K161N displayed greatly reduced chain synthesis on both mitochondria and on CISD1 and MFN2 (Figure 1A, Figure 3A–C, Figure S3A), indicating that translocation is required for efficient substrate ubiquitylation. The behavior of PARKINR275W was unique in that it is efficiently recruited to mitochondria (Figure 2A) but fails to promote UB chain synthesis (Figure 3B,C), suggesting a defect in catalytic activity (see below).
Figure 3. Mitochondrial UB chain synthesis by patient and structure-based PARKIN mutants and reversal by USP30.
(A) MFN2 and CISD1 ubiquitylation by PARKIN mutants in response to depolarization was performed as described in Figure 1B.
(B,C) Total UB (B) and the normalized UB chain linkage increase (C) on purified mitochondria was determined for cells expressing the indicated PARKIN proteins after depolarization with AO (1h). Error bars represent triplicate measurements (+/− SEM).
(D) Ectopic USP30 expression reverses PARKIN and depolarization-dependent TOMM20 ubiquitylation in HeLa cells. HeLa Flp-In T-REx PARKIN (WT or C431S, C/S) cells ectopically expressing WT or catalytically defective (C77A, C/A) USP30 were either left untreated or depolarized with AO for 1 h prior to immunoblotting of lysates.
(E,F) Effect of ectopic USP30 on the total abundance of mitochondrial UB (E) and the normalized UB chain linkage increase measured by AQUA (F). Error bars represent triplicate measurements (+/− SEM).
See also Figure S3.
UB Chain Linkage Type Independent Reversal of Depolarization-dependent Mitochondrial Poly-UB by USP30
Ectopic expression of USP30 can reverse ubiquitylation of a subset of PARKIN substrates, including TOMM20, thereby attenuating PARKIN function and mitophagy (Bingol et al., 2014). However, the ability of USP30 to remove multiple chain linkage types assembled by PARKIN has not been investigated systematically. As expected, expression of WT USP30, but not a catalytically inactive mutant (C77A), reversed TOMM20 ubiquitylation, but did not affect MFN2 or CISD1 ubiquitylation (Figure 3D, S3B). This finding is consistent with a significant level of target specificity for USP30 (Bingol et al., 2014). In the presence of USP30C77A, depolarization led to the expected increase in total UB associated with mitochondria (Figure 3E, S3C). However, expression of WT USP30 largely reversed this increase in ubiquitylation, independent of both chain linkage type and p-S65 phosphorylation (Figure 3E,F, S3C,D). Thus, despite lack of activity toward a subset of PARKIN substrates, USP30 appears, at least when overexpressed, to promiscuously remove UB chains of various linkage types from a major population of mitochondrial PARKIN substrates and does not appear to be influenced by the presence of p-UB within chains in vivo.
Direct Activation of PARKIN by PINK1 Promotes Synthesis of Canonical and Non-Canonical UB Chains in vitro
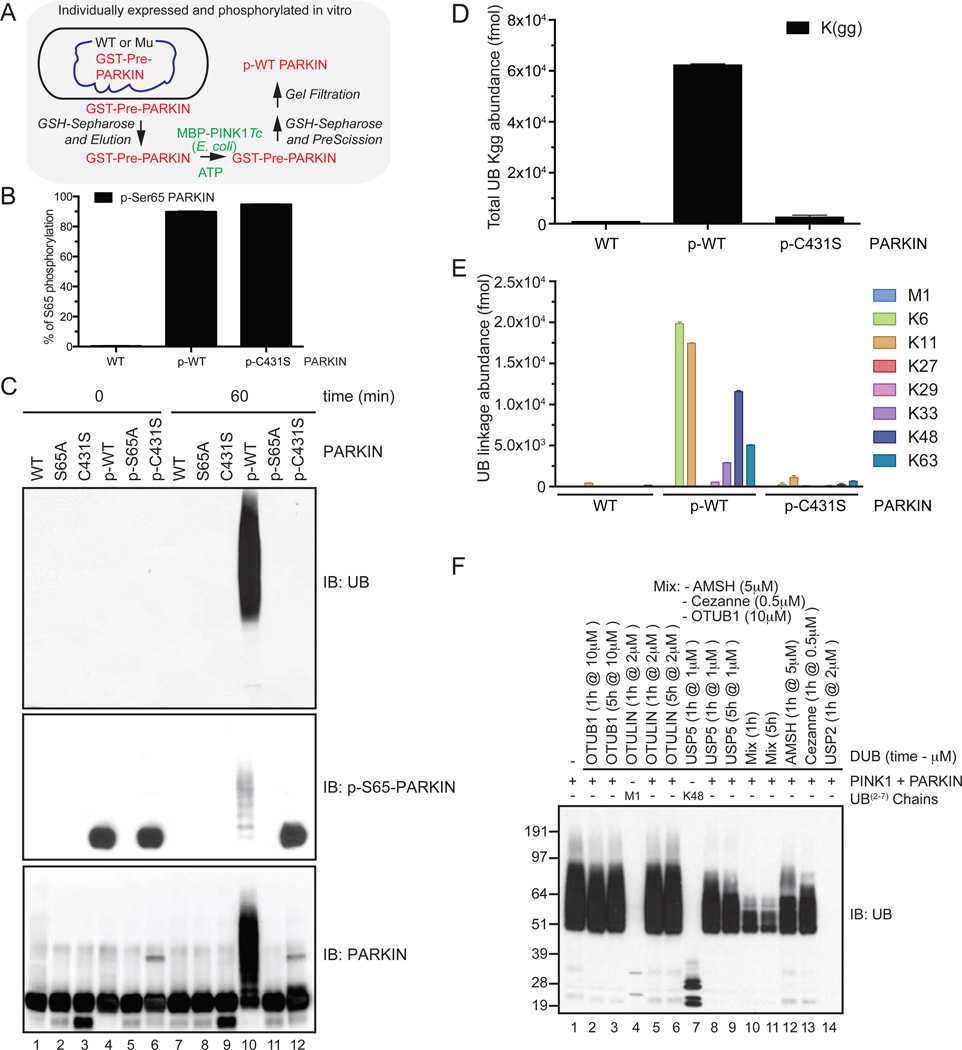
Our unexpected finding that PARKIN promoted the assembly of 4 types of UB chain linkages on mitochondria in vivo raised the question of whether PARKIN itself can promote multiple chain linkage types. We therefore sought to systematically and quantitatively examine the chain linkage types produced by PARKIN in vitro. The vast majority of papers examining PARKIN’s UB ligase activity in vitro have used forms of PARKIN that are not activated by PINK1, often employing fragments of PARKIN that lack the UBL domain phosphorylated by PINK1 (Chaugule et al., 2011; Lim et al., 2013; Matsuda, 2006; Trempe et al., 2013; Wauer and Komander, 2013), and in cases where PINK1-dependent activity has been monitored with purified proteins, the reported stoichiometry of phosphorylation at S65 in PARKIN was very low (0.08) (Kazlauskaite et al., 2014a). Thus, the relationship between the activities measured in previous studies and that of near stoichiometrically phosphorylated PARKIN, including the types of UB chains assembled, remain poorly defined.
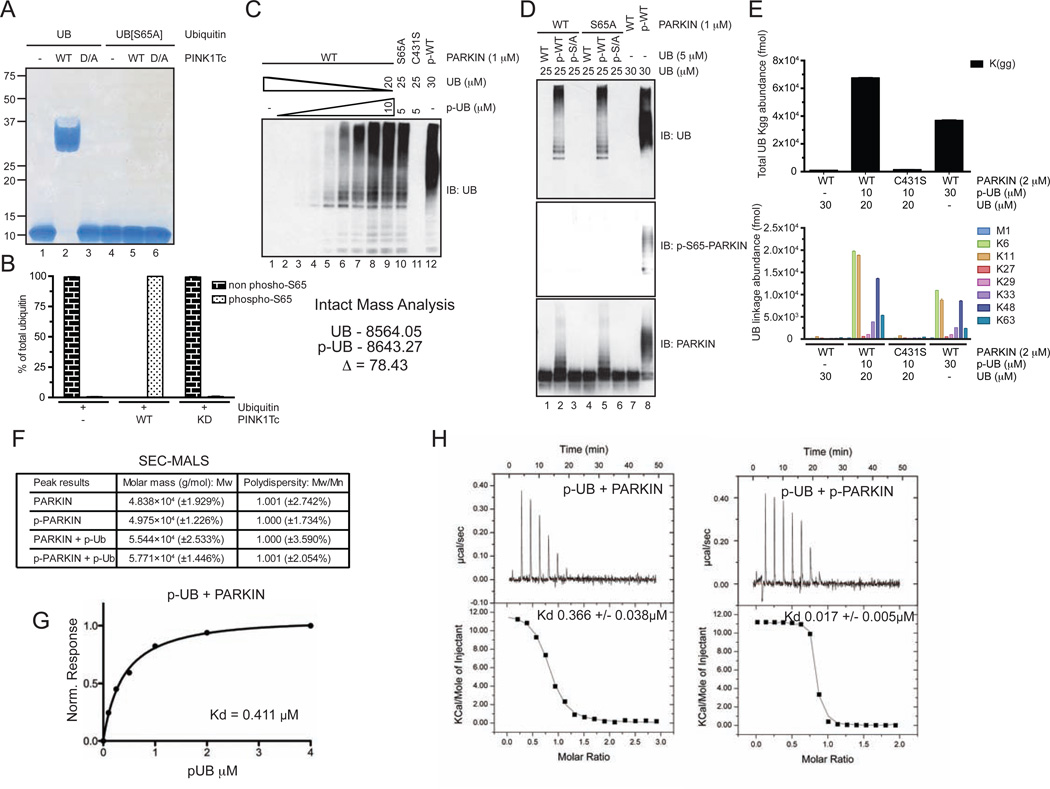
We therefore developed a robust method for production of near stoichiometrically phosphorylated Sf9 cell-derived PARKIN using Tribolium castaneum PINK1 (PINK1Tc, produced in bacteria (Kondapalli et al., 2012) and then removed PINK1Tc (Figure 4A, and Supplemental Experimental Procedures, Method 1). S65 phosphorylation was verified using an antibody produced by single chain variable fragment phage display (Koerber et al., 2013) (Figure 4C, Figure S3B and Supplemental Experimental Procedures) and the stoichiometry of S65 phosphorylation was 0.9 based on AQUA proteomics (Figure 4B). We did not detect phosphorylation of other sites in the purified p-PARKIN by LC-MS/MS (data not shown). For simplicity, throughout the paper we refer to PARKIN proteins that have been treated with PINK1Tc and ATP, and subsequently purified away from PINK1Tc, as p-WT, p-S65A, and p-C431S PARKIN. While unphosphorylated PARKIN proteins were devoid of UB chain synthesis activity (Figure 4C, lanes 7–9), p-WT PARKIN synthesized robust UB conjugates, and this activity absolutely required the catalytic Cys residue and S65 in the UBL domain (Figure 4C, lanes 10–12). UB chain synthesis occurred at least partially on p-WT PARKIN, as revealed by probing reaction products with α-PARKIN and α-p-S65 antibodies (Figure 4C).
Figure 4. UB chain synthesis by PINK1-phosphorylated PARKIN in vitro.
(A) Schematic for preparation of PINK1Tc-phosphorylated PARKIN free of PINK1Tc.
(B) The stoichiometry of PARKIN phosphorylation at S65 was determined by AQUA proteomics.
(C) UB chain synthesis by WT, S65A, or C431S PARKIN with or without phosphorylation by PINK1Tc followed by removal of PINK1Tc was measured by immunoblotting with α-UB antibodies. Control blots were probed with α-PARKIN or α-p-S65 (AVI-25) (see Extended Experimental Procedures).
(D,E) UB-AQUA of total UB linkages (Kgg) (D) and individual linkage types (E) for reaction mixtures (45 min) from the indicated PARKIN proteins.
(F) UB chain editing of PARKIN ubiquitylation products assembled in vitro. DUBs were incubated with UB chains assembled by p-WT PARKIN and the products analyzed by immunoblotting. As controls, linear or K48 UB chains were digested with the indicated DUBs.
See also Figure S4.
p-WT PARKIN displayed a dramatic increase in diGLY linkage assembly when compared with both unphosphorylated PARKIN and p-C431S PARKIN and produced primarily K6, K11, K48, and K63 linkages in vitro (Figure 4D,E, S4C), although the preference for synthesizing K48 and K63 chains in vitro was relatively lower than their production on mitochondria in vivo. Similar results were observed with PARKIN purified from bacteria and activated by PINK1Tc (Figure S4D). To validate these results, we performed a UB chain editing analysis (Mevissen et al., 2013) using chain-specific DUBs (Figure 4F). Treatment of UB chains produced by p-WT PARKIN with Cezanne (K11 specific) or AMSH (K63 specific) resulted in significant loss of conjugates as detected by immunoblotting, whereas OTUB1 (K48 specific) had a more modest effect and OTULIN (M1 specific) had no obvious effect on UB chain density. A mixture of OTUB1, Cezanne, and AMSH disassembled the majority of UB chains. The remaining UB conjugates may represent either K6 chains or mono-ubiquitylation of PARKIN itself, as this signal is lost completely upon treatment with the non-specific DUB USP2 (Figure 4F). Moreover, partial reduction of the UB immunoblot signal with USP5 indicates that a substantial fraction of the PARKIN reaction products are free UB chains (Figure 4F). Several different E2s have been proposed to be important for PARKIN-dependent mitophagy in vivo or chain assembly in vitro (Lazarou et al., 2013; Hasson et al., 2014; Geisler et al., 2014), leading us to test their ability to activate PARKIN in vitro and the chain linkage types produced. The E2 enzymes UBE2D2, UBE2E1, UBE2J2, and UBE2L3 – but not UBE2N/UBE2V1 or UBE2S (a result distinct from that reported by Kazlauskaite et al., 2014a) – promoted the robust assembly of K6, K11, K48 and K63 chains (Figure S4E–G), consistent with the idea that chain specificity is independent of the E2 used to charge PARKIN’s active site cysteine. Thus, p-WT PARKIN produces canonical and non-canonical chain linkages in vitro.
Activation of PARKIN by p-UB in vitro Promotes Assembly of Canonical and Non-canonical UB chains
Having identified PINK1-dependent phosphorylation of S65 in UB in vivo (Figure 1C, S1C–F, and Table S1), we addressed the effect of this modification on PARKIN activity in vitro. UB or UBS65A was treated with WT or kinase dead (D359A) PINK1Tc, and only WT UB underwent a mobility shift on phos-tag gels, reflecting phosphorylation (Figure 5A). Purified PINK1Tc-phosphorylated UB was validated by intact mass spectrometry as a single major species with a mass of 79.43 Da larger than unmodified UB, reflecting a single phosphorylation event, and by AQUA proteomics, the stoichiometry of S65 phosphorylation was 99.9% (Figure 5B). We will refer to PINK1-phosphorylated UB as p-UB and PINK1-treated UBS65A as p-UBS65A. We found that adding homogeneous p-UB, but not p-UBS65A, led to robust in vitro UB chain synthesis by PARKIN (either WT or S65A) in the absence of PINK1, with maximum activation at 2.5–5 µM p-UB and produced K6, K11, K48, and K63 linked chains based on AQUA analysis of reaction products (Figure 5C–E). While the ability of p-UB to activate WT PARKIN was reported while this manuscript was in preparation (Kane et al., 2014; Kazlauskaite et al., 2014b; Koyano et al., 2014), our results differ from that of Kazlauskaite et al., 2014b in that they concluded that for p-UB to fully activate PARKIN, phosphorylation of PARKIN on S65 is required. The basis for this discrepancy is not clear, but could reflect the low stoichiometry of in vitro PARKIN S65 phosphorylation in prior studies. In addition, the majority of chains elicited by p-UB appeared to be unanchored chains, whereas p-WT PARKIN appears to preferentially self-modify as indicated by gel mobility (Figure 5D). When p-UB is the sole source of UB in the reaction, the relative linkage abundance of chains produced is similar to that seen in the presence of UB but the rates of chain synthesis are 2-fold lower (Figure 5E), indicating that p-UB is not used as efficiently as UB in chain building. While UB and the UBL domain of PARKIN have ~30% identity and ~50% similarity in sequence homology (Figure S1C), we found that p-UBL has a relatively low ability compared to p-UB in activating PARKIN ligase activity in vitro (Figure S5A), indicating that they cannot substitute efficiently for each other. We also found that p-UBL domain from PARKIN as well as p-UB weakly activate PARKINΔUBL in trans (Figure S5B), suggesting that full activation preferentially occurs when the UBL is contiguous with the catalytic domains of PARKIN. Our data indicate that the ability of PARKIN to generate canonical and noncanonical chain linkages is independent of activation mode
Figure 5. UB chain synthesis by and biophysical analysis of p-UB-PARKIN complexes in vitro.
(A) UB or UBS65A was incubated with PINK1Tc and reaction products analyzed using Phos-Tag gel electrophoresis.
(B) Stoichiometry of S65 phosphorylation was determined by AQUA. The intact mass of purified p-UB is also shown.
(C,D) p-UB but not p-UBS65A activates unphosphorylated PARKIN UB ligase activity in vitro. The concentrations of p-UB used in panel C were 0, 0.1, 0.25, 0.5, 1, 2, 3.5, 5, 10 µM and PARKIN was 1 µM. The total UB concentration was 30 µM.
(E) p-UB promotes the assembly of K6, K11, K48, and K63 chains by unphosphorylated PARKIN.
(F) Inactive and active forms of PARKIN are monomeric and PARKIN binds a single molecule of p-UB. Mass determination was performed by SEC-MALS.
(G) Binding of p-UB (99% phosphorylated) to PARKIN was measured using Bio-layer interferometry.
(H) Isothermal calorimetry of the indicated protein pairs.
See also Figure S5.
PARKIN phosphorylation by PINK1 promotes formation of a heterodimeric complex with p-UB
Previous studies suggest that PARKIN can self-associate on mitochondria (Lazarou et al., 2013). In order to examine whether phosphorylation of PARKIN itself or of UB contribute to activation through self-association, we examined the interaction between different combinations of phosphorylated and unphosphorylated forms of PARKIN and UB. In order to avoid artifacts of mixed protein populations due to low phosphorylation stoichiometry, we prepared PARKIN and UB that were 99% phosphorylated on their respective S65 residues based on AQUA proteomics (Figure 5B and S5C). Size-exclusion chromatography-multi-angle light scattering (SEC-MALS) demonstrated that both unphosphorylated PARKIN and p-PARKIN from Sf9 cells are monomeric (48.4 KDa and 49.7 KDa, respectively; expected PARKIN mass is 51.4 KDa) (Figure 5F, S5D). Moreover, addition of p-UB to either species led to a ~7 KDa increase in mass, consistent with 1:1 association of PARKIN or p-WT PARKIN and p-UB (Figure 5F, S5D). Thus, activation of PARKIN by phosphorylation or by p-UB binding does not lead to overt PARKIN oligomerization in vitro. PARKIN associates with p-UB with a Kd of ~400 nM, with similar values obtained by isothermal calorimetry and bio-layer interferometry (366 and 411 nM, respectively) (Figure 5G,H), but did not detectably bind UB (data not shown). While previous structural analysis of auto-inhibited PARKIN led to the hypothesis that the side-chains of K161, R163, and K211 form a potential phosphate-binding site in the UPD domain (Figure S5E) (Wauer and Komander, 2013), these residues are not individually required for binding to p-UB (Kd~140–158 nM; Figure S5F). Remarkably, p-WT PARKIN displayed a 21-fold increase in affinity for p-UB (Kd~17 nM) (Figure 5H), indicating that the binding site for p-UB is more readily accessible when PARKIN is phosphorylated. Thus, activated PARKIN can exist as a monomer and associates with p-UB in a manner that is greatly stimulated by phosphorylation of its UBL on S65.
Rates of PARKIN-dependent UB Chain Synthesis via UB-AQUA Proteomics
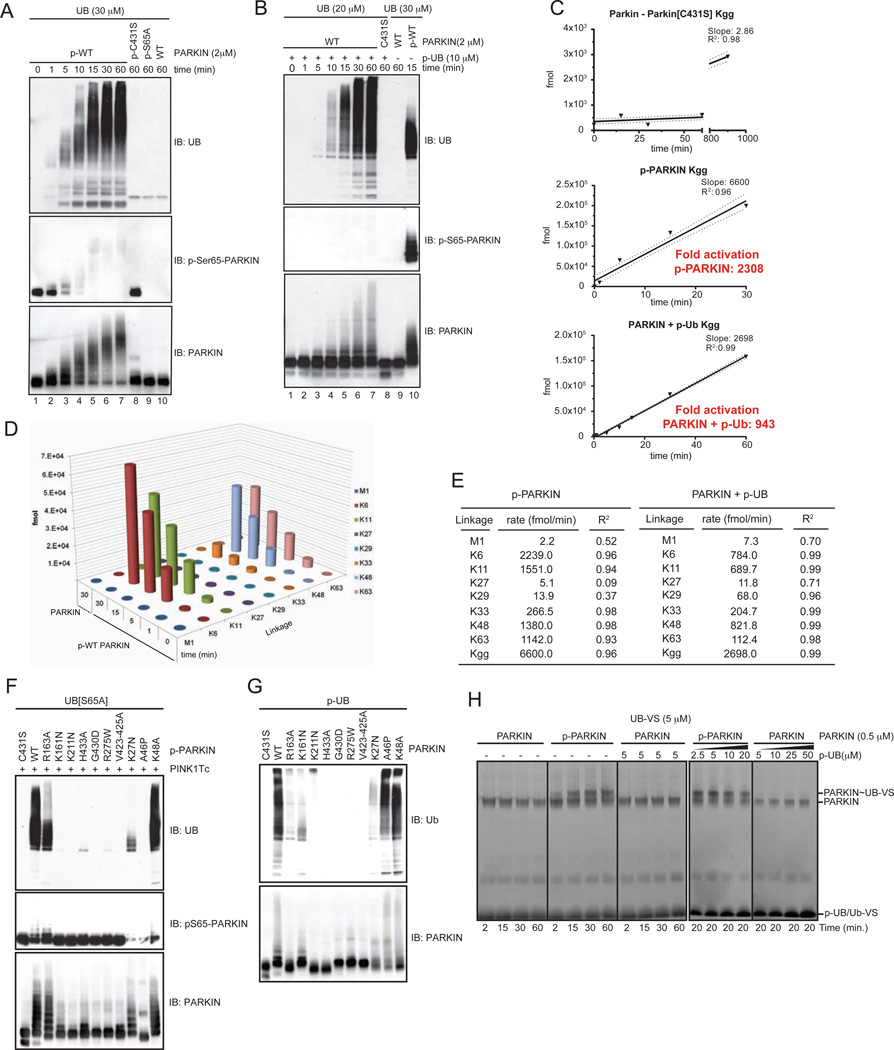
We next sought to rigorously compare UB chain synthesis by p-PARKIN and p-UB-activated PARKIN. p-WT PARKIN (purified from PINK1 and with a phosphorylation stoichiometry of 90%, Figure 4B) and unphosphorylated PARKIN activated by homogeneous p-UB both synthesized UB chains in a time-dependent manner, as observed by immunoblotting with α-UB antibodies, whereas p-C431S and S65A mutants treated with PINK1 in the same manner was p-WT PARKIN did not (Figure 6A,B). Given the inherent limitations in comparing UB ligase activities using immunoblotting, we developed a quantitative methodology based on UB-AQUA to compare the initial rates of UB chain synthesis for each chain linkage type simultaneously (Figure 6C–E, S6A–E). This analysis revealed several features of PARKIN activation. First, the formation of individual chain linkage types were linear over the 60 min time course (Figure S6C–E), allowing ligation rates to be rigorously determined for each linkage type (Figure 6E). Second, by carefully analyzing background chain synthesis rates (Figure 6C, S6B), we determined the absolute fold activation afforded by phosphorylation of PARKIN on S65 (2308-fold activation) versus activation of unphosphorylated PARKIN by p-UB (943-fold activation), as measured by the cumulative rates of each chain linkage (Figure 6C, S6B). Third, the rates of K6, K11, and K48 synthesis by p-WT PARKIN were similar and about 3-fold higher than that observed for K63 synthesis (Figure 6E). The rate constants for p-UB activated PARKIN were 1.5–10-fold lower than with p-PARKIN, depending on the linkage type (Figure 6E, S6C–E). Thus, while p-UB is capable of activating PARKIN in trans, the extent of activation is significantly lower than with PINK1-activated PARKIN across canonical and non-canonical chain linkages.
Figure 6. Kinetic analysis of activated PARKIN via AQUA proteomics.
(A,B) UB chain assembly by the indicated p-PARKIN proteins (90% phosphorylated) (panel A) or unphosphorylated PARKIN associated with p-UB (99% phosphorylated) was analyzed by immunoblotting.
(C,D) Samples from panels A and B were subjected to UB-AQUA proteomics (Figure S6B–E). Background reaction rates as determined in Extended Experimental Procedures were used to calculate the fold activation for p-WT PARKIN and for p-UB-PARKIN based on the sum of all di-GLY linkages. In panel D, the histogram shows the fmol of each UB linkage identified for p-WT PARKIN versus unphosphorylated PARKIN.
(E) Rates of synthesis of each chain linkage type determined by UB-AQUA (see Figure S6D,E for primary data).
(F) The indicated PARKIN mutants were phosphorylated with PINK1Tc (as determined by blotting with α-p-S65 (AVI-25) and subjected to UB chain synthesis reactions using UBS65A, thereby avoiding activation of PARKIN by p-UB. Reaction mixtures were subjected to immunoblotting with α-UB antibodies.
(G) The indicated PARKIN mutants in an unphosphorylated form were incubated with p-UB (99% phosphorylated) and UB chain synthesis reactions performed. Reaction products were immunoblotted with α-UB antibodies.
(H) PARKIN (with or without p-UB) or p-WT PARKIN (99% phosphorylated (Figure S5C)) was incubated with UB-VS for the indicated times and reaction products examined by SDS-PAGE and stained with SYPRO-Ruby.
See also Figure S6.
Differential Activation of PD Patient and Structure-based PARKIN Mutants by Direct Phosphorylation and by p-UB
Patient-derived PARKIN mutants display diverse phenotypes with respect to mitochondrial translocation and UB chain synthesis on mitochondria (reviewed in Youle and Narendra, 2011) (Figure 2 and Figure 3), but the extent to which these phenotypes reflect PARKIN or UB phosphorylation is unknown. We found that K161N, K211N, G430D, R275W, and K27N mutants were largely inactive in chain assembly regardless of the mode of activation, while the ubiquitylation site mutant K48A (Sarraf et al., 2013) was fully active regardless of the mode of activation (Figure 6F,G, S6F). The structure-based mutant R163 displayed partial activity when activated directly with PINK1 but was not efficiently activated by p-UB, while the opposite was found with the patient mutant A46P (Figure 6F,G). All PARKIN proteins, except K27N and A46P located in the UBL domain, were efficiently phosphorylated by PINK1Tc in vitro (Figure 6F). Thus, the inability of PINK1Tc to directly activate PARKINA46P likely reflects poor phosphorylation of S65, but this defect can be by-passed by p-UB, analogous to the finding with PARKINS65A (Figure 5D). Thus, PARKIN mutants display common and distinct activation properties in vitro.
PARKIN Phosphorylation, but not p-UB, Activates Catalytic Cysteine Reactivity in vitro
Several prior observations suggested that PINK1 may be linked to reactivity of PARKIN’s active site C431, but whether this reflected PARKIN phosphorylation by PINK1 or an independent step remained unclear. For example, PARKINC431S undergoes depolarization-dependent and PINK1-dependent formation of a stable oxy-ester with UB in human cells as well as in a cell free system (Lazarou et al., 2013; Zheng and Hunter, 2013). Furthermore, the catalytic C431 is buried due to an autoinhibitory conformation of bacterial PARKINΔUBL, which is inactive (Wauer and Komander, 2013), and that mutationally destroying autoinhibitory intramolecular protein interactions can lead to increased reactivity of the active site cysteine as measured using UB-vinyl sulfone (UB-VS) which makes a covalent adduct with the activates catalytic cysteine of PARKIN (Riley et al., 2013; Wauer and Komander, 2013). We found that unphosphorylated full-length PARKIN also failed to react with UB-VS, consistent with auto-inhibition due to burial of the active site (Figure 6H). In contrast, PINK1-phosphorylated PARKIN rapidly reacted with UB-VS to form a more slowly migrating species in a manner that depends on C431 (Figure 6H, S6G), indicating that phosphorylation of PARKIN’s UBL is sufficient to increase the reactivity of the active site cysteine. However, addition of saturating levels of p-UB to unphosphorylated PARKIN failed to promote reaction of UB-VS with the active site cysteine and did not block reaction of UB-VS with phosphorylated PARKIN (Figure 6H). These data suggest that PARKIN phosphorylation and p-UB binding activate PARKIN in vitro by distinct mechanisms.
PINK1 Promotes PARKIN Association with Poly-UB Chains by Phosphorylating Both PARKIN and Poly-UB
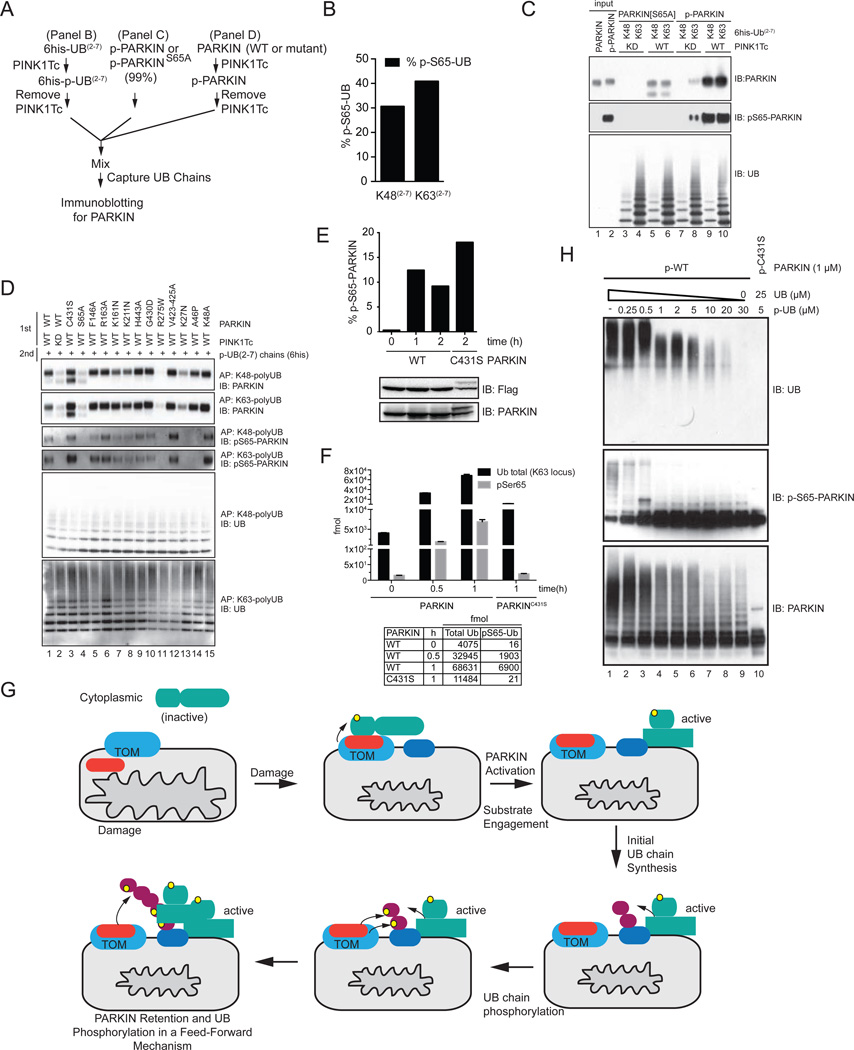
The finding that phosphorylation of PARKIN’s UBL by PINK1Tc promotes binding to monomeric p-UB (Figure 5H) together with a report that PARKIN associates with K63-linked UB chains in cell extracts through an unknown mechanism (Zheng and Hunter, 2013) led us to examine whether PARKIN could associate with UB chains directly in vitro and whether phosphorylation of PARKIN or UB chains by PINK1Tc promoted the interaction. Treatment of 6his-tagged K48 or K63 poly-UB(2–7) chains with PINK1Tc led to S65 phosphorylation with a stoichiometry of 0.3 and 0.4, respectively (Figure 7A,B), indicating that PINK1Tc can phosphorylate poly-UB chains in addition to monomeric UB. After removing PINK1Tc, poly-p-UB chains were incubated with homogeneous p-WT PARKIN or p-S65A PARKIN and captured through the 6his-tag. Co-association was examined by immunoblotting for PARKIN (Figure 7A,C). Treating S65A PARKIN with PINK1Tc did not lead to association with poly-UB chains, although the mutant associated weakly with poly-p-UB chains (Figure 7C; compare lanes 3 and 4 with 5 and 6). In contrast, p-WT PARKIN associated weakly with poly-UB K63 chains (lane 8) but showed a dramatic increase in association with poly-p-UB K48 and poly-p-K63 chains (Figure 7C, lanes 9 and 10). Thus phosphorylation of S65 on PARKIN’s UBL greatly stimulates PARKIN’s ability to associate with phosphorylated poly-UB chains, and phosphorylation of poly-UB promoted association with both unphosphorylated PARKIN and p-WT PARKIN. These results are consistent with the finding that phosphorylation of PARKIN promotes binding to monomeric p-UB as revealed by isothermal calorimetry (Figure 5H). Moreover, among our panel of PARKIN mutant proteins, there was a strong correlation between the ability to be phosphorylated by PINK1 on S65 and the ability to associate efficiently with poly-p-UB chains (Figure 7D). Thus, efficient association of PARKIN with UB chains requires phosphorylation of both PARKIN and UB on S65 by PINK1. In addition, this data indicates that particular mutants that are normally unable to stably localize on mitochondria in response to depolarization or build chains on mitochondria (Figures 2 and 3) are nevertheless able to bind efficiently to poly-p-UB chains, suggesting that their defect in translocation occurs upstream of UB chain binding.
Figure 7. PINK1-dependent phosphorylation of PARKIN’s UBL domain promotes association with PINK1-phosphorylated poly-UB chains.
(A) Scheme for examining phosphorylation-dependent binding of PARKIN to UB chains.
(B) Stoichiometry of PINK1Tc-dependent phosphorylation of K48 and K63 UB(2–7) chains used in panel C and D measured by AQUA proteomics.
(C) Phosphorylation dependent binding of PARKIN to phosphorylated UB chains. p-WT and p-S65A PARKIN were incubated with the indicated poly-UB chains as shown in panel A and UB chains captured with Ni-NTA beads prior to immunoblotting. The p-WT PARKIN used in this experiment was 99% phosphorylated on S65 as determined by AQUA (Figure S5C).
(D) Binding of a panel of PD patient and structure-based mutants of PARKIN to poly-p-UB chains. PARKIN proteins were phosphorylated with PINK1Tc, the PINK1Tc removed, and the proteins incubated with K48 and K63 UB(2–7) chains previously phosphorylated with PINK1Tc and the PINK1Tc removed (panel A). UB chains were captured and association with PARKIN examined by immunoblotting.
(E) Phosphorylation of WT PARKIN and PARKINC431S on S65 in response to depolarization was measured by AQUA proteomics.
(F) Absolute levels of UB and p-S65 UB on mitochondria from HeLa Flp-In T-REx cells expressing WT PARKIN or PARKINC431S, in the presence or absence of depolarization measured by AQUA proteomics (derived from primary data in Figure 1K).
(G) A feed forward model for PARKIN activation and retention on the mitochondrial surface in response to depolarization. See text for details.
(H) Effect of p-UB (99% phosphorylated) on UB chain synthesis by p-WT PARKIN (99% phosphorylated). The total UB concentration was 30 µM.
Phosphorylation of PARKIN on S65 by PINK1 Occurs Independently of Stable Localization on Mitochondria
While it is clear that PINK1 promotes PARKIN phosphorylation at S65 on its UBL and that PARKINC431S can undergo PINK1-dependent oxy-ester formation with UB in cells and in a cell-free system (Lazarou et al., 2013), whether this reflects PINK1-dependent phosphorylation of S65 in PARKIN and the order of this modification with respect to localization and UB chain assembly has not been investigated previously. We employed AQUA proteomics to determine the extent of PARKIN S65 phosphorylation in response to depolarization. Cells expressing WT or C431S PARKIN were depolarized, PARKIN was then immunoprecipitated, and the extent of S65 phosphorylation in the PARKIN UBL assessed by AQUA proteomics (Figure 7E). As expected, WT PARKIN was found to be phosphorylated on S65 and as much as 10–15% of PARKIN was phosphorylated 1–2h post-depolarization; we consider this a lower limit, as the extent of dephosphorylation during isolation is unknown. Remarkably, we found that an even larger fraction of PARKINC431S (18%) was phosphorylated on S65 (Figure 7E) despite its inability to stably localize on mitochondria (Figure 2B,C). A comparable fraction of PARKINC431S was shifted to a slower mobility form indicative of formation of an oxy-ester with UB, as demonstrated to occur previously in response to depolarization (Lazarou et al., 2013; Zheng and Hunter, 2013). Thus, stable association of PARKIN with mitochondria in vivo can be dissociated from the ability of activated PINK1 to phosphorylate S65 in PARKIN, indicating that PARKIN phosphorylation occurs upstream of its stable association with mitochondria, and upstream of UB chain synthesis on mitochondria.
A Feed-forward Model of PARKIN Activation and Translocation to Depolarized Mitochondria
Taken together, these results demonstrate a feed-forward mechanism for PARKIN activation and its subsequent accumulation on mitochondria (Figure 7G). We posit that cytoplasmic PARKIN present in an auto-inhibited form is continuously sampling the mitochondrial surface via a diffusion-controlled process. When PARKIN encounters activated PINK1 associated with the TOM complex on damaged mitochondria (Lazarou et al., 2012; Sarraf et al., 2013), it becomes phosphorylated on S65 (Figure 7E), leading to activation of its UB chain assembly function by more than 2000-fold based on our quantitative analysis of chain assembly in vitro (Figure 6C). We further posit that this activation event promotes the initial ubiquitylation of local substrates (Sarraf et al., 2013), possibly including TOMM20, which has been recently suggested to be a critical PARKIN substrate for mitophagy (Bingol et al., 2014). As the density of UB chains increases on mitochondria, PINK1 can then promote phosphorylation of these newly assembled chains (see below). This, in turn, promotes strong association of S65-phosphorylated PARKIN with poly-p-UB chains and retention on the mitochondria allowing phosphor-PARKIN and PINK1 to cooperatively amplify further the signal (Figure 7G).
Key findings that frame our model are based on our quantitative analysis of UB chains on mitochondria in response to depolarization (Figure 7F). In the absence of depolarization, we detected 4,075 fmol of poly-UB on mitochondria of which 16 fmol was phosphorylated on S65. After 1h of depolarization, the total abundance of poly-UB on mitochondria increased to 68,631 fmol of which 6,900 fmol was phosphorylated on S65. In striking contrast, cells expressing PARKINC431S display a small increase in total poly-UB chains on mitochondria in response to depolarization (11,484 fmol), but there was essentially no increase in the extent of UB phosphorylation (21 fmol of p-S65 UB) (Figure 7F). These data provide strong evidence that PINK1-dependent UB phosphorylation in HeLa cells is negligible on pre-existing mitochondrial poly-UB chains. Instead, we find that p-S65 UB accumulates in a manner that correlates with extensive PARKIN-dependent UB conjugation on mitochondria.
Our model is distinct from those proposed in previous studies (Kane et al., 2014; Kazlauskaite et al., 2014b; Koyano et al., 2014) in that it does not involve a major direct role for p-UB in activation of PARKIN’s E3 activity but instead posits a primary role for poly-p-UB chains in recruitment of activated PINK1-phosphorylated PARKIN. We found that at equimolar levels of homogeneous p-UB and p-WT PARKIN (in excess of their Kd), the UB chain assembly activity remained essentially unchanged (Figure 7H). However, increasing the ratio of p-UB to unphosphorylated UB is inhibitory to chain assembly by p-WT PARKIN (Figure 7H). This, together with our finding that phosphorylation of PARKIN on S65A is required for UB chain synthesis in vivo (Figure 1H) but p-UB fully activates PARKINS65A in vitro (Figure 5D), is inconsistent with the idea that accumulation of monomeric p-UB would promotes PARKIN activation in vivo.
Concluding Remarks
A key outcome of our model is that highly active phosphorylated PARKIN becomes localized on poly-p-UB chains through a feed-forward process. This feature could explain the ability of PARKIN to ubiquitylate dozens of proteins on the mitochondrial surface with no apparent substrate specificity (Sarraf et al., 2013). Our model is consistent with all but one of the patient and structure-based PARKIN mutants analyzed here, the exception being PARKINR275W, which translocates efficiently to mitochondria but fails to build UB chains. However, previous studies have shown that this mutant binds tightly to substrates and is apparently not released efficiently from MOM substrates (Johnson et al., 2012), potentially explaining its unusual biochemical behavior. An unanswered question is whether free p-UB is generated in vivo at endogenous PINK1 levels, and if so, whether it contributes to PARKIN activation or UB chain synthesis given the much higher concentration of unphosphorylated UB in cells. Indeed, our data suggest p-UB is not used as efficiently as unphosphorylated UB to build chains in vitro (Figure 5E) and is inhibitory to p-WT PARKIN at high concentration (Figure 7H). In addition, although we define PARKIN’s ability to generate canonical and non-canonical chain linkages in vivo and in vitro, we do not know whether PARKIN also generates mixed chains, akin to those produced in the IL-1 pathway (Emmerich et al., 2013), but with potentially several different linkage types, and a variety of phosphorylations all on a single substrate. Further studies are also needed to determine whether UB phosphorylation may affect other steps of the UB cascade, including chain disassembly by DUBs, although this was not seen here in the context of USP30.
Feed-forward loops are frequently used in the context of kinase driven transcriptional circuits. However, to our knowledge, this is the first demonstrated feedforward mechanism involving phosphorylation-driven activation of a UB cascade. This mechanism could be important for controlling both the timing of PARKIN recruitment to mitochondria and subsequent steps of mitophagy, and could be under negative control by USP30 through antagonism of PARKIN-dependent UB chain assembly (Bingol et al., 2014). Future studies showing the structural basis of PARKIN binding to poly-p-UB are required to mechanistically uncouple initial PARKIN activation from the poly-p-UB feedforward step. We anticipate that the power of AQUA proteomics to deconvolute individual steps in complex phosphorylation-driven UB cascades, as demonstrated here, will be useful for understanding additional intertwined signaling-ubiquitylation pathways in the future.
EXPERIMENTAL PROCEDURES
Mitochondrial UB AQUA proteomics
Inducible HeLa Flp-In T-REx cells expressing the PARKIN proteins were depolarized with Antimycin A (10 µM) and Oligomycin A (5 µM), and isolated mitochondria used for poly-UB capture with immobilized Halo-UBAUBQLN1 prior to AQUA proteomics with a library of 13C/15N-labeled reference peptides (see Figure S1G) (Phu et al., 2011). Ubiquitylation site identification by mass spectrometry was performed as described (Kim et al., 2011; Sarraf et al., 2013).
PARKIN translocation assays and live-cell microscopy analysis
Flp-In T-REx HeLa cells inducibly expressing GFP-PARKIN (and mutants) were infected with Mitochondria-RFP BacMan 2.0 (Life Technologies) and imaged using the depolarization, image capture routines, and correlation profiling method describe in Supplemental Experimental Procedures.
Protein and in vitro ubiquitylation analysis
GST-PARKIN from Sf9 cells was liberated from GST using PreScission protease, leaving a Gly-Pro N-terminal extension. Phosphorylation of PARKIN and UB by PINK1Tc (Kondapalli et al., 2012) was performed as described in Supplemental Experimental Procedures. Ubiquitylation reactions were performed as described in Supplemental Experimental Procedures and analyzed by SDS-PAGE/immunoblotting or AQUA proteomics.
Supplementary Material
Highlights.
-
*
PINK1-PARKIN pathway is dissected via quantitative proteomics and live cell imaging
-
*
PINK1-activated PARKIN synthesizes multiple chain linkage types in vitro and in vivo
-
*
PINK1 phosphorylates PARKIN to activate E3 and poly-UB to promote their interaction
-
*
The data suggest feed-forward PINK1 PARKIN activation and recruitment to mitochondria
ACKNOWLEDGMENTS
We thank M. Muqit and P. Cohen (MRC Protein Phosphorylation Unit, University of Dundee), K. Shokat (UCSF), B. Raught (Ontario Cancer Institute), V. Dixit (Genentech), and J. Shen (Brigham and Women’s Hospital) reagents, R. Kunz and C. Braun for mass spectrometry assistance, S. Harrison and Y. Dimitrova (Harvard Medical School) for SEC-MALS assistance, and the Nikon Imaging Center at Harvard Medical School for imaging support. This work was supported by NIH grants R37 NS083524 to J.W.H., RO1 GM069530 to B.A.S., and RO1 GM067945 to S.P.G. B.A.S. is an Investigator of the Howard Hughes Medical Institute. J.W.H. is a consultant for Millennium: the Takada Oncology Company and Biogen-Idec.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.O., S.A.S., and J.W.H. conceived the study. A.O. created cell lines, performed AQUA proteomics, purified proteins, performed in vitro UB chain synthesis assays and in vitro poly-UB binding assays, and SEC-MALS. S.A.S. created cell lines and performed live-cell imaging and analysis with T.X. who created the co-incidence algorithm. D.D., J.L.O., V.S. purified proteins and performed biophysical interaction measurements under the direction of B.A.S. J-M.H. created PINK1−/− Flp-In T-REx HeLa cells and with M.J. and S.P.G. performed the MEF phosphoproteomics experiments. J.T.K. and J.A.W. created α-p-S65 PARKIN antibodies by phage display. S.A.B. constructed AQUA peptides. J.W.H. and A.O. wrote the paper with input from B.A.S.
REFERENCES
- Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;509:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- Birsa N, Norkett R, Wauer T, Mevissen TE, Wu HC, Foltynie T, Bhatia K, Hirst WD, Komander D, Plun-Favreau H, et al. K27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem. 2014;289:14569–14582. doi: 10.1074/jbc.M114.563031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci U S A. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Geisler S, Vollmer S, Golombek S, Kahle PJ. The ubiquitin-conjugating enzymes UBE2N, UBE2L3 and UBE2D2/3 are essential for Parkin-dependent mitophagy. J Cell Sci. 2014;127:3280–3293. doi: 10.1242/jcs.146035. [DOI] [PubMed] [Google Scholar]
- Hasson SA, Kane LA, Yamano K, Huang CH, Sliter DA, Buehler E, Wang C, Heman-Ackah SM, Hessa T, Guha R, Martin SE, Youle RJ. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BM, Berger AK, Cortese GP, anf Lavoie MJ. The ubiquitin E3 ligase parkin regulates the proapoptotic function of Bax. Proc Natl Acad Sci U S A. 2012;109:6283–6288. doi: 10.1073/pnas.1113248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kelly V, Johnson C, Baillie C, Hastie CJ, Peggie M, Macartney T, Woodroof HI, Alessi DR, Pedrioli PG, et al. Phosphorylation of Parkin at Serine65 is essential for activation: elaboration of a Miro1 substrate-based assay of Parkin E3 ligase activity. Open Biol. 2014a;4:130213. doi: 10.1098/rsob.130213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014b;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber JT, Thomsen ND, Hannigan BT, Degrado WF, Wells JA. Nature-inspired design of motif-specific antibody scaffolds. Nat. Biotechnol. 2013;31:916–921. doi: 10.1038/nbt.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GG, Chew KC, Ng XH, Henry-Basil A, Sim RW, Tan JM, Chai C, Lim KL. Proteasome inhibition promotes Parkin-Ubc13 interaction and lysine 63-linked ubiquitination. PLoS One. 2013;8:e73235. doi: 10.1371/journal.pone.0073235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, Tanaka K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem. 2006;281:3204–3209. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phu L, Izrael-Tomasevic A, Matsumoto ML, Bustos D, Dynek JN, Fedorova AV, Bakalarski CE, Arnott D, Deshayes K, Dixit VM, et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003756. M110 003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin CA, Galeva NA, Bae K, Ahmad MN, Witte TM, Richter ML. Isolated RING2 domain of parkin is sufficient for E2-dependent E3 ligase activity. Biochemistry. 2014;53:225–234. doi: 10.1021/bi401378p. [DOI] [PubMed] [Google Scholar]
- Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4 doi: 10.1038/ncomms2982. 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf S RM, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Scientific reports. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Inoshita T, Hattori N, Imai Y. PINK1-mediated phosphorylation of Parkin boosts Parkin activity in Drosophila. PLoS Genet. 2014;10:e1004391. doi: 10.1371/journal.pgen.1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat Methods. 2012;9:303–309. doi: 10.1038/nmeth.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol Cell. 2012;47:797–809. doi: 10.1016/j.molcel.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UbcH7-reactivity profile reveals Parkin and HHARI to be RING/HECT hybrids. Nature. 2011 doi: 10.1038/nature09966. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 2013;23:886–897. doi: 10.1038/cr.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.