Abstract
Bacillus thuringiensis subsp. israelensis (Bti) has been widely for the biological control of mosquito populations. However, the mechanism of Bti toxins is still not fully understood. To further elucidate the mechanism of Bti toxins, we developed an Aedes aegypti resistant strain that shows high-level resistance to Cry11Aa toxin. After 27 selections with Cry11Aa toxin, the larvae showed a 124-fold resistance ratio for Cry11Aa (strain G30). G30 larvae showed cross-resistance to Cry4Aa (66-fold resistance), less to Cry4Ba (13-fold), but not to Cry11Ba (2-fold). Midguts from these resistant larvae did not show detectable difference in the processing of the Cry11Aa toxin compared to that in susceptible larvae (WT). Brush border membrane vesicles (BBMV) from resistant larvae bound slightly less Cry11Aa compared to WT BBMV. To identify potential proteins associated with Cry11A resistance, not only transcript changes in the larval midgut were analyzed using Illumina sequencing and qPCR, but alterations of previously identified receptor proteins were investigated using immunoblots. The transcripts of 375 genes were significantly increased and those of 208 genes were down regulated in the resistant larvae midgut compared to the WT. None of the transcripts for previously identified receptors of Cry11Aa (Aedes cadherin, ALP1, APN1, and APN2) were altered in these analyses. The genes for the identified functional receptors in resistant larvae midgut did not contain any mutation in their sequences nor was there any change in their transcript expression levels compared to WT. However, ALP proteins were expressed at reduced levels (~40%) in the resistant strain BBMV. APN proteins and their activity were also slightly reduced in resistance strain. The transcript levels of ALPs (AAEL013330 and AAEL015070) and APNs (AAEL008158, AAEL008162) were significantly reduced. These results strongly suggest that ALPs and APNs could be associated with Cry11Aa resistance in Ae. aegypti.
Keywords: Bacillus thuringiensis, Cry11Aa toxin, cadherin, ALP, APN, receptor, resistant mosquito, RNA-seq
Introduction
Aedes aegypti is an important vector of human diseases such as dengue, chikungunya and yellow fevers that are transmitted through blood feeding by the mosquito (Ligon, 2005; Ligon, 2006; Tomori, 2004). An approach used to decrease the prevalence of these diseases is by controlling the vector mosquito utilizing Bacillus thuringiensis subsp. israelensis (Bti), which also has high toxicity to Simulium species, a vector for onchocerciasis (Bravo et al., 2011; Gill et al., 1992; Margalith and Ben-Dov, 2000). The high insecticidal activity and the low toxicity to other organisms led to increased use of Bti for the control of mosquito and black fly populations. However, its mechanism is still not fully understood, in part because Bti produces a number of mosquitocidal toxins.
This bacterium contains a megaplasmid, pBtoxis, which encodes for the Cry4Aa, Cry4Ba, Cry10Aa, Cry11Aa, Cyt1Aa, Cyt1Ca and Cyt2Ba proteins (Berry et al., 2002). Of these, Cry4Aa, Cry4Ba and Cry11Aa, have been identified as the primary active toxins against mosquitoes (Chilcott and Ellar, 1988), while Cyt1Aa has low mosquitocidal activity. Cyt1A, however, synergizes Cry toxins activity by apparently acting as a surrogate receptor for these toxins in the mosquito midgut (Perez et al., 2005; Perez et al., 2007).
Among the Bti toxins, Cry11Aa is important for the control of Ae. aegypti because it is one of the more active toxin (Chilcott and Ellar, 1988), and shows high affinity to the brush border membrane vesicles (BBMV) of Ae. aegypti (Chen et al., 2013). Cry11Aa binds a number of BBMV proteins (Fernandez et al., 2006). Among them are the Aedes cadherin (AAEL018140) and two aminopeptidases N (APNs) (AAEL012778 and AAEL008155), all binding Cry11Aa with high affinity (Chen et al., 2009a; Chen et al., 2009b; Chen et al., 2013). These proteins are also present in the posterior midgut epithelial cells and/or the gastric caeca (Chen et al., 2009b; Chen et al., 2013). RNAi-mediated silencing of cadherin and of APN (AAEL012783) increased tolerance for Cry11Aa (Lee et al., 2014; Rodríguez-Almazán et al., 2012) and Cry4Ba toxicity (Saengwiman et al., 2011), respectively. These data collectively suggest that cadherins and APNs may be functionally important for Cry toxicity in mosquitoes.
In addition, ALPs have also been observed to bind a number of Cry toxins, as in Manduca sexta, Heliothis virescens and Spodoptera frugiperda (Flores-Escobar et al., 2013; Jurat-Fuentes and Adang, 2004; McNall and Adang, 2003). Importantly, ALP expression was significantly reduced in resistant strains of H. virescens and S. frugiperda implying that ALPs mediate Cry1 resistance in these insects (Jurat-Fuentes and Adang, 2004; Jurat-Fuentes et al., 2011). Further, RNAi-mediated silencing of ALP in M. sexta led to decreased Cry1Ab toxin binding (Flores-Escobar et al., 2013).
Ae. aegypti ALPs also play a role in Cry toxicity. ALP (AAEL009077) was shown to bind Cry11Aa, and moreover, it is expressed in the same midgut regions to which Cry11Aa binds – microvilli of gastric caeca and the posterior midgut (Fernandez et al., 2009). This ALP also binds Cry4Ba (Jiménez et al., 2012), but other ALPs also bind this toxin. Proteomic analysis showed Cry4Ba toxin was localized in lipid rafts from larval BBMV and bound three ALPs (AAEL003298, AAEL003313, and AAEL015070) (Bayyareddy et al., 2009; Bayyareddy et al., 2012). Moreover, the ALP (AAEL015070) bound Cry4Ba with high affinity and mediated Cry4Ba toxicity in Sf9 cells expressing the ALP (Dechklar et al., 2011; Thammasittirong et al., 2011). Thus, these ALPs are functionally important for Cry toxicity to mosquito larvae since RNAi-mediated silencing of ALP (AAEL009077) in the midgut resulted in reduced Cry11Aa and Cry4Ba toxicity (Jiménez et al., 2012).
While Bti has been used for more than three decades, field resistance to Bti has not been reported, but resistance to other Bt toxins has been observed in a number of lepidopteran insects (Bravo and Soberon, 2008; Tabashnik et al., 2008). Therefore, to further elucidate the mechanism of action of Bti toxins, resistant lab strains have been developed. As a laboratory-selected mosquito, Culex quinquefasciatus was selected with single or multiple Bti toxins (Georghiou and Wirth, 1997). After 28 generations, the strain selected with a single toxin, Cry11Aa, showed the highest resistance ratio (at least 1,000 fold), but selection with four toxins, including Cyt1Aa, resulted in the lowest resistance level (3.2 fold). The cross-resistance patterns of these strains were observed with combinations of mosquitocidal Cry toxins, and all four strains showed cross resistance to Bti toxins as well as Cry11Ba from Bacillus thuringiensis subsp. jegathesan (Btj) (Cheong et al., 1997; Wirth et al., 1998). For example, the Cry11Aa-resistant strain revealed a high resistance to Cry4Aa and Cry4Ba (41.6 fold), but very low resistance to Bti (1.1 fold) and Btj (2.8 fold) strains. These results suggest that single Cry toxins trigger the development of more rapid resistance, but Cyt1Aa prevents resistant development because it apparently functions as a surrogate receptor for Bti Cry toxins in mosquitoes (Perez et al., 2005).
A field collected Ae. aegypti strain that was selected with toxic leaf litter containing Bti for 30 generations (LiTOX strain) showed low resistance ratio (3.5 fold) to a Bti mixture (Tetreau et al., 2012). But this strain showed 67-fold, 9-fold, and 9-fold resistance for Cry4Aa, Cry4Ba, and Cry11Aa compared to susceptible Ae. aegypti. Transcript changes and differential protein expression analyses with LiTOX showed ALP (AAEL003298) and APN (AAEL012776) in LiTOX larvae midgut were down-regulated. These results imply that ALP and APN are possible receptors mediating Cry toxin resistance in mosquitoes.
In this study we describe a strain of Ae. aegypti selected for resistance to the Cry11Aa toxin. We show that alkaline phosphatases and aminopeptidases N play a role in the Cry11Aa resistance observed, but the proteins identified are different from those observed previously (Tetreau et al., 2012).
Materials and Methods
Rearing Cry11Aa-treated Aedes aegypti
Adult Ae. aegypti were reared in a 29°C, 8:12 h light:dark cycle at 50% humidity and fed 10% sugar water. After blood feeding, eggs were collected on moist filter paper. Larvae were reared in deoxygenated tap water at the same photoperiod and fed a mixture of dog food and yeast (3:1 w/w).
Selection of a Cry11A resistant mosquito colony and bioassays
Adult Ae. aegypti males (1560) were fed sugar water for 24 h containing 10 mM ethyl methansulfonate (EMS) to cause random mutations, thereby increasing heterogeneity. The EMS-treated male adults were mated with untreated virgin females (1750), which after mating were allowed to blood feed and lay eggs.
To monitor the development of Cry11Aa resistance and to determine Cry11Aa concentrations needed for selection, bioassays were performed for each generation selected. In brief, 25 early fourth-instar larvae were placed in 200 ml tap water containing Cry11Aa at different concentrations for 24 h. Bioassay results were analyzed by Probit (EPA) or Origin program (Origin Lab, Northampton, MA). Then early fourth-instar larvae were treated at the levels that were determined to be the LC90 for that generation. Selection was performed using crude spore/crystal Cry11Aa prepared as described below. Approximately 8,000 larvae of the original generation (G0), 4,000 larvae of G1 to G10, and 2,000 larvae of G11 to G29 were treated. Larvae that survived toxin treatments were transferred to fresh water and reared until the next generation. Survival levels varied from 5–30% but usually were around 20% for most generations.
Cry toxins preparation
B. thuringiensis strains producing Cry4Aa, Cry4Ba, Cry11Aa, or Cry11Ba (Chang et al., 1993; Delecluse et al., 1995) were grown in nutrient broth sporulation medium containing erythromycin (25 μg/ml) at 30°C for 4–5 days for cell autolysis to occur (Lereclus et al., 1995). For crude Cry toxin preparations, spores and crystal inclusions were harvested, washed twice with sterilized water at 10,000×g for 10 min at 4°C, and stored in water at −80°C until used. For purified Cry toxins, the inclusion bodies for Cry11Aa were isolated as previously reported using NaBr gradients (Cowles et al., 1995). The purified Cry11A inclusions were washed, solubilized in 50 mM Na2CO3 pH 10.5 buffer, and activated by trypsin (1:20 w/w) at 37°C (Dai and Gill, 1993). For biotin-labeled Cry11Aa, the activated Cry11Aa was biotinylated and then purified using a Sephadex G25 column following the manufacturer’s protocol (GE Healthcare Life Science, Pittsburgh, PA).
Preparation of Brush Border Membrane Vesicles (BBMV)
BBMV were isolated from dissected midguts of early fourth-instar Ae. aegypti larvae as reported (Nielsen-Leroux and Charles, 1992). Briefly, frozen midguts were resuspended and homogenized in ice-cold buffer A (0.3 M mannitol, 0.5 M EGTA, 20 mM Tris-Cl, pH 7.4) including a protease inhibitor cocktail (Roche, Madison, WI) and 1 mM phenylmethylsulfonyl fluoride (PMSF). MgCl2 (final concentration 12 mM) was added to the homogenate and then kept on ice for 20 min. The mixture was centrifuged at 3,000 × g for 15 min at 4°C and the supernatant was collected and kept on ice. The pellet was resuspended in ice-cold buffer A and treated as above for the first homogenization. The combined supernatants were then centrifuged at 14,000 × g for 60 min at 4°C. The pellet was resuspended in buffer A and protein concentration was quantified using the BCA assay (Pierce, Rockford, IL). BBMV were prepared and used the same day.
In vitro processing of Cry11Aa
The processing of Cry11Aa toxin was determined in wild-type (WT) and resistant (G30) strains using published methods (Forcada et al., 1996). Briefly, early fourth-instar larvae were dissected to obtain midguts including gastric caeca, but excluding the foregut and hindgut. Approximately 200 dissected midguts were resuspended and homogenized in ice-cold extraction buffer (50 mM Tris-Cl, pH 7.4). After centrifugation at 10,000 × g for 15 min at 4°C, the supernatant was transferred to a fresh tube and quantified using the BCA assay (Pierce) following the manufacturer’s protocol. The Cry11Aa protoxin (10 μg) was mixed with 0.1 μg midgut extract in 50 mM Na2CO3, pH 10 including 10 mM dithiothreitol (DTT) and incubated for 5, 10, 15, 20 min at 37°C. A control was incubated with buffer only for 20 min at 37°C. Proteolytic processing was stopped by adding SDS-PAGE sample buffer and heating the samples for 5 min at 100°C. Digested samples were analyzed in SDS polyacrylamide gel (10%). SDS polyacrylamide gels were stained in Coomassie stain solution (0.1% Coomassie brilliant blue, 10% acetic acid, and 40% methanol) and subsequently incubated in destain solution (10% acetic acid and 20% methanol). The stained band densities were measured and quantified with Image J software (NIH) and analyzed with Origin program (Origin Lab).
Alkaline phosphatase and aminopeptidase activity
Alkaline phosphatase (ALP) and aminopeptidase N (APN) enzymatic activities were measured using p-nitrophenyl phosphate and leucine-p-nitroanilide (Sigma, St. Louis, MO) as substrates following previous methods (Chen et al., 2009b; Fernandez et al., 2006; Jurat-Fuentes and Adang, 2004). Freshly prepared BBMVs (5 μg) were mixed with ALP buffer (100 mM Tris/HCl, pH 9.5, 100 mM NaCl, 5 mM MgCl2, 1.25 mM p-nitrophenyl phosphate) or APN buffer (20 mM Tricine, pH 8.0, 0.4% methanol, 0.005% bovine serum albumin, 0.18 mM leucine-p-nitroanilide) in a final volume of 200 μl. The same concentration of bovine serum albumin (BSA) in ALP or APN buffer was used as a background control. Enzymatic activities were monitored as the change in the absorbance at 405 nm for three min at room temperature in a microplate reader (Molecular Devices, Sunnyvale, CA).
Cry11Aa toxin binding to BBMV
The kinetics of Cry11Aa binding were performed with BBMV prepared above in a 96-well format as previously described (Likitvivatanavong et al., 2011; Chen et al., 2013). In brief, 4 μg BBMVs in 50 mM NaHCO3 pH 9.6 coating buffer were added into each well of a 96-well plate (Thermo Scientific, Lafayette, CO) and incubated overnight at 4°C. The plate was then washed three times with PBST (PBS and 0.1% Tween 20) and blocked with PBST for 1 h at 37°C. For total binding, biotin-labeled Cry11Aa (0.1–100 nM) in 100 μl binding buffer (PBST and 0.1% bovine serum albumin) was added into the BBMV-coated plates and incubated for 2 h at 37°C. For nonspecific binding, parallel wells were incubated under identical conditions, except in the presence of 10 μM cold Cry11Aa. The plates were washed with PBST three times and then incubated with streptavidin-horseradish peroxidase (HRP) conjugate (1:1,500, GE Healthcare Life Science) for 1 h at 37°C. After washing three times with PBST, HRP activity was detected with a luminol substrate (Thermo Scientific) and the plate exposed to an X-ray film in a darkroom. The spot densities were measured and quantified with Image J software (NIH) and analyzed with Origin program (Origin Lab). Specific binding was obtained as total binding minus nonspecific binding, and the dissociation constant (Kd) was obtained from the concentration corresponding to half the saturation response of specific binding.
Transcriptome sequencing and bioinformatics
Transcript changes in the midgut of WT and resistant (G30) strains were analyzed by RNA-seq (BGI Americas, Cambridge, MA). Early fourth-instar larvae were dissected to obtain midguts including gastric caeca. Three hundred midguts of WT and G30 strains (100 midguts for each repeat) were sent to BGI for RNA-seq analysis. Total RNA was extracted by BGI from dissected midguts and mRNA was isolated with magnetic beads. The mRNA was fragmented into short fragments, and then the cDNA was synthesized using the mRNA fragments as templates. The cDNA was purified, resolved for end repair and single nucleotide A (adenine) addition, and connected with adapters. Suitable cDNA were selected for the PCR amplification as templates. The samples were quantified and assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and ABI StepOnePlus Real-Time PCR System (Invitrogen, Grand Island, NY). The library was sequenced using Illumina HiSeq™ 2000 (Illumina, San Diego, CA) for 30 million paired-end reads (~100 bp paired end) for each sample. Three biological repeats were performed for each of the two strains. All sequence reads were assembled using de novo transcriptome assembly (Grabherr et al., 2011) and the identified genes were functionally annotated using blastX (Evalue < 0.0001) with a variety of databases including Gene Ontology, Nr, KeGG, SwissProt, COG as well as the Ae. aegypti genome, Aedes-aegypti-Liverpool_TRANSCRIPTS_AaegL1.3.fa.gz (https://www.vectorbase.org) (Nene et al., 2007). Gene expression levels were calculated using the FPKM method (Fragments Per kb per Million fragments) (Mortazavi et al., 2008) and differential gene expression analyzed with significantly expressed genes (Qvalue > 0.8) between each group using NOISeq method (Tarazona et al., 2011). In order to avoid library size bias, NOISeq method corrects the counts by a factor closely related to the sequencing depth (SD); the number of counts per million reads (the number of read counts for each gene × 106/SD). To screen gene mutations of Aedes cadherin (AAEL018140) and aminopeptidase Ns (APN1, AAEL012778 and APN2, AAEL008155), the identified genes were clustered and aligned to other similar genes (more than 70% identity) and the number of gaps quantified.
Mutation screening with direct sequencing
Aedes cadherin (AAEL018140) and alkaline phosphatase (ALP1, AAEL009077) were directly sequenced to screen for mutations (Institute for Integrative Genome Biology, University of California, Riverside, CA). Primers were designed based on gene and transcript sequences of the Ae. aegypti genome (Vectorbase, http://www.vectorbase.org). gDNA was extracted from G30 larvae with DNAzol (Molecular Research Center, Cincinnati, OH) following the manufacture’s protocol. Exons regions of ALP in G30 were amplified and sequenced for comparison of WT and resistant larvae midgut genes. Because the Aedes cadherin gene sequence was not completely annotated (Vectorbase, http://www.vectorbase.org) at the time of this study, mutation screening of the Aedes cadherin was performed on the cDNA, which was prepared from fourth-instar larvae midgut using TRIzol and SuperScript III (Invitrogen). Five fragments covering the whole Aedes cadherin sequence were amplified, sequenced and compared.
Quantitative Real-time PCR (qPCR)
Total RNA was extracted from WT and resistant larvae midgut using TRIzol and analyzed by Nanodrop ND-1000 Spectrophotometer (Institute for Integrative Genome Biology, UCR). For each sample cDNA was synthesized from total RNA with SuperScript III (Invitrogen), diluted, and 5-μl aliquots were used as template for qPCR. Respective primers specific to the target genes (Aedes cadherin, alkaline phosphatases, aminopeptidase Ns) were designed to have similar properties in terms of nucleotide length and %GC content (Table S1). As a reference gene, actin (Actin-S and Actin-A, AAEL011197) was used as a reference gene for quantification and 40S ribosomal S7 gene (S7-S and S7-A, AAEL009496) that spanned a 114-bp intron was analyzed on every run to check for gDNA contamination in RNA preparations. Our microarray data showed the actin (AAEL011197) and 40S ribosomal S7 (AAEL009496) expressions were not changed (Actin: −0.05, −0.06. and −0.07 fold change; 40S ribosomal S7: 0.17, −0.08, or 0.09 fold change) in Cry11Aa-treated larvae midgut at LC10, LC50, LC90 compared to untreated larvae midgut. PCR conditions, including the template cDNA, primer concentrations and annealing temperatures, were adjusted for amplification efficiencies (efficiency 90 – 110%) for all genes. Optimized PCR master mix (20 μl) contained the following components: 10 μl iQ SYBR Green supermix (Bio-Rad), 5 μl cDNA and 10 μM each primer. The qPCR was performed using CFX Real-time PCR (Bio-Rad). Optimized thermal program consisted of: one cycle of 95°C/1 min and 40 cycles of 95°C/1 min, 62°C/1 min, and 72°C/1 min, followed by a final extension of one cycle 72°C/5 min. Following qPCR, the homogeneity of the PCR product was confirmed by melting curve analysis. Quantification of the transcript levels or relative copy number of the genes was conducted according to the Pfaffl method (Pfaffl, 2001): ratio = (Etarget)ΔCttarget(Control-Sample)/(Eref)ΔCtref(Control-Sample) where, Etarget, amplification efficiency of the target gene, Eref; amplification efficiency of Actin; Ct, threshold cycle. Quantitative PCR was performed three times using independently prepared midgut cDNA.
Western blotting
Antibodies to the cadherin peptide (Aedes cadherin-specific antibody), and ALP and APN proteins were previously reported (Chen et al., 2009a; Chen et al., 2009b; Fernandez et al., 2009). The cadherin peptide antibody is specific for Aedes cadherin, but the ALP (AAEL009077) antibody cross reacts with a number of other ALPs (Jiménez et al., 2012). Freshly prepared BBMVs from WT and resistant larvae midgut were quantified by the BCA assay (Pierce). BBMVs (10 μg) were heated at 70°C for 5 min, separated by SDS polyacrylamide gels and electrotransferred to nitrocellulose membranes. The membranes were blocked with blocking solution (PBST and 5% Skim milk) for 1 h at room temperature and then washed with PBST. The blocked membranes were incubated overnight at 4°C with primary antibody (1:3,000), washed with PBST, and then subsequently incubated with anti-rabbit HRP (1:5,000) secondary antibody (Sigma) for 1 h at room temperature. After washing with PBST, the HRP activity was revealed with a luminol substrate (Thermo Scientific) and exposed to an X-ray film in a darkroom. The band densities of target proteins were measured and quantified using Image J software (NIH) and analyzed with Origin program (Origin Lab).
Results
Ae. aegypti developed high-level resistance to Cry11A
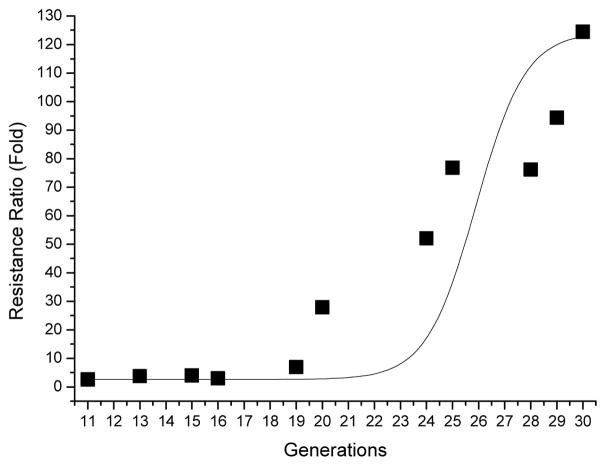
An Ae. aegypti lab colony that included populations that were collected from Malaysia was further treated with EMS to increase heterogeneity. Early fourth-instar Ae. aegypti larvae were then selected with Cry11Aa toxin at the LC50 level with G0 generation and then at the LC90 concentration levels in subsequent generations. For each selection, a bioassay was performed with susceptible (WT) and resistant larvae to determine the LC50 and LC90 values. After 20 generations, resistant larvae developed at least a 30-fold resistance ratio compared to WT larvae at the LC50 level (Figure 1). A few of the generations (G17, G22 and G26) were not selected to maintain a robust population. After 30 generations (G30), a 124-fold resistance was observed compared to WT at the LC50 level (Figure 2B).
Figure 1.
Aedes aegypti developed a high level of Cry11Aa resistance after twenty generations. Early fourth-instar larvae were selected at the LC90 concentration, except for generations 17, 22 and 26 that were not selected to maintain a health colony. Resistance ratios were obtained by comparing the LC50 values of the resistant strain to that obtained with susceptible larvae (WT). Bioassays showed the G30 larvae had a 124-fold resistant ratio.
Figure 2.
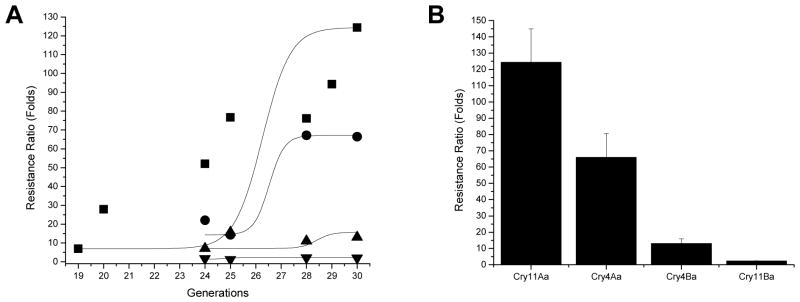
Aedes larvae developed resistance to Cry11Aa and Cry4Aa but not to Cry4Ba and Cry11Ba. Resistance ratios for individual toxins from mosquitocidal Bacillus thuringiensis israelensis (Bti) and Bacillus thuringiensis jegathesan (Btj) were tested at four generations during resistance development to Cry11Aa (A). G30 showed 124-, 66-, 13-, and 2-fold resistance ratio to Cry11Aa, Cry4Aa, Cry4Ba, and Cry11Ba (B). Cry11Aa (■), Cry4Aa (●) and Cry4Ba (▲) from Bti, and Cry11Ba (▼) from Btj. Resistance ratios for individual toxins were obtained by comparing the LC50 of the resistant strain to that of susceptible WT.
The Cry11Aa resistance strain showed cross resistance to Cry4Aa and Cry4Ba
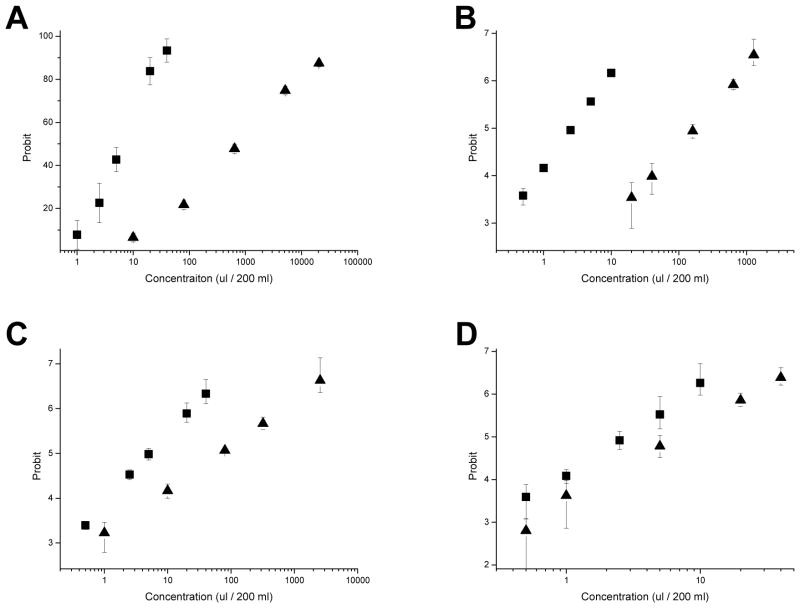
To investigate if the Cry11Aa-resistant larvae were cross resistant to other Bti toxins, the Cry11A-resistant larvae were bioassayed with the Cry4Aa, Cry4Ba, and Cry11Ba toxins (Figure 2). G30 larvae showed resistance ratios of 124, 66, 13, and 2 fold for Cry11Aa, Cry4Aa, Cry4Ba, and Cry11Ba toxins, respectively (Figure 2B). Thus, the Cry11Aa-resistant larvae showed substantial cross resistance at the LC50 level for Cry4Aa, a lesser amount to Cry4Ba, but not to Cry11Ba. Dose-response bioassay also showed that G30 larvae have increased tolerance to Cry11Aa and Cry4Aa toxins, but lower tolerance to Cry4Ba (Figure 3). Notably the LC50 values for Cry4Ba and Cry11Ba do not change much from generations 24 to 30, but do for Cry11Aa and Cry4Aa (Figure 2A).
Figure 3.
Probit analysis of Cry11Aa and Cry4Aa toxins showed significant differences in the toxicity to WT (■) and the G30 resistant strains (▲) but these were not observed with Cry4Ba and Cry11Ba. Resistance levels of G30 to mosquitocidal Cry toxins were determined with individual Cry toxins; Cry11Aa, Cry4Aa, and Cry4Ba from Bacillus thuringiensis israelensis and Cry11Ba from Bacillus thuringiensis jegathesan. (A) Cry11Aa, (B) Cry4Aa, (C) Cry4Ba, (D) Cry11Ba.
In vitro processing of Cry11Aa in resistant larval mosquito midgut
In order to investigate if resistance was associated with altered toxin processing, cleavage of the Cry11Aa protoxin with midgut homogenates was determined. Total midgut extracts were obtained from WT and resistant larvae midgut, and incubated with Cry11Aa protoxin for 5–20 min. The band densities of three Cry11Aa proteins (72, 36, 32 kDa) were measured using Image J program. No substantial difference in the cleavage patterns of the Cry11Aa toxin was observed between WT and resistant larvae (Figure 4, Table S2). Moreover, the sum of three proteins was similar at each time point implying Cry11Aa protoxin degradation was not significantly changed between WT and resistant larvae (Table S2).
Figure 4.
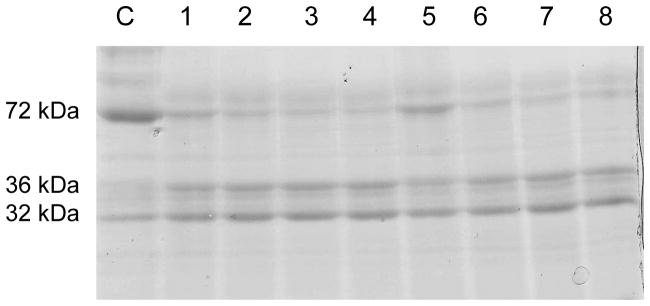
The in vitro processing of the Cry11Aa toxin was the same in WT and resistant G30 larvae midgut. Cry11Aa protoxin was incubated with midgut protein extract that included midgut proteases and then analyzed in SDS-PAGE gel. Lanes: C - control; lanes - 1, 2, 3 and 4: 5, 10, 15 and 20 min with WT midgut extract, respectively; lanes – 5, 6, 7, and 8: 5, 10, 15 and 20 min with G30 resistant strain midgut extract, respectively. The stained band densities (the sum of the gray values of all the pixels in the selection divided by the number of pixels) were measured and quantified with Image J software (NIH) and analyzed with Origin program (Origin Lab). Band densities of the 72 kDa protoxin incubated at same time were similar; lane 1 (28) and 5 (30) of 5 min, 2 (23) and 6 (20) of 10 min, 3 (21) and 7 (17) of 15 min, 4 (20) and 8 (18) of 20 min (Table S2).
Cry11A binding affinity to BBMV is slightly altered in resistant Aedes aegypti
A competitive binding assay was performed to measure Cry11Aa binding affinity to the larval midgut BBMV. To determine specific binding affinity, BBMVs from WT and resistant larvae were incubated with biotin-labeled Cry11Aa in the absence or presence of unlabeled Cry11Aa. Dissociation constant (Kd) values were calculated corresponding to half the saturation value. The resistant larvae showed reduced binding affinity (Kd = 30.5 nM) for Cry11Aa compared to that of the WT midgut (Kd = 13.4 nM) (Table 1).
Table 1.
Cry11Aa has a lower affinity to midgut membrane prepared from the G30 larvae than those from wild type.
| WT midgut | G30 midgut | |
|---|---|---|
| Kd (nM)a | 13.4 | 30.5 |
Binding affinity of WT and G30 was tested with early fourth-instar larvae midguts. Specific binding of biotin-labeled Cry11Aa to BBMV was obtained from total binding minus nonspecific binding. Cry11Aa binding affinity for G30 BBMV was slightly reduced. Kd values were obtained from the concentration corresponding to half the saturation response of specific binding.
The Kd values reported are the mean of three separate experiments.
Analysis for mutations in known receptors
To analyze if any of the previously identified Cry11Aa receptors were mutated, we analyzed these genes by direct sequencing and analyzing transcripts by Illumina and Sanger sequencing. In RNA-seq analysis, one gap (three nucleotides insertion) in the Aedes cadherin (AAEL018140) transcript of resistant larvae was observed but not in APN1 (AAEL012778) and APN2 (AAEL008155) transcripts (Table 2). However, Sanger sequencing of resistant larval midgut cDNA showed there was no mutation in the cadherin transcript. Surprisingly, RNA-seq did not detect the ALP1 (AAEL009077) sequence but its presence was observed in qPCR. Further, Sanger sequencing of the exons of these genes did not show any mutation that would lead to a change in protein sequence.
Table 2.
Previously identified receptors do not contain any mutations in the G30 larvae. Mutations in target genes were screened using RNA seq and direct sequencing.
| Name | Aedes ID | Gap in illumina sequencinga | Mutation screeninga |
|---|---|---|---|
| AaeCad | AAEL018140 | 1 | X |
| ALP1 | AAEL009077 | NI | X |
| APN1 | AAEL012778 | 0 | ND |
| APN2 | AAEL008155 | 0 | ND |
NI: non-identified gene in Illumina sequencing. X: no mutation in screened gene. ND: not-determined.
Transcriptome analysis of Cry11A resistant Aedes aegypti
To determine if there were any changes in midgut transcript expression in the resistant Aedes larvae, RNA-seq was performed with the WT and G30 strains. Differentially expressed genes were identified by comparing WT and resistant larvae transcripts, and the significantly up-regulated or down-regulated genes in resistant larvae midgut were determined (Tables S3 and S4). A total of 375 genes in the resistant larvae midgut were significantly up-regulated compared to WT, and 191 of these genes were functionally annotated (Table S3). A total of 208 genes in resistant larvae midgut were significantly down-regulated compared to WT, and 137 of these genes were functionally annotated (Table S4). Among them, proteins possibly linked to Cry11Aa resistance in mosquitoes are listed in Table 3. Two ALPs (AAEL013330, AAEL015070) and two APNs (AAEL008158, AAEL008162) were significantly down-regulated in resistant larvae and one ALP (AAEL003286) was significantly up-regulated. Two ABC transporters (AAEL012700, AAEL013833) were reduced and two kinases (AAEL009937, AAEL001442) were significantly up-regulated in resistant larvae midgut. In addition, a caspase (AAEL014658) and three serine proteases (AAEL005753, AAEL006627, and AALE007938) were up-regulated and a number of trypsins, peptidases, and serine-type proteases were down-regulated. However, none of the known receptor proteins for Cry11Aa were altered.
Table 3.
Genes of interest that show significant transcript level changes in the resistant larvae midgut.
| Gene ID | Length (bp) | Aedes ID | Annotation a | Log2b | Qvalue |
|---|---|---|---|---|---|
| ALP | |||||
| Unigene20119 | 1702 | AAEL003286-RA | alkaline phosphatase | 5.16 | 0.84 |
| Unigene13559 | 1839 | AAEL015070-RA | alkaline phosphatase | −3.06 | 0.84 |
| CL1409.Contig1 | 1754 | AAEL013330-RA | alkaline phosphatase | −4.23 | 0.86 |
|
| |||||
| APN | |||||
| CL4769.Contig1 | 3393 | AAEL008162-RA | protease m1 zinc metalloprotease | −2.26 | 0.81 |
| CL226.Contig1 | 3059 | AAEL008158-RA | protease m1 zinc metalloprotease | −2.33 | 0.81 |
| ABC transporter | |||||
|
| |||||
| CL1116.Contig2 | 269 | AAEL013833-RA | ATP-binding cassette transporter | −3.25 | 0.84 |
| CL3352.Contig1 | 436 | AAEL012700-RA | ATP-binding cassette sub-family A member 3 | −3.38 | 0.81 |
|
| |||||
| Kinase | |||||
| Unigene12551 | 327 | AAEL009937-RB | calcium/calmodulin-dependent serine protein kinase membrane-associated guanylate kinase | 12.5 | 0.94 |
| Unigene1877 | 467 | AAEL001442-RA | map-kinase activating death domain protein | 5.06 | 0.85 |
|
| |||||
| Protease | |||||
| Unigene17070 | 263 | AAEL005753-RA | serine protease | 12.3 | 0.93 |
| Unigene5390 | 509 | AAEL007938-RA | serine-type enodpeptidase | 4.33 | 0.88 |
| CL78.Contig1 | 1654 | AAEL006627-RA | serine-type enodpeptidase | 2.47 | 0.82 |
| CL850.Contig2 | 2524 | AAEL014658-RA | caspase-1 | 2.47 | 0.81 |
| Unigene15049 | 1148 | AAEL005616-RA | trypsin | −2.32 | 0.81 |
| Unigene17749 | 681 | AAEL008093-RA | trypsin | −2.97 | 0.81 |
| Unigene17807 | 986 | AAEL015036-RA | protease S51 alpha-aspartyl dipeptidase | −3.39 | 0.82 |
| CL2452.Contig1 | 867 | AAEL008782-RA | serine-type enodpeptidase | −4.55 | 0.83 |
A total of 583 genes were found to be significantly up-regulated (375 genes) or down-regulated (208 genes). See table S4 for additional details.
All sequence reads were assembled using de novo transcriptome assembly and the identified genes were functionally annotated using blastX (Evalue < 0.0001) with a variety of databases including Gene Ontology, Nr, KeGG, SwissProt, COG as well as the Ae. aegypti genome, Aedes-aegypti-Liverpool_TRANSCRIPTS_AaegL1.3.fa.gz (https://www.vectorbase.org) (Nene et al., 2007).
Gene expression levels were calculated from RNA seq data using the FPKM method (Fragments Per kb per Million fragments): FPKM = (1,000,000*C)/(N*L*1,000) where, FPKM(A) to be the expression of gene A; C to be number of fragments that uniquely aligned to gene A; N to be total number of fragments that uniquely aligned to all genes; L to be number of bases on gene A. Differential gene expression were analyzed with significantly expressed genes (Qvalue > 0.8) between each groups using the NOISeq method .
Cadherin, ALP, and APN transcript changes and protein expression
Changes in cadherin transcripts were determined in resistant larvae midgut using RNA-seq analysis and qPCR. Four cadherins were analyzed, but their expression levels were unchanged in resistant larvae midgut (Table 4). Expression of Aedes cadherin (AAEL018140) was also analyzed by qPCR. Aedes cadherin expression was unchanged when compared to WT (Table 4). This result was confirmed by immunoblot (Figure 5).
Table 4.
Transcriptome analysis of cadherins, ALPs, and APNs in resistant larvae midgut.
| Name | Aedes ID | Log2a | Qvalue | RT-qPCR (Log2)b |
|---|---|---|---|---|
| Cadherin | ||||
| AAEL012421 | 0.13 | 0.12 | ||
| AAEL011164 | −0.11 | 0.13 | ||
| AAEL007299 | −0.29 | 0.14 | ||
| AaeCadc | AAEL018140 | −0.61 | 0.41 | 0.53±0.22 |
|
| ||||
| ALP | ||||
| ALP1 | AAEL009077 | - | - | 0.27±0.37 |
| AAEL003286 | 5.16 | 0.84 | ||
| AAEL003905 | 0.73 | 0.32 | ||
| AAEL003309 | 0.23 | 0.22 | ||
| AAEL003298 | 0.05 | 0.10 | ||
| AAEL003313 | −0.53 | 0.38 | ||
| AAEL003297 | −1.43 | 0.46 | ||
| AAEL003289 | −1.63 | 0.69 | ||
| ALP5 | AAEL015070 | −3.06 | 0.84 | −3.03±0.54 |
| ALP4 | AAEL013330 | −4.23 | 0.86 | −3.88±0.12 |
|
| ||||
| APN | ||||
| AAEL012217 | 2.27 | 0.74 | ||
| AAEL012774 | 1.33 | 0.67 | ||
| AAEL003227 | 0.71 | 0.40 | ||
| APN1 | AAEL012778 | 0.37 | 0.30 | 0.50±0.45 |
| AAEL012781 | 0.36 | 0.27 | ||
| AAEL012779 | 0.27 | 0.23 | ||
| AAEL012776 | 0.24 | 0.22 | ||
| AAEL011292 | 0.08 | 0.10 | ||
| AAEL012099 | 0.05 | 0.09 | ||
| APN2 | AAEL008155 | 0.58 | 0.40 | 0.90±0.28 |
| AAEL008163 | −0.11 | 0.15 | ||
| AAEL009108 | −0.41 | 0.31 | ||
| AAEL012783 | −0.73 | 0.47 | ||
| AAEL013899 | −0.76 | 0.48 | ||
| APN5 | AAEL008162 | −2.26 | 0.81 | |
| APN4 | AAEL008158 | −2.33 | 0.81 | |
| AAEL012110 | −2.81 | 0.76 | ||
Log2 and Q values were obtained as described in Table 3.
Log2ratio = Log2{(Etarget)ΔCttarget(Control-Sample)/(Eref)ΔCtref(Control-Sample)}where, Etarget, amplification efficiency of target gene, Eref; amplification efficiency of Actin; Ct, threshold cycle.
One ALP (AAEL003286) was significantly up-regulated, two ALPs (AAEL013330 and AAEL015070) were significantly down-regulated, and two APNs (AAEL008158 and AAEL008162) were significantly down-regulated compared to WT.
Previously identified receptor proteins of Cry11Aa toxin are bold and underlined.
Figure 5.
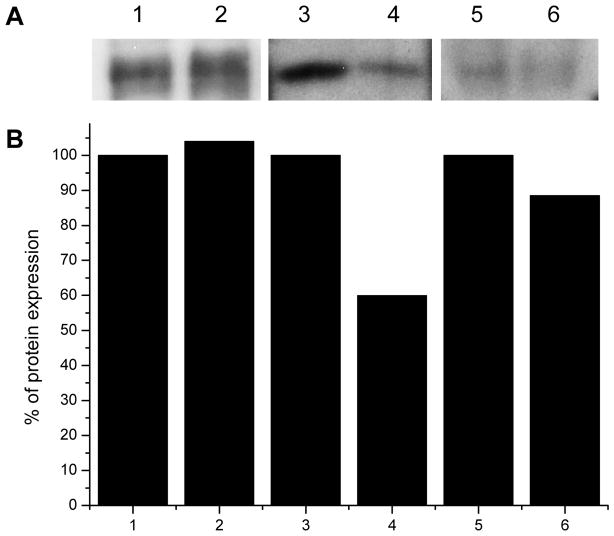
ALPs and APNs show reduced levels in the resistant larvae midgut compared to that in the wild-type strain. However, Aedes cadherin is not changed between the resistant and WT. (A) To determine the expression of target proteins in WT and G30 larvae midgut, BBMVs were extracted, loaded (10 μg) and separated in SDS-PAGE gel, transferred to a membrane and then incubated with the anti-AaeCad peptide antibody (lanes 1 and 2), anti-ALP polyclonal antibody (lanes 3 and 4), and anti-APN polyclonal antibody (lanes 5 and 6). (B) The band densities of target proteins were measured and quantified using Image J software and compared using Origin program. Lanes and columns 1, 3 and 5 – wild type and 2, 4 and 6 –resistant G30 strains. The expression of ALP (~40%) in G30 was reduced (lane and column 4) compared to that in WT (lane and column 3). The expression of APN in G30 (lane and column 6) was also reduced but less (~11%).
RNA-seq analysis revealed 9 ALPs and 17 APNs were expressed in the midgut. Comparing levels of these transcripts revealed that two ALPs (AAEL013330 and AAEL015070) in resistant larvae were significantly down-regulated compared to that in WT by Illumina sequencing and these data were confirmed using qPCR (Table 4). One ALP (AAEL003286) was highly up-regulated in resistant larvae midgut, but ALP1 (AAEL009077), a receptor protein for Cry11Aa (Fernandez et al., 2009), was not significantly changed in qPCR analysis (Table 4). Differential expression of ALPs resulted in no change of total ALP activity between WT (306 ± 41.7 μM × ml−1 × min−1/μg) and G30 (285 ± 44.7 μM × ml−1 × min−1/μg) larvae midgut. However, immunoblot assays with anti-ALP polyclonal antibody showed that a 65 kDa band, the size for ALPs, in the resistant larvae was greatly reduced (−40%) compared to WT (Figure 5). In case of APNs, two APNs (AAEL008158 and AAEL008162) were significantly reduced in resistant larvae (Table 4). However, previously identified APN receptors for Cry11Aa (APN1, AAEL012778 and APN2, AAEL008155) (Chen et al., 2009b; Chen et al., 2013) were not significantly changed or only slightly up-regulated and these results were confirmed by qPCR (Table 4). Total APN activity was slightly decreased in the midgut BBMV of the G30 resistant strain (195 ± 35.2 μM × ml−1 × min−1/μg) compared to that in the WT (273 ± 76.2 μM × ml−1 × min−1/μg). Additionally, 140 kDa bands with anti-APN polyclonal antibody were detected and slightly lower protein levels (−11%) were observed in the G30 strain compared to that in the WT.
Discussion
Ae. aegypti larvae selected with Cry11Aa toxin for 30 generations showed a 124-fold resistance ratio at the LC50 level compared to susceptible larvae (WT). However, this resistant strain (G30) had lower resistance levels to the other Bti mosquitocidal Cry toxins, namely Cry4Aa, Cry4Ba, and Cry11Ba, a homologous Cry11 toxin from Btj. These results are consistent with previous research with Cx. quinquefasciatus (Georghiou and Wirth, 1997). Cry11Aa-resistant Cx. quinquefasciatus showed high resistance to Cry11Aa (at least 1,000 fold) and to a mixture of Cry4Aa and Cry4Ba (41.6 fold), but low resistance to Cry11Ba (6.8–9.2 fold) (Cheong et al., 1997; Wirth et al., 1998; Wirth et al., 2010). These results suggest that Ae. aegypti and Cx. quinquefasciatus appear to have similar mechanisms for Cry11Aa toxin action. Further, Cry11Aa and Cry4Aa likely share some common mechanisms of toxicity in Ae. aegypti because of substantial cross-resistance. The low cross resistance to Cry4Ba suggests that although some mechanisms may be similar, there are likely differences in how Cry11Aa and Cry4Ba exert their toxicity. In contrast, Cry11Ba showed negligible cross-resistance, implying that even though the toxins are quite homologous, some of their key molecular mechanisms of toxicity likely differ. Thus competition binding data that we and others have generated does not fully explain the mechanisms of these toxins in vivo.
To determine the resistance mechanisms involved, we analyzed the processing of the Cry11Aa toxin and its ability to bind midgut BBMV from WT and G30 larvae midgut. Proteases have been demonstrated to play a role in Cry toxin resistance in some insect species (Forcada et al., 1996; Oppert et al., 1997). However, the resistant and WT strains showed similar protoxin processing patterns implying toxin processing was probably not involved in Cry11Aa resistance observed here. In contrast, there was a two-fold decrease in Cry11Aa binding affinity in the midgut of the resistant strain compared to that of the WT. This minor difference likely does not explain the resistance observed, but implies that with higher Cry11Aa resistance levels, toxin binding may be reduced in Ae. aegypti Cry11Aa resistant strains. The most common mechanism of resistance observed in lepidopteran involved changes in binding affinities of toxin receptors (Bravo and Soberon, 2008). Cry toxin resistance in Lepidoptera is associated with mutations in receptor proteins such as cadherin, ALP, APN or ABCC transporter (Atsumi et al., 2012; Gahan et al., 2001; Herrero et al., 2005; Jurat-Fuentes and Adang, 2004). Based on these data, we focused on finding a receptor protein linked to Cry11Aa resistance.
Previously identified receptor proteins of Cry11Aa in Aedes are cadherin (AAEL018140), APN1 (AAEL012778), APN2 (AAEL08155), and ALP1 (AAEL009077) (Chen et al., 2009a; Chen et al., 2009b; Chen et al., 2013; Fernandez et al., 2009). These receptors were further analyzed to determine if there were any mutations or if receptor expression was altered. In addition, RNA-seq was used to determine if the transcripts of these receptors or other previously unidentified receptors were altered in the resistant strain. These analyses showed that the expression of ALPs and APNs was significantly changed in the Cry11Aa resistant strain. Fernandez et al previously showed ALPs bound Cry11Aa, and these ALPs are involved in Cry11Aa toxicity in Ae. aegypti (Fernandez et al., 2006; Fernandez et al., 2009). Likewise, APNs were previously shown to bind the Cry11Aa toxin (Chen et al., 2009b; Chen et al., 2013). Thus, reduced ALP and APN expression in G30 resistant larvae suggests that both of these protein families could be associated with Cry11Aa resistance in Ae. aegypti. It has been previously shown that in a H. virescens Cry1A resistant strain, ALP alteration was linked to reduced Cry toxin binding, decreased pore formation, and finally increased resistance to Cry1 toxins (Jurat-Fuentes and Adang, 2004; Jurat-Fuentes et al., 2011). Also, a deletion mutant in an APN1 was associated with Cry1Ac resistance in Helicoverpa armigera (Zhang et al., 2009).
Whole genome sequence identified 14 alkaline phosphatases in Ae. aegypti (Nene et al., 2007). Previous studies investigated three ALPs as a receptor of Bti Cry toxins and found that ALP1 (AAEL009077) is a functional receptor of Cry11Aa and Cry4Ba (Fernandez et al., 2009; Rodríguez-Almazán et al., 2012). In transcriptome analysis, AAEL013330 and AAEL015070 are likely putative Cry11Aa receptors, since both of these ALPs were significantly reduced in the resistant larvae midgut. In addition, AAEL015070 was previously found in Aedes lipid rafts where Cry4Ba localized (Bayyareddy et al., 2012) and was identified as a functional receptor of Cry4Ba (Dechklar et al., 2011; Thammasittirong et al., 2011). Because the G30 strain shows high resistance to Cry11Aa and low cross-resistance to Cry4Ba, this data implies the ALPs AAEL009077 and AAEL015070 may not be associated with Cry11Aa resistance in the G30 strain, since both of these ALPs bind Cry4Ba. Further, our transcript analysis and mutation screen also showed no alteration in this ALP in this strain. On the other hand, AAEL01330 is a newly identified putative receptor of Cry toxin in mosquitoes. It is still unknown if AAEL01330 could be a functional receptor of Cry11Aa and Cry4Aa. We continue to investigate the functional role of AAEL013330 in Cry11Aa and Cry4Aa toxicity.
There were no changes in the transcript levels of APN1 AAEL008155 and APN2 AAEL012778 in the resistant strain. These were shown to be binding proteins for Cry11Aa (Chen et al., 2009b; Chen et al., 2013). However, it is possible that other APNs may be involved in Cry11Aa resistance because APN enzymatic activity and protein levels were slightly reduced in the G30 strain. In transcriptome analysis, AAEL008158 and AAEL008162 were significantly reduced in the resistant larvae midgut, implying these APNs are likely putative receptors of Cry11Aa. However, AAEL008162 may not be associated with Cry11Aa resistance in the G30 strain, since this APN bound Cry4Ba in Aedes lipid rafts (Bayyareddy et al., 2012) and only low cross resistance to Cry4Ba was observed here. On the other hand, AAEL008158 is a newly found putative receptor of Cry toxin and could be involved in Cry11Aa and Cry4Aa toxicity in mosquitoes. However, future investigations are required for both APNs.
Based on Bt resistance mechanisms in Lepidoptera, we further investigated candidate genes that could possibly be linked to Cry11Aa resistance in Ae. aegypti. The most recently identified are ABC transporters that are associated with Cry1A resistance in lepidopterans, including Heliothis virescens, Bombyx mori, and Plutella xylostella (Atsumi et al., 2012; Gahan et al., 2010; Hernández-Martínez et al., 2012; Tanaka et al., 2013). We show here that two ABC transporters (AAEL012700 and AAEL013833) were significantly reduced in G30 larvae midgut even though neither of these ABC transporters contained any mutation. However, further investigation is needed to determine if an ABC transporter is a functional receptor of Cry11Aa. Further, several proteases are differentially expressed in the G30 resistant larval midgut. In Lepidoptera, proteases trigger proteolytic activity of Cry toxin and altered protease expressions are linked in Bt resistance in Lepidoptera (Choma et al., 1990; Forcada et al., 1996; Oppert et al., 1997). However, we did not observe any difference in the processing of the Cry11Aa toxin in the G30 strain. Additionally, in a previous report, Cry toxin binding to cadherin was shown to activate a G protein and adenylyl cyclase increasing cAMP levels, which turns on a kinase A that finally causes cell death (Zhang et al., 2005; Zhang et al., 2006). In the Aedes resistant larvae midgut, two kinases, (AAEL009937 and AAEL001442), were highly up-regulated. Alteration of kinase expression could contribute to Cry11Aa resistance by regulating cell apoptosis, but additional investigation is needed.
In summary, Cry11Aa resistance in the Ae. aegypti G30 strain was associated with decreased expression of ALPs, APNs and ABC transporters, all proteins known to mediated Cry toxicity in a number of insect species. However, future investigations are needed to determine if the genes identified here are indeed involved in the toxicity of Cry11Aa.
Supplementary Material
Supplementary Table S1. Primers used for qPCR.
Supplementary Table S2. Processing of Cry11Aa in the larval midgut of WT and G30 larvae was unchanged.
Supplementary Table S3. Transcript expression of a number of genes was significantly up-regulated in the midgut of the G30 strain compared to that in the WT strain (Qvalue > 0.8).
Supplementary Table S4. Transcript expression of a number of genes was significantly down-regulated in the midgut of the G30 strain compared to that in the WT strain (Qvalue > 0.8).
Aedes aegypti developed high-level resistance to Cry11A and shows high cross-resistance to Cry4Aa, but less to Cry4Ba
Cry11A toxin processing in resistant Aedes aegypti was unchanged.
Cry11A binding affinity to midgut membrane was slightly altered in resistant Aedes aegypti.
Previously identified Cry11Aa binding proteins were not altered in resistant mosquitoes.
Transcript changes and protein expressions of alkaline phosphatases and aminopeptidase N were reduced in resistant Aedes larvae midgut.
Acknowledgments
This research was funded in part through grants from the National Institutes of Health, 1R01 AI066014 and the University of California Agricultural Experiment Station. The technical assistance of Jianwu Chen, Valeree Pons, Maria Ramirez, and Jacaqueline Ledezma is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atsumi S, Miyamoto K, Yamamoto K, Narukawa J, Kawai S, Sezutsu H, Kobayashi I, Uchino K, Tamura T, Mita K, Kadono-Okuda K, Wada S, Kanda K, Goldsmith MR, Noda H. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc Natl Acad Sci. 2012;109:E1591–E1598. doi: 10.1073/pnas.1120698109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayyareddy K, Andacht TM, Abdullah MA, Adang MJ. Proteomic identification of Bacillus thuringiensis subsp. israelensis toxin Cry4Ba binding proteins in midgut membranes from Aedes (Stegomyia) aegypti Linnaeus (Diptera, Culicidae) larvae. Insect Biochem Mol Biol. 2009;39:279–286. doi: 10.1016/j.ibmb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Bayyareddy K, Zhu X, Orlando R, Adang MJ. Proteome analysis of Cry4Ba toxin-interacting Aedes aegypti lipid rafts using geLC-MS/MS. J Proteome Res. 2012;11:5843–5855. doi: 10.1021/pr3006167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MT, Harris D, Zaritsky A, Parkhill J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2002;68:5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Likitvivatanavong S, Gill S, Soberón M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem Mol Biol. 2011;41:423–431. doi: 10.1016/j.ibmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Soberon M. How to cope with insect resistance to Bt toxins? Trends Biotechnol. 2008;26:573–579. doi: 10.1016/j.tibtech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Chang C, Yu YM, Dai SM, Law SK, Gill SS. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Aimanova KG, Fernandez LE, Bravo A, Soberon M, Gill SS. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem J. 2009a;424:191–200. doi: 10.1042/BJ20090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Aimanova KG, Pan S, Gill SS. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem Mol Biol. 2009b;39:688–696. doi: 10.1016/j.ibmb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Likitvivatanavong S, Aimanova KG, Gill SS. A 104 kDa Aedes aegypti aminopeptidase N is a putative receptor for the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Insect Biochem Mol Biol. 2013;43:1201–1208. doi: 10.1016/j.ibmb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Dhesi RK, Gill SS. Marginal cross-resistance to mosquitocidal Bacillus thuringiensis strains in Cry11A-resistant larvae: presence of Cry11A-like toxins in these strains. FEMS Microbiol Lett. 1997;153:419–424. doi: 10.1111/j.1574-6968.1997.tb12605.x. [DOI] [PubMed] [Google Scholar]
- Chilcott C, Ellar DJ. Comparative Toxicity of Bacillus thuringiensis var. israelensis Crystal Proteins in vivo and in vitro. J Gen Microbiol. 1988;134:2551–2558. doi: 10.1099/00221287-134-9-2551. [DOI] [PubMed] [Google Scholar]
- Choma CT, Surewicz WK, Carey PR, Pozsgay M, Raynor T, Kaplan H. Unusual proteolysis of the protoxin and toxin from Bacillus thuringiensis: structural implications. Eur J Biochem. 1990;189:523–527. doi: 10.1111/j.1432-1033.1990.tb15518.x. [DOI] [PubMed] [Google Scholar]
- Cowles EA, Yunovitz H, Charles JF, Gill SS. Comparison of toxin overlay and solid-phase binding assays to identify diverse CryIA(c) toxin-binding proteins in Heliothis virescens midgut. Appl Environ Microbiol. 1995;61:2738–2744. doi: 10.1128/aem.61.7.2738-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SM, Gill SS. In vitro and in vivo proteolysis of the Bacillus thuringiensis subsp. israelensis CryIVD protein by Culex quinquefasciatus larval midgut proteases. Insect Biochem Mol Biol. 1993;23:273–283. doi: 10.1016/0965-1748(93)90008-g. [DOI] [PubMed] [Google Scholar]
- Dechklar M, Tiewsiri K, Angsuthanasombat C, Pootanakit K. Functional expression in insect cells of glycosylphosphatidylinositol-linked alkaline phosphatase from Aedes aegypti larval midgut: a Bacillus thuringiensis Cry4Ba toxin receptor. Insect Biochem Mol Biol. 2011;41:159–166. doi: 10.1016/j.ibmb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Delecluse A, Rosso ML, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microbiol. 1995;61:4230–4235. doi: 10.1128/aem.61.12.4230-4235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LE, Aimanova KG, Gill SS, Bravo A, Soberon M. A GPI-anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochemical J. 2006;394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LE, Martinez-Anaya C, Lira E, Chen J, Evans A, Hernandez-Martinez S, Lanz-Mendoza H, Bravo A, Gill SS, Soberon M. Cloning and epitope mapping of Cry11Aa-binding sites in the Cry11Aa-receptor alkaline phosphatase from Aedes aegypti. Biochemistry. 2009;48:8899–8907. doi: 10.1021/bi900979b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Escobar B, Rodríguez-Magadan H, Bravo A, Soberón M, Gómez I. Differential role of Manduca sexta aminopeptidase-N and alkaline phosphatase in the mode of action of Cry1Aa, Cry1Ab, and Cry1Ac toxins from Bacillus thuringiensis. Appl Environ Microbiol. 2013;79:4543–4550. doi: 10.1128/AEM.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcada C, Alcicer E, Garceri MD, Martinez R. Differences in the midgut proteolytic activity of two Heliothis virescens strains, one susceptible and one resistant to Bacillus thuringiensis toxins. Arch Insect Biochem Physiol. 1996;31:257–272. [Google Scholar]
- Gahan LJ, Gould F, Heckle DG. Identification of a gene associated with Bt resistance in Heliothis virescens. Science. 2001;293:857–860. doi: 10.1126/science.1060949. [DOI] [PubMed] [Google Scholar]
- Gahan LJ, Pauchet Y, Vogel H, Heckel DG. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010;6:e1001248. doi: 10.1371/journal.pgen.1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georghiou GP, Wirth MC. Influence of exposure to single versus multiple toxins of Bacillus thuringiensis subsp. israelensis on development of resistance in the mosquito Culex quinquefasciatus (Diptera: Culicidae) Appl Environ Microbiol. 1997;63:1095–1101. doi: 10.1128/aem.63.3.1095-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Cowles EA, Pietrantonio PV. The mode of action of Bacillus thuringiensis endotoxins. Annu Rev Entomol. 1992;37:615–636. doi: 10.1146/annurev.en.37.010192.003151. [DOI] [PubMed] [Google Scholar]
- Grabherr M, Haas B, Yassour M, Levin J, Thompson D, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren B, Nusbaum C, Lindblad-Toh, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Martínez P, Hernández-Rodríguez C, Krishnan V, Crickmore N, Escriche B, Ferré J. Lack of Cry1Fa binding to the midgut brush border membrane in a resistant colony of Plutella xylostella moths with a mutation in the ABCC2 locus. Appl Environ Microbiol. 2012;78:6759–6761. doi: 10.1128/AEM.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero S, Gechev T, Bakker PL, Moar WJ, de Maagd RA. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four Aminopeptidase N genes. BMC Genomics. 2005;6:96. doi: 10.1186/1471-2164-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez AI, Reyes EZ, Cancino-Rodezno A, Bedoya-Pérez LP, Caballero-Flores GG, Muriel-Millan LF, Likitvivatanavong S, Gill SS, Bravo A, Soberón M. Aedes aegypti alkaline phosphatase ALP1 is a functional receptor of Bacillus thuringiensis Cry4Ba and Cry11Aa toxins. Insect Biochem Mol Biol. 2012;42:683–689. doi: 10.1016/j.ibmb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurat-Fuentes JL, Adang MJ. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur J Biochem. 2004;271:3127–3135. doi: 10.1111/j.1432-1033.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- Jurat-Fuentes JL, Karumbaiah L, Jakka SR, Ning C, Liu C, Wu K, Jackson J, Gould F, Blanco C, Portilla M, Perera O, Adang M. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS ONE. 2011;6:e17606. doi: 10.1371/journal.pone.0017606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chen J, Aimanova K, Gill S. Aedes cadherin mediates the in vivo toxicity of the Cry11Aa toxin to Aedes aegypti. Peptides. 2014 doi: 10.1016/j.peptides.2014.07.015. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D, Agaisse H, Gominet M, Chaufaux J. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Biotechnology (N Y) 1995;13:67–71. doi: 10.1038/nbt0195-67. [DOI] [PubMed] [Google Scholar]
- Ligon B. Dengue fever and dengue hemorrhagic fever: a review of the history, transmission, treatment, and prevention. Semin Pediatr Infect Dis. 2005;16:60–65. doi: 10.1053/j.spid.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Ligon BL. Reemergence of an unusual disease: the chikungunya epidemic. Semin Pediatr Infect Dis. 2006;17:99–104. doi: 10.1053/j.spid.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likitvivatanavong S, Chen J, Evans AM, Bravo A, Soberon M, Gill SS. Multiple receptors as targets of Cry toxins in mosquitoes. J Agric Food Chem. 2011;59:2829–2838. doi: 10.1021/jf1036189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalith Y, Ben-Dov E. Biological Control by Bacillus thuringiensis subsp. israelensis. In: Rechicgl JE, Rechcigl NA, editors. Insect Pest Management: Techniques for Environmental Protection. CRC Press; 2000. pp. 243–301. [Google Scholar]
- McNall RJ, Adang MJ. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem Mol Biol. 2003;33:999–1010. doi: 10.1016/s0965-1748(03)00114-0. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams B, McCue K, LS, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen-Leroux C, Charles JF. Binding of Bacillus sphaericus binary toxin to a specific receptor on midgut brush-border membranes from mosquito larvae. Eur J Biochem. 1992;210:585–590. doi: 10.1111/j.1432-1033.1992.tb17458.x. [DOI] [PubMed] [Google Scholar]
- Oppert B, Kramer KJ, Beeman RW, Johnson D, McGaughey WH. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J Biol Chem. 1997;272:23473–23476. doi: 10.1074/jbc.272.38.23473. [DOI] [PubMed] [Google Scholar]
- Perez C, Fernandez LE, Sun J, Folch JL, Gill SS, Soberon M, Bravo A. Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci U S A. 2005;102:18303–18308. doi: 10.1073/pnas.0505494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C, Munoz-Garay C, Portugal LC, Sanchez J, Gill SS, Soberon M, Bravo A. Bacillus thuringiensis ssp. israelensis Cyt1Aa enhances activity of Cry11Aa toxin by facilitating the formation of a pre-pore oligomeric structure. Cell Microbiol. 2007;9:2931–2937. doi: 10.1111/j.1462-5822.2007.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Almazán C, Reyes EZ, Zúñiga-Navarrete F, Muñoz-Garay C, Gómez I, Evans AM, Likitvivatanavong S, Bravo A, Gill SS, Soberón M. Cadherin binding is not a limiting step for Bacillus thuringiensis subsp. israelensis Cry4Ba toxicity to Aedes aegypti larvae. Biochem J. 2012;443:711–717. doi: 10.1042/BJ20111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengwiman S, Aroonkesorn A, Dedvisitsakul P, Sakdee S, Leetachewa S, Angsuthanasombat C, Pootanakit K. In vivo identification of Bacillus thuringiensis Cry4Ba toxin receptors by RNA interference knockdown of glycosylphosphatidylinositol-linked aminopeptidase N transcripts in Aedes aegypti larvae. Biochem Biophys Res Commun. 2011;407:708–713. doi: 10.1016/j.bbrc.2011.03.085. [DOI] [PubMed] [Google Scholar]
- Tabashnik BE, Gassmann AJ, Crowder DW, Carriere Y. Insect resistance to Bt crops: evidence versus theory. Nat Biotechnol. 2008;26:199–202. doi: 10.1038/nbt1382. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Miyamoto K, Noda H, Jurat-Fuentes J, Yoshizawa Y, Endo H, Sato R. The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS J. 2013;280:1782–1794. doi: 10.1111/febs.12200. [DOI] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreau G, Bayyareddy K, Jones CM, Stalinski R, Riaz MA, Paris M, David JP, Adang MJ, Després L. Larval midgut modifications associated with Bti resistance in the yellow fever mosquito using proteomic and transcriptomic approaches. BMC Genomics. 2012;13:248. doi: 10.1186/1471-2164-13-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammasittirong A, Dechklar M, Leetachewa S, Pootanakit K, Angsuthanasombat C. Aedes aegypti membrane-bound alkaline phosphatase expressed in Escherichia coli retains high-affinity binding for Bacillus thuringiensis Cry4Ba toxin. Appl Environ Microbiol. 2011;77:6836–6840. doi: 10.1128/AEM.05775-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomori O. Yellow fever: the recurring plague. Crit Rev Clin Lab Sci. 2004;41:391–427. doi: 10.1080/10408360490497474. [DOI] [PubMed] [Google Scholar]
- Wirth MC, Delecluse A, Federici BA, Walton WE. Variable cross-resistance to Cry11B from Bacillus thuringiensis subsp. jegathesan in Culex quinquefasciatus (Diptera: Culicidae) resistant to single or multiple toxins of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 1998;64:4174–4179. doi: 10.1128/aem.64.11.4174-4179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth MC, Walton WE, Federici BA. Evolution of resistance to the Bacillus sphaericus Bin toxin is phenotypically masked by combination with the mosquitocidal proteins of Bacillus thuringiensis subspecies israelensis. Environ Microbiol. 2010;12:1154–1160. doi: 10.1111/j.1462-2920.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Cheng H, Gao Y, Wang G, Liang G, Wu K. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Mol Biol. 2009;39:421–429. doi: 10.1016/j.ibmb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, Candas M, Griko NB, Rose-Young L, Bulla LA., Jr Cytotoxicity of Bacillus thuringiensis Cry1Ab toxin depends on specific binding of the toxin to the cadherin receptor BT-R1 expressed in insect cells. Cell Death Differ. 2005;12:1407–1416. doi: 10.1038/sj.cdd.4401675. [DOI] [PubMed] [Google Scholar]
- Zhang X, Candas M, Griko NB, Taussig R, Bulla LAJ. A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc Natl Acad Sci USA. 2006;103:9897–9902. doi: 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Primers used for qPCR.
Supplementary Table S2. Processing of Cry11Aa in the larval midgut of WT and G30 larvae was unchanged.
Supplementary Table S3. Transcript expression of a number of genes was significantly up-regulated in the midgut of the G30 strain compared to that in the WT strain (Qvalue > 0.8).
Supplementary Table S4. Transcript expression of a number of genes was significantly down-regulated in the midgut of the G30 strain compared to that in the WT strain (Qvalue > 0.8).