Abstract
Histone H2Ba of Toxoplasma gondii was expressed as recombinant protein (rH2Ba) and used to generate antibody in mouse that is highly specific. Antibody recognizing rH2Ba detects a single band in tachyzoite lysate of the expected molecular weight (12 kDa). By indirect immunofluorescence (IFA) in in vitro grown tachyzoites and bradyzoites, the signal was detected only in the parasite nucleus. The nucleosome composition of H2Ba was determined through co-immunoprecipitation assays. H2Ba was detected in the same immunocomplex as H2A.X, but not with H2A.Z. Through chromatin immunoprecipitation (ChIP) assays and qPCR, it was observed that H2Ba is preferentially located at promoters of inactive genes and silent regions, accompanying H2A.X and opposed to H2A.Z/H2B.Z dimers.
Keywords: Toxoplasma, epigenetics, canonical histones, nucleosome, ChIP, chromatin
1. INTRODUCTION
Toxoplasma gondii is an obligate intracellular protozoan parasite belonging to the phylum Apicomplexa, along with the genera Plasmodium, Cryptosporidium, Eimeria, Neospora and other economically and medically important parasites. T. gondii is the agent causing toxoplasmosis, a significant human and animal disease, impacting billions of people worldwide [1]. Toxoplasmosis is incurable because of the parasite´s ability to form latent cysts containing bradyzoites that are able to reconvert to infective tachyzoites when the immune system is compromised [2]. Consequently, AIDS patients or organ transplant recipients are at serious risk [3–6]. The parasites can also cause abortion or congenital birth defects if the mother becomes infected for the first time during pregnancy [7, 8]. Changes in gene expression are tightly controlled throughout the lytic and developmental cycles of T. gondii [9]. As in other species, a key contributor to gene expression regulation in T. gondii involves epigenetic control, including post-translational modifications (PTMs) of histones as well as histone variant replacement [10].
The basic unit of chromatin wraps 147 bp of DNA around the nucleosome, an octamer of histones that is comprised of two dimers containing histones H2A-H2B and an (H3-H4)2 tetramer [11]. These histones may be canonical or variants and can be dynamically interchanged by specific enzymes. In addition, histones are altered by a number of enzymes that introduce a wide variety of PTMs including phosphorylation, acetylation, methylation, and sumoylation [12, 13]. In T. gondii, two H2A variants are present in addition to the canonical (H2A1), H2A.X and H2A.Z, which have been characterized previously by Dalmasso et al. [14]. Regarding H2B, there are two canonical forms, H2Ba and H2Bb of which only H2Ba is transcribed in the tachyzoite stage; there is one variant H2B called H2B.Z, which has also been described by Dalmasso [15]. H2B.Z is the major H2B histone of T. gondii, in contrast to most species that have no H2B variants with the exception of tissue specific ones [15–20].
Histone H2B.Z was found in the same nucleosome with H2A.Z, while H2A.X is not present in this nucleosome particle [14]. We have also observed that these variants are enriched in specific locations in T. gondii genome. H2A.X is associated with silent gene promoters while H2A.Z and H2B.Z are enriched in active promoters [14]. In order to fully characterize the nucleosome composition of T. gondii H2A variants, we generated recombinant histone rH2Ba to obtain specific antibody. On the other hand, we transfected parasites with a tagged version of H2A.X, and obtained clones that stably express this histone. With these new tools, we were able to analyze the interactions of canonical H2B histone with the other variants in the T. gondii nucleosome and map its location in the parasite genome.
2. MATERIALS AND METHODS
2.1. Parasite culture and manipulation
RHΔhxgprt strain was used in all cases and grown in standard tachyzoite conditions in vitro: human foreskin fibroblast (HFF) monolayers were infected with tachyzoites and incubated with Dulbecco’s modified Eagle medium (DMEM, Gibco BRL) supplemented with 1% fetal bovine serum at 37°C and 5% CO2.
2.2. Source of the cDNA clone, sequencing and sequence analysis
The predicted open reading frame of H2Ba cDNA (TGME49_305160) was amplified by PCR using specific primers: sense, 5’ GGATCCATGGTGGCCAAGAAGTCCGC; anti-sense, 5’ GGTACCGAAGTGTAAACTGCCGAGACTAC. BamHI and KpnI recognition sites were included in the sequence (underlined). These fragments were cloned in pRSET-A plasmid and transformed in E. coli strain BL21pLys to express the recombinant protein under induction with 0.1 mM IPTG overnight at 37°C and purified through a Ni2+ affinity column (Qiagen) under denaturing conditions following the manufacturer's instructions [14].
2.3. Generation of anti-H2Ba antibody
Mice were immunized with rH2Ba (100 µg per dose). Three boosters of each antigen emulsified with incomplete Freund's adjuvant at intervals of 2 weeks followed a primary immunization performed with complete Freund's adjuvant. Samples of pre-immune serum were collected from each animal before antigenic stimulation [14]. Similarly, rabbit antiserum was obtained with few modifications (200 µg per dose).
2.4. Western-blot (WB) analysis
Tachyzoites were collected, filtered and counted. Recombinant histones were quantified by absorbance at 280 nm assay (Perkin Elmer Lamba 25 UV/VIS spectrometer). In all cases, 1×107 parasites, or 1.5 µg of recombinant protein were loaded per well and resolved by 15% SDS-PAGE. Proteins were transferred to PVDF membrane for 1h at 100V. Western blot was then performed as described [21]. The primary antibodies: αrH2BZ [14], was used at 1/5000 for 1 h at room temperature, whereas α-rH2Ba was used at 1/100. Appropriate secondary antibodies were used: phosphatase alkaline-conjugated goat anti-mouse or anti-rabbit (Sigma) along with the NBT and BCIP (Promega) detection system, or horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit employed along with the ECL detection system (Amersham-GE).
2.5. Immunofluorescence assay (IFA)
Extracellular tachyzoites RHΔhxgprt strain were fixed in cover slips using cold methanol 100% for 8 min and blocked with 1% BSA. Primary antibody αrH2Ba (rabbit) and α-IMC1 (mouse) (kindly provided by Dr. Gary Ward, University of Vermont, USA) diluted 1/50 or 1/500 respectively with 0.5% BSA were incubated for 1 h at room temperature. IMC1 is a cytoskeletal protein that allows visualization of daughter cells during division. After several washes with PBS cover slips were incubated with secondary antibodies Alexa fluor goat anti-mouse 488 and anti-rabbit 594 (Invitrogen). DAPI was used to stain nuclei. To generate bradyzoite cysts in vitro, 500 PRU strain tachyzoites were inoculated onto confluent HFF monolayers grown on glass coverslips. Four hours post infection, culture medium was replaced with alkaline medium (pH8.1), which was replaced daily. After five days infected monolayers were fixed in ice-cold methanol, and bradyzoite cysts were visualized by indirect fluorescent-antibody assay (IFA) following staining with fluorescein isothiocyanate (FITC)-conjugated Dolichos biflorus lectin (1:200, FL-1031; Vector Laboratories).
2.6. Co-Immunoprecipitation (Co-IP)
Approximately 5×108 RHΔhxgprt tachyzoites were used for each immunoprecipitation. Tachyzoites were treated with micrococcal endonuclease (MNase) in order to obtain mononucleosomes as described [14] and Co-IP was performed as described in Dalmasso et al [14] with minor modifications. Mononucleosomes were incubated with the antibody of interest (20 µl) overnight at 4°C. On the following day, 40 µl of Protein A/G (Santa Cruz) was added and incubated for 2 h. Immunocomplexes were washed twice with washing buffer 1 (50 mM Tris, pH8, 200 mM NaCl and 0.05 % Igepal100), twice with washing buffer 2 (50 mM Tris, pH8, 300 mM NaCl and 0.05 % Igepal100), and twice with buffer TE (10 mM Tris, pH8, 1 mM EDTA); then resuspended in 60 µl of SDS-PAGE loading buffer. Samples were boiled for 5 min and 20 µl was loaded per well in a 15% SDS-PAGE gel for immunoblotting. The absence of contaminating proteins was corroborated by Western blot with murine anti-SAG1antibody [22].
2.7. Generation of Myc-H2A.X
To generate the MycH2AX construct, the open reading frame (ORF) was amplified using the primers F: 5’-ATGCATATGTCCGCCAAAGGTGCAGG-3’ and R: 5’- TTAATTAAAGGATAGCCATTCTGTCTGGTG-3’. NsiI and PacI restriction sites (underlined) were engineered into the 5’ end of primers F and R, respectively, to clone into T. gondii expression vectors. The resulting PCR product of the expected size was removed using a Qiaex II Gel Extraction kit (Qiagen), cloned in pGEM-T vector (Promega) and sequenced employing the ABIPRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (PerkinElmer Applied Biosystems). The cloned ORF was digested with NsiI and PacI and ligated into the plasmid pTUB8MycGFPPf.myc tailTy-1/HX (kindly provided by D. Soldati, Université de Genéve, Switzerland). The final plasmid was linearized with NotI and electroporated into RH strain tachyzoites lacking hypoxanthine–xanthine guanine phosphoribosyl transferase (RHΔhxgprt). Transgenic parasites were selected in mycophenolic acid (25µg/ml) and xanthine (50µg/ml) and cloned by limiting dilution.
2.8. Chromatin immunoprecipitation (ChIP)
ChIP was performed as described in Dalmasso et al [14] with minor modifications. Samples for ChIP consisted of ~5 × 107 parasites that were subjected to sonication at 4°C, 30% amplitude, during 5 min 20 pulses (with 30 s stop between each 15 s pulse) in a Q500 Sonicator with water bath cup horn (QSonica). Ten µl of the rabbit sera (αrH2A.X, αrH2A.Z, αrH2B.Z ), or 25 µl of the mouse sera (αrH2Ba) were added for overnight incubation, after pre-incubation with 80 µl salmon sperm for 30 min at 4°C, for pre-clearing. Sixty µl of salmon sperm DNA/protein-A agarose was added and incubated for 2 h to collect the antibody-protein-DNA complex. The resulting complex was washed once with 1.0 ml of each of the following buffers: Low Salt wash buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl, 150mM NaCl, pH 8.1); High Salt wash buffer (0.1% SDS, 1% Triton-X 100, 2mM EDTA, 20mM Tris-HCl, 500mM NaCl, pH 8.1). Samples were washed 2× in TE (10 mM Tris, pH8, 1 mM EDTA) prior to elution in 250 µl elution buffer (1% SDS, 0.1M NaHCO ) for 15 min at room temperature with agitation. After a spin at 1,000 rpm for 1 min, a second elution step was performed. Crosslinks were reversed from the combined eluates and input samples with 20 µl 5M NaCl and 65°C overnight. Samples were then treated with proteinase K and DNA was precipitated with isopropanol, after extraction with phenol/chloroform. DNA samples (1.0 µl) were used as a template for qPCR to detect specific targets. Amplification was performed in a 10 µl final volume containing SYBR Green PCR Master Mix (Roche) and 0.5 µM of each forward and reverse primers previously described [14, 23]. Also a set of primers designed to amplify TASL TGME49_chrIV: 2179605–2207233 (F: CAAGCCAGTGTCGTCAAAGGAG; R: TACGGGTTGCCTACTGTGAAGC) was used.
3. RESULTS AND DISCUSSION
3.1. Generation of antibody recognizing canonical T. gondii histone H2Ba
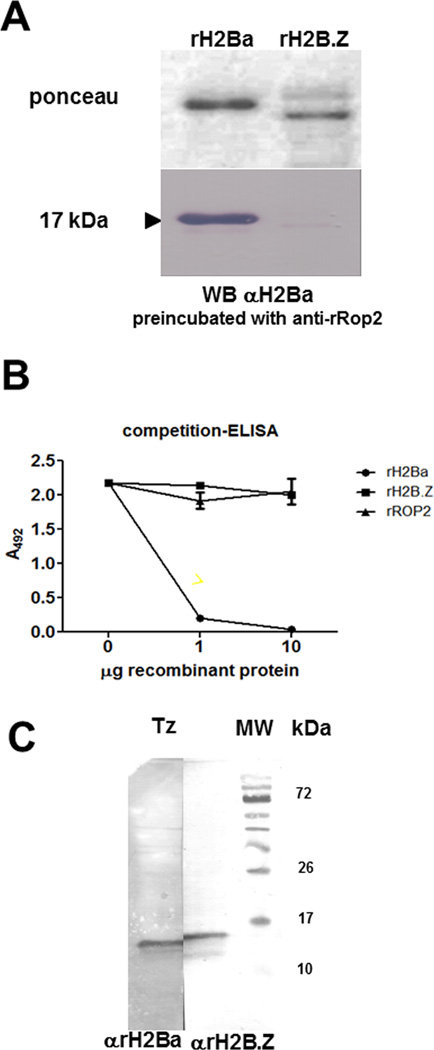
To obtain antibodies against T. gondii canonical H2Ba, we generated recombinant H2Ba (rH2Ba) by cloning the complete open reading frame in pRSET-A plasmid from a PCR product. Mouse anti T. gondii rH2Ba (αrH2Ba) serum was generated by immunization of mice and tested by Western-blot to determine its specificity. Since both rH2Ba and rH2B.Z share the 6xHis tag, the serum was pre-incubated with rROP2 [24], another 6xHis tag recombinant protein unrelated to the histones, in order to avoid cross reaction due to this tag. The purified anti-rH2Ba antibody reacted with a single band against rH2Ba, but did not cross-react with rH2B.Z (Fig 1A). To confirm the specificity of anti-rH2Ba antibody, a competition ELISA assay was performed (Fig 1B). The ELISA plate was sensitized with 1 µg/ml of rH2Ba and incubated with anti-rH2Ba serum sample pre-incubated with different quantities of rH2Ba, rH2B.Z [15] and rROP2 [24] recombinant proteins. After that the plate was incubated with secondary antibody labeled with horseradish peroxidase, revealed with OPD and the absorbance at 492 nanometers was measured. Only the treatment with rH2Ba displayed a competition effect for the immobilized rH2Ba, suggesting that the anti-rH2Ba antibody is highly specific for rH2Ba (Fig 1B). Since, H2Ba and H2Bb (TGME49_251870) share a 99% of identity at amino acid level, it is highly probable that the anti-rH2Ba antibody recognizes both histones, being an anti-canonical H2B antibody. However, only h2ba could be amplified using mRNA form tachyzoites and bradyzoites as template [15]. By contrast, despite the high conservation between canonical H2Bs and H2B.Z (68–70%), mainly at globular region, we have observed no cross reaction between these two types of histone. Similar results with other conserved histones such as H2A.X and canonical H2A were obtained [14] . Histones are also highly conserved in mammals, so we suspect that the immune response is mainly targeted to the highly polymorphic region on the histones that are far less conserved within T. gondii histone families.
Fig. 1.
Expression and characterization of T. gondii canonical H2B. (A) Recombinant H2Ba (rH2Ba) and rH2Bv (renamed H2B.Z) were resolved in SDS-PAGE, blotted to nitrocellulose membrane (stained with Ponceau red), and analyzed by Western blot against mouse rH2Ba (αrH2Ba). (B) Competition ELISA was performed with αrH2Ba serum incubated with increasing quantities of rH2Ba against recombinant proteins. The panel corresponds to the affinity of the antibody, indicated as the title, respect to the specific recombinant protein immobilized (1 µg/ml of rH2Ba). The anti-rH2Ba serum sample was pre-incubated with different quantities of rH2Ba, rH2B.Z and rROP2 recombinant proteins. (C) Whole tachyzoite lysate (Tz) was analyzed by Western blot using αrH2Ba and αrH2B.Z antibodies. The αrH2Ba recognized a single band at expected size, with a lower MW than H2B.Z.
A Western-blot analysis was performed with αrH2Ba and αrH2B.Z against tachyzoite lysate obtained in vitro. Consistent with the high specificity of the αrH2Ba antibody, a single band of ~ 12 kDa was detected, slightly smaller than the main band obtained with αrH2B.Z (Fig 1C). This result is in agreement with the expected molecular weight of both histones: 12.6 (H2Ba) and 13.3 kDa (H2B.Z).
3.2. Cellular localization of H2Ba in replicating and non-replicating parasites
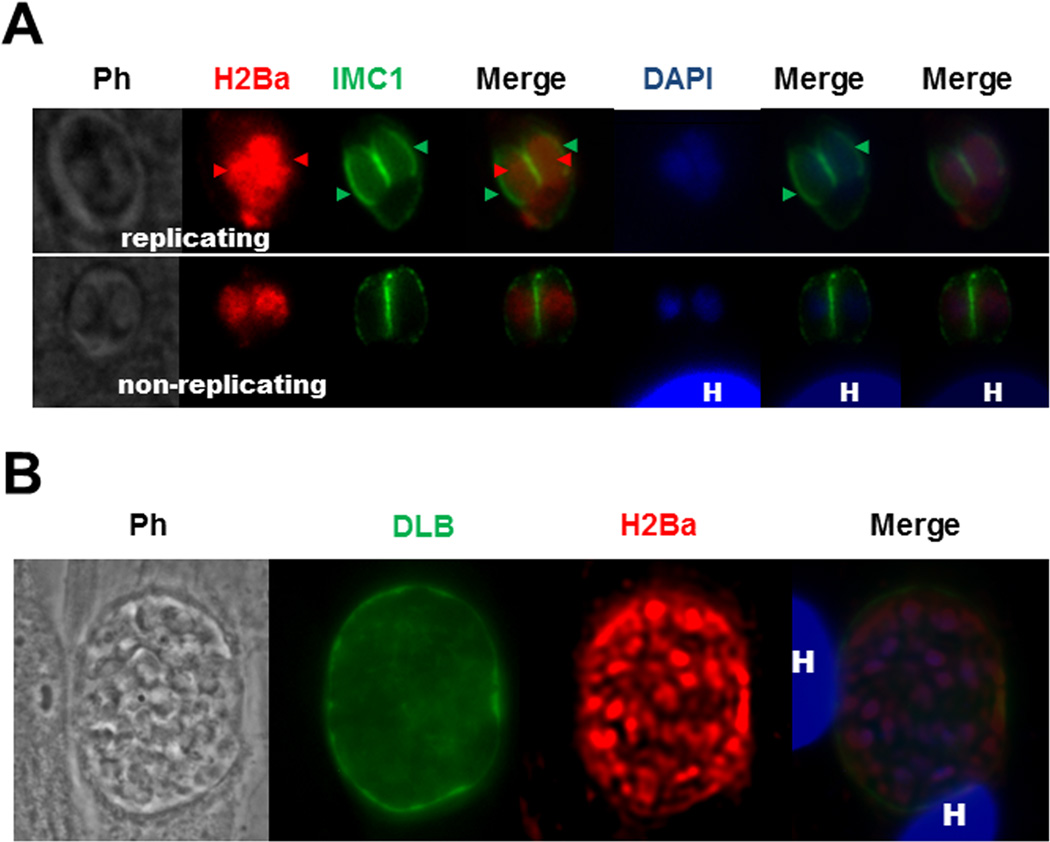
The cellular localization of H2Ba in the parasite was determined by immunofluorescence assay (IFA). This assay was performed in intracellular tachyzoites, grown in normal conditions in vitro, fixed with methanol and permeabilized with Triton X-100. Nuclear DNA was stained with DAPI. The replicating state of the parasites was analyzed by using α-IMC antibody. The parasite IMC (for inner membrane complex) appears surrounding the new tachyzoites during daughter cell segregation, whereas mother cell structure is conserved [25]. A clear signal for canonical H2B was detected only in the nucleus (yellow arrows), co-localizing with DAPI, both in replicating or non-replicating parasites (Fig 2 A). We also found H2Ba signal co-localizing with DAPI in in vitro generated bradyzoites (Fig. 2 B). This is in accordance with the previous observations in Dalmasso [15] in which h2ba gene is expressed in bradyzoites, although at a lower rate in comparison to tachyzoites.
Fig. 2.
Immunolocalization of H2Ba. (A) Non-replicating tachyzoites and replicating intracellular tachyzoites. The αrH2B antibody labeled the nucleus of tachyzoites in IFA assay, as expected for a histone protein. H2Ba stained with Alexa Fluor 594 (red) was detected in segregating nuclei (red arrowheads) and nuclei of non-replicating tachyzoites. IMC1 stained with Alexa Fluor 488 (green) shows the inner membrane complex both of the mother cell and also the daughter cells (green arrowheads). DAPI was used to stain nuclei. H: host cell nucleus. Ph: phase contrast. Merge panels, left to rigth: combination of H2Ba/IMC1, DAPI/IMC1 and H2Ba/DAPI/IMC1 images, respectively. (B) Bradyzoites. The αrH2B antibody labeled the nucleus of in vitro generated bradyzoites in IFA assay, as expected for a histone protein. H2Ba stained the nuclei inside the cyst, stained with Alexa Fluor 594 (red). Fluorescein isothiocyanate (FITC)-conjugated Dolichos biflorus lectin (DBL) (green) shows the cyst wall. DAPI was used to stain nuclei. H: host cell nucleus. Ph: phase contrast.
3.3. Nucleosome composition and position of histone H2Ba in the genome
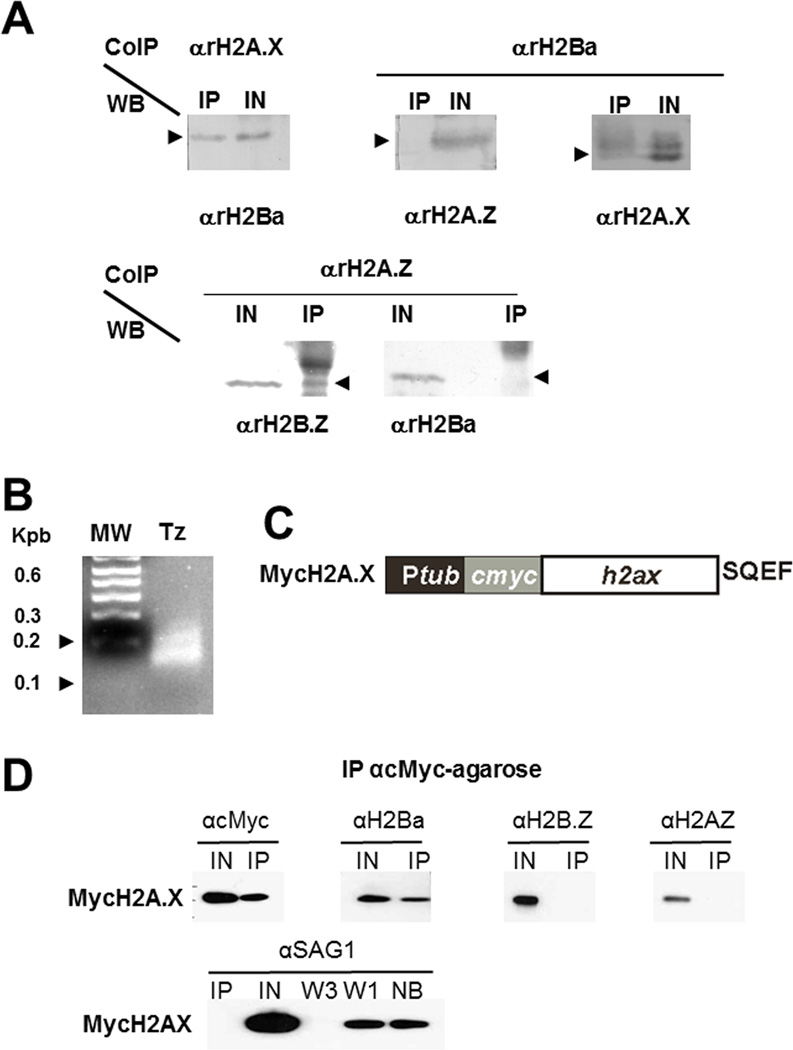
Previously we established that H2A.Z forms dimers with H2B.Z and H2A.X does not [14], however the nucleosome composition of H2A.X remained to be elucidated. Using the αrH2Ba antibody, a series of co-immunoprecipitation (Co-IP) assays were performed. Figure 3 shows that T. gondii H2A.X was able to pull down H2Ba whereas H2A.Z did not form detectable dimers with this histone. The reciprocal co-IP assay showed the same results: H2Ba was able to pull down H2A.X but not H2A.Z (Fig. 3 A). Figure 3B shows that the sample is enriched in mononucleosomes by MNase digestion. H2Ba interaction with H2A.X was further validated based on the stable expression of H2A.X fused to a c-Myc tag (Fig 3 C), where it is also evident that Myc-H2A.X is not able to immunoprecipitate H2B.Z (Fig 3 D). These data suggest that the nucleosomes detected in T. gondii with these antibodies are predominantly either H2A.X/H2Ba or H2A.Z/H2B.Z (Fig 5).
Fig. 3.
Nucleosome composition. (A) H2Ba. Immunoprecipitation was carried out using αrH2Ba, αrH2A.X or αrH2A.Z antibodies. The complexes were analyzed by Western blot using αrH2Ba, αrH2A.X or αrH2A.Z antibodies. H2A.X co-immunoprecipitates with H2Ba but H2A.Z did not immunoprecipitate with H2Ba. (B) MNase digestion of DNA control. DNA was reduced mainly to fragments between 100 and 200 pb. (C) Generation of stable lines of T. gondii expressing H2A.X fused to c-Myc epitope at N-terminal end. Diagram of the construction. (D) MycH2A.X. Immunoprecipitation was carried out using anti-cMyc (αcMyc) antibody. The complexes were analyzed on Western blots using αcMyc, αrH2Ba, αrH2B.Z or αrH2A.Z. Anti-SAG1 antibody, which detects surface antigen 1, was used as a control to identify contaminants in the immunoprecipitate. IN, input; IP, immunoprecipitate; NB, not bound; W1, wash 1; W3, wash 3
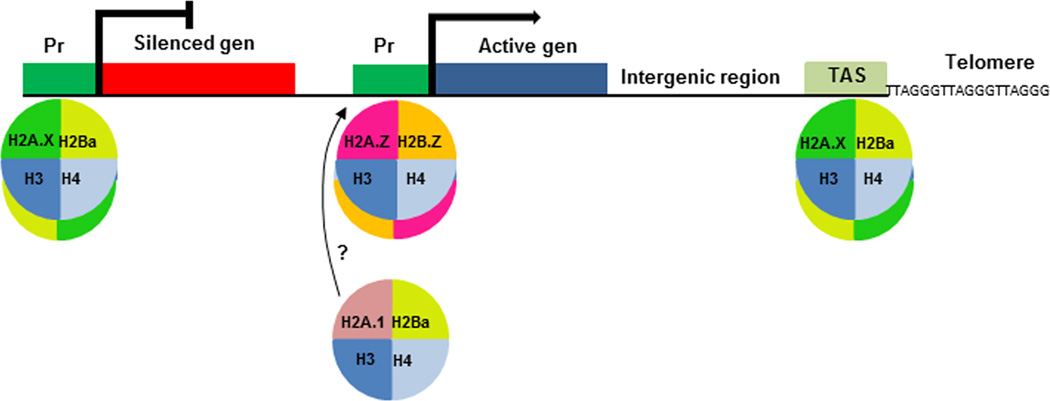
Fig. 5.
Model of nucleosome composition and genome localization in T. gondii. Nucleosomes composed of H2A.X/H2Ba histones would be present in promoters of inactive genes, as well as telomeric regions; H2A.Z/H2B.Z nucleosomes would be present at promoters of active genes. Pr: promoter; TAS: telomeric associated sequence
To study the genomic position of this histone, Chromatin IP (ChIP) experiments performed with αrH2A.X, αrH2A.Z, αrH2B.Z, and αrH2Ba antibodies were analyzed by qPCR with primers that amplify the following regions: upstream regions of constitutively active genes (β-tubulin, DHFR and elF4) and two tachyzoite-specific genes (SAG1 and SAG2A) as well as two bradyzoite-specific genes that are repressed during the tachyzoite stage (LDH2 and BAG1). In addition, two silenced gene-free genomic regions representative of telomeric associated sequences (TAS), named tgire and TASL_IV [14, 26] were also analyzed. Results indicate that H2A.X and H2Ba are enriched upstream of repressed genes LDH2 and BAG1 compared to the active genes, β-tubulin, DHFR, ElF4 and SAG1 (Fig 2B). They are also enriched in TAS regions (Fig. 4) as had been already seen before with H2A.X [26]. H2Ba but not H2A.X seems to be enriched also in SAG2A, a promoter of a tachyzoite active gene [23]. On the contrary, H2A.Z and H2B.Z are enriched at active genes relative to the inactive genes, confirming previous results [14, 26] (Fig. 4). These data suggest that H2A.Z and H2A.X have opposing roles in their regulation of chromatin, and that H2B.Z accompanies H2A.Z in its role, whereas H2Ba complements H2A.X (Fig. 5). The enrichment of H2Ba in sag2a promoter could indicate that canonical H2B may occasionally be also located in the same nucleosomes or near to H2A.Z/H2B.Z containing nucleosomes, in this case maybe by dimerization with canonical H2A. This should be confirmed by further analysis.
Fig. 4.
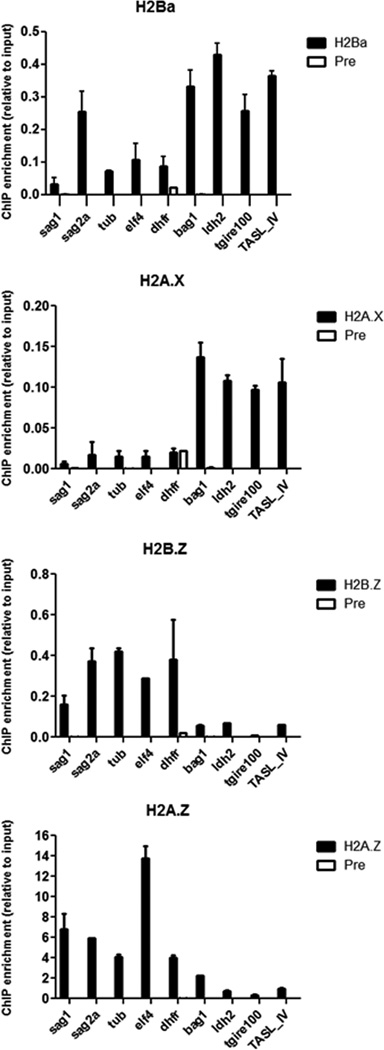
Genome localization. Chromatin immunoprecipitation (ChIP) was carried out using αrH2A.X, αrH2A.Z, αrH2B.Z or αrH2A.X or αrH2Ba antibodies, and preimmune serum (Pre). Immunoprecipitated DNA was then analyzed by qPCR with primers of SAG1, SAG2A, β-tubulin (tub), DHFR, ElF4, BAG1 and LDH2 promoters as well as TgIRE100 and TASL_IV, both from different regions on TAS element. In the figure ChIP enrichment relative to the input DNA (taken as 1) is shown.
This data, considered with our previous findings, makes it clear that T. gondii has a peculiar nucleosome composition. The presence of a unique H2B variant histone (H2B.Z) throughout Apicomplexa, apart from canonical H2B, suggests that this nucleosome composition is likely to exist in other species in the phylum. These findings reveal that the nucleosomal arrangement in protozoa may exhibit intriguing differences relative to their mammalian counterparts.
The model shown in Fig. 5 predicts that H2A.Z and H2B.Z are associated at least with promoters of active genes, whereas H2A.X and H2Ba are predominately positioned at silenced promoters and constitutive heterochromatic regions such as TAS. Our study only focused on promoter regions and two regions inside T. gondii TAS, near 30 kb genomic domain. Since T. gondii H2A.X was also enriched in other regions inside TAS in comparison to H2A.Z and H2B.Z [26], it is expected that canonical H2B is positioned along this subtelomeric sequence. However, we have not tested the nucleosome composition of intergenic and gene regions, which will be important to expand our understanding of the role of each of these histones in chromatin modulation. In addition, the enrichment of H2Ba, but not H2A.X, together with H2A.Z/H2B.Z in the promoter of tachyzoite active gene sag2a, opens the question if canonical H2B could be localized in or near active promoters of some genes.
In general, the model envisages that canonical H2B/H2A.X and the double variant nucleosome formed by H2A.Z/H2B.Z present opposite genome localization, which should be confirmed by a genome wide analysis. Up to now, a genome wide analysis on Plasmodium falciparum, another apicomplexa protozoan parasite, further confirms that H2A.Z and H2B.Z form the same double variant nucleosome occupying AT rich promoters, intergenic regions and var gene promoters [27, 28]. Despite var genes, which are absent in T. gondii, are located in clusters within the heterochromatic regions at plasmodial TAS, H2A.Z-H2B.Z nucleosomes are present in single var gene that escapes silencing and is abundantly transcribed [28, 29]. Since P. falciparum does not have a H2A.X variant, it remains to be elucidated the nucleosome composition, concerning H2B and H2A histones, along P. falciparum TAS .
The opposing genomic localization of H2A.Z/H2B.Z and H2A.X/H2Ba is not surprising considering the PTMs present in these histones are quite different. H2A.Z and H2B.Z have been shown to be hyper-acetylated in the N-terminal [30]. Acetylation is one of the most prominent modifications influencing gene expression [31, 32] as neutralization of the positively charged histone N-terminal tails leads to chromatin decondensation, thereby enhancing transcriptional activity [32]. In other organisms, as well as T. gondii, H2A.Z with its hyperacetylated N-terminal region, is preferentially present in regions of the genome correlating with transcriptional activation [33], and this should also be true for H2B.Z, where the N-terminal tail is acetylated on lysines 3, 8, 13, 14, and 18 [30]. Similar results were also observed in P. falciparum [34, 35]. In contrast, H2A.X and canonical H2Ba have a striking lack of PTMs at the N-terminus compared to H2A.Z and H2B.Z. On H2Ba, apart from an acetylation mark on K4, only methylation of certain residues were found along with some phosphorylation [30]. This is in agreement with our results showing localization of these histones in the same nucleosome, positioned in silenced regions of the genome.
The localization of H2A.X and H2Ba in TAS regions may link these histones to roles in subtelomere silencing and/or DNA damage control. It is interesting that some mutations in yeast H2B have been shown to produce significant changes in telomeric and subtelomeric compactation, interaction with silenced DNA, or DNA damage checkpoint associated factors such as silent information regulator, sirtuin (Sir) complex or disruptor of telomeric silencing (Dot1) proteins [36–38]. In addition, the distribution of H2A.X and γH2A.X histones in the replicating genome is enriched near chromosomal ends (telomeres and subtelomeres) [39]. Sub-telomeres are exposed to replication-mediated DSB, maybe because during highly replicative stages DNA contains many replication forks in sub-telomeric regions [40, 41]. Elucidation of the role of H2A.X/H2Ba histones in the TAS region of T. gondii is an interesting subject for future studies.
Highlights.
We developed an antibody against recombinant histone H2Ba of Toxoplasma gondii
Co-IP using α-rH2Ba showed that H2Ba shares nucleosome with H2A.X, not with H2A.Z
We obtained a stable line of tachyzoites expressing Myc-H2AX
Co-IP assays using α-c-Myc confirmed that H2Ba shares nucleosome with H2A.X
We determined the genomic location of H2Ba in promoters of silent genes and TAS
Acknowledgements
SO Angel (Researcher), S Bogado (Fellow), L Vanagas (Post-doctoral Fellow) are members of National Research Council of Argentina (CONICET). A Ganuza is member of Scientific Research Comission (CIC, provincia Buenos Aires). This work was supported by ANPCyT PICT 1864 (to L.V.), ANPCyT PICT 0623 (to S.O.A.), NIH-NIAID R01AI083162 (to S.O.A. and W.J.S.), R01 AI077502 (to W.J.S.) and R01AI087625 (to K.K.), and RC4AI092801 (to K.K. and W.J.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. International journal for parasitology. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyons RE, McLeod R, Roberts CW. Toxoplasma gondii tachyzoite-bradyzoite interconversion. Trends in parasitology. 2002;18:198–201. doi: 10.1016/s1471-4922(02)02248-1. [DOI] [PubMed] [Google Scholar]
- 3.Nissapatorn V. Toxoplasmosis in HIV/AIDS: a living legacy. The Southeast Asian journal of tropical medicine and public health. 2009;40:1158–1178. [PubMed] [Google Scholar]
- 4.Luft BJ, Remington JS. Toxoplasmic encephalitis in AIDS. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1992;15:211–222. doi: 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 5.Derouin F, Pelloux H. Prevention of toxoplasmosis in transplant patients. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2008;14:1089–1101. doi: 10.1111/j.1469-0691.2008.02091.x. [DOI] [PubMed] [Google Scholar]
- 6.Moncada PA, Montoya JG. Toxoplasmosis in the fetus and newborn: an update on prevalence, diagnosis and treatment. Expert review of anti-infective therapy. 2012;10:815–828. doi: 10.1586/eri.12.58. [DOI] [PubMed] [Google Scholar]
- 7.Kravetz J. Congenital toxoplasmosis. Clinical evidence. 2013;2013 [PMC free article] [PubMed] [Google Scholar]
- 8.Halonen SK, Weiss LM. Toxoplasmosis. Handbook of clinical neurology. 2013;114:125–145. doi: 10.1016/B978-0-444-53490-3.00008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bougdour A, Braun L, Cannella D, Hakimi MA. Chromatin modifications: implications in the regulation of gene expression in Toxoplasma gondii. Cellular microbiology. 2010;12:413–423. doi: 10.1111/j.1462-5822.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan WJ, Jr, Naguleswaran A, Angel SO. Histones and histone modifications in protozoan parasites. Cellular microbiology. 2006;8:1850–1861. doi: 10.1111/j.1462-5822.2006.00818.x. [DOI] [PubMed] [Google Scholar]
- 11.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 12.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 13.Karch KR, Denizio JE, Black BE, Garcia BA. Identification and interrogation of combinatorial histone modifications. Frontiers in genetics. 2013;4:264. doi: 10.3389/fgene.2013.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalmasso MC, Onyango DO, Naguleswaran A, Sullivan WJ, Jr, Angel SO. Toxoplasma H2A variants reveal novel insights into nucleosome composition and functions for this histone family. Journal of molecular biology. 2009;392:33–47. doi: 10.1016/j.jmb.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalmasso MC, Echeverria PC, Zappia MP, Hellman U, Dubremetz JF, Angel SO. Toxoplasma gondii has two lineages of histones 2b (H2B) with different expression profiles. Molecular and biochemical parasitology. 2006;148:103–107. doi: 10.1016/j.molbiopara.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Yuan G, Zhu B. Histone variants and epigenetic inheritance. Biochimica et biophysica acta. 2012;1819:222–229. doi: 10.1016/j.bbagrm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Aul RB, Oko RJ. The major subacrosomal occupant of bull spermatozoa is a novel histone H2B variant associated with the forming acrosome during spermiogenesis. Developmental biology. 2001;239:376–387. doi: 10.1006/dbio.2001.0427. [DOI] [PubMed] [Google Scholar]
- 18.Churikov D, Siino J, Svetlova M, Zhang K, Gineitis A, Morton Bradbury E, Zalensky A. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics. 2004;84:745–756. doi: 10.1016/j.ygeno.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Churikov D, Zalenskaya IA, Zalensky AO. Male germline-specific histones in mouse and man. Cytogenetic and genome research. 2004;105:203–214. doi: 10.1159/000078190. [DOI] [PubMed] [Google Scholar]
- 20.Talbert PB, Ahmad K, Almouzni G, Ausio J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, Cross GA, Cui L, Dimitrov SI, Doenecke D, Eirin-Lopez JM, Gorovsky MA, Hake SB, Hamkalo BA, Holec S, Jacobsen SE, Kamieniarz K, Khochbin S, Ladurner AG, Landsman D, Latham JA, Loppin B, Malik HS, Marzluff WF, Pehrson JR, Postberg J, Schneider R, Singh MB, Smith MM, Thompson E, Torres-Padilla ME, Tremethick DJ, Turner BM, Waterborg JH, Wollmann H, Yelagandula R, Zhu B, Henikoff S. A unified phylogeny398 based nomenclature for histone variants. Epigenetics & chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Echeverria PC, Matrajt M, Harb OS, Zappia MP, Costas MA, Roos DS, Dubremetz JF, Angel SO. Toxoplasma gondii Hsp90 is a potential drug target whose expression and subcellular localization are developmentally regulated. Journal of molecular biology. 2005;350:723–734. doi: 10.1016/j.jmb.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 22.Laguia-Becher M, Martin V, Kraemer M, Corigliano M, Yacono ML, Goldman A, Clemente M. Effect of codon optimization and subcellular targeting on Toxoplasma gondii antigen SAG1 expression in tobacco leaves to use in subcutaneous and oral immunization in mice. BMC biotechnology. 2010;10:52. doi: 10.1186/1472-6750-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ, Jr, Cesbron-Delauw MF, Hakimi MA. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Molecular and cellular biology. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nigro M, Gutierrez A, Hoffer AM, Clemente M, Kaufer F, Carral L, Martin V, Guarnera EA, Angel SO. Evaluation of Toxoplasma gondii recombinant proteins for the diagnosis of recently acquired toxoplasmosis by an immunoglobulin G analysis. Diagnostic microbiology and infectious disease. 2003;47:609–613. doi: 10.1016/s0732-8893(03)00156-1. [DOI] [PubMed] [Google Scholar]
- 25.Hu K, Mann T, Striepen B, Beckers CJ, Roos DS, Murray JM. Daughter cell assembly in the protozoan parasite Toxoplasma gondii. Molecular biology of the cell. 2002;13:593–606. doi: 10.1091/mbc.01-06-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalmasso MC, Carmona SJ, Angel SO, Aguero F. Characterization of Toxoplasma gondii subtelomeric-like regions: identification of a long-range compositional bias that is also associated with gene-poor regions. BMC genomics. 2014;15:21. doi: 10.1186/1471-2164-15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Francoijs KJ, Treeck M, Gilberger TW, Stunnenberg HG, Bartfai R. H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Molecular microbiology. 2013;87:1061–1073. doi: 10.1111/mmi.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petter M, Selvarajah SA, Lee CC, Chin WH, Gupta AP, Bozdech Z, Brown GV, Duffy MF. H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum. Molecular microbiology. 2013;87:1167–1182. doi: 10.1111/mmi.12154. [DOI] [PubMed] [Google Scholar]
- 29.Petter M, Lee CC, Byrne TJ, Boysen KE, Volz J, Ralph SA, Cowman AF, Brown GV, Duffy MF. Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter. PLoS pathogens. 2011;7:e1001292. doi: 10.1371/journal.ppat.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nardelli SC, Che FY, Silmon de Monerri NC, Xiao H, Nieves E, Madrid-Aliste C, Angel SO, Sullivan WJ, Jr, Angeletti RH, Kim K, Weiss LM. The histone code of Toxoplasma gondii comprises conserved and unique posttranslational modifications. mBio. 2013;4 doi: 10.1128/mBio.00922-13. e00922-00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 32.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nature reviews. Molecular cell biology. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 34.Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. Journal of proteome research. 2009;8:3439–3450. doi: 10.1021/pr9000898. [DOI] [PubMed] [Google Scholar]
- 35.Cui L, Miao J. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryotic cell. 2010;9:1138–1149. doi: 10.1128/EC.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang CY, Hua CY, Hsu HE, Hsu CL, Tseng HY, Wright DE, Hsu PH, Jen CH, Lin CY, Wu MY, Tsai MD, Kao CF. The C-terminus of histone H2B is involved in chromatin compaction specifically at telomeres, independently of its monoubiquitylation at lysine 123. PloS one. 2011;6:e22209. doi: 10.1371/journal.pone.0022209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai J, Hyland EM, Norris A, Boeke JD. Yin and Yang of histone H2B roles in silencing and longevity: a tale of two arginines. Genetics. 2010;186:813–828. doi: 10.1534/genetics.110.118489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyriss MN, Jin Y, Gallegos IJ, Sanford JA, Wyrick JJ. Novel functional residues in the core domain of histone H2B regulate yeast gene expression and silencing and affect the response to DNA damage. Molecular and cellular biology. 2010;30:3503–3518. doi: 10.1128/MCB.00290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo J, Kim SC, Lee HS, Kim JK, Shon HJ, Salleh NL, Desai KV, Lee JH, Kang ES, Kim JS, Choi JK. Genome-wide profiles of H2AX and gamma-H2AX differentiate endogenous and exogenous DNA damage hotspots in human cells. Nucleic acids research. 2012;40:5965–5974. doi: 10.1093/nar/gks287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 41.Zschenker O, Kulkarni A, Miller D, Reynolds GE, Granger-Locatelli M, Pottier G, Sabatier L, Murnane JP. Increased sensitivity of subtelomeric regions to DNA double-strand breaks in a human cancer cell line. DNA repair. 2009;8:886–900. doi: 10.1016/j.dnarep.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]