Abstract
Poliomavirus JC replicates in glial cells in the brain, and causes the fatal demyelinating disease, progressive multifocal leukoencephalopathy (PML). PML is usually seen in patients with underlying immunocompromised conditions, notably among AIDS patients and those on chronic immunosuppressive regimens. The late leader sequence of JC virus contains an open reading frame encoding a small regulatory protein called agnoprotein. Agnoprotein contributes to progressive viral infection by playing significant roles in viral replication cycle. Here, we demonstrate that agnoprotein can be detected in cell-free fractions of glial cultures infected with JCV, transfected with expression plasmids or transduced with adenovirus expression system. We also provide evidence that extracellular agnoprotein can be taken up by uninfected neighboring cells. These studies have revealed a novel phenomenon of agnoprotein during the viral life cycle with a potential of developing diagnostic and therapeutic interventions.
Keywords: JC virus, agnoprotein, release, uptake, secretion, viral pathogenesis, biomarker
Introduction
JC virus (JCV) is a human polyomavirus that infects greater than 50% of the human population during childhood, and establishes a latent/persistent infection for the rest of the life in healthy individuals (Weber T., 2008; Moens et al., 2008). Replication of the neurotropic strain of JCV in glial cells causes the fatal demyelinating disease of the central nervous system, progressive multifocal leukoencephalopathy (PML), which is seen in patients with underlying immunocompromised conditions, notably HIV-1/AIDS (Safak et al., 2005; Berger et al., 1995; Miller et al., 1982). PML is the only viral demyelinating disease of the human brain characterized by lytic infection of oligodendrocytes (Safak et al., 2005; Berger et al., 1995; Padget et al., 1971). Over the past few years, exogenous immunosuppressive treatments such as natalizumab, efalizumab, and rituximab have also been associated with PML in patients with autoimmune diseases, including Multiple Sclerosis, Crohn’s Disease, Psoriasis, and Lupus (Frenzczy et al., 2012; Tavazzi et al., 2011). Like other polyomaviruses, the genome of JCV is composed of a double-stranded circular DNA genome of approximately 5 kb in size with a bi-directional non-coding control region that is located between the early and late coding sequences (Frenzczy et al., 2012). The early coding region is responsible for the expression of large T antigen (T-Ag), small t antigen (t-Ag), and a group of T′ proteins, which are produced upon alternative splicing of the early primary transcript. Similarly, alternative splicing of the late transcript results in production of the viral capsid proteins VP1, VP2, and VP3 which are essential for completion of the viral lytic cycle and formation of viral particles.
In addition to the capsid proteins, JCV encodes a small (71 aa long), regulatory, phosphoprotein, agnoprotein, from the late viral transcript. Agnoprotein forms highly stable dimers and oligomers (Saribas et al., 2011 and 2013) and has an important role in viral DNA replication by enhancing T-Ag binding to the origin of replication (Saribas et al., 2012). The expression pattern of agnoprotein in tissue sections from PML shows localization to the cytoplasmic and perinuclear regions of infected glial cells (Okada et al., 2002). Recent observations also suggest that agnoprotein localizes to the endoplasmic reticulum, interacts with lipid membranes and may function as a viroporin (Suzuki et al., 2010 and 2013). Furthermore, agnoprotein expression is required for the successful completion of JC virus life cycle, because mutant JC virus with a deletion in the agno gene is unable to propagate (Ellis et al., 2013, Sariyer et al., 2006, and 2011a).
Because of its highly basic structure, co-localization with endoplasmic reticulum at the perinuclear area and its association with intracellular lipid membranes, we sought to investigate possible release of agnoprotein by infected cells. Our results has revealed the presence of extracellular agnoprotein in cell free supernatant fractions of infected cultures as well as in glial cell lines expressing agnoprotein in the absence of viral lytic infection.
Results
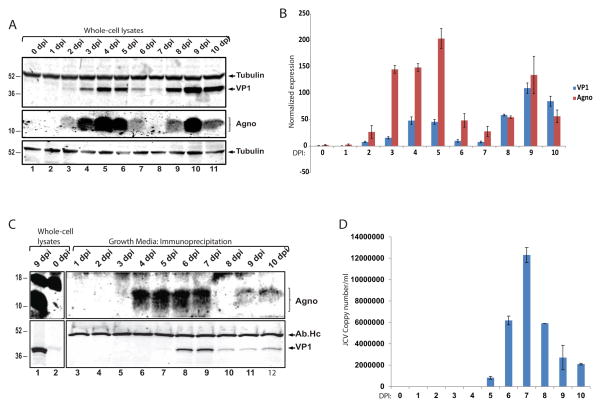
To determine the possible secretion of JC virus agnoprotein from infected cells, we first infected SVG-A human glial cell line with Mad-1 strain of JC virus. SVG-A cells were transfected with viral genome to initiate a uniform infection cycle and whole cell protein lysates were collected at 24h intervals up to 10 days post-infection (dpi). Protein samples were processed for SDS-PAGE, transferred to nitrocellulose membranes and expression of VP1 and agnoprotein were determined by Western blot. As shown in Fig. 1A and B, VP1 expression was started at the second day post-infections, reached a peak at 4 dpi, and showed a dramatic decrease at 6 and 7 dpi that corresponded to the time of the completion of the first replication cycle (Sariyer et al., 2009). Similar to the VP1, agnoprotein expression was detectable as early as 2 dpi, reached a peak at 4 dpi, and stayed high until 6 dpi. Consistent with the expression of VP1, agnoprotein levels were barely detectable at 6 and 7 dpi. Interestingly, agnoprotein expression levels came back to peak levels at 9 dpi, followed by another sharp reduction in expression at 10 dpi. These experiments suggested that unlike the VP1 expression, agnoprotein showed a dynamic expression pattern during the replication cycle of the virus in glial cells. Next, we asked whether agnoprotein could be released by infected cells and its expression in the cells correlate with its release pattern during the course of viral replication cycle. The growth media from the cells were collected from the same infection studies presented in Fig. 1A, and processed for agnoprotein detection by immunoprecipitation followed by Western blot. Whole cell protein lysates from 9 dpi and 0 dpi (uninfected) were loaded as positive and negative controls of agnoprotein expression. As shown in Fig. 1C (upper panel), unlike to its cellular expression, agnoprotein was not detected in growth media earlier than 4 dpi (compare Fig. 1A and Fig. 1C). Surprisingly, a significant amount of agnoprotein was present in the growth media at 4 dpi when there were still peak levels of cellular expression (compare Fig. 1A lane 5 with Fig. 1C lane 6). At 6 and 7 dpi when agnoprotein cellular expression levels started to decrease, agnoprotein was still present in the growth media in significant amounts (compare Fig. 1A lanes 7 and 8 with Fig. 1C lanes 8 and 9). Interestingly, there were no detectable agnoprotein in growth media at 8 dpi when agnoprotein expression began to reappear in the cells. Agnoprotein was again present in the growth media in parallel to its peak expression at 9 dpi and stayed at a constant level in contrast to a drop in its cellular expression level at 10 dpi. To determine the time of viral release, the major capcid protein VP1 was also immunoprecipitated from the growth media of infected cells and analyzed by Western blotting (Fig. 1C lower panel). Unlike agnoprotein, VP1 appeared in the growth media at 6 and 7 dpi when its cellular expression levels was dropping (compare Fig. 1A and C), and showed a lower but constant level at 8, 9, and 10 dpi. In addition, Q-PCR analysis of the viral copy numbers in the growth medium further demonstrated that most viral particles were released at 6 and 7 dpi (Fig. 1D). Thus, these infection studies have revealed the presence of extracellular agnoprotein which was released into the growth media by infected cells before the completion of viral lytic life cycle.
Figure 1. Agnoprotein is detected in cell-free fractions of glial cells infected with JCV.
A. SVG-A cells were transfected/infected with Mad-1 JCV as described previously (23, 24). Whole cell protein samples were prepared from cellular pellets with 24 h intervals up to 10 dpi. Expression of VP1 and agnoprotein were analyzed by western blot. In lane 1, protein samples from uninfected SVG-A cells were loaded as negative control (0 dpi). Membranes were stripped and re-probed for β-tubulin as a loading control. DPI is an abbreviation for “days post infection”. B. Bar graph representation of normalized expression of VP1 and agnoprotein during JCV infection in glial cells. C. Immunoprecipitation of agnoprotein and VP1 from the growth medium of cells infected with JC virus. Immune complexes were separated by SDS-PAGE and processed for Western blot of agnoprotein and VP1. In lanes 1 and 2, whole-cell protein lysates from JCV-infected SVG-A cells (9 dpi) and from uninfected cells (0 dpi) were loaded as positive and negative controls of agnoprotein and VP1 expression, respectively. D. Q-PCR analysis of JCV copy number in growth medium. The growth medium of JCV-infected cells used in IP studies (panel C) were also subjected to Q-PCR analysis for the detection of viral loads as described in materials and methods. All experiments were carried out in triplicate. Images depict representative data.
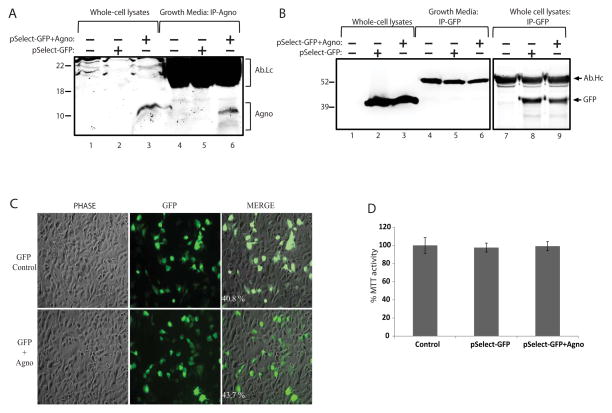
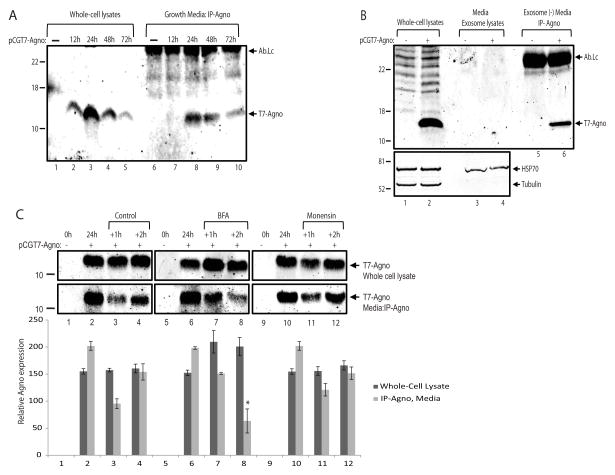
One can argue that the presence of viral regulatory proteins in the growth medium of infected cultures could be due to the mechanical disruption of the cell membrane by the viral particles at the stage of their release from infected cells. In order to investigate this possibility, agnoprotein was cloned into a mammalian expression vector pSELECT-GFPzeo (InvivoGen, USA). The final plasmid construct contained two transcription units, the first drove the expression of agnoprotein and the second drove the expression of GFP as described by the manufacturer. T98G glioblastoma cell lines were transfected with this construct and growth medium was collected in parallel to the whole cell protein lysates at 48hrs post-transfection. As shown in Figure 2A, agnoprotein was clearly detected in growth media as well as in whole cell protein lysates (lane 6 and 3, respectively). In parallel to the agnoprotein, GFP was also immunoprecipitated from the same set of experiments. Unlike agnoprotein, GFP was not detected in growth media of the cells expressing either GFP together with agnoprotein (Fig. 2B, lane 6) or GFP alone from control plasmid transfections (Fig. 2B, lane 5). Of note, GFP was sufficiently immunoprecipitated by the anti-GFP antibody from whole cell protein lysates suggesting its efficiency for IP studies (Fig. 2B, lanes 6 - 8). The possible toxicity of agnoprotein expression for the cells was also assessed by live-fluorescein imaging of GFP (Fig. 2C) and MTT viability (Fig. 2D) assays, and found no significant effect on cellular morphology and viability, respectively. Since both agnoprotein and GFP were expressed from the same plasmid backbone, detection of extracellular agnoprotein with no GFP contamination suggested that agnoprotein was possibly secreted by glial cells. In order to investigate this possibility, agnoprotein was cloned into a mammalian expression vector pCGT7 as a fusion protein to T7-tag. The 11 amino acid T7-tag allowed us to utilize a commercially available monoclonal anti-T7 antibody for the detection of agnoprotein. T98G glioblastoma cell lines were transfected with this construct and growth medium was collected in parallel to whole cell protein lysates at 12, 24, 48, and 72 hours post-transfections. As shown in Figure 3A, agnoprotein expression started at 12hrs, reached the highest level at 24hrs and gradually decreased at 48 and 72hrs post-transfections in the cellular lysates (lanes 1 – 5). In parallel, immunoprecipitation of agnoprotein from the growth media was performed at the same time points. Interestingly, agnoprotein appeared in the growth medium at 24hrs and the signal was decreased in correlation with its expression pattern in the cells (Fig. 3A, lanes 6 – 10). These data, once again, suggested that agnoprotein may be released into the extracellular matrix in the absence of viral lytic infection. One possible mechanism for agnoprotein release could be the involvement of exosomes. To clarify whether exosomes were involved in agnoprotein release, T98G cells were transfected with pCGT7-Agno expression plasmid and conditioned media was collected at 24hrs post-transfections. Exosomes were precipitated using ExoQuick exosome precipitation solution (System Biosciences, address) according to the manufacturer’s instructions, and lysed with TNN buffer containing 1% NP40. As shown in Fig. 3B, exosome lysates from cells expressing agnoprotein contained Hsp70 (bottom panel, lanes 3 and 4), a well described internal exosome marker (Lancaster and Febbraio, 2005) with no detectable levels of agnoprotein (lane 4). In addition, agnoprotein was clearly detected by immunoprecipitation in exosome-depleted growth media suggesting that extracellular agnoprotein may not be associated with exosomes. Many secretory proteins are secreted from cells by using the endoplasmic reticulum to Golgi pathway (conventional secretion pathway), which can be blocked by Brefeldin-A (BFA). To determine the possible role of ER to Golgi transport in agnoprotein release, T98G cells were transfected with pCGT7-agno expression plasmid, and growth medium of the cells was changed with fresh media containing BFA or Monensin at 24hrs post-transfection. Whole cell protein lysates were prepared at 24hrs (no treatment), 24+1hrs, and 24+2hrs post-treatment. In parallel, growth medium from cells was collected at the same time points and processed for immunoprecipitation for agnoprotein. As shown in Fig. 3C, agnoprotein was readily detectable in the growth medium at 24hrs post-transfections (lanes 2, 6, and 10). As expected, the level of extracellular agnoprotein was reduced at 24+1hrs in control, BFA, and Monensin treatments (lanes, 3, 7, and 11). Interestingly, only BFA but not Monensin caused a significant reduction in the levels of extracellular agnoprotein at 24+2hrs post-treatments (compare lane 8 with lane 4 and 12), suggesting possible involvement of ER to Golgi transport pathway in agnoprotein release into the extracellular space.
Figure 2. Agnoprotein is released by glial cells.
A. Immunoprecipitation of agnoprotein in growth media. T98G cells were transfected with pSELECT-GFPzeo control plasmid encoding GFP or with pSELECT-GFPzeo-agno expression plasmid which contained two transcription units, the first drove the expression of agnoprotein and the second drove the expression of GFP. Growth medium was collected at 48h post-transfections simultaneously along with the whole cell protein extracts. Agnoprotein was immunoprecipitated in the growth medium, and analyzed by western blotting (lanes 6 – 10). Whole cell protein extracts were also processed and loaded in parallel for the cellular expression of agnoprotein (lanes 1 – 5). Ab-Lc points the light chain of agnoprotein antibody. B. Immunoprecipitation of GFP in growth medium of T98G cells. GFP was immunoprecipitated in the growth media from the same set of experiments presented in panel A and analyzed by western blot (lanes 6 – 10). Whole cell protein extracts were also processed and loaded in parallel for the cellular expression of GFP (lanes 1 – 5). Efficiency of anti-GFP antibody for immunoprecipitating GFP protein was also assessed in whole cell protein lysates and shown (right panel, lanes 7–9). Ab-Hc indicates the IgG heavy chain of the antibody. C. T98G cells were transfected with expression plasmids encoding either GFP alone or GFP and agnoprotein together as described above. Live-fluorescein images of GFP expression was taken at 48 hrs posttransfections. Transfection efficiencies were determined as ~40% for GFP control and ~43% for GFP + agnoprotein by calculating the percentile of GFP positive cells over total number of cells. D. T98G cells were transfected with expression plasmids encoding either GFP alone or GFP and agnoprotein together as described in panel A and B. Forty eight hours post transfections, MTT activities were detected. MTT activity of untransfected cells is presented as 100%, and MTT activity of the transfected groups is presented proportionally.
Figure 3. Agnoprotein release may involve conventional pathway.
A. Immunoprecipitation of agnoprotein in growth media from T98-G cells. Cells were transfected with an expression polasmid encoding T7-tagged agnoprotein (pCGT7-Agno) and growth media was collected at 0, 12, 24, 48, and 72h post-transfections simultaneously along with whole cell extracts. Agnoprotein was immunoprecipitated from the growth media, and analyzed by western blotting (lanes 6 – 10). Whole cell protein extracts were also processed and loaded in parallel for the cellular expression of agnoprotein (lanes 1 – 5). Ab-Lc points the light chain of T7 tag antibody. B. The secreted agnoprotein is not associated with extracellular vesicles. Extracellular vesicles were isolated from growth media of either control cells or cells expressing agnoprotein by ExoQuick exosome precipitation solution (System Biosciences, SBI) according to the manufacturer’s instructions. The precipitated exosomes were lysed with TNN buffer containing 1% NP40, and analyzed by Western blot for the detection of agnoprotein (upper panel, lanes 3 and 4) and Hsp70 (bottom panel, lanes 3 and 4) proteins. The exosome depleted growth media was subjected to immunoprecipitation for agnoprotein (lanes 5 and 6) by using an anti-T7 tag monoclonal antibody. Whole cell protein extracts were also processed in parallel for the cellular expression of agnoprotein (lanes 1 and 2). C. Agnoprotein secretion is suppressed by Brefeldin A. T98G cells were transfected with pCGT7-Agno expression plasmid and grouped as 24h, +1h, and +2h. At 24h post-transfection, growth media and whole cell protein extracts were obtained from the first 24h group for the cellular and secreted expression of agnoprotein. Meanwhile, the growth media of +1h and +2h groups were aspirated, cells were washed twice with PBS, and treated with Brefeldin A (10 μg/ml) or Monensin (1 μM) in fresh growth media. At 1h and 2h post-treatment, growth medium was collected and processed for immunoprecipitation of agnoprotein (lower panel). In parallel, whole cell protein extracts were also prepared and analyzed by Western blot for its cellular expression (top panel). Bar graph represents the relative expression of agnoprotein in whole cell lysates and growth medium presented in the upper panels. All experiments were carried out in triplicate.
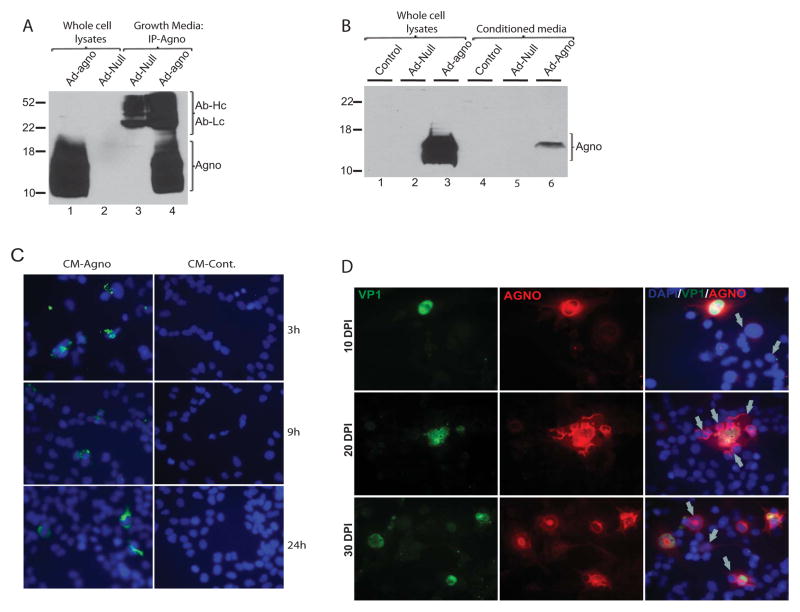
In an alternative approach, agnoprotein release was also analyzed by adenoviral transduction (Fig. 4A). Tc620 oligodendroglioma cells were transduced with aneno-null and adeno-agno constructs and whole cell protein lysates and growth media were collected at 48hrs post-transduction. Consistent with the infection and transfection studies, agnoprotein was once again detected in the cell-free supernatants of cells transduced with adeno-agno construct (Fig. 3A, lane 4). Next, we asked whether secreted agnoprotein could be taken up by glial cells. First, we collected growth media and whole cell protein lysates from Tc620 cells either transduced with adeno-null or adeno-agnoprotein constructs. In the second step of the experiment, another set of Tc620 cells were only incubated with the conditioned media collected from control cells and cells transduced with either adeno-null or adeno-agno (50% of culture volume). At 48h post-incubation, cells were washed three times with PBS, and whole cell protein extracts were prepared. Both direct and indirect protein extracts were processed for agnoprotein detection by Western blotting. As expected, agnoprotein was detected in the whole cell protein lysates (Fig. 3B, lane 3). Interestingly, a decent amount of agnoprotein was also detected in extracts prepared from cells treated with conditioned media from adeno-agno transduced cells (Fig. 4B, lane 6), suggesting that extracellular agnoprotein in the conditioned media was possibly taken up by the second set of Tc620 cells. We also analyzed the cellular uptake and subcellular localization of agnoprotein by immunocytochemistry. We treated TC620 cells with conditioned media from cells either transduced with adeno-agno (CM-Agno) or adeno-null (CM-control). Cells were fixed at various time points and processed for immunostaining as described in materials and methods. A signal corresponding to the presence of agnoprotein was detected within the cultured cells as early as 3h post-treatments (Fig. 3C, upper panel). At 9 and 24hrs post-treatments, the characteristic cytoplasmic localization of agnoprotein was observed. In the next set of experiments, we asked whether agnoprotein secreted during the infection was taken up by neighboring uninfected cells. To analyze this, SVG-A cells were infected with Mad1 strain of JCV and cells were fixed at 10, 20, and 30 day post-infection. Immunocytochemical detection of agnoprotein and VP1 was performed by using a mouse monoclonal antibody to VP1 (Pab597) and a rabbit polyclonal antibody to agnoprotein (Pab7903). As shown in Fig. 4D, VP1 showed an exclusive nuclear expression (green) and agnoprotein (red) showed its characteristic cytoplasmic and perinuclear localization within the infected glial cells. Interestingly, merge analysis of green (VP1) and red (agnoprotein) fluorescein channels revealed the presence of agnoprotein positive but VP1 negative cells within the infected SVG-A cultures (depicted by arrows). Since both VP1 and agnoprotein are expressed from the late viral transcript, the presence of agnoprotein in neighboring cells with no detectable VP1 expression may suggest its possible uptake from the culture medium.
Figure 4. Agnoprotein is taken up by glial cells.
A. TC620 cells were transduced with either a null adenovirus or adenovirus encoding agnoprotein. Growth medium was collected in parallel to whole cell protein lysates at 48 h post-transduction. Agnoprotein was immunoprecipitated from growth medium and analyzed by Western blot (lanes 3 and 4) as described in Fig. 1C. Whole cell protein lysates were also loaded as positive and negative controls of expression (lanes 1 and 2, respectively). B. Western blot analysis of whole cell extracts from cells expressing agnoprotein (lanes 1–3) or from cells treated with conditioned medium obtained from control, adeno-null, and adeno-agno transduced cells. C. TC620 cells were seeded in two-well chamber slides and treated with growth medium from agnoprotein-expressing cells or control cells. At 3, 9, and 24 h post-treatment, cells were washed 3X with PBS and fixed with cold acetone/methanol (1/1). Immunocytochemistry was performed to detect agnoprotein as described in materials and methods. D. SVGA cells were transfected/infected with Mad-1 JCV and cells were fixed on 2-well chamber slides at 10, 20, and 30 days post-infection. Expression and co-localization of VP1 and agnoprotein were analyzed by immunocytochemistry. Nuclei were also labeled with DAPI (blue) and merged with the agno (red) and VP1 (green). Arrows point cells positive for agno (red) and negative for VP1 (green) fluorescein.
Discussion
Many eukaryotic viruses encode small secretory proteins which provide essential functions for the replication cycle and the pathogenesis of the diseases (Lucas and McFadden, 2004). Agnoprotein is a small regulatory protein of JC virus and it is not incorporated into the virions therefore it should be present in cells actively replicating the virus. However, our results have revealed that agnoprotein is also present in cell-free fractions of infected glial cultures. This detection of agnoprotein in growth medium was not only due to the lytic destruction of the infected cell membrane during virion release of viral particles. Analysis of viral copy numbers and detection of VP1 in growth media suggest that completion of first round of JCV infection cycle starts around 5 dpi. Detection of agnoprotein in growth media of cells at 4 dpi with no detectable levels of VP1 protein and viral genome clearly suggest that agnoprotein secretion may occur prior to virion release. In addition, detection of agnoprotein in cell-free fractions of T98G cells transfected with a dual-promoter expression plasmid encoding agnoprotein and GFP has further confirmed that agnoprotein is indeed released by transfected cells with no detectable GFP contamination. Moreover, glial cell lines transduced with an adenovirus encoding agnoprotein also resulted in its release.
The presence of agnoprotein in the extracellular matrix may suggest an important novel role of agnoprotein in the molecular pathogenesis of JCV infection in the brain. One possible role for the secreted agnoprotein could be an extracellular immunomodulatory function to facilitate the invasion of the virus within the infected tissue. Many DNA viruses encode immunomodulatory proteins that are secreted into the extracellular matrix (Lucas and McFadden, 2004). Among these, myxomavirus encodes a small 37-kDa secretory protein (M-T7) which binds to a broad spectrum of C/CC/CXC chemokines and inhibits leukocyte chemotaxis into virus-infected tissue (Liu et al., 2000 and 2004). Some strains of vaccinia virus encode vaccinia complement control protein (VCP), which has been shown to interact with complement components and limit the complement cascade (Kotwal and Moss, 1988; Murthy et al., 2001). Whether secreted agnoprotein has an immunomodulatory function that is similar to the myxoma and vaccinia virus proteins remains to be determined.
PML is usually a fatal disease and little is known about the progression of the disease from latency to the pathologic stage of the disease. Most PML cases are diagnosed at very late stage of disease progression when neurological symptoms appear and this increases the morbidity and mortality of the disease. Currently, PML is diagnosed by combination of JCV DNA detection in cerebrospinal fluid or brain biopsy specimen and by the observation of multifocal non-enhancing lesions in MRI. There is no established diagnostic early marker applicable to the clinic that indicates reactivation of JCV. Since the agnoprotein is only expressed by cells actively replicating the virus and released into the extracellular matrix, detection of agnoprotein in clinical samples may serve as such a marker. This would be a significant advance in clinical diagnosis. Whether agnoprotein could be a biomarker for the detection of the early stages of JCV reactivation and the progression of PML remains to be investigated.
In conclusion, our results suggest that agnoprotein is released by infected glial cells during the course of viral propagation, and present in the extracellular matrix. We have also provided evidence that agnoprotein may also be taken up by neighboring uninfected cells that may play important roles in the pathogenesis and progression of the disease, PML. These results open new possibilities for research to understand the molecular pathogenesis of JCV.
Materials and Methods
JCV genomic strains, plasmid constructs, and adenovirus vectors
The wild type Mad1 strain of JCV was linearized by BamH1 digestion, and cloned into the pBlueScript KS (+) vector as previously described (Sariyer and Khalili, 2011b). The pCGT7-Agno expression vector was created as following. The full-length coding region for the agnoprotein was amplified by using Agno-forward-(Xba1) 5′-attctatctagagttcttcgccagctgtcacgt-3′ and Agno-reverse-(BamH1) 5′-attctaggatccctatgtagcttttggttcaggca-3′ primers by using the Mad1 genome as template. The amplification products were subjected to Klenow reaction, and were blunt-end ligated into pBlueScript KS (+) vector. The agnoprotein coding sequence was removed from pBlueScript KS (+) background by Xba1/BamH1 digestion and cloned into pCGT7 vector. The final construct was sequenced to confirm the integrity and sequence frame before using in the experiments. The pSELECT-GFPzeo-agno expression vector was created as following. The full-length coding region for the agnoprotein was amplified by using Agno-forward-(SalI) 5′-accttccagtcgacatggttcttcgccagctgtcacgtaa-3′ and Agno-reverse-(BamH1) 5′-attctaggatccctatgtagcttttggttcaggca-3′ primers by using the Mad1 genome as template. The amplification products were subjected to Klenow reaction, and were blunt-end ligated into pBlueScript KS (+) vector. The agnoprotein coding sequence was removed from pBlueScript KS (+) background by SalI/BamH1 digestion and cloned into pCGT7 vector. The final construct was sequenced to confirm the integrity and sequence frame before using in the experiments. Adenovirus construct expressing JCV agnoprotein (Ad-Agno) was described previously (Merabova et al, 2008).
JCV infection
Transfection/infection of cells with JCV Mad-1 genome was described previously (Sariyer and Khalili, 2011b; Uleri et al, 2013). Briefly, pBlue.script-Mad1-WT plasmid was digested with BamH1 enzyme to remove the complete viral genome from pBlue.Script KS (+) plasmid. SVG-A cells were plated in 10 different T75-cm tissue culture flasks at a confluence of 0.5 × 106 cells per T75-cm tissue culture flask, and transfected/infected with the Mad1 genomic DNA (10 μg/flask) using Fugene-6 transfection reagent as indicated by the manufacturer (Roche). For the time course analysis of JC virus infection cycle, growth media of the cells and whole cell protein lysates were collected at 24hrs intervals up to 10 days post-infection (dpi). Of note, the growth media of the cells were never removed from the cells during the course of 10 day infection. Instead, 5ml of fresh media was added to the cells at 4dpi and 8dpi for the late time points of the infections. Whole cell protein extracts were prepared to analyze the expression of viral proteins. The growth media of the cells were also obtained in parallel to whole cell protein extracts to analyze the presence of viral proteins (agnoprotein and VP1) by immunoprecipitation and to measure viral load by Q-PCR analysis.
Immunoprecipitation of agnoprotein from growth media
Growth media of cells expressing agnoprotein was collected at different time points as indicated in the experimental settings. Before subjecting to immunoprecipitation, growth media was centrifuged at 3000 rpm for at least 15 minutes to remove cellular debris, and diluted with TNN buffer containing 1% NP-40 at a ratio of 1:1. Immunoprecipitations were performed with a Dynabeads® Protein G immunoprecipitation kit (Invitrogen). Since the growth media of the cells was supplemented with 10% fetal bovine serum (FBS) which contained massive amount of immunoglobulins, we followed two steps to avoid the saturation of the beads with FBS originated immunoglobulins. In the first step, we depleted the immunoglobulins from media by incubating them with protein G beads. Secondly, anti-agno antibody (#7903) was pre-loaded on dynabeads before subjecting to immunoprecipitations.
Q-PCR analyses of JCV copy numbers in growth media
Q-PCR analysis of JCV copy numbers in growth media was performed as described previously (Uleri et al., 2013). Briefly, the culture medium (containing viral particles) were collected and incubated at 95°C for 10 minutes to inactivate virus. Ten microliters of the medium was then used as a template in Q-PCR reactions. The standard curve was obtained after serial dilution of pJCV, a plasmid containing the whole genome of the JCV Mad-1 strain. The standard curve was then used to extrapolate the viral load of each sample. Negative and positive controls were included in each reaction and each sample was tested in triplicate. All Q-PCR analyses were done by using Light cycler 480 (Roche). Primers were JCV Q-PCR-forward: 5′-AGTTGATGGGCAGCCTATGTA-3′ and JCV Q-PCR-reverse: 5′-TCATGTCTGGGTCCCCTGGA-3′. The probe for the Q-PCR was 5′-/5HEX/CATGGA TGCTCAAGTAGAGGAGGTTAGAGTTT/3BHQ_1/-3′.
Exosome purification
Extracellular vesicles were isolated from growth media of either control cells or cells expressing agnoprotein by ExoQuick exosome precipitation solution (System Biosciences, SBI) according to the manufacturer’s instructions. Briefly, 2 ml ExoQuick exosome precipitation solution was mixed with 10 ml growth media from cells expressing agnoprotein and incubated at 4 °C for 12hrs (overnight). The mixture was centrifuged and exosomes were precipitated at 1500 ×g for 30 minutes. The supernatant (exosome depleted media) was recovered and all the trace of fluid was removed. The precipitated exosomes were lysed with TNN buffer containing 1% NP40, and analyzed by Western blot for the detection of agnoprotein and exosome marker Hsp70 (Santa Cruz, W27). The exosome depleted growth media was subjected to immunoprecipitation for agnoprotein. Whole-cell protein extracts were also processed in parallel for the cellular expression of agnoprotein.
Brefeldin A and Monensin treatments
T98G cells were plated in 60mm tissue culture dishes at a density of %80, and transfected with pCGT7-Agno expression plasmid. At 24h post-transfections, cells were washed 3X with PBS and fresh growth media were added with BFA or Monensin. The growth media of the cells were collected at +1h and +2h of the 24h post-transfection. In parallel, whole cell protein lysates were also prepared to analyze the cellular & released agnoprotein ratios. Whole cell protein extracts were processed for western blotting using an anti-T7 monoclonal antibody. In parallel, agnoprotein was immunoprecipitated from growth media with Dynabeads® Protein G immunoprecipitation kit (Invitrogen # 10007D) as described above.
Immunocytochemistry
In order to show cellular uptake of agnoprotein, Tc620 cells were seeded in two-well chamber slides and treated with conditioned media obtained from agnoprotein expressing cells. At 3, 9, and 24 h post-treatments, cells were washed 3X with PBS, and fixed with cold acetone/methanol (1/1). Slides were blocked with a 10% BSA solution, followed by incubation with anti-agno (7903) polyclonal antibody. Cells were then incubated with FITC-conjugated secondary antibody, mounted with aqueous mounting medium, and examined under immunoflourescence microscope. To analyze the expression of agnoprotein in infected cells, SVG-A cells were infected with JC virus and fixed with cold acetone/methanol (1/1) at 10, 20, and 30 day postinfections (dpi). After treatment with a 10% BSA solution, samples were incubated with JCV VP1-specific monoclonal and anti-agno (7903) polyclonal antibodies followed by incubation with FITC and Rhodamine-conjugated secondary antibodies. Samples were mounted with aqueous mounting media with DAPI, and examined under fluorescein microscope.
Highlights.
Agnoprotein is detected in cell-free fractions of glial cells infected with JC virus.
Agnoprotein is released by glial cells.
Agnoprotein release may involve conventional secretion pathway.
Agnoprotein is taken up by neighboring glial cells.
Acknowledgments
The authors thank the past and present members of the Department of Neuroscience/Center for Neurovirology for sharing their ideas and reagents, particularly Drs. Mahmut Safak, Martyn K. White and Jennifer Gordon. We would like to also thank Kasra Houshmand and Rahsan Sariyer for their contributions. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI101192. The funding organization played no role in the design of the study, in the collection, analysis, and interpretation of the data and in the decision to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berger JR, Concha M. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J Neurovirology. 1995;1(1):5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- 2.Ellis LC, Norton E, Dang X, Koralnik IJ. Agnogene deletion in a novel pathogenic JC virus isolate impairs VP1 expression and virion production. PLoS One. 2013;12;8(11):e80840. doi: 10.1371/journal.pone.0080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferenczy MW, Marshall LJ, Nelson CD, Atwood WJ, Nath A, Khalili K, Major EO. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 2012;25(3):471–506. doi: 10.1128/CMR.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotwal GJ, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 5.Lancaster GI, Febbraio MA. Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem. 2005;280(24):23349–55. doi: 10.1074/jbc.M502017200. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Lalani A, Dai E, Seet B, Macauley C, Singh R, Fan L, McFadden G, Lucas A. The viral anti-inflammatory chemokine-binding protein M-T7 reduces intimal hyperplasia after vascular injury. J Clin Invest. 2000;105:1613. doi: 10.1172/JCI8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, Dai R, Miller L, Seet B, Lalani A, Macauley C, Li X, Virgin HW, Bunce C, Turner P, Moyer R, Macauley C, Lucas A. Viral chemokine-binding proteins inhibit inflammatory responses and aortic allograft transplant vasculopathy in rat models. Transplantation. 2004;77:1652. doi: 10.1097/01.tp.0000131173.52424.84. [DOI] [PubMed] [Google Scholar]
- 8.Lucas A, McFadden G. Secreted immunomodulatory viral proteins as novel biotherapeutics. J Immunol. 2004;15;173(8):4765–74. doi: 10.4049/jimmunol.173.8.4765. [DOI] [PubMed] [Google Scholar]
- 9.Merabova N, Kaniowska D, Kaminski R, Deshmane SL, White MK, Amini S, Darbinyan A, Khalili K. JC virus agnoprotein inhibits in vitro differentiation of oligodendrocytes and promotes apoptosis. J Virol. 2008;82(3):1558–69. doi: 10.1128/JVI.01680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JR, Barrett RE, Britton CB, Tapper ML, Bahr GS, Bruno PJ, Marquardt MD, Hays AP, McMurtry JG, 3rd, Weissman JB, Bruno MS. Progressive multifocal leukoencephalopathy in a male homosexual with T-cell immune deficiency. N Engl J Med. 1982;307(23):1436–8. doi: 10.1056/NEJM198212023072307. [DOI] [PubMed] [Google Scholar]
- 11.Moens U, Johannessen M. Human polyomaviruses and cancer: expanding repertoire. J Dtsch Dermatol Ges. 2008;6(9):704–8. doi: 10.1111/j.1610-0387.2008.06810.x. [DOI] [PubMed] [Google Scholar]
- 12.Murthy KH, Smith SA, Ganesh VK, Judge KW, Mullin N, Barlow PN, Ogata CM, Kotwal GJ. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell. 2001;104:301. doi: 10.1016/s0092-8674(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 13.Okada Y, Sawa H, Endo S, Orba Y, Umemura T, Nishihara H, Stan AC, Tanaka S, Takahashi H, Nagashima K. Expression of JC virus agnoprotein in progressive multifocal leukoencephalopathy brain. Acta Neuropathol (Berl) 2002;104:130–136. doi: 10.1007/s00401-002-0526-8. [DOI] [PubMed] [Google Scholar]
- 14.Padgett BL, Zu Rhein GM, Walker DL, Echroade R, Dessel B. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet. 1971;i:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 15.Safak M, Major E, Khalili K. Human polyomavirus, JC virus, and progressive multifocal encephalopathy. In: Howard IG, Gendelman E, Everall Ian Paul, Lipton Stuart A, Swindells Susan, editors. The Neurology of AIDS. Oxford University Press; New York: 2005. pp. 461–474. [Google Scholar]
- 16.Saribas AS, Arachea BT, White MK, Viola RE, Safak M. Human polyomavirus JC small regulatory agnoprotein forms highly stable dimers and oligomers: implications for their roles in agnoprotein function. Virology. 2011;10;420(1):51–65. doi: 10.1016/j.virol.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saribas AS, Abou-Gharbia M, Childers W, Sariyer IK, White MK, Safak M. Essential roles of Leu/Ile/Phe-rich domain of JC virus agnoprotein in dimer/oligomer formation, protein stability and splicing of viral transcripts. Virology. 2013;443(1):161–76. doi: 10.1016/j.virol.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saribas AS, White MK, Safak M. JC virus agnoprotein enhances large T antigen binding to the origin of viral DNA replication: evidence for its involvement in viral DNA replication. Virology. 2012;10;433(1):12–26. doi: 10.1016/j.virol.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, Safak M. Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. J Virol. 2006;80(8):3893–903. doi: 10.1128/JVI.80.8.3893-3903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sariyer IK, Safak M, Gordon J, Khalili K. Generation and characterization of JCV permissive hybrid cell lines. J Virol Methods. 2009;159(1):122–6. doi: 10.1016/j.jviromet.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sariyer IK, Saribas AS, White MK, Safak M. Infection by agnoprotein- negative mutants of polyomavirus JCV and SV40 results in release of virions that are deficient In DNA content. Virol J. 2011a;8:255. doi: 10.1186/1743-422X-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sariyer IK, Khalili K. Regulation of human neurotropic polyomavirus, JCV, by alternative splicing factor, SF2/ASF, in glial cells. PLoS One. 2011b;10(1):e14630. doi: 10.1371/journal.pone.0014630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, Nagashima K, Hall WW, Sawa H. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 2010;12;6(3) doi: 10.1371/journal.ppat.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki T, Orba Y, Makino Y, Okada Y, Sunden Y, Hasegawa H, Hall WW, Sawa H. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc Natl Acad Sci U S A. 2013;110(46):18668–73. doi: 10.1073/pnas.1311457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavazzi E, Ferrante P, Khalili K. Progressive multifocal leukoencephalopathy: an unexpected complication of modern therapeutic monoclonal antibody therapies. Clin Microbiol Infect. 2011;17(12):1776–80. doi: 10.1111/j.1469-0691.2011.03653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uleri E, Regan P, Dolei A, Sariyer IK. SF2/ASF binding region within JC virus NCCR limits early gene transcription in glial cells. Virol J. 2013;14(10):147. doi: 10.1186/1743-422X-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber T. Progressive Multifocal Leukoencephalopathy. Neurol Clin. 2008;26:833–854. doi: 10.1016/j.ncl.2008.03.007. [DOI] [PubMed] [Google Scholar]