Abstract
The epithelium of gastrointestinal (GI) mucosa is a rapidly self-renewing tissue in the body, and its homeostasis is preserved through strict regulation of cell proliferation and apoptosis. Epithelial cells originate from a small number of pluripotent stem cells, which divide to either renew themselves or become committed crypt cells. RNA-binding proteins (RBPs) and microRNAs (miRNAs) regulate gene expression at the posttranscriptional level and are recently shown to modulate GI mucosal growth and repair after injury. Here we highlight the roles of RBPs HuR, CUG-binding protein 1, AU-binding factor 1, and several GI epithelial-specific miRNAs in gut mucosal homeostasis and diseases and also further analyze the mechanisms through which RBPs and miRNAs modulate the stability and translation of target mRNAs.
Keywords: posttranscriptional regulation, mRNA stability and translation, cellular polyamines, cell proliferation, apoptosis
Introduction
Maintenance of the GI epithelial integrity in response to stressful environments requires epithelial cell decisions that regulate signaling networks controlling expression of different genes. Under biological conditions, undifferentiated epithelial cells continuously replicate in the proliferating zone within the crypts and differentiate as they migrate up the luminal surface of the colon and the villous tips in the small intestine [1]. To maintain stable numbers of enterocytes, cell division is counterbalanced by apoptosis that occurs in the crypt area and at the luminal surface of the colon and villous tips in the small intestine. This rapid dynamic turnover rate of the intestinal epithelium is highly regulated and critically controlled by numerous factors. Although the exact roles and mechanisms of transcriptional gene regulation in GI epithelial homeostasis have been extensively studied for decades, the essential contribution of posttranscriptional events is becoming increasingly recognized recently. Posttranscriptional processes, particularly altered mRNA turnover and translation, are major mechanisms by which mammalian cells control gene expression [2,3*]. Changes in mRNA stability and translation are governed by two types of trans-acting factors: RBPs and miRNAs that directly bind to the cis-elements located at the 3′-untranslated regions (3′-UTRs) of target mRNAs and regulate gene expression synergistically or antagonistically.
Over the past ten years, an increasing body of evidence indicates that control of the stability and translation of mRNAs encoding growth/apoptosis-associated factors such as p53, JunD, nucleophosmin (NPM), X chromosome-linked inhibitor of apoptosis protein (XIAP), mitogen-activated protein kinase kinase-1 (MEK-1), activating transcription factor-2 (ATF-2), Smad ubiquitin regulatory factor 2 (Smurf2), cyclin-dependent kinase 4 (CDK4), CDK2, and c-Myc is crucial for maintaining normal gut mucosal homeostasis and epithelial integrity and its disruption is related to pathogenesis of GI mucosal diseases. Here we highlight the roles of RBPs and several epithelial tissue-specific miRNAs in the regulation of GI epithelial homeostasis and their involvement in mucosal growth-related diseases. We also further analyze in some detail the mechanisms through which RBPs and miRNAs modulate the stability and translation of target mRNAs.
RBPs in Gut Epithelial Homeostasis and Diseases
After transcription from their genes, mRNAs are subjected to multiple processing and regulatory steps that are coordinated by RBPs. Through these processes, their product proteins are efficiently produced to meet the need of the cell. U- and AU-rich elements (AREs) are the best-characterized cis-acting sequences located in 3′-UTR of many labile mRNAs and function as elements controlling mRNA exportation, decay, and translation [4,5]. Several RBPs, including AUF1, BRF1, TTP, and KSRP, are shown to promote ARE/mRNA decay, whereas Hu/ELAV family of RBPs, which consists of three primary neuronal members HuB, HuC, HuD, and one ubiquitous member HuR, stabilize and stimulate translation of their target mRNAs [6]. Several RBPs are implicated in human diseases, because there are significant changes in their binding affinity to target mRNAs, defect in binding region, and dysregulation of RBP expressions in various pathologic conditions. A well-known example is that fragile X mental retardation results from loss of expression of the RBP FMR1 [7]. In addition, CUG-binding protein 1 (CUGBP1) dysfunction was found to contribute to the pathogenesis of myotonic dystrophy; HuR is intimately involved in the processes of different cancers and aging. The study investigating RBPs in gut epithelial homeostasis is still in its early stage, but several RBPs are shown to play an important role in the regulation of intestinal epithelial cell (IEC) proliferation, apoptosis, migration, and cell-to-cell interactions.
HuR
HuR is one of the most studied RBPs and exhibits a high affinity for AREs. HuR is predominantly located in the nucleus in unstimulated cells, but it rapidly translocates to the cytoplasm, where it directly interacts with and regulates the stability and/or translation of target mRNAs in response to specific stimuli. In IECs, HuR is highly expressed and its subcellular distribution and binding affinity are tightly regulated and dependent on the levels of cellular polyamines. Polyamines (spermidine, spermine, and their precursor putrescine) are ubiquitous polycationic molecules found in all eukaryotic cells and are implicated in many aspects of cellular functions [8–10]. We [11–13] and others [14] have demonstrated that normal cell proliferation in the intestinal mucosa depends on the supply of polyamines to the dividing cells and that decreasing cellular polyamines inhibits epithelial cell proliferation. We have further shown that depletion of cellular polyamines enhances the cytoplasmic abundance of HuR, while increased levels of polyamines decrease cytoplasmic HuR; neither intervention changes whole-cell HuR levels [15]. HuR specifically binds the 3′-UTRs of several mRNAs including NPM, p53, and JunD mRNAs. HuR silencing renders these mRNAs unstable and prevents increases in the levels of NPM, p53, and JunD mRNAs and proteins in polyamine-deficient cells. These results indicate that polyamines modulate cytoplasmic HuR levels in IECs, in turn controlling the stability of NPM, p53, and JunD mRNAs and influencing levels of NPM, p53 and JunD proteins.
Polyamines modulate the subcellular localization of HuR through AMP-activated protein kinase (AMPK)-regulated phosphorylation and acetylation of importin α1 [16]. The AMPK is an enzyme involved in responding to metabolic stress and also plays a role in regulating the nuclear import of HuR [17]. Decreased levels of cellular polyamines repress AMPK activity and reduce importin α1 levels, whereas increased polyamines induce both AMPK and importin α1 levels. AMPK activation increases importin α1 but reduces the cytoplasmic levels of HuR in control and polyamine-deficient cells. IECs overexpressing wild-type importin α1 exhibit a reduction of cytoplasmic HuR abundance, while cells overexpressing importin α1 proteins bearing K22R (lacking acetylation site), S105A (lacking phosphorylation site), or K22R/S105A (lacking both sites) mutations display increased levels of cytoplasmic HuR. Ectopic expression of these importin α1 mutants also prevents the increased levels of cytoplasmic HuR following polyamine depletion. These results strongly suggest that increase in AMPK activity by polyamines triggers HuR nuclear import, whereas polyamine depletion increases the cytoplasmic levels of HuR by inactivating the AMPK-mediated importin α1 signaling.
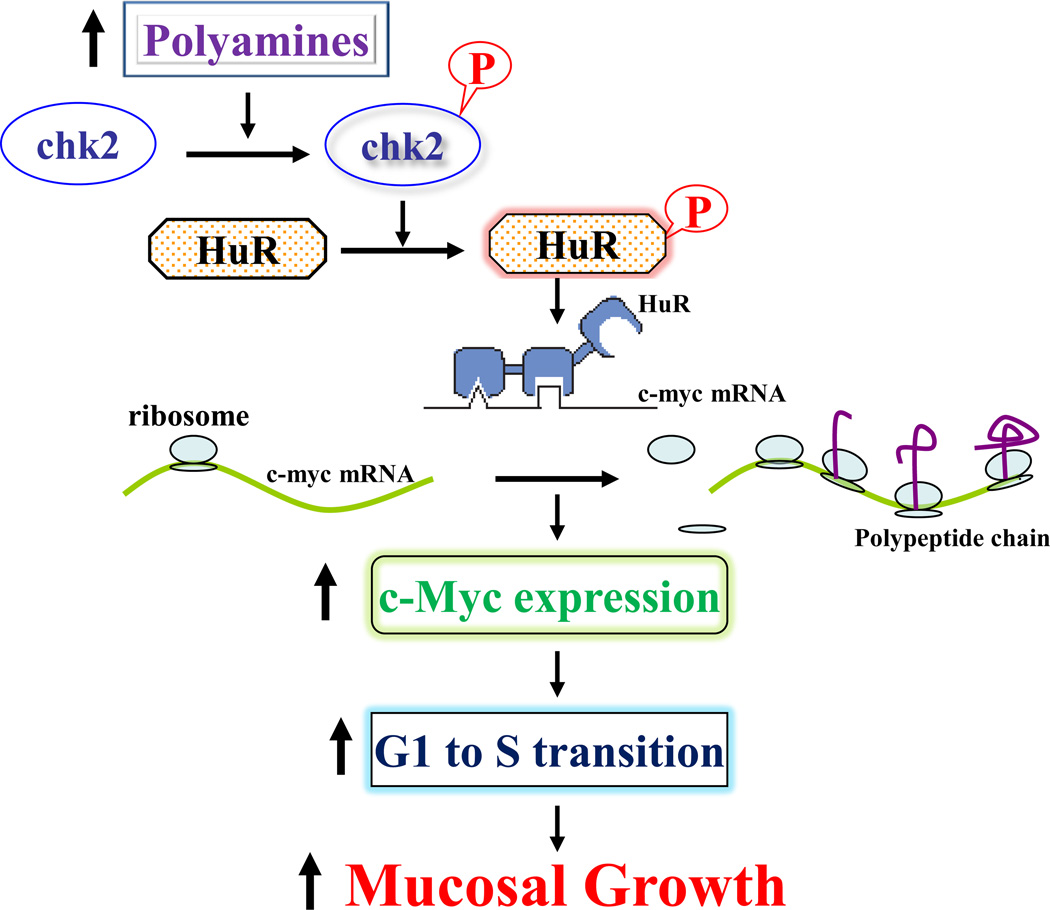
Polyamines also regulate HuR binding affinity for target mRNAs through Chk2-dependent HuR phosphorylation [18,19]. HuR is phosphorylated by Chk2 kinase in vivo as well as in vitro and this posttranslational modification influences HuR’s association with its target transcripts [20*]. We have demonstrated that Chk2 expression requires polyamines and that polyamine depletion reduces Chk2 and decreases levels of phosphorylated-HuR (p-HuR), associated with a reduction in HuR interaction with mRNAs encoding c-Myc and occludin [19–21**]. Ectopic Chk2 overexpression increases p-HuR, thus enhancing HuR association with the mRNAs of c-Myc and occludin. The levels of [HuR/c-Myc mRNA] complexes in polyamine-deficient cells are markedly higher than those observed in control cells after Chk2 overexpression, as polyamine-deficient cells show higher cytoplasmic HuR levels. Moreover, polyamines enhance c-Myc mRNA translation by increasing HuR phosphorylation by Chk2. Since c-Myc plays an important role in the regulation of the cell cycle and normal gut mucosal growth and because inhibition of c-Myc expression represses IEC proliferation and delays mucosal healing, we propose a model delineating the role of HuR-induced c-Myc expression following increased polyamines in intestinal mucosal renewal (Fig. 1). In this model, increased polyamines stimulate Chk2 and increase HuR phosphorylation, in turn triggering c-Myc translation and enhancing IEC proliferation. In contrast, polyamine depletion inhibits Chk1 and decreases c-Myc translation, thus repressing mucosal growth. In addition, HuR modulates intestinal epithelial homeostasis by regulating expression of genes involved in apoptosis [22–25]. These genes includes: XIAP, MEK-1, and ATF-2. The XIAP mRNA is a direct target of HuR, and increased level of [HuR/XIAP mRNA] complex stabilizes the XIAP mRNA and increases cellular abundance of XIAP, thus desensitizing IECs to apoptosis. HuR also displays a strong affinity to the ATF-2 and MEK-1 mRNAs. The binding of HuR to the ATF-2 mRNA primarily increases the stability of ATF-2 mRNA, whereas HuR association with the MEK-1 mRNA not only increases the stability of MEK1 mRNA but also enhances its translation.
Figure 1.
Schematic diagram of polyamine-induced c-Myc translation in the regulation of gut mucosal growth. Increased levels of cellular polyamines induce HuR phosphorylation by activating Chk2, promote HuR association with the c-Myc mRNA, and enhance c-Myc translation, thus stimulating intestinal epithelial cell proliferation.
Several recent studies show that HuR also regulates gut permeability by altering expression of tight junction (TJ) proteins such as occludin [20*, 21**]. HuR interacts with the occludin mRNA via its 3′-UTR and this association enhances occludin translation. HuR association with the occludin mRNA depends on Chk2-dependent HuR phosphorylation, since reduced HuR phosphorylation by Chk2 silencing decreases HuR binding to the occludin mRNA and represses occludin translation. In mice exposed to septic stress, Chk2 levels in the intestinal mucosa decrease dramatically, which is associated with an inhibition of occludin expression and gut barrier dysfunction. Recently, we have also reported that HuR regulates early intestinal mucosal restitution after injury by stabilizing the mRNA of Stromal interaction molecule 1 (STIM1) and that increased STIM1 by HuR enhances TRPC1-mediated Ca2+ influx and stimulates IEC migration over wounded area [26**].
The binding regions and regulatory effects of HuR are transcript-specific. As pointed out above, HuR selectively binds to the mRNAs of NPM, p53, ATF-2, MEK-1, c-Myc, and occludin via their 3′-UTRs, but it interacts with the XIAP mRNA through both coding region (CR) and 3′-UTR. HuR primarily regulates the stability of mRNAs encoding NPM, p53, JunD, ATF-2, and XIAP, but it enhances expression of MEK-1, c-Myc, and occludin at the translation level. Importantly, HuR association with its transcripts is determined by the crosstalk with other RBPs. For example, HuR and AUF1 competitively bind to the JunD mRNA and regulate the stability of the JunD mRNA in opposite directions [27]. Moreover, polyamines regulate the stability of the JunD mRNA by modulating the competitive binding of the JunD mRNA with HuR and AUF1.
CUGBP1
CUGBP1 binds to GC-rich elements (GREs) rather than AREs of target mRNAs. The interaction of CUGBP1 with its target mRNAs commonly enhances mRNA decay and represses translation, although in some instances CUGBP1 promotes mRNA translation [28,29]. In normal IECs, CUGBP1 interacts with the CDK4 mRNA and represses CDK4 translation. CUGBP1 binds to the CDK4 mRNA via both its CR and 3′-UTR, enhances the CDK4 mRNA association with argonaute (Ago)-containing complexes, and increases the recruitment of CDK4 mRNA to processing bodies (P-bodies), where mRNAs are sorted for degradation and/or translation repression [30]. Interestingly, polyamines promote the translation of CDK4 by inhibiting CDK4 mRNA association with CUGBP1, since depletion of cellular polyamines increases cytoplasmic CUGBP1 abundance and induces its associations with the CDK4 mRNA. In contrast, increasing the levels of cellular polyamines decreases CDK4 mRNA interaction with CUGBP1 and induces CDK4 expression. Furthermore, decreasing the levels of CUGBP1 by polyamines is associated with an increase in IEC proliferation, which is prevented by ectopic CURBP overexpression and CDK4 silencing. These findings indicate that polyamines stimulate IEC proliferation at least partially by enhancing CDK4 expression via down-regulation of CUGBP1. On the other hand, CUGBP1 also regulates intestinal epithelial homeostasis by modulating IEC apoptosis [31**]. Increased levels of cellular CUGBP1 desensitize IECs to apoptosis, whereas CUGBP1 silencing increases the sensitivity of IECs to apoptosis. The mechanism underlying this process is that CUGBP1 represses expressions of IAP proteins such as c-IAP1 and c-IAP2, thereby enhancing caspase-3 signaling pathway.
Recently, CUGBP1 is shown to play an important role in regulation of TJ expression and gut barrier function [21**]. Ectopic CUGBP1 overxpression represses occludin expression but fails to alter the levels of claudin-2, claudin-3 and claudin-5, ZO-1 and β-catenin. This inhibitory effect of CUGBP1 on occludin expression occurs at the translation level and is mediated through its 3′-UTR rather than CR and 5′-UTR. Consistently, increased CUGBP1 also impairs the epithelial barrier function as indicated by an increase in epithelial paracellular permeability. Interestingly, CUGBP1 and HuR compete for association with the same occludin 3′-UTR and regulate occludin translation competitively and in opposite directions [21**]. CUGBP1 overexpression decreases HuR binding to occludin mRNA and represses occludin translation, whereas HuR overexpression inhibits CUGBP1 association with occludin mRNA and promotes occludin translation. Studies using purified GST-HuR or GST-CUGBP1 fusion proteins have revealed that the occludin 3′-UTR association with HuR is progressively increased when increasing concentrations of GST-HuR in the binding reaction mixture, but its interaction with CUGBP1 is reduced with increasing GST-HuR levels. These results indicate that HuR and CUGBP1 competitively bind to the occludin 3′-UTR and that CUGBP1 represses occludin mRNA translation by displacing HuR.
AUF1
AUF1 is the first identified RBP and displays high affinity to ARE-containing RNAs [32,33]. Experiments using cDNA cloning and immunoblotting assays show that there are four isoforms of AUF1 family with molecular weights of 37 (p37), 40 (p40), 42 (p42), and 45 (p45) kDa. With varied RNA-binding affinities and specificities, AUF1 isoforms permit cells to differentially control mRNA decay in a cell-type specific manner. AUF1 is a general RNA destabilizer, but recent studies have found that AUF1 also acts as an enhancer of mRNA stability and translation in some instances [34,35].
The expression of AUF1 family proteins is abundant in IECs, with the isoform p42 as the dominant protein. We have demonstrated that AUF1 associates with and destabilizes the JunD mRNA and that this process is regulated by polyamines and HuR [27]. Polyamine depletion stabilizes JunD mRNA without affecting its transcription [36], and increased JunD/AP-1 transcriptional activity contributes to an inhibition of IEC proliferation by repressing CDK4 [37] through dimerization with ATF-2 [38]. Our studies show that JunD mRNA is a target of HuR and AUF1 and that polyamines modulate JunD mRNA degradation by altering the competitive binding of HuR and AUF1 to the JunD 3′-UTR. Polyamine depletion enhances HuR binding to JunD mRNA and decreases the levels of JunD transcript associated with AUF1, thus stabilizing JunD mRNA. The silencing of HuR increases AUF1 binding to the JunD mRNA, decreases the abundance of [HuR/JunD mRNA] complexes, renders the JunD mRNA unstable, and prevents increases in JunD mRNA and protein in polyamine-deficient cells.
TIAR
Gut mucosa also highly expresses the RBP TIAR (TIA-1 related protein) that is implicated in intestinal epithelial homeostasis. TIAR acts generally as a translational repressor and regulates its target gene expression negatively [39]. TIAR binds to mRNAs via its RNA recognition motif domains, and AREs are common cis-elements of TIAR. TIAR was shown to directly bind the mRNA encoding the membrane-associated TJ protein ZO-1 via its 3′-UTR, and increased [TIAR/ZO-1 mRNA] complex represses ZO-1 translation [40], thus disrupting the intestinal epithelial barrier function. Moreover, JunD regulates ZO-1 expression by altering the interaction of the ZO-1 mRNA with TIAR.
miRNAs in GI Epithelial Homeostasis
miRNAs are small noncoding RNAs of ∼22 nucleotides, which posttranscriptionally repress the expression of target genes [41]. Although the exact functions of miRNAs in human development and physiology remain largely unknown, differential expression of given miRNAs during disease progression suggests an especially significant role for miRNAs in human pathologic conditions. For example, miRNA-21, miRNA-26, miRNA-31, miRNA-141, miRNA-145, miRNA-196, and miRNA-200 are shown to play a critical role in the regulation of migration and invasiveness of colon cancer cells, while miRNA-493 inhibits the settlement of colon cancer cells in the liver parenchyma. In addition, the mRNA and protein levels of Dicer1, which is essential for miRNA biogenesis, are reduced during disease progression in gastric cancer. We have profiled global miRNA expression in normal intestinal mucosa and IECs and revealed that several miRNAs, including miRNA-29b, miRNA-222, miRNA-503, and miR-195, are highly expressed in the intestinal epithelium and their expression levels are affected rapidly in response to food starvation and polyamine depletion [30,42**]. Further studies show that control of these intestinal epithelial tissue-specific miRNAs is crucial for maintenance of normal intestinal epithelial integrity.
miRNA-222
miRNA-222 functions as a regulator of cell survival and migration in lung cancer, prostate carcinoma, hepatocellular cancer, and breast cancer [43,44], but its importance in normal intestinal epithelial homeostasis has not been elucidated until recently. Our studies show that miR-222 binds to the CDK4 mRNA and this association is inhibited by polyamines. Depletion of cellular polyamines increases cellular abundance of miR-222, induces its association with the CDK4 mRNA, and inhibits CDK4 translation, whereas increasing the levels of cellular polyamines decreases CDK4 mRNA interaction with miR-222, thereby inducing CDK4 expression. miR-222 and CUGBP1 jointly regulate CDK4 expression and they repress CDK4 mRNA translation synergistically [30]. Moreover, miR-222 and CUGBP1 associate with the CDK4 mRNA through distinct non-overlapping binding site, and there are no common sites for both miR-222 and CUGBP1 in the CDK4 mRNA. CUGBP1 binds both the CDK4 CR and its 3′-UTR, but miR-222 only interacts with the CDK4 CR. Although the CDK4 mRNA does not contain any high-affinity CUGBP1-binding site such as the canonical GRE (defined as UGUUUGUUUGU), there are several GU repeats and CUG repeats in the 3′-UTR and CR of the CDK4 transcript, which are also recently recognized as the GRE and interact with CUGBP1. On the other hand, there are two predicted miR-222 binding sites within the CDK4 CR, but only one sequence (spanning positions 264 to 284) is functional.
It is intriguing that increased miR-222 expression causes growth arrest in normal IECs, but it is required in certain cancer cells to inhibit p27 expression and may induces cancer cell proliferation [45]. The reasons for this distinct effect remain to be fully understood, but they are related to: 1) expression and function of miRNAs are tissue-specific and development-specific in general; and 2) changes in the accessibility of miR-222 to its target transcripts. For example, upon growth factor stimulation, increased binding of the RBP Pumilio-1 to the p27 mRNA 3′-UTR in cancer cells induces the partial change in the p27 mRNA structure that favors its association with miRNA-222. Induced binding of miR-222 to the p27 mRNA suppresses p27 expression, thus promoting cancer cells entry into S phase of the cell cycle. On the other hand, basal levels of p27 and Pumilio are low in normal IECs. It is likely that under the stress of growth inhibition, interaction of miRNA-222 with p27 is decreased as a result of target site hindrance. In contrast, [miRNA-222/CDK4 mRNA] complex increases at the same time, thus leading to growth arrest in normal IECs.
miRNA-503
miR-503 is an abundant miRNA which is conserved among many species and is clustered with miR-322 on chromosome X. miR-503/322 have been studied previously in myogenesis, where they were shown to promote cell cycle quiescence and differentiation by down-regulating Cdc25 [46]. Recent studies show that miR-503/322 regulates the vascular smooth muscle cell phenotype and neointimal formation and that miR-322 and its target Tob2 control osteogenesis through modulation of Osx mRNA degradation [47]. We have recently demonstrated that small intestinal mucosal atrophy following polyamine depletion or fasting is associated with a decrease in the levels of miR-322/503 [31]. Overexpression of a miR-503 precursor (pre-miR-503) reduces newly synthesized CUGBP1 protein levels, whereas inhibiting miR-503 enhances CUGBP1 biosynthesis and elevates its abundance [31]. Studies using heterologous reporter constructs have revealed a greater repressive effect of miR-503 through the CUGBP1 CR sites than through the single CUGBP1 3′-UTR target site. CUGBP1 mRNA levels in P-bodies increases in cells transfected with pre-miR-503, while silencing P-body resident proteins Ago2 or RCK decreases miR-503-mediated repression of CUGBP1 expression. This effect is tightly regulated by polyamines, since decreasing the levels of cellular polyamines reduces endogenous miR-503 levels but promotes CUGBP1 expression, which is prevented by ectopic miR-503 overexpression. Furthermore, repression of CUGBP1 by miR-503 in turn alters the expression of CUGBP1 target mRNAs such as c-IAP1 and c-IAP2 and thus increases the sensitivity to apoptosis.
Smurf2 is an E3 ubiquitin ligase that regulates TGF-β/Smad signaling and is implicated in gut epithelial homeostasis. Our group has reported that miR-503/322 are novel factors that regulate Smurf2 expression posttranscriptionally [48]. Both miR-503 and miR-322 interact with Smurf2 mRNA via its 3′-UTR and repress Smurf2 translation but do not affect total Smurf2 mRNA levels. miR-503/322-induced repression of Smurf2 translation is mediated through a single binding site in the Smurf2 3′-UTR. Increased levels of endogenous Smurf2 by antagonization of miR-503/322 inhibit TGF-β-induced Smad2 activation by increasing the degradation of phosphorylated Smad2. Moreover, the increase in Smurf2 in IECs expressing lower levels of miR-503/322 is associated with increased resistance to apoptosis, which is abolished by Smurf2 silencing. These findings indicate that miR-503/322 repress Smurf2 translation in IECs, thereby affecting intestinal epithelial homeostasis by altering TGF-β/Smad2 signaling and apoptosis.
miRNA-29b
miR-29b is a member of the miR-29 family and modulates a variety of cellular functions. miR-29b regulates DNA methylation-related reprogramming events by targeting dnmt3a and dnmt3b [49], and promotes fibrosis by altering expression of collagen isoforms [50]. miR-29b modulates cell proliferation and apoptosis and also plays a role in the development of abdominal aortic aneurysm [51*]. The abnormal expression of miR-29b is associated with tumorigenesis and cancer progression, and miR-29b also alters the tumor microenvironment to repress metastasis. Patients with advanced liver cirrhosis show lower levels of miR-29a in their serum when compared with healthy controls or patients with early fibrosis. Disrupted expression of miR-29 family contributes to the pathogenesis of type-1 diabetes. miR-29 modulates innate and adaptive immunity by targeting interferon-γ and also regulates gut permeability in patients with irritable bowel syndrome [52].
We have investigated the role of miR-29b in normal intestinal mucosal growth and shown that mucosal atrophy in the small intestine induced by fasting or polyamine depletion is associated with increased expression of miR-29b [42**]. The simple systemic delivery of locked nucleic acid-modified antimiR-29b oligonucleotides reduces endogenous miR-29b levels in the small intestinal mucosa, increases CDK2 expression, and stimulates mucosal growth. In contrast, overexpression of the miR-29b precursor represses CDK2 expression and results in growth arrest in G1 phase. miR-29b represses CDK2 translation through direct interaction with the cdk2 mRNA via its 3′-UTR, whereas point-mutation of miR-29b binding-site in the cdk2 3′-UTR prevents miR-29b-induced repression of CDK2 translation. These results indicate that miR-29b inhibits intestinal mucosal growth by repressing CDK2 translation.
miR-195
The evolutionarily conserved miR-195 is highly abundant in normal GI mucosa, but its levels decrease significantly in cancer tissues. miR-195 inhibits cell proliferation by targeting CDK4, cyclin D1, CDK6, and WEE1, promotes apoptosis by down-regulating Sirt1, and affects cell invasion by modulating ActRIIA expression. We have found that miR-195 competes with HuR to modulate STIM1 mRNA stability and regulate early rapid intestinal epithelial restitution after wounding [26**]. STIM1 functions as a sensor of Ca2+ within stores and plays an essential role in the activation of store-operated Ca2+ entry (SOCE). Lowering Stim1 levels reduces SOCE and inhibits intestinal epithelial repair. Our results show that cellular STIM1 abundance is controlled posttranscriptionally via factors that associate with 3′-UTR of stim1 mRNA. miR-195 and HuR compete for association with the stim1 3′-UTR and regulate stim1 mRNA decay in opposite directions. Interaction of miR-195 with the stim1 3′-UTR destabilizes stim1 mRNA, whereas the stability of stim1 mRNA increases with HuR association. Interestingly, ectopic miR-195 overexpression increases the colocalization of tagged stim1 RNA with P-bodies and this translocation of stim1 mRNA is abolished by HuR overexpression. Moreover, decreased levels of STIM1 by miR-195 overexpression inhibit cell migration over the denuded area after wounding, which is rescued by increasing HuR levels.
In addition, several miRNAs, including miR-192, let-7, miR-122a, miR-155, miR-9, miR-374, and miR-145, are implicated in the regulation of TJ expression and gut permeability under physiological and pathological conditions. These interesting observations have been summarized and discussed in our recent publication [53].
Conclusions
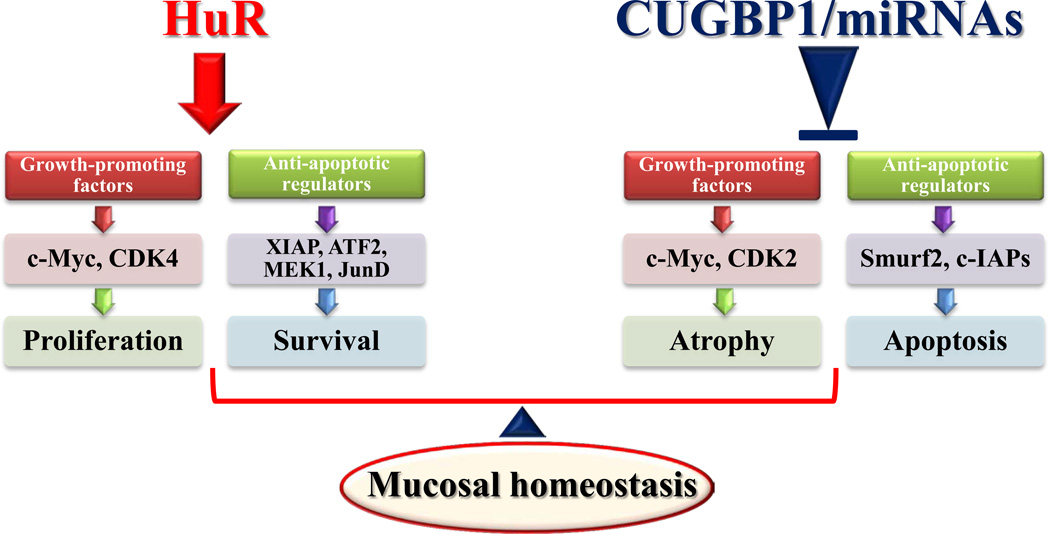
Although the gene expression programs that regulate the gut epithelial integrity are strongly regulated at the transcription level, the essential contribution of posttranscriptional events is becoming increasing recognized. The experimental data summarized here provide novel evidence indicating the importance of several RBPs and miRNAs in posttranscriptional gene regulation. Based on our studies and others, we propose a model delineating the posttranscriptional control of IEC proliferation/apoptosis and subsequent epithelial homeostasis by RBPs and miRNAs (Fig. 2). In this model, expression of growth-promoting factors such as c-Myc, CDK2, and CDK4 and anti-apoptotic regulators such as XIAP, ATF-2, MEK-1, JunD, and Smurf1 is controlled by two factors competing for influence on their mRNA stability and translation: the positive factor HuR and negative regulators including CUGBP1, AUF1, TIAR, and miRNAs. HuR stabilizes and/or promotes translation of mRNAs encoding these factors, thus up-regulating their expression and protecting the epithelial integrity or resulting in mucosal hyperplasia. In contrast, CUGBP1, AUF1, TIAR, and miRNAs destabilize and/or inhibit translation of mRNAs encoding growth-promoting factors and anti-apoptotic regulators, leading to mucosal atrophy. Importantly, normal intestinal epithelial homeostasis depends on the balance between HuR and negative regulators including CUGBP1, AUF1, TIAR, and miRNAs.
Figure 2.
Schematic diagram of depicting posttranscriptional regulation of intestinal epithelial homeostasis by RBPs and miRNAs. HuR stimulates expression of growth-promoting and anti-apoptotic factors by increasing the stability and translation of their mRNAs, thus up-regulating cell proliferation and survival. In contrast, CUGBP1, AUF1, TIAR, and given miRNAs inhibit expression of growth-promoting and anti-apoptotic factors posttranscriptionally and function as negative regulators of intestinal epithelial renewal. Normal epithelial homeostasis is dependent on the balance between the positive factor HuR and negative RBPs/miRNAs.
However, there are still many critical issues that remain to be addressed regarding this model. First, the molecular processes responsible for the control of RBP activation and miRNA biogenesis in response to stress remain largely unknown. Second, it is unclear how altered RBPs and miRNAs modulate the stability and translation of mRNAs in the intestinal epithelium. Third, experiments defining the in vivo functions of RBPs and miRNAs by using intestinal epithelial tissue-specific RBP and miRNA knockout or transgenic mouse models are badly needed and will provide a better understanding of physiological functions of RBPs/miRNAs. Finally, translational studies investigating the roles of RBPs/miRNAs in the pathogenesis of gut mucosal hyperplasia and atrophy in patients are still limited and should be emphasized in the future study.
Highlights.
Gut epithelium is a self-renewing tissue and its homeostasis is tightly regulated
RBPs and miRNAs play an important role in regulating GI mucosal growth and repair
The roles of RBPs and miRNAs in gut mucosal homeostasis and diseases are discussed
Acknowledgments
This work was supported by a Merit Review Grant from the Department of Veterans Affairs and by NIH Grants DK-57819, DK-61972, and DK-68491. J-Y. Wang is Senior Research Career Scientist, Medical Research Service, US Department of Veterans Affairs.
ABBREVIATIONS
- miRNA
microRNA
- RBPs
RNA-binding proteins
- IEC
intestinal epithelial cell
- UTRs
untranslated regions
- CR
coding region
- GI
gastrointestinal
- AREs
AU-rich elements
- P-bodies
processing bodies
- AMPK
AMP-activated protein kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Simons BD, Clevers H. Stem cell self-renewal in intestinal crypt. Exp Cell Res. 2011;317:2719–2724. doi: 10.1016/j.yexcr.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 3.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005.Emerging principles of miRNA in stress signaling pathways and concepts of miRNAs in disease are reviewed; the challenges and opportunities associated with the mechanistic dissection of miRNA function and miRNA-based therapeutics are also highlighted.
- 4.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 6.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo X, Rao JN, Liu L, Zou TT, Keledjian KM, Boneva D, Marasa BS, Wang JY. Polyamines are necessary for synthesis and stability of occludin in intestinal epithelial cells. Am J Physiol. 2005;288:G1159–G1169. doi: 10.1152/ajpgi.00407.2004. [DOI] [PubMed] [Google Scholar]
- 9.Guo X, Rao JN, Liu L, Rizvi M, Turner DJ, Wang JY. Polyamines regulate β-catenin tyrosine phosphorylation via Ca2+ during intestinal epithelial cell migration. Am J Physiol. 2002;283:C722–C734. doi: 10.1152/ajpcell.00054.2002. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Rao JN, Zou T, Liu L, Marasa BS, Xiao L, Zeng X, Turner DJ, Wang JY. Polyamines are required for expression of Toll-like receptor 2 modulating intestinal epithelial barrier integrity. Am J Physiol. 2007;293:G568–G576. doi: 10.1152/ajpgi.00201.2007. [DOI] [PubMed] [Google Scholar]
- 11.Timmons J, Chang E, Wang JY, Rao JN. Polyamines and gut mucosal homeostasis. J Gastrointest digest sys. 2012;S:7. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JY. Polyamines and mRNA stability in regulation of intestinal mucosal growth. Amino Acids. 2007;33:241–252. doi: 10.1007/s00726-007-0518-z. [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Santora R, Rao JN, Guo X, Zou TT, Zhang HM, Turner DJ, Wang JY. Activation of TGF-β-Smad signaling pathway following polyamine depletion in intestinal epithelial cells. Am J Physiol. 2003;285:G1056–G1067. doi: 10.1152/ajpgi.00151.2003. [DOI] [PubMed] [Google Scholar]
- 14.Gerner EW, Meyskens FL., Jr. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 15.Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 16.Zou T, Liu L, Rao JN, Marasa BS, Chen J, Xiao L, Zhou H, Gorospe M, Wang JY. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1. Biochem J. 2008;409:389–398. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Fan J, Yang X, Furer-Galban S, de Silanes IL, von Kobbe C, Guo J, Georas SN, Foufelle F, Hardie DG, Carling D, Gorospe M. AMP-activated kinase regulates cytoplasmic HuR. Mol Cell Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdelmohsen K, Pullmann R, Jr., Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell. 2009;20:4885–4898. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu TX, Wang PY, Rao JN, Zou T, Liu L, Xiao L, Gorospe M, Wang JY. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2012;39:8472–8487. doi: 10.1093/nar/gkr567. This is the first study showing the role of the RBP HuR in TJ expression and gut epithelial barrier function. HuR interacts with the occludin 3′-UTR and stimulates occludin translation through Chk2-dependent HuR phosphorylation.
- 21.Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. CUGBP1 and HuR compete for association with the occludin 3′-UTR and regulate occludin translation in opposite directions. CUGBP1 represses occludin translation, whereas HuR promotes occludin transaltion.
- 22.Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, Turner DJ, Gorospe M, Wang JY. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem J. 2009;426:293–306. doi: 10.1042/BJ20091459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol Biol Cell. 2007;18:4579–4590. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009;37:7623–7637. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda K, Abdelmohsen K, Kim MM, Srikantan S, Lee EK, Tominaga K, Selimyan R, Martindale JL, Yang X, Lehrmann E, Zhang Y, Becker KG, Wang JY, Kim HH, Gorospe M. Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation. EMBO J. 2011;30:1040–1053. doi: 10.1038/emboj.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013;41:7905–7919. doi: 10.1093/nar/gkt565. This study shows that interaction of miR-195 with the stim1 3′-UTR destabilizes stim1 mRNA, whereas the stability of stim1 mRNA increases with HuR association.
- 27.Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3’ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HH, Gorospe M. GU-rich RNA: expanding CUGBP1 function, broadening mRNA turnover. Mol Cell. 2008;29:151–152. doi: 10.1016/j.molcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, Wang JY. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22:3055–3069. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui YH, Xiao L, Rao JN, Zou T, Liu L, Chen Y, Turner DJ, Gorospe M, Wang JY. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2012;23:151–162. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 33.Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–6. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 35.Sela-Brown A, Silver J, Brewer G, Naveh-Many T. Identification of AUF1 as a parathyroid hormone mRNA 3′-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J Biol Chem. 2000;275:7424–7429. doi: 10.1074/jbc.275.10.7424. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Liu L, Rao JN, Esmaili A, Strauch ED, Bass BL, Wang JY. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through p21 after polyamine depletion. Gastroenterology. 2002;123:764–779. doi: 10.1053/gast.2002.35386. [DOI] [PubMed] [Google Scholar]
- 37.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Passaniti A, Wang JY. Induced JunD in intestinal epithelial cells represses CDK4 transcription through its proximal promoter region following polyamine depletion. Biochem J. 2007;403:573–581. doi: 10.1042/BJ20061436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao L, Rao JN, Zou T, Liu L, Yu TX, Zhu XY, Donahue JM, Wang JY. Induced ATF-2 represses CDK4 transcription through dimerization with JunD inhibiting intestinal epithelial cell growth after polyamine depletion. Am J Physiol Cell Physiol. 2010;298:C1226–1234. doi: 10.1152/ajpcell.00021.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dean JL, Sully G, Clark AR, Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol Biol Cell. 2008;19:3701–3712. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 42.Xiao L, Rao JN, Zou T, Liu L, Cao S, Martindale JL, Su W, Chung HK, Gorospe M, Wang JY. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell. 2013;24:3038–3046. doi: 10.1091/mbc.E13-05-0287. miR-29b silencing enhances CDK2 expression and stimulates mucosal growth in the small intestine; miR-29b represses CDK2 translation through interaction with the CDK2 mRNA.
- 43.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G, Chiariello M, Croce CM. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, Burow ME, Ivan M, Croce CM, Nephew KP. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3’ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 46.Sarkar S, Dey BK, Dutta A. MiR-322/424 and −503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamez B, Rodriguez-Carballo E, Bartrons R, Rosa JL, Ventura F. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J Biol Chem. 2013;288:14264–14275. doi: 10.1074/jbc.M112.432104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao S, Xiao L, Rao JN, Zou T, Liu L, Zhang D, Turner DJ, Gorospe M, Wang JY. Inhibition of Smurf2 translation by miR-322/503 modulates TGF-beta/Smad2 signaling and intestinal epithelial homeostasis. Mol Biol Cell. 2014;25:1234–1243. doi: 10.1091/mbc.E13-09-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takada S, Berezikov E, Choi YL, Yamashita Y, Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA. 2009;15:1507–1514. doi: 10.1261/rna.1418309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, Tacke F, Trautwein C, Luedde T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2010;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 51.Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, Raaz U, Schoelmerich AM, Raiesdana A, Leeper NJ, McConnell MV, Dalman RL, Spin JM, Tsao PS. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J Clin Invest. 2012;122:497–506. doi: 10.1172/JCI61598. Abdominal aortic aneurysm (AAA) development associates with decreased aortic expression of miR-29b, whereas miR-29b silencing results in a significant reduction in AAA progression.
- 52.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–9. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 53.Yang H, Rao JN, Wang JY. Posttranscriptional regulation of intestinal epithelial tight junction barrier by RNA-binding proteins and microRNAs. Tissue Barriers. 2014;1(1):e28320. doi: 10.4161/tisb.28320. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]