Abstract
Cells evolve to actively coordinate nutrient availability with cellular activity in order to maintain metabolic homeostasis. In addition, active pathways to repair DNA damage are crucial to avoid deleterious genomic instability. In recent years, it has become increasingly clear that availability of intermediate metabolites may play an important role in DNA repair, suggesting that these two seemingly distant cellular activities may be highly coordinated. The sirtuin family of proteins now described as deacylases (they can also remove acyl groups other than acetyl moieties), it appears to have evolved to control both metabolism and DNA repair. In this review, we discuss recent advances that lay the foundation to understanding the role of sirtuins in these two biological processes, and the potential crosstalk to coordinate them.
Introduction
Sirtuins are members of a family of evolutionarily conserved enzymes with NAD+-dependent deacylase activity. Since the discovery of Sir2 (silencing information regulator 2) in the budding yeast Saccharomyces cerevisiae as a transcriptional silencer of the mating-type loci more than 20 years ago [1], many studies have demonstrated diverse biological roles for sirtuins, such as in genome stability, cellular metabolism, and lifespan regulation [2,3]. Mammalian sirtuins have seven isoforms (SIRT1–7), each one with unique subcellular localization and distinct functions [4]. SIRT1 and SIRT2 can be found in both nucleus and cytoplasm, SIRT6 and SIRT7 are almost exclusively nuclear and SIRT3, SIRT4, and SIRT5 are located in the mitochondria [5]. Studies on sirtuin biology have shown great progress in the past two decades, emphasizing the critical importance of these enzymes in human biology and disease.
Due to their NAD+ dependency, it had been speculated that sirtuins play a crucial role in modulating energy metabolism. Indeed, sirtuins are broadly recognized as critical regulators of multiple metabolic pathways, including glucose, glutamine, and lipid metabolism [6]. For cells to thrive, energy and metabolic demands have to be carefully coordinated with nutrients availability. As sensors of energy and redox status in cells, these protein deacylases can directly modulate activity of key metabolic enzymes -by posttranslational modifications- as well as regulate transcription of metabolic genes. In addition, several sirtuins play additional roles in metabolic homeostasis. For instance, both SIRT1 and SIRT2 control autophagy responses under various nutrient stress conditions, as modulators of FOXO signaling pathway [7]. Autophagy will be covered in detail in an accompanying article in this issue.
Nuclear sirtuins have also evolved as regulators of genome integrity. Our cells experience ~ 1×104−1×105 DNA lesions per day [8], hence they have developed repair machineries to avoid detrimental outcomes from oxidative and genotoxic stress. In the past decade, the roles of sirtuins in maintaining genomic stability have been described, as regulators of DNA repair pathways [9], chromatin structure [10], and telomere maintenance [11,12].
Based on the fact that sirtuins possess dual roles in metabolism and DNA repair, sirtuins can serve as nodal points in regulating both processes. Intriguingly, new studies have started to appreciate that DNA damage can directly trigger adaptive metabolic responses [13,14], indicating that these two seemingly separate biological entities may function in a highly coordinated fashion. In this review, we will focus on recent progress in understanding the roles of sirtuins in both metabolism and DNA repair, and the possible crosstalk between these two phenomena.
Sirtuins in metabolism
Glucose and glutamine metabolism
Since glucose is a primary nutrient for cell survival and proliferation, systemic glucose levels should be tightly regulated throughout tissues. Crucial organs such as liver, muscle, and pancreas are main modulators of glucose homeostasis. At the cellular level, once glucose enters a cell, it is converted into pyruvate in the cytoplasm through glycolysis in a multi-enzyme, strictly regulated process. In most cells, pyruvate will then enter the TCA cycle to generate energy through oxidative phosphorylation (OXPHOS) in a highly efficient process (34–36 mols of ATP per mol of glucose). However, in specific cases, pyruvate will be diverted in the cytoplasm to produce lactate, a less efficient way to produce ATP, but a critical adaptive mechanism in cells where OXPHOS is impeded (hypoxia, for instance) or to produce intermediate metabolites for biomass in highly proliferating cells.
Extensive studies have previously shown that SIRT1 can modulate both gluconeogenesis and glycolysis by regulating important metabolic factors, including PGC1α and FOXO [15]. More recently, intracellular levels of NAD+ has been shown to regulate SIRT1 deacetylase activity, affecting high fat diet (HFD)-induced obesity and aging, as discussed below [reviewed in 16].
SIRT3 is a major mitochondrial protein deacetylase [17], regulating multiple metabolic proteins such as the TCA cycle protein isocitrate dehydrogenease 2 (IDH2) [18] and key proteins in the electron transfer chain (ETC) [19–21]. In skeletal muscle, SIRT3 plays an important role in regulating metabolic adaptive responses. Decreased levels of SIRT3 cause increasing oxidative stress and insulin resistance [22] and recent studies showed that active deacetylation of pyruvate dehydrogenase (PDH) E1α by SIRT3 provides metabolic flexibility under nutrient stress conditions [23]. Wang and his colleagues discovered that SIRT3 can deacetylate FOXO3a, in turn enhancing FOXO3a activity and increased expression of its targets, including antioxidant genes. In this way, SIRT3 protects mitochondria from oxidative stress [24]. Since SIRT3 actively modulate carbohydrate metabolism and ROS production, the role of SIRT3 in cancer metabolism has been highlighted [25]. Gius et al. first described that SIRT3 acts as a tumor suppressor by maintaining intact mitochondria in breast cancer [26]. Later, two studies provided mechanistic proof that HIF-1α (hypoxia inducible factor-1α) stabilization following mitochondrial ROS generation is critical to sustain cancer-prone metabolic reprogramming in SIRT3-deleted tumors [27,28].
SIRT4 is mostly known for its role in glutamine metabolism. In proliferating cells, glutamine is the main source to replenish the TCA cycle as a source of α-ketoglutarate (α-KG) [29]. Two different groups recently reported new roles for SIRT4 in glutamine metabolism. Jeong et al. described that SIRT4 inhibits glutamine entry to the TCA cycle under genotoxic stress, preventing dysregulated proliferation and genomic instability [14]. Although SIRT4 appears to work by inhibiting GDH activity, how SIRT4 does so mechanistically remains to be fully understood. Notably, Csibi et al. found that the mTORC1-CREB2 axis can regulate SIRT4 transcription under various nutrient stress conditions, thereby affecting glutamine anaplerosis into the TCA cycle and cell proliferation [30], further confirming an important role for this sirtuin in glutamine metabolism.
SIRT5 has recently been defined as a lysine demalonylase and desuccinylase [31]. The global analysis of lysine succinylation (“succinylome”) in the context of SIRT5 demonstrated that this posttranslational modification has a regulatory effect on glucose metabolism by modulating the activities of PDH, SDH and mitochondrial respiration in mouse liver and MEFs [32]. The pioneering work of the Lin laboratory provided the first proof that sirtuins can work by removing non-acetyl acyl groups, defining sirtuins as “protein deacylases” and opening a whole new field in enzymology and biochemistry.
Previous work defined SIRT6 as a critical epigenetic regulator of glucose metabolism [33]. SIRT6 knockout (KO) mice exhibited a fatal hypoglycemic phenotype, which leads to death few weeks after birth [34]. The hypoglycemia resulted mainly from increased glucose uptake in muscle and brown adipose tissue. Mechanistically, SIRT6 negatively regulates HIF-1α-dependent transcription by deacetylating H3K9Ac at the promoter of several metabolic genes such as glucose transporter 1 (GLUT1), lactate dehydrogenase A (LDHA), and PDH kinase 1 (PDHK1), thereby augmenting glucose uptake and glycolysis even under normoxia [35]. Such phenotype of aerobic glycolysis (also known as “Warburg effect” [36]), led to the hypothesis that SIRT6 could play a crucial role as a tumor suppressor. Indeed, ablation of SIRT6 enhanced tumor growth both in vitro and in vivo in models of colorectal cancer, [37]. More strikingly, treatment with the PDHK1 inhibitor dichloroacetate (DCA), reversed the tumorigenic phenotype in the context of SIRT6-deleted tumors, demonstrating that metabolic reprogramming is a driver of tumorigenesis. Two additional studies support the idea that SIRT6 acts as a tumor suppressor. Wagner and his colleagues reported that decreased level of SIRT6 plays a key role in AP-1-driven liver tumor by increasing H3K9Ac at the promoter of survivin and thus promoting cell survival [38]. This event is specific to tumor initiation, working in a c-Jun-dependent manner, thus implicating SIRT6 in liver tumor initiation. Another study reported that decreased level of SIRT6 is associated with poor clinical consequences in hepatocellular carcinoma (HCC) [39]. Taking into account that SIRT6 acts as well as a negative regulator of gluconeogenesis in liver via GCN5-dependent PGC-1α activation [40], it will be of particular interest to dissect how different metabolic outputs may contribute to liver tumorigenesis in a SIRT6 dependent manner.
Lipid metabolism
Lipids play fundamental roles as cellular membrane constituents and energy source, whose synthesis, storage, and expenditure are tightly regulated by different physiological cues, including fasting and nutrients availability. Excess nutrients from glucose, lipid and protein metabolism stimulate lipid synthesis, primarily in liver, in order to store energy inside white adipose tissue (WAT). Fatty acid (FA) synthesis occurs in the cytoplasm by using malonyl-CoA as an adaptor molecule and acetyl-CoA as a substrate of FA synthase (FAS), yielding acyl-CoA. On the other hand, FA oxidation happens in the mitochondrial matrix where β-oxidation produces acetyl-CoA, a key molecule in the TCA cycle to generate ATP. As energy/redox sensors, sirtuins actively modulate both FA synthesis and oxidation via transcriptional or posttranslational regulation [41]. Depending on the subcellular localization of sirtuins, they preferentially regulate either FA synthesis (cytoplasm) or FA oxidation (mitochondria).
SIRT1 deacetylates and suppresses sterol-response element-binding protein 1c (SREBP1c)-dependent transcription, targeting triglyceride synthesis in the liver [42,43]. SIRT1 also plays a key role in hepatic FA utilization during fasting [44] or HFD [45], mediating the transcriptional activation of PPARα/PGC-1α-dependent genes. Li and his group further demonstrated that liver-specific genetic ablation of SIRT1 caused hepatic steatosis in vivo [46]. In skeletal muscle, a SIRT1/PGC-1α complex is activated via cAMP/PKA signaling cascade from adrenergic stimuli to increase FA oxidation [47]. Interestingly, oleic acid among long-chain free FA (LCFFA) specifically stimulates FA utilization in skeletal muscle through a PKA-SIRT1-PGC-1α pathway [48], suggesting that a single LCFFA evolved the capability to regulate FA metabolism. Whether this represents a highly specialized feedback mechanism and the physiological relevance of these effects remain to be determined.
SIRT3 and SIRT4 play as well important roles in FA oxidation. Genetic ablation of SIRT3 alters acetylation status of several metabolic enzymes including long-chain acyl-CoA dehydrogenase (LCAD), decreasing FA oxidation in liver mitochondria [49] and predisposing to metabolic syndrome [50]. Recently, the precise lysine sites in LCAD targeted by SIRT3 (K318/K322) were identified [51]. Given that hundreds of mitochondrial proteins are hyperacetylated in SIRT3−/− mitochondria, future work will be required to fully grasp the functional and physiological consequences of such modifications. Although SIRT4 has been known to regulate FA oxidation in liver and skeletal muscle [52], only recently we learned SIRT4 as a repressor of malonyl-CoA decarboxylase (MCD) [53]. MCD is a core enzyme to balance the levels of malonyl-CoA and acetyl-CoA in mitochondria, and thus it is a key module of lipid anabolism and catabolism. Through deacetylation and inhibition of MCD activity, SIRT4 favors FA synthesis over FA oxidation in fed condition and deletion of SIRT4 has a protective role in HFD-induced obesity.
SIRT6 KO mice presents complete loss of subcutaneous fat in addition to its hypoglycemic phenotype, indicating a potential role for SIRT6 in lipid metabolism [34]. Indeed, Kim et al. observed that liver-specific deletion of SIRT6 facilitates fatty liver formation by increasing triglyceride (TG) synthesis [54]. SIRT6 represses transcription of lipid metabolism-related genes including acetyl-CoA carboxylase (ACC) and FAS by H3K9 deacetylation. Recently, SIRT6 role in lipid metabolism was further investigated as a regulator of LDL (low-density lipoprotein) and cholesterol [55,56]. SIRT6 form a complex with FOXO3a, regulating H3K9Ac and H3K56Ac levels in the promoter of the Pcsk9 (proprotein convertase subtilisin/kexin type 9) gene, in turn repressing LDLR (LDL receptor) expression, an important membrane receptor for LDL and cholesterol internalization in liver [55]. Notably, SIRT6 overexpressing mice exhibited protective effect from HFD-induced LDL and cholesterol increase in the blood. In a separate study, the same group also reported that SREBP-2 is another key regulator in cholesterol homeostasis in a FOXO3/SIRT6-dependent manner [56]. Using one of the first models of SIRT6 overexpression, Cohen and his colleagues deciphered further mechanistic insights on SIRT6 regulating SREBP-1/2 in liver [57], following their original study demonstrating extension of lifespan in SIRT6 transgenic mice [58]. In addition to transcriptional repression of SREBP-1/2, SIRT6 modulates SREBP-1/2 by proteolytic cleavage and phosphorylation of SREBP-1 via activation of AMPK (AMP kinase). In a reciprocal manner, the microRNAs miR33a and miR33b, expressed from the introns of SREBP-2 and -1 respectively, down-regulate SIRT6 level. Such roles for SIRT6 explained the protection against hypercholesterolemia following HFD treatment in SIRT6 transgenic mice. These studies provide multi-layered regulation of lipid metabolism by SIRT6, confirming a critical role for SIRT6 in lipid metabolism and metabolic syndrome related disorders.
Surprisingly, SIRT6 has shown very weak in vitro deacetylase activity, making biochemical analysis challenging. This in vitro observation led to two possible hypotheses: one was that SIRT6 needs a certain biological context to fully act as a deacetylase. The other postulated a novel enzymatic activity. Astonishingly, it appears that both hypotheses were right. Similar to the desuccinylase and demalonylase activity defined for SIRT5 [31], the same group demonstrated that SIRT6 possesses a novel enzymatic activity as a LCFA deacylase, working as demyristoylase and depalmitoylase in vitro [59]. Supported by in vivo results, the study found that SIRT6 demyristoylate TNFα, stimulating its secretion in macrophages. On the other hand, Denu and colleagues discovered that in vitro SIRT6 deacetylase activity is stimulated a thousand fold by free FA (FFA), performing as robust as any of the other sirtuins [60]. This study provided a unique biochemical basis to define a novel regulatory loop, where FFAs in cells can act as allosteric regulators to stimulate SIRT6 activity, which in turn will tune FA metabolism to bring back homeostasis.
Although much less is known about SIRT7, a recent study showed that it alleviates HFD-induced hepatosteatosis by co-repressing Myc transcriptional activity and thus decreasing ER stress in liver [61]. In vivo genetic ablation and overexpression of SIRT7 confirmed that this protective effect of SIRT7 is Myc-dependent. Although a previous study defined SIRT7 as an H3K18 deacetylase [62], future investigations will uncover by which mechanism(s) SIRT7 regulates Myc-dependent transcription in lipid metabolism.
NAD+ and Metabolism
It has long been postulated that modulation of NAD levels could serve as a mean to regulate sirtuin activity, influencing metabolism. A first proof for such hypothesis came from work by the Imai lab, where they showed that treatment of mice with the NAD precursor nicotinamide mononucleotide (NMN) ameliorated glucose intolerance and diabetes in both HFD-treated and aged animals [63]. Such effects were partly dependent on SIRT1. Supporting these studies, recent work demonstrated that supplementation with another NAD+ precursor, nicotinamide riboside (NR), increases intracellular and mitochondrial NAD+ levels, thus activating SIRT1 and SIRT3, and subsequently enhancing oxidative metabolism both in vitro and in vivo [64]. This study also showed that NR supplementation protected against HFD-induced obesity. Remarkably, Sinclair and colleagues discovered that nuclear NAD+ levels affect SIRT1 activity to regulate mitochondrial OXPHOS and overall homeostasis in mice [65]. When nuclear NAD+ levels are significantly reduced, as seen with aging, SIRT1 activity is compromised and mitochondrial metabolism is severely impaired through HIF-1α-, c-Myc- and PGC-1α-mediated mechanisms, causing a pseudohypoxic state. Furthermore, interventions to increase NAD+ levels by calorie restriction and supplementation with NMN partially restored mitochondrial homeostasis and metabolism, further defining NAD+ as a key modulatory factor in metabolism-associated aging phenotypes. Even though all these studies provided evidence to support a role for SIRT1 and SIRT3 downstream of NAD availability, whether other sirtuins may as well being involved in those phenotypes remains to be established.
Sirtuins in DNA repair
Our cells are constantly exposed to genomic insults and four major pathways evolved in eukaryotes to resolve DNA damage; homologous recombination (HR), non-homologous end joining (NHEJ), base-excision repair (BER), and nucleotide-excision repair (NER) [66]. For single-strand breaks (SSB), BER and NER are major repair mechanisms to repair the nucleotides using the sister strand as a template. ROS-mediated SSBs preferentially undergoes BER repair, while bulky adducts and UV-induced thymidine dimers are prone to be repaired by NER. For double-strand breaks (DSB), more detrimental to the genome, cells choose either HR or NHEJ to repair the damaged DNA. If cells find a homologous DNA region from a sister chromatid in proximity to the DNA damage, HR serves as a repair mechanism to rebuild the whole damaged area using the template chromatid (indeed, HR is the dominant repair pathway during S phase). In contrast, in non-dividing cells, ligation of two damaged DNA ends with little homology occurs via NHEJ, an error-prone DDR pathway. Notably, sirtuins have evolved to modulate multiple repair pathways. As explained in detail below, some of them modulates activity of DNA repair factors through deacetylation, others influence chromatin accessibility to enhance recruitment of repair factors, while others influence repair by preventing DNA damage indirectly, by means of modulating the cell cycle and preventing oxidative stress.
SIRT1
SIRT1 null mice present embryonic lethality mainly due to impaired DDR and chromosomal abnormalities [67]. Indeed, SIRT1 regulates the activity of several proteins important for HR repair, such as NBS1 [68], Rad51 [69], and the DSB sensing protein WRN [70]. Recent studies show that SIRT1 also regulates NHEJ via cooperative action with ATM and HDAC1 in postmitotic neurons [71]. On one hand, SIRT1 sustains prolonged activity of ATM, and on the other hand, it also stimulates HDAC1 activity by deacetylating this enzyme at sites of DSBs. SIRT1 also plays key roles in NER via deacetylation and recruitment of XPA [72] and XPC [73] (Xeroderma Pigmentosum A and C) to the sites of damage. All together, these results indicate that SIRT1 evolved to perform multiple functions in different DNA repair pathways, highlighting its critical role in protecting against genomic instability.
SIRT2
Initial studies demonstrated a potential role for SIRT2 in cell cycle progression, especially during mitosis, given that levels of SIRT2 drastically varies throughout the cell cycle [74]. Recently, two studies highlighted novel roles for SIRT2 in replication stress and genomic integrity [75,76]. The replication stress response (RSR) is one of the DDR signaling pathways to keep genome integrity. SIRT2 can relieve RS through deacetylation and activation of CDK9 [75]. Vaquero and his group discovered that deacetylation of H4K16Ac by SIRT2 facilitates H4K20 methylation by the PR-Set7 methyltransferase, regulating mitotic entrance [76]. Notably, loss of SIRT2 facilitated tumor formation in a model of skin squamous cell carcinoma [76], indicating a potential role for SIRT2 in protecting against genomic instability, thereby preventing tumorigenesis.
Mitochondrial sirtuins
Considering their exclusive localization in the mitochondria, it is reasonable to think that SIRT3, 4 and 5 play no direct role on nuclear DNA repair. However, they are of critical importance to prevent accumulation of mitochondrial ROS (reactive oxygen species) in turn preventing DNA damage. Kim et al. first demonstrated that SIRT3 deletion leads to an increased level of superoxide and genomic instability under stress conditions, enhancing tumor development in mammary glands [25]. Several groups showed that high ROS level results from failure to activate SOD2 (MnSOD, manganese superoxide dismutase) via deacetylation by SIRT3 [77–79]. SIRT3 also regulates glutathione-mediated redox balance [18] and ROS generation from complex III [26], further supporting a protective role for SIRT3 against oxidative stress. As mentioned above, SIRT4 levels are increased by DNA damage, acting as a glutamine gatekeeper to regulate anaplerosis towards the TCA cycle, coordinating DDR and metabolism [14]. SIRT4 reduces entrance of glutamine to the TCA cycle by reducing glutamate dehydrogenase (GDH) activity. Inhibition of glutamine metabolism causes cells to arrest, providing sufficient time for these cells to repair DNA. Cells lacking SIRT4 continue to proliferate unabated following DNA damage, in turn accumulating genomic instability. In this context, SIRT4 acts as a tumor suppressor in models of breast and lung cancer [13,29].
SIRT6
Extensive studies cemented a role for SIRT6 in numerous DNA repair pathways. SIRT6 KO cells exhibit hypersensitivity to genotoxic agents and genomic instability [33]. In that original study, SIRT6 was proposed to work on BER, by mechanisms that remain poorly understood. Chua and her group first illustrated that SIRT6 is necessary for efficient DNA DSB repair as well, mainly by stabilizing DNA-PK (DNA-dependent protein kinase) at DSB sites in turn promoting NHEJ repair [80]. Moreover, SIRT6 protects telomeric chromatin from DNA damage and genomic instability, acting as an H3K9 and H3K56 deacetylase [12,81]. Deacetylation of H3K9 by SIRT6 promotes the stable association of the WRN protein at telomere regions, important for processing telomeres in S phase [12]. Another study found CtIP (CtBP interacting protein) as a novel substrate of SIRT6, which facilitates the resection of the DSBs and DNA repair by HR [82]. PARP1 (poly-[adenosine diphosphate (ADP)–ribose] polymerase 1) is the first target for mono- ADP ribosylation by SIRT6, resulting in enhanced DSB repair both by NHEJ and HR specifically following oxidative stress [83]. Interestingly, a recent study found that chromatin remodeling by SIRT6 also plays a crucial role in DSB repair [84]. SIRT6 is recruited to sites of DSBs, recruiting SNF2h, an ATP-dependent chromatin remodeler, to open chromatin, providing proper docking sites for further recruitment of downstream DDR factors, allowing efficient repair. This study is particularly meaningful, providing in vivo data that SIRT6 is critical for SNF2h recruitment to chromatin following DDR in brain and pancreas. Taken together, all these studies demonstrate that SIRT6 plays multiple roles at different layers during DNA repair. How such roles are coordinated, and what are the unique determinants that modulate specificity, remain yet to be discovered.
Concluding remarks: the metabolism-DNA repair connection
Considering that the repair of DNA needs energy as well as particular metabolic intermediates for signaling, cells may have evolved specific mechanistic crosstalks to coordinate metabolic activities for efficient DNA repair responses. As we discussed above, sirtuins play significant roles both in metabolism and DNA repair, implicating that sirtuins may work as a hub to coordinate these two seemingly different cellular processes. One interesting perspective is that metabolic intermediates are necessary for a series of enzymatic functions in DDR. Indeed, several enzymes in DDR are regulated by sirtuin-dependent acetylation/deacetylation, such as NBS1 [68] and WRN [70]. Given that acetyl-CoA is necessary for acetylation of proteins, and NAD+ is a cofactor of both sirtuins and the DNA repair factor PARP, one could argue that changes in availability of acetyl-CoA and NAD+ could play critical limiting steps for proper DDR. Further, given the role of chromatin dynamics in DNA repair, changes in chromatin structure that depends on histone acetylation and methylation could directly impinge on efficient DNA repair. Therefore, we could infer that availability of acetyl-coA and methyl groups from one-carbon metabolism could directly influence genetic stability. Although such scenarios have recently been elegantly discussed from a theoretical point of view [85,86], such hypotheses remain to be experimentally tested. Given the extensive roles (discussed above) in both metabolism and DNA repair, sirtuins could be coordinating such efforts. For example, DNA damage-dependent increase in SIRT4 dampens glutamine metabolism, providing a proliferation checkpoint to ensure proper DNA repair, as implicated in Jeong et al. [14]. The recent findings defining activation of SIRT6 by FFA [60], suggest that limiting availability of acetyl-CoA could determine levels of FFA in cells, in turn modulating SIRT6 activity. SIRT6 regulates, at the transcriptional level, genes involved in lipolysis, while at the same time influences DNA repair through its deacetylase activity. Finally, acetyl-groups removed from histones in the nucleus could be shuffle back into the cellular pools to restore metabolic balance [85]. While such crosstalks are debated in a theoretical arena, active research in these areas is likely to provide experimental evidence in the near future.
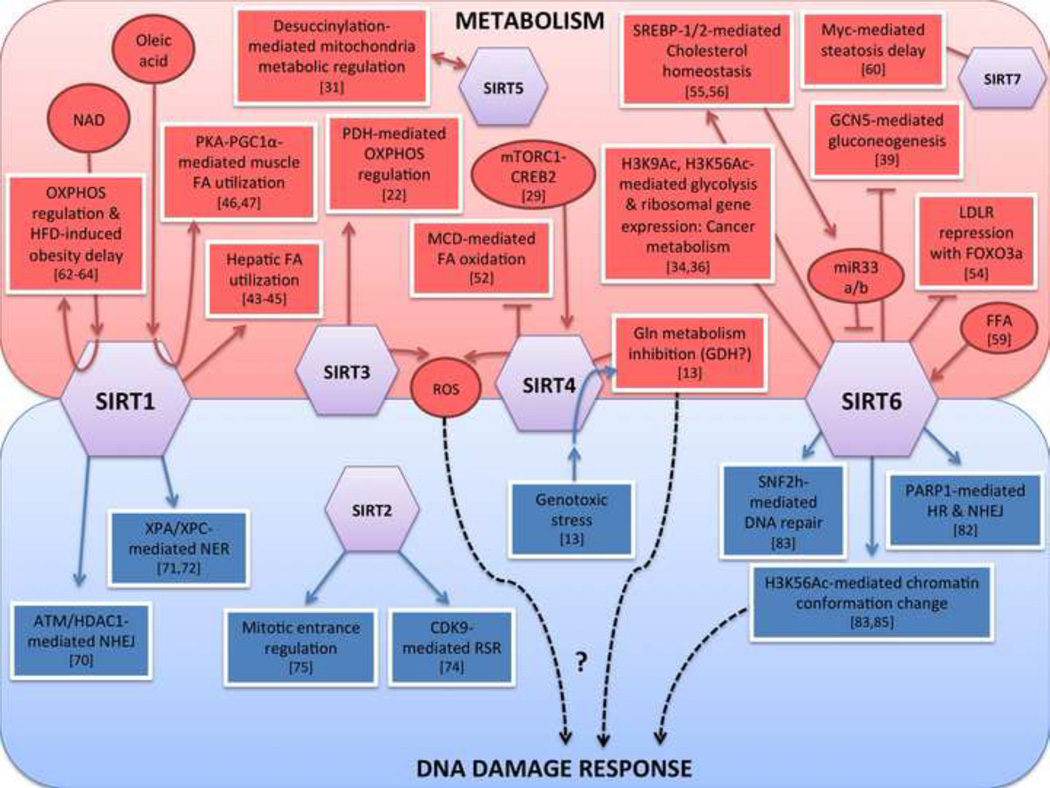
Figure 1. Sirtuins functions in metabolism and DNA repair.
A diagram depicting the different functions for the mammalian sirtuins in cellular metabolism (red) and DNA repair (blue). Specific targets and biological roles are summarized.
Acknowledgments
Work in the Mostoslavsky lab is supported in part by NIH grants GM093072-01, DK088190-01A1 and the National Pancreatic Foundation. RM is the Kristine and Bob Higgins MGH Research Scholar, a Howard Goodman Awardee and the Andrew Warshaw Institute for Pancreatic Cancer Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haigis MC, Sinclair DA. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu. Rev. Pathol. Mech. Dis. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sebastian C, Satterstrom FK, Haigis MC, Mostoslavsky R. From Sirtuin Biology to Human Diseases: An Update. Journal of Biological Chemistry. 2012;287:42444–42452. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finkel T, Deng C-X, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michishita E. Evolutionarily Conserved and Nonconserved Cellular Localizations and Functions of Human SIRT Proteins. Molecular Biology of the Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nature Reviews Molecular Cell Biology. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng F, Tang BL. Sirtuins' modulation of autophagy. J. Cell. Physiol. 2013;228:2262–2270. doi: 10.1002/jcp.24399. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T. Instability and deacy of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 9.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 10.Vaquero A. The conserved role of sirtuins in chromatin regulation. Int. J. Dev. Biol. 2009;53:303–322. doi: 10.1387/ijdb.082675av. [DOI] [PubMed] [Google Scholar]
- 11.Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. Journal of Cell Biology. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TLA, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. The EMBO Journal. 2010;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeong SM, Xiao C, Finley LWS, Lahusen T, Souza AL, Pierce K, Li Y-H, Wang X, Laurent G, German NJ, et al. SIRT4 Has Tumor-Suppressive Activity and Regulates the Cellular Metabolic Response to DNA Damage by Inhibiting Mitochondrial Glutamine Metabolism. Cancer Cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. This study is one of the first to demonstrate that specific metabolic adaptations (namely, rerouting of glutamine) are an integral part of the DNA damage response.
- 15.Cantó C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol. Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdin E. The Many Faces of Sirtuins: Coupling of NAD metabolism, sirtuins and lifespan. Nature Medicine. 2014;20:25–27. doi: 10.1038/nm.3447. [DOI] [PubMed] [Google Scholar]
- 17. Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. This study is the first comprehensive analysis of the broad regulation of protein acetylation in the mitochondria by SIRT3 and calorie restriction.
- 18.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 Mediates Reduction of Oxidative Damage and Prevention of Age-Related Hearing Loss under Caloric Restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao J, Scott I, Lu Z, Pang L, Dimond CC, Gius D, Sack MN. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radical Biology and Medicine. 2010;49:1230–1237. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimen H, Han M-J, Yang Y, Tong Q, Koc H, Koc EC. Regulation of Succinate Dehydrogenase Activity by SIRT3 in Mammalian Mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finley LWS, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, Haigis MC. Succinate Dehydrogenase Is a Direct Target of Sirtuin 3 Deacetylase Activity. PLoS ONE. 2011;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proceedings of the National Academy of Sciences. 2011;108:14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jing E, O'Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, et al. Sirt3 Regulates Metabolic Flexibility of Skeletal Muscle Through Reversible Enzymatic Deacetylation. Diabetes. 2013;62:3404–3417. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng AHH, Shieh S-S, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radical Biology and Medicine. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Haigis MC, Deng CX, Finley LWS, Kim HS, Gius D. SIRT3 Is a Mitochondrial Tumor Suppressor: A Scientific Tale That Connects Aberrant Cellular ROS, the Warburg Effect, and Carcinogenesis. Cancer Research. 2012;72:2468–2472. doi: 10.1158/0008-5472.CAN-11-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H-S, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 Is a Mitochondria-Localized Tumor Suppressor Required for Maintenance of Mitochondrial Integrity and Metabolism during Stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell EL, Emerling BM, Ricoult SJH, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finley LWS, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Csibi A, Fendt S-M, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, et al. The mTORC1 Pathway Stimulates Glutamine Metabolism and Cell Proliferation by Repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. This manuscript is the first to describe a non-deacetylase, novel deacylase activity for a sirtuin, opening a whole new field in enzymology and biochemistry.
- 32.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BMM, Skinner ME, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Molecular Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong L, Mostoslavsky R. SIRT6: a master epigenetic gatekeeper of glucose metabolism. Transcription. 2010;1:17–21. doi: 10.4161/trns.1.1.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic Instability and Aging-like Phenotype in the Absence of Mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 35.Zhong L, Urso AD, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1a. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warburg O. On the Origin of Cancer Cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 37. Sebastian C, Zwaans BMM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. The Histone Deacetylase SIRT6 Is a Tumor Suppressor that Controls Cancer Metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. This study describes SIRT6 as a tumor suppressor modulating cancer metabolism, representing one of the first demonstration that metabolism may play a role as a driver, rather than a late event, during tumorigenesis.
- 38.Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, Chen L, Scheuch H, Zheng H, Qin L, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nature Cell Biology. 2012;14:1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- 39.Marquardt JU, Fischer K, Baus K, Kashyap A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, et al. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology. 2013;58:1054–1064. doi: 10.1002/hep.26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominy JE, Jr, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, Ruan H-B, Feldman J, Pierce K, Mostoslavsky R, et al. The Deacetylase Sirt6 Activates the Acetyltransferase GCN5 and Suppresses Hepatic Gluconeogenesis. Molecular Cell. 2012;48:900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomb DJ, Laurent G, Haigis MC. Sirtuins regulate key aspects of lipid metabolism. Biochim. Biophys. Acta. 2010;1804:1652–1657. doi: 10.1016/j.bbapap.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 42.Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 Deacetylates and Inhibits SREBP-1C Activity in Regulation of Hepatic Lipid Metabolism. Journal of Biological Chemistry. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker AK, Yang F, Jiang K, Ji J-Y, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes & Development. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proceedings of the National Academy of Sciences. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) Activity Leads to Liver Steatosis in the SIRT1+/− Mice: A Role of Lipid Mobilization and Inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metabolism. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerhart-Hines Z, Dominy JE, Blättler SM, Jedrychowski MP, Banks AS, Lim J-H, Chim H, Gygi SP, Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Molecular Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim JH, Gerhart-Hines Z, Dominy JE, Lee Y, Kim S, Tabata M, Xiang YK, Puigserver P. Oleic Acid Stimulates Complete Oxidation of Fatty Acids through Protein Kinase A-dependent Activation of SIRT1-PGC1 Complex. Journal of Biological Chemistry. 2013;288:7117–7126. doi: 10.1074/jbc.M112.415729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stančáková A, Goetzman E, Lam MM, Schwer B, et al. SIRT3 Deficiency and Mitochondrial Protein Hyperacetylation Accelerate the Development of the Metabolic Syndrome. Molecular Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, et al. Sirtuin 3 (SIRT3) Protein Regulates Long-chain Acyl-CoA Dehydrogenase by Deacetylating Conserved Lysines Near the Active Site. Journal of Biological Chemistry. 2013;288:33837–33847. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 Regulates Fatty Acid Oxidation and Mitochondrial Gene Expression in Liver and Muscle Cells. Journal of Biological Chemistry. 2010;285:31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laurent G, German NJ, Saha AK, de Boer VCJ, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, et al. SIRT4 Coordinates the Balance between Lipid Synthesis and Catabolism by Repressing Malonyl CoA Decarboxylase. Molecular Cell. 2013;50:686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim H-S, Xiao C, Wang R-H, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong W-I, Park O, Ki SH, et al. Hepatic-Specific Disruption of SIRT6 in Mice Results in Fatty Liver Formation Due to Enhanced Glycolysis and Triglyceride Synthesis. Cell Metabolism. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao H, Xiong X, DePinho RA, Deng CX, Dong XC. FoxO3 Transcription Factor and Sirt6 Deacetylase Regulate Low Density Lipoprotein (LDL)-cholesterol Homeostasis via Control of the Proprotein Convertase Subtilisin/Kexin Type 9 (Pcsk9) Gene Expression. Journal of Biological Chemistry. 2013;288:29252–29259. doi: 10.1074/jbc.M113.481473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tao H, Xiong X, DePinho RA, Deng CX, Dong XC. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. The Journal of Lipid Research. 2013;54:2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elhanati S, Kanfi Y, Varvak A, Roichman A, Carmel-Gross I, Barth S, Gibor G, Cohen HY. Multiple Regulatory Layers of SREBP1/2 by SIRT6. Cell reports. 2013;4:905–912. doi: 10.1016/j.celrep.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 59.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Feldman JL, Baeza J, Denu JM. Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins [Internet] J. Biol. Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. After years of discussion trying to understand why SIRT6 deacetylase activity in vitro was weak, Denu and colleagues demonstrate that free fatty acids (FFAs) directly activate SIRT6, pointing to a potential mechanism by which lipid metabolism could modulate SIRT6, which in turn may regulate metabolism to bring back homeostasis.
- 61.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, et al. SIRT7 Represses Myc Activity to Suppress ER Stress and Prevent Fatty Liver Disease. Cell reports. 2013;5:654–665. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2013;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoshino J, Mills KF, Yoon MJ, Imai S-I. Nicotinamide Mononucleotide, a Key NAD+ Intermediate, Treats the Pathophysiology of Diet- and Age- Induced Diabetes in Mice. Cell Metabolism. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. This study is the first to demonstrate that treatment in vivo with a small molecule intermediate in NAD metabolism, NMN, is sufficient to increase NAD levels and modulate metabolism at the organismal level, protecting against diabetes. As such, it represents one of the first proofs supporting the central role of NAD in organismal metabolism.
- 64.Cantó C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. The NAD+ Precursor Nicotinamide Riboside Enhances Oxidative Metabolism and Protects against High-Fat Diet-Induced Obesity. Cell Metabolism. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gomes AP, Price NL, Ling AJY, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD+ Induces a Pseudohypoxic State Disrupting Nuclear-Mitochondrial Communication during Aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. This study demonstrates for the first time, that NAD levels decrease with age, affecting expression of mitochondrial OXPHOS subunits in a SIRT1 dependent manner. Increasing NAD levels was sufficient to partially restore mitochondrial function in aged tissue.
- 66.Hoeijmakers J-HJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 67.Wang R-H, Sengupta K, Li C, Kim H-S, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Molecular Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park S-K, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 Redistribution on Chromatin Promotes Genomic Stability but Alters Gene Expression during Aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, Ge Q, Gu W, Orren D, Luo J. Regulation of WRN Protein Cellular Localization and Enzymatic Activities by SIRT1-mediated Deacetylation. Journal of Biological Chemistry. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 71.Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao P-C, Qiu Y, Zhao Y, et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci. 2013;16:1008–1015. doi: 10.1038/nn.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fan W, Luo J. SIRT1 Regulates UV-Induced DNA Repair through Deacetylating XPA. Molecular Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 73.Ming M, Shea CR, Guo X, Li X, Soltani K, Han W, He Y-Y. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proceedings of the National Academy of Sciences. 2010;107:22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for Human SIRT2 NAD-Dependent Deacetylase Activity in Control of Mitotic Exit in the Cell Cycle. Molecular and Cellular Biology. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, Park S-H, Pantazides BG, Karpiuk O, Warren MD, Hardy CW, Duong DM, Park S-J, Kim H-S, Vassilopoulos A, et al. SIRT2 directs the replication stress response through CDK9 deacetylation. Proceedings of the National Academy of Sciences. 2013;110:13546–13551. doi: 10.1073/pnas.1301463110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Serrano L, Martínez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes & Development. 2013;27:639–653. doi: 10.1101/gad.211342.112. This is one of the first proofs for a nuclear function for SIRT2, a sirtuin long thought to work mainly in the cytoplasm.
- 77.Tao R, Coleman MC, Pennington JD, Ozden O, Park S-H, Jiang H, Kim H-S, Flynn CR, Hill S, Hayes McDonald W, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metabolism. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 79.Chen Y, Zhang J, Lin Y, Lei Q, Guan K-L, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO reports. 2011;12:534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, et al. SIRT6 stabilizes DNA-dependent Protein Kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1:109. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 Promotes DNA Repair Under Stress by Activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. SIRT6 Recruits SNF2H to DNA Break Sites, Preventing Genomic Instability through Chromatin Remodeling. Molecular Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martinez-Pastor B, Cosentino C, Mostoslavsky R. A Tale of Metabolites: The Cross-Talk between Chromatin and Energy Metabolism. Cancer Discovery. 2013;3:497–501. doi: 10.1158/2159-8290.CD-13-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J, Kim J, Oberdoerffer P. Metabolic modulation of chromatin: implications for DNA repair and genomic integrity. Frontiers in genetics. 2013;4:1–11. doi: 10.3389/fgene.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]