Abstract
Cyanobacterial blooms have an impact on the aquatic ecosystem due to the production of toxins (e.g. microcystins, MCs), which constrains fish health or even cause fish death. However the toxicokinetics of the most abundant toxin, microcystin-LR (MC-LR), are not yet fully understood. To investigate the uptake mechanism, the novel Oatp1d1 in rainbow trout (rtOatp1d1) was cloned, identified and characterized. The cDNA isolated from a clone library consisted of 2772 bp containing a 2115 bp open reading frame coding for a 705 aa protein with an approximate molecular mass of 80 kDa. This fish specific transporter belongs to the OATP1 family and has most likely evolved from a common ancestor of OATP1C1. Real time PCR analysis showed that rtOatp1d1 is predominantly expressed in the liver, followed by the brain while expression in other organs was not detectable. Transient transfection in HEK293 cells was used for further characterization. Like its human homologs OATP1A1, OATP1B1 and OATP1B3, rtOatp1d1 displayed multi-specific transport including endogenous and xenobiotic substrates. Kinetic analyses revealed a Km value of 13.9 μM and 13.4 μM for estrone-3-sulfate and methotrexate, respectively and a rather low affinity for taurocholate with a Km value of 103 μM. Furthermore, it was confirmed that rtOatp1d1 is a MC-LR transporter and therefore most likely plays a key role in the susceptibility of rainbow trout to MC intoxications.
Keywords: microcystin, Oatp, rainbow trout, toxin transport
INTRODUCTION
Cyanobacterial blooms occur worldwide in fresh and coastal water. Since a huge amount of toxins can be released during or after the breakdown of the bloom, toxic cyanoblooms have been associated with fish kills all over the globe (Albay et al., 2003; Bürgi and Stadelmann, 2002; Rodger and Turnbull, 1994). Fish kills may be the result of numerous simultaneously occurring factors including e.g. oxygen depletion, alkaline pH, and cyanotoxins. It is not surprising that cyanobacterial blooms have deleterious effects on fish populations given that approximately 75 % of the blooms contain toxins (Lawton and Codd, 1991) while at the same time fish, in comparison to terrestrial animals, have limited possibilities in avoiding exposure to the bloom and its toxins. In fact it could be shown that bony fish from surface waters experiencing blooms also incorporated the most abundant cyanobacterial toxin present: microcystin (MC) (Krishnamurthy et al., 1986; Magalhães, 2001; Sipiä et al., 2001). Given that there is a fish species-dependent sensitivity towards MC (Kotak et al., 1996; Råbergh et al., 1991; Tencalla, 1995; Tencalla et al., 1994), these species differences in toxin susceptibility will lead to profound effects on the species diversity within a given ecosystem. In addition, as discussed previously in various studies (Peng et al., 2010; Poste et al., 2011), accumulation of MC in edible fish is likely to pose a threat to human health. However, despite the serious implications described above only limited information is available e.g. the underlying mechanisms (toxicodynamics) of MC toxicity in bony fish, while the disposition of MC i.e. MC uptake and systemic distribution in fish remains completely unclear. Investigation of the latter however could help to understand why certain species appear more likely to accumulate MC and whether edible parts of the fish are potentially more contaminated with toxin than others.
Indeed, experimental applications of either MC containing bloom material or purified MC in fish resulted in liver-, kidney-, gut- and gill-pathology as well as associated effects e.g. decreased hemoglobin, increased serum liver enzyme activities and ROS formation (Fischer and Dietrich, 2000; Li et al., 2004; Tencalla, 1997). These pathological alterations are explained by the fact that MCs inhibit cytosolic as well as nucleosolic serine (Ser) and threonine (Thr) protein phosphatases (PP). Ser/Thr-PP-inhibition leads to protein hyperphosphorylation and as a consequence to altered signal transduction pathways, cytoskeletal disintegration and oxidative stress via mitochondrial toxicity, finally leading to apoptosis and cell necrosis. MCs are structurally complex molecules with a molecular weight around 1000 Da. Obviously these amphiphilic to lipophilic cyclic peptides do not readily pass through the cell membrane to enter into a hydrophilic environment e.g. the cytosol. Indeed, MCs require active uptake from the cells ambient media and carrier mediated transport across the membrane into the cell, which was demonstrated in humans, mice, rats and in the little skate to occur via organic anion transporting polypeptides (OATPs/Oatps) (Fischer et al., 2010; Fischer et al., 2005; Hagenbuch and Gui, 2008; Lu et al., 2008; Meier-Abt et al., 2005).
Organic anion transporting polypeptides (OATPs in mammals/Oatps in fish) (Hagenbuch and Stieger, 2013) mediate the uptake of various amphipathic endogenous and exogenous organic compounds. So far, ≥ 300 OATPs/Oatps have been annotated, thereby representing ≥ 40 animal species. Based on their amino acid identity OATPs can be divided into six families (OATP1-6, with ≥ 40 % amino acid identity) and several subfamilies (with ≥60 % amino acid identity) (Hagenbuch and Meier, 2004). OATPs can be expressed in an ubiquitous or in an organ specific manner (Hagenbuch and Stieger, 2013). Typical substrates are bile salts, eicosanoids, steroids and steroid conjugates, thyroid hormones, prostaglandins, oligo-peptides as well as pharmaceuticals and toxins, e.g. the phalloidins and amanitins of the highly toxic amanita species (death cap) and the cyanobacterial microcystins (König, 2011; Roth et al., 2012). In humans MC organ/cellular uptake is mediated by the liver specific OATP1B1 and OATP1B3 as well as by OATP1A2 (Fischer et al., 2010; Fischer et al., 2005; Monks et al., 2007). In rodents OATP1B2 was identified as a specific MC transporter (Fischer et al., 2005; Lu et al., 2008).
Based on the knowledge in mammals we hypothesize that an ortholog of the OATP1 subfamily is expressed in bony fish as well, fulfilling a similar function in the liver and playing a key role in detoxification. We propose that this ortholog transports MC which would help to understand the kinetics of MC. We furthermore aim to investigate if the expression profile of this Oatp can explain the often observed pathologies and accumulations in various fish tissue including the muscle as most edible tissue.
In cartilaginous fish a transporter belonging to the OATP1 family has been identified and was demonstrated to transport [3H]-demethylphalloidin and [3H]-dehydro-MC-LR (Cai et al., 2002; Meier-Abt et al., 2005). In addition, several Oatps have been annotated for zebrafish in a phylogenetic analysis (Popovic et al., 2010). Amongst these, the zebrafish Oatp1d1 was further characterized regarding its functional and structural properties using specific OATP substrates, albeit without specifically characterizing its MC transport capabilities (Popovic et al., 2013). As primarily salmonids appeared to be involved in many toxic cyanobacterial bloom associated fish kills reported, the widespread salmonid aquaculture, and the reported MC intoxication of salmonids in aquacultures (net-pen disease) (Andersen et al., 1993), MC contaminated fish may pose an important route of MC exposure for humans. Therefore the focus for identifying potential Oatp orthologs of the human liver OATP1 members was placed on salmonids, in this case rainbow trout. Thus, a rainbow trout liver cDNA library was screened for a putative Oatp that could be functionally expressed in vitro and then characterized with regard to Oatp substrate specificity and MC transport.
METHODS
Reagents and materials
[3H] taurocholic acid (TCA) (0.21 Tera Becquerel (TBq)/mmol), [3H] estrone sulfate ammonium salt (E3S) (2 TBq/mmol), [3H] methotrexate disodium salt (MTX) (1.8 TBq/mmol), [3H] estradiol-17-β-D-glucuronide (E17βG) (1.8 TBq/mmol), [3H] bromosulfophthalein [BSP] (0.5 TBq/mmol), [3H] [D-penicillamine 2,5]encephalin (DPDPE) (1.7 TBq/mmol), [3H] dehydroepiandrosterone sulfate sodium salt (DHEAS) (2.9 TBq/mmol), [3H] ritonavir (0.04 TBq/mmol), [3H] paclitaxel (1.7 TBq/mmol), [3H] docetaxel (2.2 TBq/mmol), [3H] pravastatin (0.6 TBq/mmol), [3H] ouabain (0.6 TBq/mmol), [3H] oleic acid (1.2 TBq/mmol) and [3H] digoxin (1.4 TBq/mmol) were purchased from Perkin Elmer Inc. (Waltham, USA), American Radiolabeled Chemicals Inc. (Saint Louis, USA), Moravek Biochemicals (Brea, USA) and Hartmann Analytic GmbH (Braunschweig, Deutschland). MC-LR was from Enzo Life Science, Inc. (New York, USA). Reverse-transcription and PCR reagents were purchased from New England Biolabs (Ipswich, UK) and cell culture material from PAA Laboratories (Cölbe, Germany) unless indicated otherwise. All other chemicals and antibodies, unless indicated otherwise, were from Sigma-Aldrich (Taufkirchen, Germany). Live rainbow trout were purchased from a local hatchery (Riebel, Reichenau, Germany).
Isolation of Oatp cDNA and phylogenetic analysis
A rainbow trout liver cDNA library was constructed in the vector pCMV-Sport6 with the Gateway Super Script Plasmid Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The library was screened with a 32P dCTP labeled PCR-fragment of a rainbow trout sequence, which was amplified using degenerate primers binding to OATP/Oatp consensus sequences as suggested in (Cai et al., 2002). The resulting PCR product was sequenced and identified as Oatp via blast analysis, and gene specific primers were designed. Amplification with these specific primers resulted in a 377 bp amplicon which was used for screening of the library under high stringency conditions (final two wash steps: 0.5 × SSC/0.1 % SDS; 65 °C; 20 min). After two rounds of screening a single clone with a 2772 bp insert was identified. It contained the full-length open reading frame of a rainbow trout liver Oatp (rtOatp) flanked by 5′- and 3′-untranslated sequences. The DNA sequence was determined for both strands (Eurofins MWG GmbH, Ebersberg, Germany) and was submitted to NCBI (http://www.ncbi.nlm.nih.gov/projects/geo) with Acc# KJ831065. After aligning OATP sequences with ClustalW, phylogenetic analyses were carried out using MEGA 6.06 software (statistical method: Neighbor-Joining, Jones-Taylor-Thornton model, pairwise deletions). Branch strength was tested using bootstrap methodology (5000 replicates). Representative OATP amino acid sequences from vertebrates including fish were used as listed in Supplemental table 1. According to the above analyses the cloned rtOatp was classified as rtOatp1d1. The tree was constructed using iTOL (Letunic and Bork, 2007).
Organ distribution of rtOatp1d1 via PCR and real time PCR
To determine tissue specific expression of rtOatp1d1, rainbow trout were dissected immediately after cervical dislocation and tissues stored in RNAlater at −80 °C. For isolation of total RNA, tissue samples were disrupted in TriFast (Peqlab Biotechnologie GmbH, Erlangen, Germany) using a tissue lyser II (Quiagen, Hilden, Germany). RNA was extracted using a Quiagen Mini Kit according to manufacturer’s instructions. One microgram of RNA was used to prepare cDNA using 200 U of SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA), 2.5 μM oligo (dT), 2.5 μM random hexamers, 500 μM dNTP, 10 mM DTT and 40 U Rnase Inhibitor. The reverse transcription reaction was incubated at 42 °C for 5 min followed by 45 min at 50 °C. The reaction was stopped by 15 min incubation at 72 °C. RNA was degraded by incubation with 5 U RNase H for 20 min at 37 °C. PCR and real-time PCR was carried out on 5 individual rainbow trout. PCR with subsequent gel-electrophoresis was carried out with Taq polymerase and the specific primers for Oatp1d1 as listed in Supplemental table 2. Elongation factor 1α (ef1α) was used as reference gene. For semi-quantitative real time PCR a 1:5 dilution of the cDNA was used whereby two reference genes, ef1α and 18S-rRNA, served as normalization controls as a similar expression amongst organs could be shown for the two reference genes (Supplemental figure 1 F). SYBR Green (Bioline, London, UK) was used according to manufacturer’s instructions. RNA melting curve analysis was conducted following real-time PCR to ensure lack of secondary products (Supplemental figure 1 A–C). Standard curves for the primers used were analyzed in a preliminary study with technical triplicates, whereby efficiency of the amplification was 76 % for 18s-rRNA and nearly 100 % for ef1α and rtOatp1d1 (Supplemental figure 1 D–E). Data were analyzed according to the Δct method.
Expression of rtOatp1d1 in HEK293 cells
Human embryonic kidney (HEK293) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 25 mM glucose and 4 mM L-glutamine supplemented with 10 % FBS, 100 units/ml penicillin and 100 mg/ml streptomycin at 37 °C and 5 % CO2. For transient transfections of rtOatp1d1 two different plasmids were used. One plasmid was the original pCMV-Sport6 containing the isolated cDNA. The other plasmid was constructed by cutting the cDNA with NotI and EcoRI out of pCMV-Sport6 and cloning the resulting insert after gel purification into the precut pTracer™CMV/Bsd (Invitrogen, Carlsbad, CA, USA). This construct allowed using expression of GFP to verify transfection success. Comparison of transfection with both vectors with regard to taurocholate uptake resulted in similar transport results (data not shown). The respective cDNA or the empty vector as a control (0.5 μg per well on a 24-well plate) were transfected with FuGENE (Promega) at 60–80 % HEK293 cell confluence.
Uptake studies
HEK293 cells were seeded on poly-D-lysine coated 24 well plates and transfected as described above. Substrate uptake was measured 24 h and 48 h after transfection with either pCMV-Sport6 or pTracer™CMV/Bsd, respectively. For uptake experiments cells were washed 3 times with warm uptake buffer (116.4 mM NaCl, 5.3 mM KCl, 1.6 mM NaH2PO4, 0.8 mM MgSO4, 5.5 mM D-Glucose, 20 mM Hepes, pH adjusted to 7.4 with Trizma base). Subsequently, cells were incubated with 200 μl/well of uptake buffer containing 0.3 μCi of the respective radiolabeled substrate at the following final concentrations: [3H] TCA (30.0 nM), [3H] E3S (6.6 nM), [3H] MTX (13.9 nM), [3H] E17βG (6.1 nM), [3H] BSP (20.7 nM), [3H] DPDPE (6.7 nM), [3H] DHEAS (3.8 nM), [3H] ritonavir (300.0 nM), [3H] paclitaxel (6.6 nM), [3H] docetaxel (5.0 nM) or [3H] pravastatin (40.0 nM). After 5 min of incubation substrate uptake was stopped by washing the cells 6 times with ice-cold uptake buffer. Subsequently, cells were solubilized with 300 μl 1 % Triton X-100 per well and 200 μl were used to quantify radioactivity by β-scintillation counting, while the remaining solution was used to determine total protein concentrations (triplicate analyses) using the BCA protein assay kit (Pierce Biotechnology, Rockford, USA).
For three demonstrated rtOatp1d1 substrates additional experiments were performed to determine uptake kinetics under initial linear rate conditions (30 sec) at substrate concentrations ranging from 0.1 μM to maximally 300 μM. Uptake was normalized to total protein concentration within each respective experiment. Transporter specific uptake was calculated by subtracting the counts of empty vector transfected cells from those of rtOatp1d1-transfected cells. Data points were means ± SEM of three technical replicates and a minimum of 4 experiments were performed. In one case, only 1 experiment could be performed. In that case the 3 technical replicates served to determine the means ± SEM for each time-point. A non-linear regression (Michaelis-Menten) curve was fitted to the data to obtain the Km-values.
Immunoblotting for MC-LR detection
Forty-eight hours after transfection with pTracer™CMV/Bsd HEK293 cells were incubated with 50 nM MC-LR for 1 h, 6 h or 24 h. Incubation with methanol served as solvent control, whereby methanol concentrations never exceeded 0.2 %. HEK293 cells stably expressing human OATP1B3 served as positive and empty vector transfected cells or non-transfected cells as negative controls. Proteins were isolated from cells with cold buffer containing 10 mM Tris-base, pH 7.5, 140 mM NaCl, 5 mM EDTA, 0.1 % (v/v) Triton X-100, pH 7.5 and 1 % protease inhibitor. Following centrifugation at 10,000 g, protein content in the supernatant was measured using the BCA assay kit. Fifteen μg of protein/lane were separated using 10 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at constant 200 V (Laemmli, 1970). Proteins were then transferred onto a nitrocellulose membrane (Whatman, Dassel, Germany) at 300 mA for 90 min (Towbin et al., 1992) and membranes were incubated in blocking buffer (Tris-buffered saline with1 % Tween 20 containing 1 % bovine serum albumin) for 1 h at room temperature. To detect MC-LR bound to phosphatases, nitrocellulose membranes were incubated with the MC-specific primary monoclonal anti-Adda- antibody (1:10) over night at 4 °C (clone AD4G2, (Zeck et al., 2001)). To detect the primary antibody the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (1:80,000; Sigma- Aldrich, Taufkirchen, Germany) for 1 h at room temperature. Immunopositive bands were visualized with Amersham ECL Advance™ Western Blotting Detection Kit (GE Healthcare, Amersham, UK) according to manufacturer’s protocol and the chemiluminescent signal was detected using ImageQuant LAS 4000 mini (GE Healthcare, Germany). After stripping the membranes with 0.2 M NaOH for 10 min they were re-probed with the polyclonal rabbit anti-GAPDH antibody (1:200) (Santa Cruz Biotechnologies, Heidelberg, Germany) again using overnight 4 °C incubation. GAPDH served as immunoblot as well as protein reference control. Quantification was carried out with greyscale analysis (Quantity One version 4.6.9).
Immunoblotting for rtOatp1d1
A polyclonal rabbit antibody was generated against a peptide corresponding to 13 amino acids (PRKSDCDRMFKFYM) at the C-terminal end of rtOatp1d1 by Washington Biotechnology, Inc., Baltimore, USA.
Rainbow trout liver tissue (0.5 g) was homogenized in a glass-teflon potter with 10 ml homogenization buffer (0.25 M Sucrose, 10 mM Tris-HCl, 1 % protease inhibitor and 40 μg/ml phenylmethylsulfonylfluorid). The homogenate was centrifuged for 5 min at 1,000 × g and 4 °C. The resulting supernatant was centrifuged for 5 min at 6,000 × g and 4 °C. The pellet was discarded and the supernatant was centrifuged for 2 h at 100,000 × g and 4 °C. The resulting pellet containing the membrane protein fraction, was re-suspended in 500 μl buffer (0.25 M sucrose and 20 mM HEPES at a pH 7.5) and 25 μg protein were separated using a 10 % gel. SDS-PAGE and western blotting were carried out as described above. Nitrocellulose membranes were blocked with 2 % milk powder in Tris-buffered saline with 1 % Tween 20 overnight at 4 °C followed by incubation with either a 1:10,000 dilution of the anti-rtOatp1d1 antiserum or pre-immunserum for 2 h at 4 °C. Detection and visualization was carried out after a 1 h incubation with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (1:160,000) (Sigma- Aldrich, Taufkirchen, Germany) and subsequent visualization was obtained with the Amersham ECL AdvanceTM Western Blotting Detection Kit (GE Healthcare, Amersham, UK). The chemiluminescent signal was detected using ImageQuant LAS 4000 mini (GE Healthcare, Germany).
RESULTS
Cloning and phylogenetic analysis of a cDNA encoding rtOatp1d1
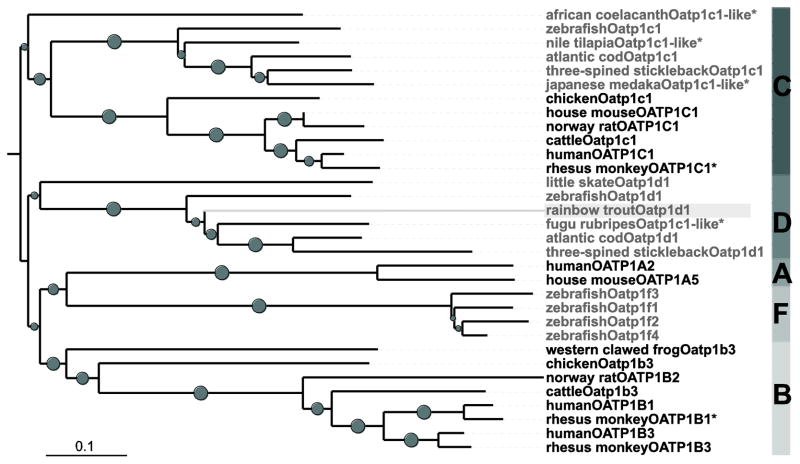
To identify and characterize a rainbow trout ortholog of the human, mouse, rat and little skate OATPs/Oatps, which have been demonstrated to transport MC, a 32P-labeled 377 bp DNA fragment was used as a probe to screen a cDNA library obtained from rainbow trout liver. A 2772 bp cDNA was isolated containing a 5′-UTR of 91 bp, an open reading frame (ORF) of 2115 bp and a 3′-UTR of 586 bp. The ORF encodes a protein with 705 amino acids and based on phylogenetic analyses clusters within the OATP1 family (Figure 1). The rainbow trout Oatp amino acid sequence shares 53–60 % identity with the annotated fish-specific Oatp1d1 subfamily but only 45–48 % identity with members of the OATP1C1 subfamily also present in fish and other vertebrates. Based on these analyses this new rainbow trout Oatp was classified as Oatp1d1, the sequence was deposited (Acc# KJ831065) and is subsequently called rtOatp1d1.
Figure 1.
Phylogenetic tree of selected vertebrates in the OATP1 family. Based on a Clustal W alignment the tree was assembled using Neighbour-Joining Method with pairwise deletion; bootstrap values (5000 replicates) > 0.5 are indicated as grey circles, while the size is proportional to the bootstrap value. * indicates predicted sequences. Fish Oatps are marked in grey, the novel rtOatp1d1 is highlighted with a light grey background.
Tissue distribution of rtOatp1d1
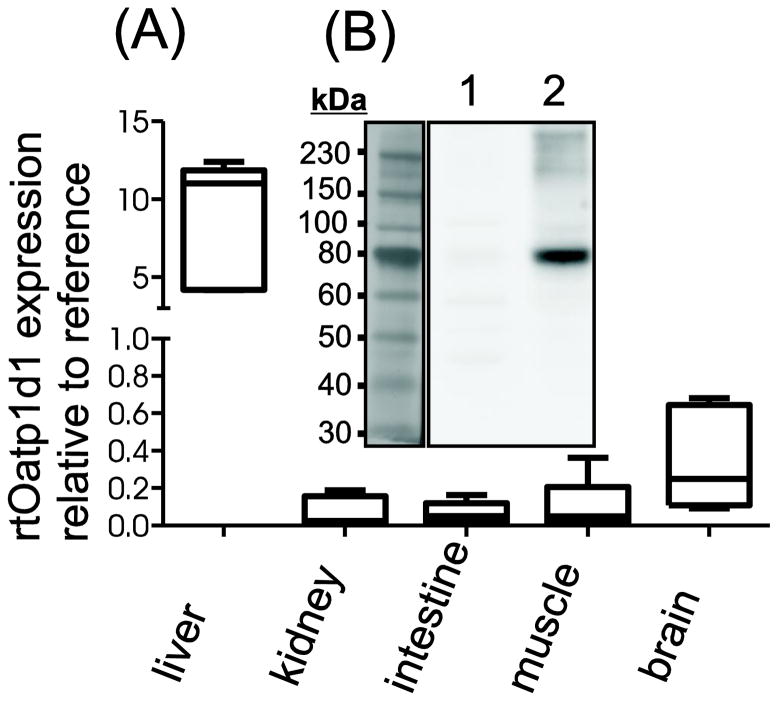
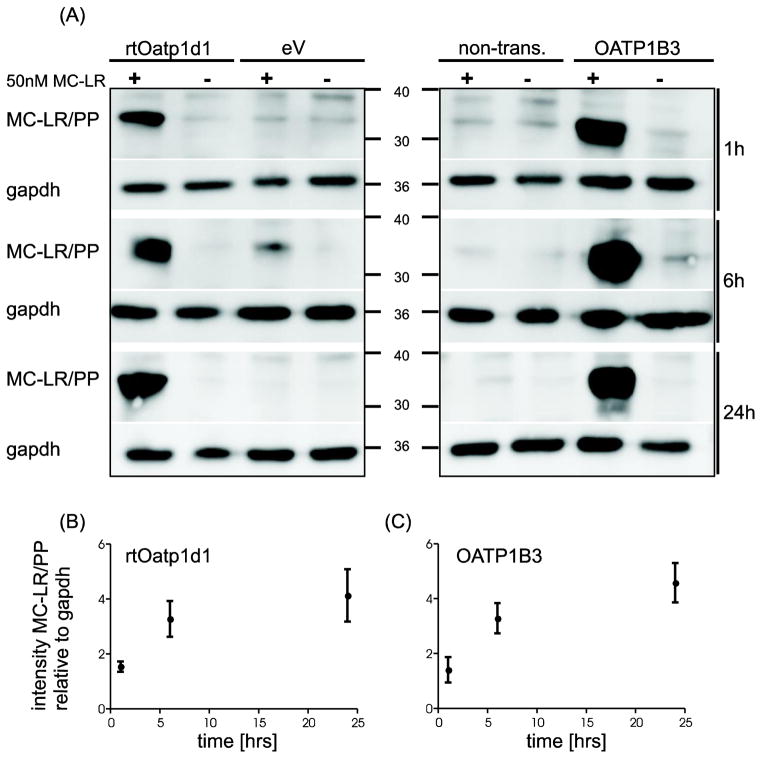
Tissue distribution of rtOatp1d1 at the RNA level was determined using semi-quantitative qRT-PCR. Data summarized in Figure 2A revealed a predominant expression of rtOatp1d1 in the liver, while its expression was moderate in the brain. No mRNA was detected in kidney, muscle and intestine, either due to absence of rtOatp1d1 or due to too low a level of expression to allow reliable detection. Western blot analyses were used to confirm that rtOatp1d1 indeed is expressed in the liver (Figure 2B). An immunopositive band at 80 kDa was detected corresponding nicely with size predictions based on the amino acid composition of rtOatp1d1 sequence (77.1 kDa). Low expression levels prevented detection of rtOatp1d1 protein in other tissues.
Figure 2.
Organ distribution of rtOatp1d1. (A) RNA expression of rtOatp1d1 in several rainbow trout tissues performed with semi-quantitative real time PCR. Expression of rtOatp1d1 was normalized to the mean expression of elongation factor 1α and 18s-RNA as reference genes. Whisker box plots with minimum to the maximum value whiskers; n = 5 fish. (B) rtOatp1d1 protein expression in rainbow trout liver homogenate. Lane 1 pre-immune serum; Lane 2: anti-rtOatp1d1 antibody.
rtOatp1d1 substrate specificity
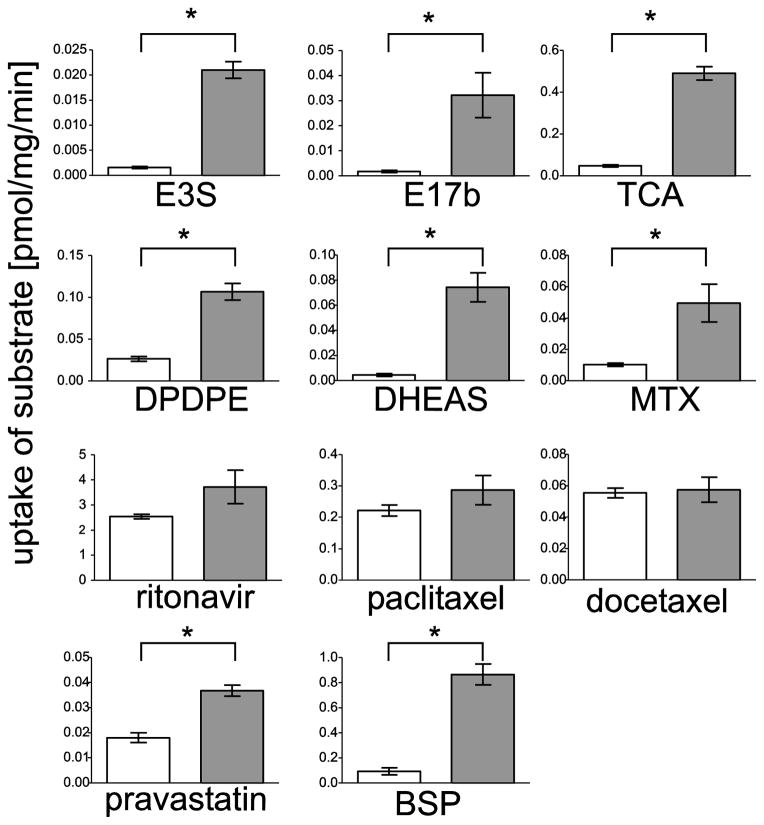
To test the substrate specificity of rtOatp1d1, transiently transfected HEK293 cells (rtOatp1d1-HEK293) were used to determine the uptake of 11 radiolabeled compounds i.e. reported substrates for human OATP1B1, OATP1B3 and OATP1A2. Figure 3 demonstrates that rtOatp1d1-HEK293 cells transported pravastatin, E3S, E17βG, TCA, BSP, DPDPE, DHEAS and MTX, whereby uptake rates ranged between 0.02 (Pravastatin) and 0.44 (TCA) pmol/mg/protein/minute. No transport was observed for docetaxel, paclitaxel and ritonavir.
Figure 3.
Functional characterization of rtOatp1d1 in HEK293 cells. 24 h after transfection, cells were incubated for 5 min with [3H] TCA (30.0 nM), [3H] E3S (6.6 nM), [3H] MTX (13.9 nM), [3H] E17βG (6.1 nM), [3H] BSP (20.7 nM), [3H] DPDPE (6.7 nM), [3H] DHEAS (3.8 nM), [3H] ritonavir (300.0 nM), [3H] paclitaxel (6.6 nM), [3H] docetaxel (5.0 nM) or [3H] pravastatin (40.0 nM). Values represent the mean ± SEM of technical triplicates. Differences in substrate uptake between rtOatp1d1-HEK293 (grey bars) and either eV-HEK293 (in case of DPDPE, DHEAS, TCA and MTX) or non-transfected cells (all others) (represented in white bars). Significant differences were statistically analyzed using a non-parametric Mann–Whitney test (* p<0.05).
E3S, TCA and MTX kinetics in rtOatp1d1-HEK293 cells
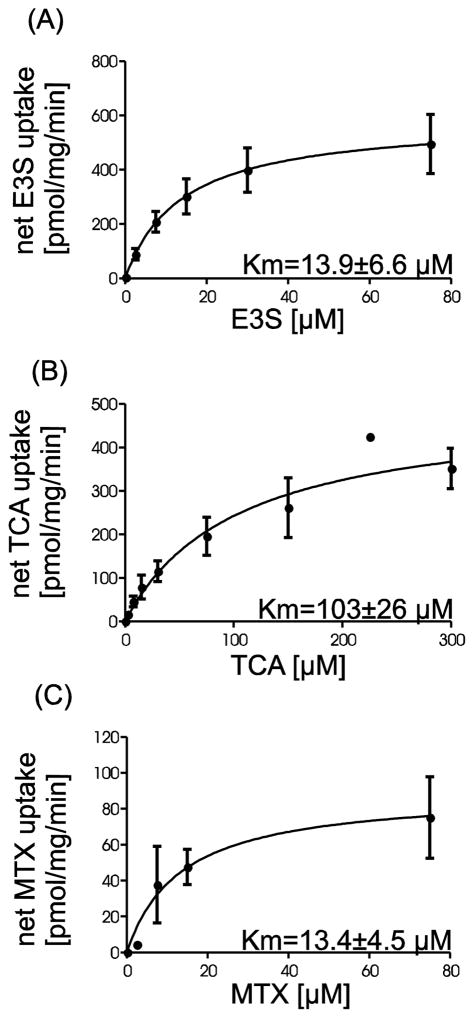
Kinetic experiments were carried out to further characterize substrate transport mediated by rtOatp1d1. Concentration dependent uptake was performed for E3S (0.1 – 75 μM), MTX (0.1 – 75 μM) and TCA (0.1 – 300 μM) under initial linear rate conditions (30 sec) (Figure 4). Nonlinear regression analyses (Michaelis-Menten) revealed Km values of 13.9 μM, 13.4 μM and 103 μM for E3S, MTX and TCA, respectively.
Figure 4.
Kinetic analyses of rtOatp1d1-mediated substrate uptake. Transport was measured within the initial linear uptake phase at 30 sec with varying concentrations between 0.1 μM and 75 μM or 300 μM respectively. (A) E3S (B) TCA and (C) MTX. Values depict mean ± SEM of five (A) or four (B) independent replicates with three technical replicates each. For MTX (C) three determinations of one replicate are shown. A nonlinear regression (Michelis-Menten) was fitted to the net uptake which was calculated by subtracting the unspecific uptake of the eV-HEK293 cells from the uptake of the rtOatp1d1-HEK293 cells.
Microcystin-LR transport by rtOatp1d1-HEK293 cells
In order to test whether rtOatp1d1 is capable of transporting MC-LR, rtOatp1d1-HEK293 cells, HEK293 cells transfected with the empty vector as a negative control, and HEK293 cells expressing human OATP1B3 as a positive control were incubated with 50 nM MC-LR for 1, 6 or 24 h. Subsequent western-blotting with the AD4G2 MC-antibody demonstrated MC covalently bound to intracellular phosphatases and thus represented the MC-LR transported by rtOatp1d1 or OATP1B3 into the HEK293 cells. Indeed, a strong MC-immuno-positive band was detected (between 30 and 40 kDa) for HEK293 cells expressing either rtOatp1d1 or human OATP1B3 (Figure 5A) which compared well with similar immunoblots of OATP1B3 transfected HEK293 cells (Fischer et al., 2010) and neuronal cells (Feurstein et al., 2009) exposed to MC-LR. As previously demonstrated by Feurstein et al. (2009) the MC-LR immunopositive bands corresponded to MC-LR covalently bound to the catalytic subunit of PP1 and PP2A, albeit no specific immuno-detection of PP1 and 2A was carried out on the same blots. Quantification of the immunoblots suggested that immunodetectable MC-LR increased with MC-LR exposure time in both, the transiently transfected rtOatp1d1-HEK293 cells (Figure 5B) and the stably transfected human OATP1B3-HEK293 cells (Figure 5C). Detectable MC-LR decreased when rtOatp1d1-HEK293 cells where co-incubated with 100 μM TCA or BSP (Figure 6). Contrary to expectations, co-incubation with 10 μM TCA or BSP did not lead to a reduced signal, suggesting a higher affinity of MC-LR than BSP or TCA for rtOatp1d1, as also suggested by kinetic experiments with TCA. As demonstrated earlier by Fischer et al. (2010) TCA did not appear to significantly compete with MC-LR uptake in OATP1B3-HEK293 cells. Co-incubation with BSP however provided for a similar reduction of detectable intracellular MC-LR in both rtOatp1d1- and OATP1B3-transfected HEK293 cells.
Figure 5.
MC-LR uptake mediated by rtOatp1d1. (A) Immunoblot showing uptake of MC-LR mediated by transiently transfected rtOatp1d1-HEK293, ev-HEK293, non-transfected cells (non-tr.) or stable transfected human OATP1B3-HEK293. Cells were incubated with 50 nM MC-LR (+), or solvent control MeOH (−) for 1, 6 and 24 h, GAPDH has been used as reference gene; (B) and (C) Densitometric greyscale analysis (n=5, mean ± SEM) was used to compare the uptake of MC-LR in (B) rtOatp1d1 and (C) human OATP1B3- transfected HEK293 cells, after subtracting greyscale signals for ev-HEK293 cells or non-transfected HEK293 cells respectively.
Figure 6.
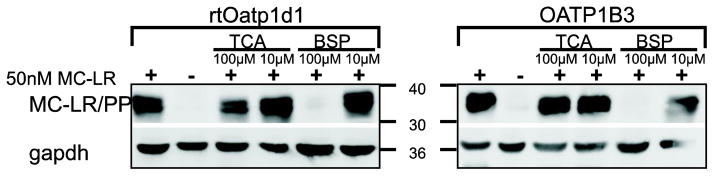
Competitive inhibition of rtOatp1d1-mediated uptake of MC-LR by TCA and BSP. Immunoblot showing uptake of 50nM MC-LR for 24 h (MC-LR +) in the absence of presence of 100 μM or 10 μM TCA or BSP. MeOH was used as solvent control (−). rtOatp1d1: transiently transfected rtOatp1d1-HEK293 cells; OATP1B3: stably transfected OATP1B3-HEK cells.
DISCUSSION
In this study a novel rainbow trout Oatp transporter (rtOatp) was identified and its functional characterization revealed that it can transport MC. Indeed, screening of a rainbow trout liver cDNA library resulted in the isolation of a 2772 bp cDNA that encodes for a 705 aa protein. Subsequent blast analysis confirmed the presumed affiliation of this sequence with the OATP1 family. The ensuing phylogenetic analysis of OATP1 subtypes from 16 vertebrates including 9 fish species added more insight and suggested that the novel rtOatp belongs to the Oatp1d1 subfamily, which to date has only been reported in fish.
Functional expression of rtOatp1d1 in HEK293 cells resulted in the transport of several of the known OATP substrates, including E17βG, E3S, TCA, DPDPE, DHEAS, MTX, pravastatin, docetaxel, paclitaxel and MC (summarized in (Hagenbuch and Stieger, 2013), suggesting that rtOatp1d1 is a multi-specific transporter comparable with the liver-specific human OATP1B1 and OATP1B3 or the human OATP1A2, primarily expressed in the brain but also in various other tissues. rtOatp1d1 affinities for the three tested substrates E3S, TCA and MTX were in general comparable to the affinities of other transporters of the OATP1 family in humans and fish, which mostly vary between 10 and 80 μM (Table 1). However, given that these results were obtained by different research groups in vastly differing expression systems, the comparison of the substrate affinities is not absolute, but can serve for orientation purposes. It appears that within fish, both little skate and rainbow trout Oatp1d1 have comparable affinities for TCA, while they vary for E3S (Table 1). Transport of E3S in rtOatp1d1 appears to be monophasic with a Km value of 13.9 μM, whereas a biphasic transport mechanism has been shown for OATP1B1 (Gui and Hagenbuch, 2009; Noé et al., 2007; Tamai et al., 2001).
Table 1.
Affinities for the tested substrates in this study compared to human and fish transporters of the Oatp1 family. Given as Km value in μM.
| subtype | species | Km [μM] | references | ||
|---|---|---|---|---|---|
| E3S | TCA | MTX | |||
| OATP1B1 | Human | 0.23/45 | 10–34 | - | (Abe et al., 1999; Cui et al., 2001; Hsiang et al., 1999; Noé et al., 2007) |
| OATP1B3 | Human | 58 | 6–42 | 25–39 | (Abe et al., 2001; Briz et al., 2006; Gui et al., 2008; Leuthold et al., 2009) |
| OATP1A2 | Human | 16–59 | 60 | 457 | (Badagnani et al., 2006; Bossuyt et al., 1996; Kullak-Ublick et al., 1995; Lee et al., 2005) |
| Oatp1d1 | Zebrafish | 1.75 | - | - | (Popovic et al., 2013) |
| Oatp1d1 | Little skate | 61 | 85 | - | (Cai et al., 2002) |
| Oatp1d1 | Rainbow trout | 13.9 | 103 | 13.4 | |
Several members of the OATP1 subfamily have been demonstrated to transport various MC congeners (Fischer et al., 2010). Based on the fact that this novel rtOatp1d1 is also a member of the OATP1 subfamily it is not surprising that also rtOatp1d1 is capable of transporting MC-LR. A similar MC-LR uptake velocity was found when comparing the time course of MC-LR uptake into HEK293 cells transiently transfected with rtOatp1d1- and stably transfected with human OATP1B3 (Figure 5B and C). Direct competition experiments using BSP and TCA to inhibit MC-LR uptake (Figure 6) suggest that MC-LR has a lower affinity for rtOatp1d1 than for OATP1B3 given that the affinities of TCA for the two transporters are similar (Table 1). In view of the lacking Km values for BSP in rtOatp1d, the affinity of BSP and thus competition with MC-LR can only be speculated, albeit the comparison of western blots (Figure 6) would suggest that BSP has a lower affinity to rtOatp1d1 when tested at equimolar competitive concentrations of MC-LR. The fact that other Oatp substrates like TCA and BSP can inhibit MC uptake conversely raises the question whether MC is also able to minimize the uptake of physiological substrates transported by Oatp1d1 with a low affinity. Since this could lead to an altered homeostasis of endogenous substrates, a reduced fitness of the fish would be expected.
In order to understand the significance of rtOatp1d1 for potential MC induced organ toxicity, expression analyses suggested that rtOatp1d1 is highly expressed in the liver followed by the brain. This distribution pattern compares well with the ortholog pattern found in the little skate Oatp1d1 (Cai et al., 2002). Thus rtOatp1d1 represents a functional fish homolog of the mammalian OATP1B1/OATP1B3 with a similar substrate spectrum i.e. including the cyanotoxin MC. Therefore a functional role of rtOatp1d1 as transporter for bile acid metabolism, steroid distribution and detoxification can be assumed.
The organ distribution pattern observed and the capability to transport MC-LR corroborate earlier findings of intra-organ/intra-cellular MC detected in Salmonidae and Cyprinidae (Fischer and Dietrich, 2000; Fischer et al., 2000). Pathologies of MC were mainly reported in the liver of several fish, suggesting that due to high expression of Oatp1d1 in the liver, the liver is primarily affected during MC intoxications. However, MC was also reported in the brain of carp (Fischer and Dietrich, 2000) and live-bearing fish (Cazenave et al., 2005), suggesting an Oatp1d1 mediated MC accumulation and neurotoxicity. Indeed, changes in fish swimming activity (Cazenave et al., 2008; Ernst et al., 2007; Tencalla et al., 1994) support a MC mediated neurotoxicity. Consequently the presence of fish Oatp1d1 in liver and brain appears to be key for MC uptake and the subsequent development of MC mediated organ pathology and thus morbidity and mortality of fish exposed.
Interestingly organs primarily affected following MC intoxication as well as the severity of pathologies/toxicity observed varies between species. Indeed, while in rainbow trout pathology was primarily observed in the liver, cyprinids presented also with kidney pathology, albeit less severe than that observed in the liver of the same animals (Carbis et al., 1996; Fischer and Dietrich, 2000; Råbergh et al., 1991). The latter observations corroborate findings of a species-specific susceptibility towards MC upon oral exposure and intraperitoneal injection and employing LD50/LC50 values for comparison (Fischer and Dietrich, 2000; Kotak et al., 1996; Råbergh et al., 1991; Tencalla, 1995; Tencalla et al., 1994). As the expression levels and MC transporting capabilities of OATPs/Oatps appear more critical than the Ser/Thr PP inhibiting capabilities of the individual MC-congeners for the development of tissue damage and thus the apical toxicity (Fischer et al., 2010), current data would suggest that also for fish the expression and expression level of Oatp1d1 is the critical factor governing morbidity and mortality of fish exposed to MC containing cyanobacterial blooms. Hence, adverse effects of toxic cyanobacterial blooms to aquatic ecosystems including a shift of species distribution largely depend on the organ distribution of Oatp1d1 and their inherent susceptibility to MCs of the respective inhabitant fish species.
It is interesting to note that oral exposure of fish to MC resulted in pathological alterations and accumulation of MC in various organs including kidney, muscle gut and gill (Fischer and Dietrich, 2000; Li et al., 2004; Tencalla, 1997). Despite the obvious transport of MCs across the gastro-intestinal wall to the portal vein, expression of rtOatp1d1 was not found in the gastro-intestinal tract of rainbow trout. A very low expression level of rtOatp1d1 and thus low uptake of MC per time unit would be in disagreement with the rapid onset of pathology in the liver as observed upon oral MC exposure in trout and carp. Likewise an accumulation of MC in muscle tissue cannot be explained by the rtOatp1d1 organ expression data shown. As most likely other Oatp subtypes are present in the fish, the presence of MC in the gastrointestinal tract and muscle can be explained by a so far unidentified transporter, capable of transporting MCs. Indeed, several studies reported the accumulation of MC in fish muscle tissue at tissue concentrations that would surpass the tolerable daily intake (TDI) value, recommended by the world health organization (WHO) (Magalhães, 2001; Poste et al., 2011; Song et al., 2007), upon normal daily fish consumption.
In summary, despite that rtOatp1d1 shows the highest phylogenetic relationship with OATP1C1, the comparable functional characteristics places it closer to the human OATP1A2, OATP1B1 and OATP1B3. Indeed, the broad spectrum of transported substrates, including bile salts, hormone steroids and xenobiotica, overlaps greatly with the human OATP1A and OATP1B subtypes suggesting that rtOatp1d1 plays a similar role in fish as OATP1A2, OATP1B1 and OATP1B3 in humans. Similar to these three human OATPs rtOatp1d1 was identified as an MC transporter, allowing for further insight into the kinetics of MC in fish. Based on the organ distribution of rtOatp1d1 in rainbow trout, adverse effects mediated by acute as well as chronic MC exposure may be expected primarily in the liver and in brain and result in the acute or slow demise of individual fish as well as whole populations in ecosystems subject to recurring toxin producing cyanobacterial blooms.
Supplementary Material
Highlights.
A new Oatp1d1 in rainbow trout (rtOatp1d1) was cloned, identified and characterized
rtOatp1d1 is predominantly expressed in the liver
rtOatp1d1 displays multi-specific transport of endogenous and xenobiotic substrates
rtOatp1d1 is a homolog of the OATP1A1, OATP1B1 and OATP1B3
rtOatp1d1 is a microcystin (MC) transporter
Acknowledgments
FUNDING
This work was enabled by financial support from a DFG Center Grant, the International Max Planck Research School for Organismal Biology (IMPRS), by a Marie Curie International Research Staff Exchange Scheme Fellowship (PIRSES-GA-2011-295223), and by grants from the National Center for Research Resources (RR021940) and the National Institute of General Medical Sciences (GM077336 and GM103549) of the National Institutes of Health.
LIST OF ABBREVIATIONS
- OATP
organic anion transporting polypeptide
- MC
Microcystin
- MC-LR
Microcystin-LR
- PP
protein phosphatases
- ROS
reactive oxygen species
- rtOatp1d1
rainbow trout Oatp1d1
- HEPES
-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- ORF
open reading frame
- TCA
taurocholic acid
- E3S
estrone sulfate ammonium salt
- MTX
methotrexate disodium salt
- E17βG
estradiol-17-β-D-glucuronide
- BSP
bromosulfophthalein
- DPDPE
[D-penicillamine 2,5]encephalin
- DHEAS
dehydroepiandro-sterone sulfate sodium salt
- LD50
lethal dose killing 50% of dosed animals
- LC50
lethal concentration killing 50 % of exposed animals
- TDI
tolerable daily intake
- WHO
World Health Organization
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Konstanze Steiner, Email: konstanze.steiner@uni-konstanz.de.
Bruno Hagenbuch, Email: bhagenbuch@kumc.edu.
Daniel R. Dietrich, Email: daniel.dietrich@uni-konstanz.de.
References
- Abe T, Kakyo M, Tokui T, Nakagomi R, Nishio T, Nakai D, Nomura H, Unno M, Suzuki M, Naitoh T, Matsuno S, Yawo H. Identification of a novel gene family encoding human liver-specific organic anion transporter LST-1. J Biol Chem. 1999;274:17159–17163. doi: 10.1074/jbc.274.24.17159. [DOI] [PubMed] [Google Scholar]
- Abe T, Unno M, Onogawa T, Tokui T, Kondo TN, Nakagomi R, Adachi H, Fujiwara K, Okabe M, Suzuki T, Nunoki K, Sato E, Kakyo M, Nishio T, Sugita J, Asano NAjopGalp, Tanemoto M, Seki M, Date F, Ono K, Kondo Y, Shiiba K, Suzuki M, Ohtani H, Shimosegawa T, Iinuma, Nagura H, Ito S, Matsuno S. LST-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology. 2001;120:1689–1699. doi: 10.1053/gast.2001.24804. [DOI] [PubMed] [Google Scholar]
- Albay M, Akcaalan R, Tufekci H, Metcalf J. Depth profiles of cyanobacterial hepatotoxins (microcystins) in three Turkish freshwater lakes. Hydrobiologia 2003 [Google Scholar]
- Andersen RJ, Luu HA, Chen DZ, Holmes CF, Kent ML, Le Blanc M, Taylor FM, Williams DE. Chemical and biological evidence links microcystins to salmon ‘netpen liver disease’. Toxicon. 1993;31(10):1315–1323. doi: 10.1016/0041-0101(93)90404-7. [DOI] [PubMed] [Google Scholar]
- Badagnani I, Castro RA, Taylor TR, Brett CM, Huang CC, Stryke D, Kawamoto M, Johns SJ, Ferrin TE, Carlson EJ, Burchard EG, Giacomini KM. Interaction of methotrexate with organic-anion transporting polypeptide 1A2 and its genetic variants. J Pharmacol Exp Ther. 2006;318(2):521–9. doi: 10.1124/jpet.106.104364. [DOI] [PubMed] [Google Scholar]
- Bossuyt X, Muller M, Meier PJ. Multispecific amphipathic substrate transport by an organic anion transporter of human liver. J Hepatol. 1996;25(5):733–738. doi: 10.1016/s0168-8278(96)80246-7. [DOI] [PubMed] [Google Scholar]
- Briz O, Romero MR, Martinez-Becerra P, Macias RI, Perez MJ, Jimenez F, San Martin FG, Marin JJ. OATP8/1B3-mediated cotransport of bile acids and glutathione: an export pathway for organic anions from hepatocytes? J Biol Chem. 2006;281(41):30326–35. doi: 10.1074/jbc.M602048200. [DOI] [PubMed] [Google Scholar]
- Bürgi H, Stadelmann P. Change of phytoplankton composition and biodiversity in Lake Sempach before and during restoration. Hydrobiologia. 2002;469:33–48. [Google Scholar]
- Cai SY, Wang W, Soroka CJ, Ballatori N, Boyer JL. An evolutionarily ancient Oatp: insights into conserved functional domains of these proteins. Am J Physiol Gastrointest Liver Physiol. 2002;282:G702–10. doi: 10.1152/ajpgi.00458.2001. [DOI] [PubMed] [Google Scholar]
- Carbis C, Rawlin G, Mitchell G, Anderson J, McCauley I. The histopathology of carp, Cyprinus carpio L exposed to microcystins by gavage, immersion and intraperitoneal administration. Journal of Fish Diseases. 1996;19(3):199–207. [Google Scholar]
- Cazenave J, Nores ML, Miceli M, Díaz MP, Wunderlin Da, Bistoni Ma. Changes in the swimming activity and the glutathione S-transferase activity of Jenynsia multidentata fed with microcystin-RR. Water Res. 2008;42:1299–307. doi: 10.1016/j.watres.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Cazenave J, Wunderlin DA, de Los Angeles Bistoni M, Amé MV, Krause E, Pflugmacher S, Wiegand C. Uptake, tissue distribution and accumulation of microcystin-RR in Corydoras paleatus, Jenynsia multidentata and Odontesthes bonariensis A field and laboratory study. Aquat Toxicol. 2005;75:178–90. doi: 10.1016/j.aquatox.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Cui Y, König J, Leier I, Buchholz U, Keppler D. Hepatic uptake of bilirubin and its conjugates by the human organic anion transporter SLC21A6. J Biol Chem. 2001;276:9626–9630. doi: 10.1074/jbc.M004968200. [DOI] [PubMed] [Google Scholar]
- Ernst B, Hoeger SJ, O’brien E, Dietrich DR. Physiological stress and pathology in European whitefish (Coregonus lavaretus) induced by subchronic exposure to environmentally relevant densities of Planktothrix rubescens. Aquat Toxicol. 2007;82:15–26. doi: 10.1016/j.aquatox.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Feurstein DJ, Holst K, Fischer A, Dietrich DR. Oatp-associated uptake and toxicity of microcystins in primary murine whole brain cells. Toxicol Appl Pharmacol. 2009;234:247–55. doi: 10.1016/j.taap.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Fischer A, Hoeger SJ, Stemmer K, Feurstein DJ, Knobeloch D, Nussler A, Dietrich DR. The role of organic anion transporting polypeptides (OATPs/SLCOs) in the toxicity of different microcystin congeners in vitro: a comparison of primary human hepatocytes and OATP-transfected HEK293 cells. Toxicol Appl Pharmacol. 2010;245(1):9–20. doi: 10.1016/j.taap.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Fischer WJ, Altheimer S, Cattori V, Meier PJ, Dietrich DR, Hagenbuch B. Organic anion transporting polypeptides expressed in liver and brain mediate uptake of microcystin. Toxicol Appl Pharmacol. 2005;203:257–63. doi: 10.1016/j.taap.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Fischer WJ, Dietrich DR. Pathological and biochemical characterization of microcystin-induced hepatopancreas and kidney damage in carp (Cyprinus carpio) Toxicol Appl Pharmacol. 2000;164:73–81. doi: 10.1006/taap.1999.8861. [DOI] [PubMed] [Google Scholar]
- Fischer WJ, Hitzfeld BC, Tencalla F, Eriksson JE, Mikhailov A, Dietrich DR. Microcystin-LR toxicodynamics, induced pathology, and immunohistochemical localization in livers of blue-green algae exposed rainbow trout (Oncorhynchus mykiss) Toxicol Sci. 2000;54:365–73. doi: 10.1093/toxsci/54.2.365. [DOI] [PubMed] [Google Scholar]
- Gui C, Hagenbuch B. Role of transmembrane domain 10 for the function of organic anion transporting polypeptide 1B1. Protein Sci. 2009;18:2298–306. doi: 10.1002/pro.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui C, Miao Y, Thompson L, Wahlgren B, Mock M, Stieger B, Hagenbuch B. Effect of pregnane X receptor ligands on transport mediated by human OATP1B1 and OATP1B3. Eur J Pharmacol. 2008;584(1):57–65. doi: 10.1016/j.ejphar.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ. Organic anion transporting polypeptides of the OATP/SLC21 family: phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Pflugers Arch. 2004;447(5):653–65. doi: 10.1007/s00424-003-1168-y. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Stieger B. The SLCO(former SLC21) superfamily of transporters. Mol Aspects Med. 2013;34:369–412. doi: 10.1016/j.mam.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang B, Zhu Y, Wang Z, Wu Y, Sasseville V, Yang WP, Kirchgessner TG. A novel human hepatic organic anion transporting polypeptide (OATP2) J Biol Chem. 1999;274:37161–37168. doi: 10.1074/jbc.274.52.37161. [DOI] [PubMed] [Google Scholar]
- König J. Uptake transporters of the human OATP family: molecular characteristics, substrates, their role in drug-drug interactions, and functional consequences of polymorphisms. Handb Exp Pharmacol. 2011:1–28. doi: 10.1007/978-3-642-14541-4_1. [DOI] [PubMed] [Google Scholar]
- Kotak BG, Semalulu S, Fritz DL, Prepas EE, Hrudey SE, Coppock RW. Hepatic and renal pathology of microcytin-LR in rainbow trout (Oncorhynchus mykiss) Toxicon. 1996;34:517–525. doi: 10.1016/0041-0101(96)00009-8. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy T, Carmichael WW, Sarver EW. Toxic peptides from freshwater cyanobacteria (blue-green algae). I Isolation, purification and characterization of peptides from Microcystis aeruginosa and Anabaena flos-aquae. Toxicon. 1986;24:865–873. doi: 10.1016/0041-0101(86)90087-5. [DOI] [PubMed] [Google Scholar]
- Kullak-Ublick GA, Hagenbuch B, Stieger B, Schteingart CD, Hofmann AF, Wolkoff AW, Meier PJ. Molecular and functional characterization of an organic anion transporting polypeptide cloned from human liver. Gastroenterology. 1995;109(4):1274–1282. doi: 10.1016/0016-5085(95)90588-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawton LA, Codd GA. Cyanobacterial (Blue-Green Algal) Toxins and their Significance in UK and European Waters. Water Environ J. 1991;5:460–465. [Google Scholar]
- Lee W, Glaeser H, Smith LH, Roberts RL, Moeckel GW, Gervasini G, Leake BF, Kim RB. Polymorphisms in human organic anion-transporting polypeptide 1A2 (OATP1A2): implications for altered drug disposition and central nervous system drug entry. J Biol Chem. 2005;280(10):9610–7. doi: 10.1074/jbc.M411092200. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Leuthold S, Hagenbuch B, Mohebbi N, Wagner CA, Meier PJ, Stieger B. Mechanisms of pH-gradient driven transport mediated by organic anion polypeptide transporters. Am J Physiol Cell Physiol. 2009;296(3):C570–82. doi: 10.1152/ajpcell.00436.2008. [DOI] [PubMed] [Google Scholar]
- Li XY, Chung IK, Kim JI, Lee JA. Subchronic oral toxicity of microcystin in common carp (Cyprinus carpio L) exposed to Microcystis under laboratory conditions. Toxicon. 2004;44:821–7. doi: 10.1016/j.toxicon.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Lu H, Choudhuri S, Ogura K, Csanaky IL, Lei X, Cheng X, Song P-z, Klaassen CD. Characterization of organic anion transporting polypeptide 1b2-null mice: essential role in hepatic uptake/toxicity of phalloidin and microcystin-LR. Toxicol Sci. 2008;103:35–45. doi: 10.1093/toxsci/kfn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães VFd. Microcystin contamination in fish from the Jacarepaguá Lagoon (Rio de Janeiro, Brazil): ecological implication and human health risk. Toxicon. 2001:1077–1085. doi: 10.1016/s0041-0101(00)00251-8. [DOI] [PubMed] [Google Scholar]
- Meier-Abt F, Mokrab Y, Mizuguchi K. Organic anion transporting polypeptides of the OATP/SLCO superfamily: identification of new members in nonmammalian species, comparative modeling and a potential transport mode. J Membr Biol. 2005;208:213–27. doi: 10.1007/s00232-005-7004-x. [DOI] [PubMed] [Google Scholar]
- Monks NR, Liu S, Xu Y, Yu H, Bendelow AS, Moscow JA. Potent cytotoxicity of the phosphatase inhibitor microcystin LR and microcystin analogues in OATP1B1- and OATP1B3-expressing HeLa cells. Mol Cancer Ther. 2007;6(2):587–98. doi: 10.1158/1535-7163.MCT-06-0500. [DOI] [PubMed] [Google Scholar]
- Noé J, Portmann R, Brun M, Funk C. Substrate-dependent drug-drug interactions between gemfibrozil, fluvastatin and other organic anion-transporting peptide (OATP) substrates on OATP1B1, OATP2B1. Drug Metab Dispos. 2007;35:1308–1314. doi: 10.1124/dmd.106.012930. [DOI] [PubMed] [Google Scholar]
- Peng L, Liu Y, Chen W, Liu L, Kent M, Song L. Health risks associated with consumption of microcystin-contaminated fish and shellfish in three Chinese lakes: significance for freshwater aquacultures. Ecotoxicology and environmental safety. 2010;73(7):1804–1811. doi: 10.1016/j.ecoenv.2010.07.043. [DOI] [PubMed] [Google Scholar]
- Popovic M, Zaja R, Fent K, Smital T. Molecular Characterization of Zebrafish Oatp1d1 (Slco1d1), a Novel Organic Anion-transporting Polypeptide. J Biol Chem. 2013;288:33894–911. doi: 10.1074/jbc.M113.518506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Zaja R, Smital T. Organic anion transporting polypeptides (OATP) in zebrafish (Danio rerio): Phylogenetic analysis and tissue distribution. Comp Biochem Physiol A Mol Integr Physiol. 2010;155:327–35. doi: 10.1016/j.cbpa.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Poste AE, Hecky RE, Guildford SJ. Evaluating microcystin exposure risk through fish consumption. Environmental science & technology. 2011;45(13):5806–5811. doi: 10.1021/es200285c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Råbergh C, Bylund G, Eriksson J. Histopathological effects of microcystin-LR, a cyclic peptide toxin from the cyanobacterium (blue-green alga) Microcystis aeruginosa on common carp (Cyprinus carpio L.) Aquat Toxicol. 1991;20(3):131–145. [Google Scholar]
- Rodger H, Turnbull T. Cyanobacterial (blue-green algal) bloom associated pathology in brown trout, Salmo trutta L., in Loch Leven, Scotland. J Fish Dis. 1994:17.
- Roth M, Obaidat A, Hagenbuch B. OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. Br J Pharmacol. 2012;165(5):1260–87. doi: 10.1111/j.1476-5381.2011.01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiä VO, Kankaanpää HT, Flinkman J, Lahti K, Meriluoto JA. Time-dependent accumulation of cyanobacterial hepatotoxins in flounders (Platichthys flesus) and mussels (Mytilus edulis) from the northern Baltic Sea. Environ Toxicol. 2001;16:330–6. doi: 10.1002/tox.1040. [DOI] [PubMed] [Google Scholar]
- Song L, Chen W, Peng L, Wan N, Gan N, Zhang X. Distribution and bioaccumulation of microcystins in water columns: A systematic investigation into the environmental fate and the risks associated with microcystins in Meiliang Bay, Lake Taihu. Water Res. 2007;41(13):2853–2864. doi: 10.1016/j.watres.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Tamai I, Nozawa T, Koshida M, Nezu J, Sai Y, Tsuji A. Functional characterization of human organic anion transporting polypeptide B (OATP-B) in comparison with liver-specific OATP-C. Pharm Res. 2001;18:1262–9. doi: 10.1023/a:1013077609227. [DOI] [PubMed] [Google Scholar]
- Tencalla F. PhD Thesis. Swiss Federal Institute of Technology; Zürich: 1995. Toxicity of cyanobacterial peptide toxins to fish. [Google Scholar]
- Tencalla F. Biochemical characterization of microcystin toxicity in rainbow trout (Oncorhynchus mykiss) Toxicon. 1997;35:583–595. doi: 10.1016/s0041-0101(96)00153-5. [DOI] [PubMed] [Google Scholar]
- Tencalla F, Dietrich D, Schlatter C. Toxicity of Microcystis aeruginosa peptide toxin to yearling rainbow trout (Oncorhynchus mykiss) Aquat Toxicol. 1994;30:215–224. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications 1979. Biotechnology. 1992;24:145–9. [PubMed] [Google Scholar]
- Zeck A, Weller MG, Bursill D, Niessner R. Generic microcystin immunoassay based on monoclonal antibodies against Adda. Analyst. 2001;126:2002–7. doi: 10.1039/b105064h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.