Abstract
Notch signaling is essential for cell-fate specification in metazoans, and dysregulation of the pathway leads to a variety of human diseases including heart and vascular defects as well as cancer. Glycosylation of the Notch extracellular domain has emerged as an elegant means for regulating Notch activity, especially since the discovery that Fringe is a glycosyltransferase that modifies O-fucose in 2000. Since then, several other O-glycans on the extracellular domain have been demonstrated to modulate Notch activity. Here we will describe recent results on the molecular mechanisms by which Fringe modulates Notch activity, summarize recent work on how O-glucose, O-GlcNAc, and O-GalNAc glycans affect Notch, and discuss several human genetic disorders resulting from defects in Notch glycosylation.
Keywords: Notch signaling, O-Glycosylation, EGF repeat
1. Introduction
Notch signaling is an evolutionarily conserved signaling pathway that is required for proper development and homeostasis [1,2,3,4]. Mutations of Notch receptors and the other components of this signaling pathway lead to congenital disorders such as Alagille syndrome and Spondylocostal dysostosis, and adult onset diseases such as CADASIL [5,6]. Activating and inactivating mutations in Notch result in a variety of hematopoietic and solid tumors, indicating that Notch can function as a tumor suppressor or tumor promoter depending on context [7]. As a result, Notch signaling has emerged as molecular target of a variety of cancers [6,8,9,10]. Thus, a thorough understanding of this pathway and how it is regulated will contribute to advancement of basic biology as well as medical science.
The basic molecular structure of the Notch receptors and ligands is well-conserved from flies to humans (Figure 1) [2,11]. Both receptors and ligands are type-I transmembrane proteins expressed at the plasma membrane. Ligand binding to the Notch extracellular domain (NECD) of Notch triggers receptor activation by inducing a conformational change of the negative regulatory region (NRR) [12,13,14]. This conformational change exposes site 2 (S2) that is cleaved by ADAM10/17 metalloproteases, followed by an S3 cleavage catalyzed by the γ-secretase complex. These successive cleavages result in liberation of the Notch intracellular domain (NICD) that moves into the nucleus and regulates transcriptional activation of Notch target genes. Notch receptor activation is regulated at multiple steps to ensure its specificity and robustness.
Figure 1. Potential glycosylation on mouse Notch1.
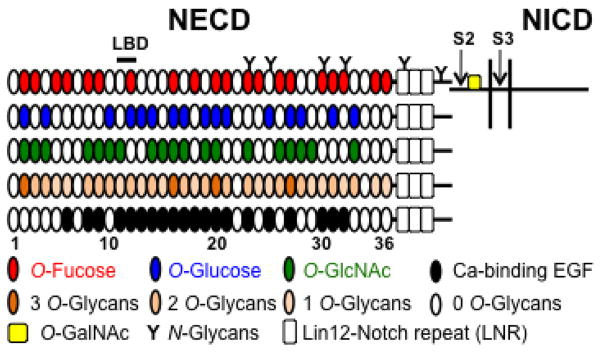
Mouse Notch1 is a heterodimer where the NECD is bound non-covalently to the transmembrane-intracellular domain (shown for the top diagram, not drawn to scale). The positions of the S2 and S3 protease cleavage sites are shown. The NICD is released upon cleavage at S3. The NECD of mouse Notch1 consists of 36 EGF repeats (ovals) and 3 Lin12-Notch repeats (LNR, open rectangles). EGF repeats 11–12 are the ligand binding domain (LBD). EGF repeats containing the consensus sequence for O-Fucose (red), O-Glucose (blue), or O-GlcNAc (green) are shown. EGF repeats with any of these O-glycans are shown in different colors according to whether they have one, two or three potential O-glycans modifications. Ca-binding EGF repeats are shown in black. O-GalNAc glycans may exist near the ADAM10/17-cleavage site 2 (S2) and thereby regulate a subsequent cleavage by γ-secretase complex at S3 within the transmembrane.
Since obtaining the sequence of the Notch receptor in 1985 [15], the presence of multiple tandem epidermal growth factor-like (EGF) repeats in the NECD has been a mystery. Recent evidence points to significance of the NECD for robustness and paralogue-specificity in Notch signaling [16]. The number of EGF repeats are different among Notch paralogues: Drosophila has a single Notch receptor with 36 EGF repeats, while humans have four Notch receptors, designated as NOTCH1-4. Both NOTCH1 and NOTCH2 have 36 EGF repeats while NOTCH3 and NOTCH4 have 34 and 29 EGF repeats, respectively [2]. Early work suggested that the 11th and 12th EGF repeats of Drosophila Notch are necessary and sufficient for ligand binding and Notch activation [17], and recent mutagenesis work has revealed specific amino acids in EGF12 involved in ligand binding [18]. Other regions of the NECD have also been implicated in ligand binding [19,20,21,22]. Endocytosis of the NECD-bound ligand into the signal sending cells generates a pulling force that relaxes the closed conformation of the NRR, which leads to the exposure of the S2 cleavage sites [12,23,24,25]. Thus, the NECD somehow links the binding of ligand and the pulling force generated from endocytosis of ligand to an alteration in the conformation of the NRR. How the NECD does this is unknown, but glycosylation of NECD has been implicated both in ligand binding and in linking ligand binding to proteolysis.
2. Types of glycosylation as post-translational modifications of Notch
The NECD is modified with different types of carbohydrates including asparagine-linked N-glycans and several serine- or threonine-linked O-glycans (Figure 1). O-Glycans are classified by types of monosaccharides that are directly attached to serine or threonine residues of proteins. O-Glycans observed on the EGF repeats of the NECD include O-fucose [26], O-glucose [26], O-GlcNAc (N-acetylglucosamine) [27], and O-xylose [28]. Recently, conventional mucin-type O-GalNAc glycans were described in the NECD, although not modifying the EGF repeats [29]. Modification of Notch proteins by glycosylation occurs during transit through the endoplasmic reticulum (ER) and Golgi apparatus where the glycosylation machinery (glycosyltransferases) work just like an assembly line, adding glycans in a progressive manner. In this section, we will review our current knowledge of carbohydrate structures and their roles for Notch activation based on existing biochemical and genetic evidence.
2-1. O-Glycan modifications of NECD EGF repeats
There are nine different glycosyltransferases that preferentially modify properly folded EGF repeats with O-glycans (Table 1). A single EGF repeat contains six cysteine residues that form three disulfide bonds in a distinct pattern, C1-C3, C2-C4, and C5-C6 where C1 indicates the first cysteine in a primary sequence [30]. EGF repeats in the NECD are linked by a short spacer (5-7 amino acids) between C6 of one EGF repeat and C1 of the next. The last four amino acids of this spacer region and a part of the EGF repeat can coordinate a calcium ion depending on their amino acid sequences [31]. Many EGF repeats in NECD are predicted to bind calcium (Figure 1), and these sites are highly conserved in Notch receptors across species [19]. Bound calcium ions are thought to enhance the rigidity of tandemly connected EGF repeats, and possibly the stability of individual EGF repeats [31].
Table 1.
Mammalian glycosyltransferases that preferentially modify EGF repeats.
| Gene | Acceptor Substrate | Donor Substrate | Subcellular Localization | Effect on Notch | |
|---|---|---|---|---|---|
| POFUT1/Ofut1 | EGF | GDP-Fuc | ER | Essential | |
| Lunatic fringe | Fuc-EGF | UDP-GlcNAc | Golgi | Notch activation | |
| Manic fringe | Fuc-EGF | UDP-GlcNAc | Golgi | Notch activation | |
| Radical fringe | Fuc-EGF | UDP-GlcNAc | Golgi | Notch activation | |
| POGLUT1/Rumi | EGF | UDP-Glc | UDP-Xyla | ER | Essential |
| GXYLT1 | Glc-EGF | UDP-Xyl | Unknown | Inhibitory | |
| GXYLT2 | Glc-EGF | UDP-Xyl | Unknown | Inhibitory | |
| XXYLT1 | Xyl-Glc-EGF | UDP-Xyl | ER | Inhibitory | |
| EOGT1 | EGF | UDP-GlcNAc | ER | ? | |
POGLUT1/Rumi can utilize UDP-Xyl when acceptor substrates have a diserine-motif (C-X-S-S-P/A-C) within the O-glucose consensus sequence.
2-1-1. O-Fucose
O-Fucose glycans were originally described as amino acid fucosides isolated from human urine in 1975 [32] and then were found on EGF repeats from urokinase plasminogen activator (PA), tissue-type PA, several blood coagulation factors, and Notch [26,33,34]. The fully extended structures of O-fucose glycans differ among species [26,35]. O-Fucose monosaccharides are elongated to a GlcNAcβ1-3Fuc disaccharide by the action of N-acetylglucosaminyltransferase Fringe in Drosophila. The disaccharide can be further elongated to the tetrasaccharide, Neu5Acα2-3/6Galβ1-4GlcNAcβ1-3Fuc, by the sequential action of several glycosyltransferases in mammals. O-Fucosylation occurs on distinct serine or threonine residues within the consensus sequence C2-X-X-X-X-(S/T)-C3 (where X is any amino acid). While database searches identify over 100 proteins containing EGF repeats with this consensus sequence [36], the Notch family of receptors has more O-fucose consensus sites than any other protein (Figure 1).
Fringe was originally named because mutants result in tissue loss of the edge, or “fringe” of fly wings, a Notch-like phenotype [37]. Subsequent work demonstrated that Fringe is a modulator of Notch activity [38]. Three Fringe homologues exist in mammals: Lunatic fringe, Manic fringe, and Radical fringe [39,40]. Demonstrating that Fringe modulates Notch activation through elongating O-fucose with a β3-linked GlcNAc provided a clear example of how altering glycan structures on a receptor could regulate a signaling pathway [41,42]. However, we and the others are still working to understand how changes in O-fucose structure regulate Notch signaling. We recently reviewed our knowledge about the significance of O-fucosylation and Fringe for Notch signaling [43,44]. In this article, we summarize these points briefly and emphasize more recent data.
Protein O-fucosyltransferase-1 (designated as Pofut1 in mammals and Ofut1 in Drosophila) solely catalyzes the addition of O-fucose to EGF repeats. The gene encoding mammalian Pofut1 was identified by conventional protein purification and molecular cloning [45]. Unlike the other known fucosyltransferases, Pofut1 is a soluble protein with a C-terminal KDEL-like ER retention signal, suggesting that O-Fucosylation of EGF repeats by Pofut1 occurs in the ER [46]. Pofut1 transfers fucose from GDP-fucose to properly folded EGF repeats containing the O-fucose consensus sequence [47,48]. Gene-targeted elimination of Pofut1 in mice result in embryonic lethality with Notch-like phenotypes that are similar to those in mice lacking core components of Notch signaling like RBP-Jk [49]. Since the Pofut1 null phenotype is more severe than that of mice lacking individual Notch paralogues, O-fucosylation by Pofut1 is thought to be involved in proper function of all Notch paralogues. RNAi-mediated knockdown of Ofut1 showed that Ofut1 is cell-autonomously required for proper Notch function in Drosophila [50]. Identification of a mutation in Ofut1, neurotic, supported this notion [51]. These data indicate the essential nature of Pofut1 for function of Notch receptors.
Fringe-mediated elongation of O-fucose glycans regulates Notch receptor activation in several distinct contexts [43,44]. In flies, Fringe regulates Notch activation cell-autonomously by making Notch more sensitive to Delta and less sensitive to Serrate, which was explained by showing that addition of GlcNAc to O-fucose by Fringe enhances Delta binding to Notch and decreases Serrate binding to Notch using Drosophila components [35]. The mammalian system is significantly more complex, with three different Fringes, four different Notch receptors and five different ligands. The mechanism by which Fringe modulates Notch in mammals is not fully understood even after significant effort [52,53,54,55]. Moreover, it is not known how individual O-fucose modification sites contribute to Notch ligand binding in any system. Early work suggested that EGF repeats 11–12 are necessary and sufficient for ligand binding [17]. Consistent with this view, bacterially expressed (and therefore unglycosylated) EGF repeats 11–13 from human Notch1 bind to Notch ligands, albeit weakly [18,20,31]. Very recently, in collaboration with the Handford and Lea labs, we found that Fringe-mediated extension of O-fucose on EGF12 of EGF11–13 from human Notch1 enhances binding to ligands from both families, Delta-like1 (Delta homologue) and Jagged1 (Serrate homologue) [56]. These results show that Fringe modification of O-fucose on EGF12 enhances Delta-like1 binding, providing at least a partial explanation for the enhancement of Delta-Notch signaling caused by Fringe. Nonetheless, these results do not explain the Fringe-mediated inhibition of Jagged-induced Notch signaling [43,44]. Thus, inhibition of Jagged-induced signaling must involve Fringe modification of other O-fucose sites. Preliminary data have suggested that O-fucose on EGF repeats outside of the ligand binding domain (e.g. EGF26 and EGF27) are also modified with Fringe and are important for Notch1 activation [57]. Several reports have suggested physical interaction between the ligand binding domain and other regions of the NECDs [58,59]. These findings may help to explain how Fringe modifications at other sites inhibit Jagged-induced signaling.
To examine whether the addition of GlcNAc to O-fucose on EGF12 induces a conformational change in the protein backbone, EGF11–13 was crystallized in the absence of O-fucose, with O-fucose monosaccharide, and with O-fucose disaccharide. Comparison of the structures revealed that addition of GlcNAc by Fringe does not alter the backbone structure of EGF repeats 11–13, suggesting that the increase in ligand binding (to both Delta-like1 and Jagged1) can be explained by direct interactions with the sugars (Figure 2) [56]. The crystal structure also revealed that the GlcNAcβ1-3Fuc disaccharide is highly ordered resulting from significant interactions with the underlying amino acids, consistent with earlier studies on O-fucose glycans on EGF12 alone [60].
Figure 2. Structure of O-fucosylated variants of human Notch1 EGF repeats 11–13.
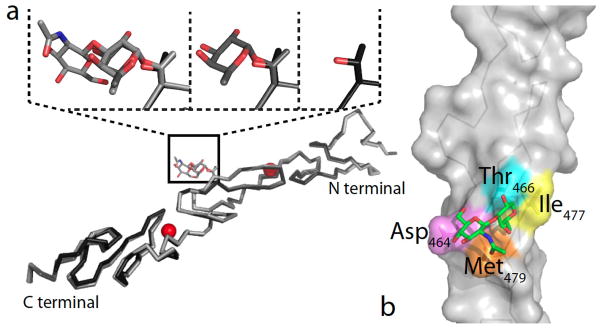
(a) Superimposed X-ray structures of the unmodified human Notch1 EGF11–13 (PDB ID code 2VJ3) and the monosaccharide and disaccharide structures with the subsequent additions to the O-fucose site (Thr466) region highlighted. Calcium ions are shown in red. (b) Details of human Notch1 EGF12 X-ray structure highlighting contacts between the C6 methyl group of the O-fucose with Ile477 (yellow) and Met479 (orange), the C6 methoxy group of GlcNAc with Asp464 (violet), and the N-acetyl group of GlcNAc with Met479 (orange). Thr466 is highlighted in cyan. Reproduced with permission from [56].
2-1-2. O-Glucose
O-Glucose glycans on EGF repeats were originally found on several coagulation factors and defined structurally as O-glucose elongated to a linear trisaccharide by the addition of two α1-3 linked xylose residues [61,62,63]. Subsequently, mouse Notch1 and Notch2 isolated from mammalian cell lines were found to be modified with O-glucose glycans [26,28,64,65,66]. Based on the amino acid sequences surrounding the serine residue modified with O-glucose glycans, a consensus sequence for O-glucosylation was proposed: C1-X-S-X-P-C2. We recently revised this to C1-X-S-X-(P/A)-C2 based on our biochemical and mass spectral analyses of O-glucose glycan modification sites on mouse Notch1 and Drosophila Notch [67]. Database searches for this consensus sequence identify over 40 proteins predicted to be O-glucosylated, although like O-fucose, the Notch family of receptors has more consensus sites than any other protein (Figure 1) [44].
O-Glucose glycans are also essential for Notch activity in both mice and flies [66,68]. The enzymatic activity responsible for addition of O-glucose to EGF repeats was initially characterized in crude cell lysates and shown to be a soluble enzyme [69]. Shortly thereafter, rumi was identified as a temperature-sensitive mutant in a forward genetic screen designed to identify novel players in the Notch signaling pathway in Drosophila [68,70]. The predicted protein product of rumi has a signal peptide, a CAP10 domain, and a KDEL ER-retention signal, indicating that Rumi would be a soluble, ER-localized protein. CAP genes are thought to be glycosyltransferases involved in the formation of capsule of Cryptococcus neoformans [71]. RNAi on Rumi in S2 cells showed Rumi is required for proper O-glucosylation on Notch. More directly, in vitro enzymatic assays showed that Rumi is a protein O-glucosyltransferase (Poglut) acting on EGF repeats with the O-glucose consensus sequence. To determine whether catalytic activity of Rumi is critical for Notch function, one allele that showed full-blown Notch phenotypes resulting from a single point mutation (G189E), rumi79, was further analyzed [68]. Rumi-G189E was expressed at normal levels in rumi79, and it was expressed in Drosophila S2 cells similarly to wild type Rumi, which indicates the G189E mutation does not alter protein expression or stability [68]. In vitro enzyme assays with Rumi-G189E showed no detectable activity, indicating that O-glucosylation is essential for Notch activation [68].
Three Rumi homologues exist in mammals. Only the one with highest homology to Drosophila Rumi (56%, termed POGLUT1) shows in vitro enzymatic activity and rescues Notch-phenotypes in Rumi mutant flies [28]. The functional significance of the other Rumi homologues (termed KDELC1 and KDELC2) is not known. Elimination of Poglut1 in mice results in embryonic lethality with many Notch-like phenotypes (e.g. somitogenesis, cardiogenesis, and vascular remodeling) but others that are distinct from Notch (e.g. defects in neural tube development) [66]. These results clearly indicate that addition of O-glucose to Notch is fundamentally required for proper Notch activation, but that O-glucosylation of other targets is also essential for mammalian development.
Elongation of O-glucose glycans by two α3-xylose residues is catalyzed by specific xylosyltransferases. Mammals have three xylosyltransferases: UDP-xylose: glucoside α1-3 xylosyltransferases 1 and 2 (GXYLT1 and GXYLT2) and UDP-xylose: xyloside α1-3 xylosyltransferase 1 (XXYLT1) [72,73]. They are all type II transmembrane proteins with the putative catalytic domain in the luminal domain. In mammalian cells, overexpressed XXYLT1 was localized in the ER although it does not bear an obvious ER-localization signal in its primary sequence.
To begin to understand how O-glucose glycans regulate Notch activation at the molecular level, we have analyzed whether predicted O-glucose consensus sequences are modified with O-glucose glycans. Multiple EGF repeats in the NECD contain O-glucose consensus sequences, and the sites are conserved relatively well in the middle region of the NECD (EGF 10–20) (Figure 1) [43]. Using mass spectral-based glycoproteomic methods, we mapped O-glucose glycans to each predicted site on mouse Notch1 [67]. Essentially all predicted O-glucose sites are modified at high stoichiometry, and the major structure at each site is the trisaccharide. Only EGF27 was not fully modified. In vitro Poglut assays revealed the EGF27 is a poorer substrate for Rumi/POGLUT1 than other EGF repeats that are efficiently modified [48]. An arginine adjacent to the modified serine in the EGF27 consensus sequence (C1DSRPC2) is at least partially responsible for making EGF27 a poor substrate, suggesting that efficiency of modification at a particular site is affected by the primary sequence of an individual EGF repeat [48]. Site mapping on Drosophila Notch has shown that most predicted sites are also modified at high stoichiometry, but interestingly elongation of O-glucose to di- or tri-saccharide is limited to EGF14–20 [74]. These results suggest that the xylosyltransferases also show preference for some EGF repeats over others.
Although we do not yet know how O-glucose glycans affect Notch activity at the molecular level, several clues have emerged during analysis of mutants. An initial clue came from the observation that Rumi mutant flies showed a temperature-sensitive Notch phenotype [68]. Since the temperature sensitivity was not due to instability of Rumi (Rumi protein null alleles also showed temperature sensitivity), we hypothesized that lack of O-glucose somehow destabilized Notch. Notch accumulates intracellularly and at the surface of cells lacking Rumi, but fails to signal [68]. Further analyses showed that O-glucose glycans are required for S2 cleavage of Notch during activation, however Delta ligand binding was not affected [68]. These results indicated that O-glucose glycans are required for proper folding and/or trafficking of Notch, thereby regulating its activation. The molecular basis for the temperature-sensitivity of the Rumi mutant was further investigated by the Jafar-Nejad group by mutating O-glucose sites [75]. No single mutation from serine to alanine causes a significant decrease in Notch activation. Rather, all of the potential sites contribute to robust Notch activation at higher temperatures, especially the EGF repeats in and around the ligand-binding domain. They also showed Rumi is required for ligand binding-independent Notch activation caused by deletion of LNR repeats. These results strongly suggested that O-glucosylation at multiple EGF repeats of Notch allow proper S2 cleavage at higher temperatures in Drosophila. We have investigated the contribution of individual O-glucose sites in mouse Notch1 to the regulation of Notch signaling by using co-culture Notch reporter assays. As in flies, elimination of single sites had little effect on Notch1 activity, except for a mutation of the O-glucose site of EGF28, which caused a statistically significant decrease in Delta1-mediated Notch1 activation in NIH3T3 cells [67]. EGF28 is not a highly conserved site, so it may play a unique role in regulation of mouse Notch1. Additional work needs to be done to resolve context-dependent differences.
The Jafar-Nejad group recently published a very interesting report suggesting that xylosyl extension of O-glucose glycans on Notch inhibits Notch activation in Drosophila [74]. RNAi knock down of the Drosophila GXYLT homologue, Shams, increases Notch activation while overexpression of human GXYLT1 decreases Notch activation in Drosophila. The overexpression of GXYLT1 reduced cell surface expression of Notch, providing a potential molecular explanation for this effect. Thus, elongation of O-glucose with xylose residues appeared to inhibit proper trafficking of Notch. This is the first data suggesting that elongation of O-glucose with xylose could modulate Notch activity much like Fringe-mediated elongation of O-fucose. Much work remains to be done to better understand the regulation of Notch activation by O-glucose glycans.
2-1-3. O-GlcNAc
Up until recently, the O-GlcNAc modification was thought to be found primarily on nuclear and cytoplasmic proteins [76], but O-GlcNAc modifications have also been found to occur between the 5th and 6th cysteine residues of EGF repeats of Drosophila Notch [27] and Dumpy [77]. Subsequently, O-GlcNAc was found in mammalian Notch [78]. Notch ligands such as Delta and Serrate are also modified with O-GlcNAc [79]. Unlike O-Fucose or O-Glucose, the stoichiometry of modification does not appear to be high based on the sites mapped to date [27]. The consensus sequence of attachment of O-GlcNAc has been proposed as C5-X-X-G-X-(S/T)-G-X-X-C6 based on experimental mapping by mass spectral methods [80]. The enzyme responsible for O-GlcNAc modification on EGF repeats has been identified and is designated as EGF-specific O-GlcNAc-transferase (Eogt) in Drosophila [77] and Eogt1 in mammals [78]. Their enzymatic activities detected in vitro are very similar. Mammalian Eogt1 can rescue blistering phenotype of the fly Eogt mutant, which strongly suggests that both fly Eogt and mammalian Eogt1 are functionally equivalent.
A role for the O-GlcNAc modification in Notch signaling is not yet clear. While loss of Eogt in flies does not cause obvious defects in Notch signaling [77], genetic interactions of Eogt with Notch pathway components have been reported [79]. Elimination of Eogt in flies results in a severe wing blistering defect which is similar to phenotypes caused by loss of Dumpy, suggesting O-GlcNAc is essential for Dumpy function [77]. There is no homologous protein to Dumpy in mammals. While elimination of mouse Eogt1 has not yet been reported, recent studies have shown that mutations in human EOGT1 cause a rare autosomal recessive disorder called Adams-Oliver syndrome, clearly demonstrating the significance of EOGT1 in human disease [81,82]. Interestingly, some forms of Adams-Oliver syndrome are caused by defects in the Notch pathway [83], raising the possibility that O-GlcNAc modifications on Notch may affect its function.
2-2. O-Glycans outside the NECD EGF repeats
In addition to the variety of O-glycans that occur on the EGF repeats of Notch described above, a mucin-type O-GalNAc glycan outside the EGF repeats was very recently reported as a novel regulator for Notch activation [29]. Mucin-type O-GalNAc glycans are widely distributed on mucins and many secreted or transmembrane glycoproteins. Addition of O-GalNAc to core proteins is initiated by a family of polypeptide N-acetygalactosaminyltransferases (GALNT) which consists of 20 isoforms in human [84]. There is no consensus sequence for O-GalNAc attachment, and the specificity of GALNT isoforms appear to be redundant in many cases. For these reasons, it is challenging to identify a biologically relevant target of a specific GALNT isoform.
The Khokha group identified a copy number deletion of GALNT11 as a candidate gene responsible for heterotaxy [85]. Heterotaxy is a congenital disorder of left-right patterning during development. Using biological assays in Xenopus, they successfully found that GALNT11 alters left-right patterning and Notch signaling. They identified a synthetic peptide (NIPYKIEAVQSETVEPPPPA) corresponding to a S2 cleavage region from human NOTCH1 that can be modified by GALNT11. Moreover, they showed that attachment of GalNAc at this distinct threonine facilitates the S2 cleavage by ADAM17. Further work will need to be done to clarify the underlying mechanism of O-GalNAc glycan-mediated Notch activation. In addition, Libisch and co-workers recently identified GALNT11 as a new molecular marker for chronic lymphocytic leukemia (CLL) [86]. Interestingly, Notch signaling is known to be oncogenic in CLL [7]. O-GalNAc glycans may also be important in this context.
2-3. N-Glycans on EGF repeats and other regions of NECD
There are multiple consensus sequences for N-glycan attachment (N-X-S/T where X is any amino acid except proline) in the Notch ECD (Figure 1), several of which are known to be modified [26]. Studies in mutant CHO cell mutants show that alterations in N-glycan structure (i.e. complex versus high-mannose structures) have little or no effect on Notch signaling [41,87]. Thus, little is known about the biological role of these modifications.
3. Human diseases caused by mutations or aberrant expression affecting Notch glycosylation
Mutations of Notch and the components of Notch signaling pathway are found in human developmental disorders such as Alagille syndrome and CADASIL and in different types of cancer [6,36]. Increasingly, defects in Notch-related glycosylation have been reported to cause human disease, confirming the importance of these modifications in human biology. The defect in GALNT11 resulting in heterotaxy described above is one such example [85]. Several others are highlighted below.
Dowling-Degos disease (DDD) is a rare autosomal-dominant form of a reticulate pigmentary disorder causing a postpubertal reticulate hyperpigmentation that is progressive and disfiguring, including small hyperkeratotic dark-brown papules [88]. No effective therapy is available. Loss-of-function mutations in KRT5 (Keratin 5) had been identified in two families of DDD patients; however, there are affected individuals who do not possess any mutations in KRT5. Genome-wide linkage analysis and exome sequencing of patients affected by generalized DDD identified a nonsense mutation (p.Glu144*) in POFUT1 and a heterozygous deletion mutation (p.Lyc161Sefs*42) in POFUT1 [89]. Knockdown of POFUT1 causes reduction in the expression of NOTCH1, NOTCH2, a Notch downstream target, HES1, and KRT5 in human keratinocyte HaCaT cells [89]. Similarly, Basmanav et al. found nine different heterozygous POGLUT1 mutations in 13 DDD-affected individuals, including a nonsense mutation located at the very beginning of the POGLUT1 protein (p.Trp4*) [88] and a point mutant (p.R279W) with no detectable enzymatic activity (HT and RSH, Unpublished). Thus, haploinsufficiency of either POFUT1 or POGLUT1 is a plausible explanation for DDD. Of note, similar skin defects have not been reported in the mice heterozygous for Pofut1 or Poglut1 [49,66]. Intriguingly, several lines of evidence have pointed out significance of Notch signaling in the context of skin homeostasis and diseases [90,91,92,93]. Future work will elucidate why partial loss of POFUT1 or POGLUT1 function results in DDD in this context.
The role of Notch signaling in breast cancers has been well studied. Early work in mice revealed that insertion of the mouse mammary tumor virus into either the Notch1 or Notch4 loci leads to constitutive Notch activation and formation of mammary tumors [94,95]. Notch1 and Notch4 are known to be upregulated in breast cancers and correlate with malignancy and poor clinical prognosis [96,97,98,99]. The role of Notch2 in breast cancer remains controversial where it may have a tumor-suppressive role [98,100]. A recent study demonstrated that deficiency in Lunatic Fringe leads to basal-like breast cancer [101]. This was important since there had not been clear indication that upregulation of Notch pathway components associated with this particular breast cancer subtype. Similarly, a tumor-suppressive role of Lunatic Fringe has also been suggested in prostate cancers [102].
In addition, an increasing number of studies have reported aberrant expression of Notch-modifying glycosyltransferases in different types of cancers. Upregulation of POFUT1 has been detected in brain tumors, hepatocellular carcinoma, colorectal cancers, and oral squamous cell carcinoma [103, 104, 105, 106]. POGLUT1 is overexpressed in primary acute myelogenous leukemia and T cell-acute lymphoblastic leukemia [107]. RNAi-mediated knockdown of POGLUT1 results in reduction in Notch activation in human myeloid leukemia U937 cells [108]. The precise roles of POFUT1 and POGLUT1 in cancers are still unclear. Both O-fucosylation and O-glucosylation are present at high stoichiometry on mouse Notch1 produced in HEK293T cells or Drosophila Notch produced in S2 cells [67,74]. If stoichiometry of O-fucosylation and O-glucosylation on Notch is low in the context of these cancers, overexpression of POFUT1 or POGLUT1 might affect Notch function through changing the levels of modifications. Alternatively, overexpression of these enzymes might increase the biosynthetic capacity of the cells and allow more Notch to be expressed on the cell surface.
4. Conclusions and perspectives
Since the discovery of O-fucose and O-glucose glycans on Notch in 2000 [26], we have learned a great deal about the significance of Notch glycosylation. The enzymes required for the addition of sugars to Notch have been identified, and mutations in several of these enzymes cause Notch phenotypes in model organisms (Table 1). More recently human diseases resulting from mutations in some of these enzymes have been described. Significant advances in the molecular details describing how these glycans affect Notch function have been made, but more work remains to be done. Site mapping has revealed that most O-fucose and O-glucose sites are modified at high stoichiometry, but that elongation past the monosaccharide is site specific. The first crystal structure of the Notch ligand-binding domain modified with a GlcNAcβ1-3Fuc disaccharide has been solved (Figure 2), and binding studies suggest that Fringe-mediated elongation of O-fucose on EGF12 enhances binding to both Delta and Jagged ligands [56]. These results nicely explain how Fringe enhances Delta-initiated Notch signaling but suggest that Fringe modification of O-fucose on other EGF repeats are responsible for Fringe inhibition of Jagged-initiated Notch signaling. Studies to identify which EGF repeats are involved in inhibition of Jagged-initiated signaling are in progress.
While our understanding of how glycosylation affects Notch function continues to grow, our understanding of how these glycans affect other proteins is in its infancy. O-Fucosyation has already been shown to be required for proper function of agrin [109], but its importance on the nearly 100 other Pofut1 predicted targets [36] remains unstudied. Similarly, the effects of O-glucose on the other 40 predicted POGLUT1 targets remain unknown. The fact that Poglut1 null mice show phenotypes that are not associated with Notch suggests that other relevant targets exist [66]. Detailed analysis of mutants in model organisms as well as phenotypes observed in patients with mutations in these enzymes should shed more light on which of these other targets are affected by glycosylation.
Highlights.
Notch signaling is regulated by glycosylation of its extracellular domain
Multiple O-linked carbohydrate modifications are found on the Epidermal Growth Factor-like repeats of Notch
Defects in glycosylation of Notch leads to a variety of human disorders
Acknowledgments
We are delighted to contribute a review to this special issue honoring Dr. William Lennarz who has frequently encouraged us in our studies on the glycosylation of Notch. We apologize that we cannot include all the relevant studies on Notch signaling in this article due to limitation of space. Primary work was supported by NIH grant GM061126 (to RSH). We thank members of the Haltiwanger lab for critical comments on this article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Developmental cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. Journal of cell science. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Seminars in cell & developmental biology. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling - are we there yet? Nature reviews Drug discovery. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 7.Ntziachristos P, Lim JS, Sage J, Aifantis I. From Fly Wings to Targeted Cancer Therapies: A Centennial for Notch Signaling. Cancer cell. 2014;25:318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groth C, Fortini ME. Therapeutic approaches to modulating Notch signaling: current challenges and future prospects. Seminars in cell & developmental biology. 2012;23:465–472. doi: 10.1016/j.semcdb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacology & therapeutics. 2013;139:95–110. doi: 10.1016/j.pharmthera.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinoza I, Pochampally R, Xing F, Watabe K, Miele L. Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. OncoTargets and therapy. 2013;6:1249–1259. doi: 10.2147/OTT.S36162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 13.Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling--a structural and biochemical perspective. Journal of cell science. 2008;121:3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 16.Kopan R, Chen S, Liu Z. Alagille, Notch, and robustness: why duplicating systems does not ensure redundancy. Pediatric nephrology. 2014;29:651–657. doi: 10.1007/s00467-013-2661-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF Repeats of Notch Mediate Interactions with Delta and Serrate: Implications for Notch as a Multifunctional Receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 18.Whiteman P, de Madrid BH, Taylor P, Li D, Heslop R, Viticheep N, Tan JZ, Shimizu H, Callaghan J, Masiero M, Li JL, Banham AH, Harris AL, Lea SM, Redfield C, Baron M, Handford PA. Molecular basis for Jagged-1/Serrate ligand recognition by the Notch receptor. The Journal of biological chemistry. 2013;288:7305–7312. doi: 10.1074/jbc.M112.428854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu A, Lei L, Irvine KD. Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J Biol Chem. 2005;280:30158–30165. doi: 10.1074/jbc.M505569200. [DOI] [PubMed] [Google Scholar]
- 20.Cordle J, Redfieldz C, Stacey M, van der Merwe PA, Willis AC, Champion BR, Hambleton S, Handford PA. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. The Journal of biological chemistry. 2008;283:11785–11793. doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto S, Charng WL, Rana NA, Kakuda S, Jaiswal M, Bayat V, Xiong B, Zhang K, Sandoval H, David G, Wang H, Haltiwanger RS, Bellen HJ. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science. 2012;338:1229–1232. doi: 10.1126/science.1228745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrawes MB, Xu X, Liu H, Ficarro SB, Marto JA, Aster JC, Blacklow SC. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. The Journal of biological chemistry. 2013;288:25477–25489. doi: 10.1074/jbc.M113.454850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meloty-Kapella L, Shergill B, Kuon J, Botvinick E, Weinmaster G. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Developmental cell. 2012;22:1299–1312. doi: 10.1016/j.devcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2757–2765. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ploscariu N, Kuczera K, Malek KE, Wawrzyniuk M, Dey A, Szoszkiewicz R. Single Molecule Studies of Force-Induced S2 Site Exposure in the Mammalian Notch Negative Regulatory Domain. The journal of physical chemistry B. 2014 doi: 10.1021/jp5004825. [DOI] [PubMed] [Google Scholar]
- 26.Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. The Journal of biological chemistry. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. The Journal of biological chemistry. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi H, Fernandez-Valdivia RC, Caswell DS, Nita-Lazar A, Rana NA, Garner TP, Weldeghiorghis TK, Macnaughtan MA, Jafar-Nejad H, Haltiwanger RS. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16600–16605. doi: 10.1073/pnas.1109696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013;504:456–459. doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang JY, Li L, Lai PH. A major kinetic trap for the oxidative folding of human epidermal growth factor. The Journal of biological chemistry. 2001;276:4845–4852. doi: 10.1074/jbc.M005160200. [DOI] [PubMed] [Google Scholar]
- 31.Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, Handford PA, Downing AK. Structural and functional properties of the human notch-1 ligand binding region. Structure (Camb) 2004;12:2173–2183. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Hallgren P, Lundblad A, Svensson S. A new type of carbohydrate-protein linkage in a glycopeptide from normal human urine. J Biol Chem. 1975;250:5312–5314. [PubMed] [Google Scholar]
- 33.Kentzer EJ, Buko AM, Menon G, Sarin VK. Carbohydrate Composition and Presence of a Fucose-Protein Linkage in Recombinant Human Pro-Urokinase. Biochem Biophys Res Commun. 1990;171:401–406. doi: 10.1016/0006-291x(90)91407-j. [DOI] [PubMed] [Google Scholar]
- 34.Harris RJ, Spellman MW. O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology. 1993;3:219–224. doi: 10.1093/glycob/3.3.219. [DOI] [PubMed] [Google Scholar]
- 35.Xu A, Haines N, Dlugosz M, Rana NA, Takeuchi H, Haltiwanger RS, Irvine KD. In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. The Journal of biological chemistry. 2007;282:35153–35162. doi: 10.1074/jbc.M707040200. [DOI] [PubMed] [Google Scholar]
- 36.Rampal R, Luther KB, Haltiwanger RS. Notch signaling in normal and disease States: possible therapies related to glycosylation. Current molecular medicine. 2007;7:427–445. doi: 10.2174/156652407780831593. [DOI] [PubMed] [Google Scholar]
- 37.Irvine KD, Wieschaus E. Fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 38.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates notch ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 39.Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, Whiting E, Ryan D, Zinyk D, Boulianne G, Hui CC, Gallie B, Phillips RA, Lipshitz HD, Egan SE. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nature Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- 40.Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 41.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 42.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi H, Haltiwanger RS. Role of glycosylation of Notch in development. Seminars in cell & developmental biology. 2010;21:638–645. doi: 10.1016/j.semcdb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rana NA, Haltiwanger RS. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Current opinion in structural biology. 2011;21:583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, Haltiwanger RS. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. The Journal of biological chemistry. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 46.Luo Y, Haltiwanger RS. O-fucosylation of Notch occurs in the endoplasmic reticulum. J Biol Chem. 2005;280:11289–11294. doi: 10.1074/jbc.M414574200. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Spellman MW. Purification and characterization of a GDP-fucose:polypeptide fucosyltransferase from Chinese hamster ovary cells. The Journal of biological chemistry. 1998;273:8112–8118. doi: 10.1074/jbc.273.14.8112. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi H, Kantharia J, Sethi MK, Bakker H, Haltiwanger RS. Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats. The Journal of biological chemistry. 2012;287:33934–33944. doi: 10.1074/jbc.M112.401315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi S, Stanley P. Protein O-fucosyltransferase I is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okajima T, Irvine KD. Regulation of notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 51.Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M, Kitagawa M, Harigaya K, Spana E, Bilder D, Perrimon N, Matsuno K. Neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development. 2003;130:4785–4795. doi: 10.1242/dev.00679. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu K, Chiba S, Saito T, Kumano K, Takahashi T, Hirai H. Manic fringe and lunatic fringe modify different sites of the notch2 extracellular region, resulting in different signaling modulation. J Biol Chem. 2001;276:25753–25758. doi: 10.1074/jbc.M103473200. [DOI] [PubMed] [Google Scholar]
- 53.Shao L, Moloney DJ, Haltiwanger R. Fringe modifies O-fucose on mouse Notch1 at epidermal growth factor-like repeats within the ligand-binding site and the Abruptex region. The Journal of biological chemistry. 2003;278:7775–7782. doi: 10.1074/jbc.M212221200. [DOI] [PubMed] [Google Scholar]
- 54.Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. The Journal of biological chemistry. 2005;280:42454–42463. doi: 10.1074/jbc.M509552200. [DOI] [PubMed] [Google Scholar]
- 55.Yang LT, Nichols JT, Yao C, Manilay JO, Robey EA, Weinmaster G. Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol Biol Cell. 2005;16:927–942. doi: 10.1091/mbc.E04-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor P, Takeuchi H, Sheppard D, Chillakuri C, Lea SM, Haltiwanger RS, Handford PA. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1319683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rampal R, Arboleda-Velasquez JF, Nita-Lazar A, Kosik KS, Haltiwanger RS. Highly conserved O-fucose sites have distinct effects on Notch1 function. J Biol Chem. 2005;280:32133–32140. doi: 10.1074/jbc.M506104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pei Z, Baker NE. Competition between Delta and the Abruptex domain of Notch. BMC Dev Biol. 2008;8:4. doi: 10.1186/1471-213X-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma A, Rangarajan A, Dighe RR. Antibodies against the extracellular domain of human Notch1 receptor reveal the critical role of epidermal-growth-factor-like repeats 25–26in ligand binding and receptor activation. The Biochemical journal. 2013;449:519–530. doi: 10.1042/BJ20121153. [DOI] [PubMed] [Google Scholar]
- 60.Hiruma-Shimizu K, Hosoguchi K, Liu Y, Fujitani N, Ohta T, Hinou H, Matsushita T, Shimizu H, Feizi T, Nishimura S. Chemical synthesis, folding, and structural insights into O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor. Journal of the American Chemical Society. 2010;132:14857–14865. doi: 10.1021/ja105216u. [DOI] [PubMed] [Google Scholar]
- 61.Hase S, Kawabata S, Nishimura H, Takeya H, Sueyoshi T, Miyata T, Iwanaga S, Takao T, Shimonishi Y, Ikenaka T. A New Trisaccharide Sugar Chain Linked to a Serine Residue in Bovine Blood Coagulation Factors VII and IX. J Biochem (Tokyo) 1988;104:867–868. doi: 10.1093/oxfordjournals.jbchem.a122571. [DOI] [PubMed] [Google Scholar]
- 62.Nishimura H, Kawabata S, Kisiel W, Hase S, Ikenaka T, Takao T, Shimonishi Y, Iwanaga S. Identification of a disaccharide (Xyl-Glc) and a trisaccharide (Xyl2-Glc) O-glycosidically linked to a serine residue in the first epidermal growth factor-like domain of human factors VII and IX and protein Z and bovine protein Z. J Biol Chem. 1989;264:20320–20325. [PubMed] [Google Scholar]
- 63.Hase S, Nishimura H, Kawabata S, Iwanaga S, Ikenaka T. The structure of (xylose)2glucose-O-serine 53 found in the first epidermal growth factor-like domain of bovine blood clotting factor IX. The Journal of biological chemistry. 1990;265:1858–1861. [PubMed] [Google Scholar]
- 64.Bakker H, Oka T, Ashikov A, Yadav A, Berger M, Rana NA, Bai X, Jigami Y, Haltiwanger RS, Esko JD, Gerardy-Schahn R. Functional UDP-xylose transport across the endoplasmic reticulum/Golgi membrane in a Chinese hamster ovary cell mutant defective in UDP-xylose Synthase. The Journal of biological chemistry. 2009;284:2576–2583. doi: 10.1074/jbc.M804394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitworth GE, Zandberg WF, Clark T, Vocadlo DJ. Mammalian Notch is modified by D-Xyl-alpha1-3-D-Xyl-alpha1-3-D-Glc-beta1-O-Ser: implementation of a method to study O-glucosylation. Glycobiology. 2010;20:287–299. doi: 10.1093/glycob/cwp173. [DOI] [PubMed] [Google Scholar]
- 66.Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS, Jafar-Nejad H. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138:1925–1934. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rana NA, Nita-Lazar A, Takeuchi H, Kakuda S, Luther KB, Haltiwanger RS. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse notch1. The Journal of biological chemistry. 2011;286:31623–31637. doi: 10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shao L, Luo Y, Moloney DJ, Haltiwanger R. O-Glycosylation of EGF repeats: identification and initial characterization of a UDP-glucose: protein O-glucosyltransferase. Glycobiology. 2002;12:763–770. doi: 10.1093/glycob/cwf085. [DOI] [PubMed] [Google Scholar]
- 70.Lee TV, Takeuchi H, Jafar-Nejad H. Regulation of notch signaling via O-glucosylation insights from Drosophila studies. Methods in enzymology. 2010;480:375–398. doi: 10.1016/S0076-6879(10)80017-5. [DOI] [PubMed] [Google Scholar]
- 71.Chang YC, Kwon-Chung KJ. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. Journal of bacteriology. 1999;181:5636–5643. doi: 10.1128/jb.181.18.5636-5643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. The Journal of biological chemistry. 2010;285:1582–1586. doi: 10.1074/jbc.C109.065409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sethi MK, Buettner FF, Ashikov A, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. Molecular cloning of a xylosyltransferase that transfers the second xylose to o-glucosylated epidermal growth factor repeats of notch. The Journal of biological chemistry. 2012;287:2739–2748. doi: 10.1074/jbc.M111.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee TV, Sethi MK, Leonardi J, Rana NA, Buettner FF, Haltiwanger RS, Bakker H, Jafar-Nejad H. Negative regulation of notch signaling by xylose. PLoS genetics. 2013;9:e1003547. doi: 10.1371/journal.pgen.1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leonardi J, Fernandez-Valdivia R, Li YD, Simcox AA, Jafar-Nejad H. Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling. Development. 2011;138:3569–3578. doi: 10.1242/dev.068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 77.Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nature communications. 2011;2:583. doi: 10.1038/ncomms1591. [DOI] [PubMed] [Google Scholar]
- 78.Sakaidani Y, Ichiyanagi N, Saito C, Nomura T, Ito M, Nishio Y, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochemical and biophysical research communications. 2012;419:14–19. doi: 10.1016/j.bbrc.2012.01.098. [DOI] [PubMed] [Google Scholar]
- 79.Muller R, Jenny A, Stanley P. The EGF repeat-specific O-GlcNAc-transferase Eogt interacts with notch signaling and pyrimidine metabolism pathways in Drosophila. PloS one. 2013;8:e62835. doi: 10.1371/journal.pone.0062835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG, 2nd, Shabanowitz J, Stanley P, Hart GW, Hunt DF, Yang F, Smith RD. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaheen R, Aglan M, Keppler-Noreuil K, Faqeih E, Ansari S, Horton K, Ashour A, Zaki MS, Al-Zahrani F, Cueto-Gonzalez AM, Abdel-Salam G, Temtamy S, Alkuraya FS. Mutations in EOGT confirm the genetic heterogeneity of autosomal-recessive Adams-Oliver syndrome. American journal of human genetics. 2013;92:598–604. doi: 10.1016/j.ajhg.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohen I, Silberstein E, Perez Y, Landau D, Elbedour K, Langer Y, Kadir R, Volodarsky M, Sivan S, Narkis G, Birk OS. Autosomal recessive Adams-Oliver syndrome caused by homozygous mutation in EOGT, encoding an EGF domain-specific O-GlcNAc transferase. European journal of human genetics: EJHG. 2014;22:374–378. doi: 10.1038/ejhg.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hassed SJ, Wiley GB, Wang S, Lee JY, Li S, Xu W, Zhao ZJ, Mulvihill JJ, Robertson J, Warner J, Gaffney PM. RBPJ mutations identified in two families affected by Adams-Oliver syndrome. American journal of human genetics. 2012;91:391–395. doi: 10.1016/j.ajhg.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Libisch MG, Casas M, Chiribao M, Moreno P, Cayota A, Osinaga E, Oppezzo P, Robello C. GALNT11 as a new molecular marker in chronic lymphocytic leukemia. Gene. 2014;533:270–279. doi: 10.1016/j.gene.2013.09.052. [DOI] [PubMed] [Google Scholar]
- 87.Chen J, Moloney DJ, Stanley P. Fringe modulation of Jagged1-induced Notch signaling requires the action of beta 4galactosyltransferase-1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13716–13721. doi: 10.1073/pnas.241398098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Basmanav FB, Oprisoreanu AM, Pasternack SM, Thiele H, Fritz G, Wenzel J, Grosser L, Wehner M, Wolf S, Fagerberg C, Bygum A, Altmuller J, Rutten A, Parmentier L, El Shabrawi-Caelen L, Hafner C, Nurnberg P, Kruse R, Schoch S, Hanneken S, Betz RC. Mutations in POGLUT1, Encoding Protein O-Glucosyltransferase 1, Cause Autosomal-Dominant Dowling-Degos Disease. American journal of human genetics. 2014;94:135–143. doi: 10.1016/j.ajhg.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li M, Cheng R, Liang J, Yan H, Zhang H, Yang L, Li C, Jiao Q, Lu Z, He J, Ji J, Shen Z, Hao F, Yu H, Yao Z. Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease. American journal of human genetics. 2013;92:895–903. doi: 10.1016/j.ajhg.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, Delmas V, Kageyama R, Beermann F, Larue L, Nishikawa S. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. The Journal of cell biology. 2006;173:333–339. doi: 10.1083/jcb.200509084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kumano K, Masuda S, Sata M, Saito T, Lee SY, Sakata-Yanagimoto M, Tomita T, Iwatsubo T, Natsugari H, Kurokawa M, Ogawa S, Chiba S. Both Notch1 and Notch2 contribute to the regulation of melanocyte homeostasis. Pigment cell & melanoma research. 2008;21:70–78. doi: 10.1111/j.1755-148X.2007.00423.x. [DOI] [PubMed] [Google Scholar]
- 92.Schouwey K, Delmas V, Larue L, Zimber-Strobl U, Strobl LJ, Radtke F, Beermann F. Notch1 and Notch2 receptors influence progressive hair graying in a dose-dependent manner. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:282–289. doi: 10.1002/dvdy.21000. [DOI] [PubMed] [Google Scholar]
- 93.Aubin-Houzelstein G, Djian-Zaouche J, Bernex F, Gadin S, Delmas V, Larue L, Panthier JJ. Melanoblasts’ proper location and timed differentiation depend on Notch/RBP-J signaling in postnatal hair follicles. The Journal of investigative dermatology. 2008;128:2686–2695. doi: 10.1038/jid.2008.120. [DOI] [PubMed] [Google Scholar]
- 94.Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes & development. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 95.Gallahan D, Callahan R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4) Oncogene. 1997;14:1883–1890. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- 96.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, Rudolf M, Siziopikou K, Kast WM, Miele L. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nature medicine. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 97.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 98.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. International journal of molecular medicine. 2004;14:779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 99.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 100.O’Neill CF, Urs S, Cinelli C, Lincoln A, Nadeau RJ, Leon R, Toher J, Mouta-Bellum C, Friesel RE, Liaw L. Notch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growth. The American journal of pathology. 2007;171:1023–1036. doi: 10.2353/ajpath.2007.061029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, Wang W, Loch AJ, Deng T, Zhao W, Cardiff RD, Yoon K, Gaiano N, Ling V, Beyene J, Zacksenhaus E, Gridley T, Leong WL, Guidos CJ, Perou CM, Egan SE. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer. Cancer cell. 2012;21:626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang S, Chung WC, Wu G, Egan SE, Xu K. Tumor-Suppressive Activity of Lunatic Fringe in Prostate through Differential Modulation of Notch Receptor Activation. Neoplasia. 2014;16:158–167. doi: 10.1593/neo.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kroes RA, Dawson G, Moskal JR. Focused microarray analysis of glyco-gene expression in human glioblastomas. Journal of neurochemistry. 2007;103(Suppl 1):14–24. doi: 10.1111/j.1471-4159.2007.04780.x. [DOI] [PubMed] [Google Scholar]
- 104.Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, Zhao A, Busuttil RW, Yee H, Stein L, French DM, Finn RS, Lowe SW, Powers S. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Loo LW, Tiirikainen M, Cheng I, Lum-Jones A, Seifried A, Church JM, Gryfe R, Weisenberger DJ, Lindor NM, Gallinger S, Haile RW, Duggan DJ, Thibodeau SN, Casey G, Le Marchand L. Integrated analysis of genome-wide copy number alterations and gene expression in microsatellite stable, CpG island methylator phenotype-negative colon cancer. Genes, chromosomes & cancer. 2013;52:450–466. doi: 10.1002/gcc.22043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yokota S, Ogawara K, Kimura R, Shimizu F, Baba T, Minakawa Y, Higo M, Kasamatsu A, Endo-Sakamoto Y, Shiiba M, Tanzawa H, Uzawa K. Protein O-fucosyltransferase 1: a potential diagnostic marker and therapeutic target for human oral cancer. International journal of oncology. 2013;43:1864–1870. doi: 10.3892/ijo.2013.2110. [DOI] [PubMed] [Google Scholar]
- 107.Wang Y, Chang N, Zhang T, Liu H, Ma W, Chu Q, Lai Q, Liu L, Wang W. Overexpression of human CAP10-like protein 46 KD in T-acute lymphoblastic leukemia and acute myelogenous leukemia. Genetic testing and molecular biomarkers. 2010;14:127–133. doi: 10.1089/gtmb.2009.0145. [DOI] [PubMed] [Google Scholar]
- 108.Ma W, Du J, Chu Q, Wang Y, Liu L, Song M, Wang W. hCLP46 regulates U937 cell proliferation via Notch signaling pathway. Biochemical and biophysical research communications. 2011 doi: 10.1016/j.bbrc.2011.03.124. [DOI] [PubMed] [Google Scholar]
- 109.Kim ML, Chandrasekharan K, Glass M, Shi S, Stahl MC, Kaspar B, Stanley P, Martin PT. O-fucosylation of muscle agrin determines its ability to cluster acetylcholine receptors. Molecular and cellular neurosciences. 2008;39:452–464. doi: 10.1016/j.mcn.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]


