Abstract
OBJECTIVE
In this study, we aimed to investigate relationships between maternal prepregnancy obesity and gestational diabetes mellitus and placental leptin DNA methylation.
STUDY DESIGN
This study comprises data on 535 mother-infant dyads enrolled in the Rhode Island Child Health Study (RICHS), a prospective cohort study of healthy term pregnancies. Prepregnancy body mass index was calculated from self-reported anthropometric measures and gestational diabetes mellitus diagnoses gathered from inpatient medical records. DNA methylation of the leptin promoter region was assessed in placental tissue collected at birth using quantitative bisulfite pyrosequencing.
RESULTS
In multivariable regression analysis adjusted for confounders, infants exposed to gestational diabetes mellitus had higher placental leptin methylation (β=1.89, P=0.04), as did those demonstrating prepregnancy obesity (β=1.17, P=0.06). Using a structural equations model (SEM), we observed that gestational diabetes mellitus is a mediator of the effects of prepregnancy obesity on placental leptin DNA methylation (β =0.81, 95% CI: 0.27, 2.71).
CONCLUSIONS
Our results suggest that maternal metabolic status before and during pregnancy can alter placental DNA methylation profile at birth and potentially contribute to metabolic programming of obesity and related conditions.
Keywords: leptin, DNA methylation, maternal obesity, gestational diabetes mellitus, placenta
1. INTRODUCTION
Maternal obesity and gestational diabetes mellitus (GDM) constitute two common, often comorbid pregnancy complications.1 In line with the developmental origins of health and disease (DOHaD) hypothesis2, increasing evidence suggests that these conditions modify the intrauterine environment and augment the offspring’s risk of obesity and diabetes in adult life.3, 4 Epigenetic marks have been proposed as a mechanism for this developmental programming because they respond to environmental stimuli but they are also mitotically stable.5 Due to the high tissue-specificity of epigenetic mechanisms5, though, it is critical to appropriately focus studies in relevant tissues. The placenta, a metabolically active organ that regulates the intrauterine environment and is crucial for fetal growth and development, is such a tissue.6, 7
Leptin is an adipokine central for energy homeostasis that functions as a satiety signal. During pregnancy, leptin is produced by the placenta where it has pleiotropic functions, including regulating growth and nutrient exchange.8 Leptin gene (LEP) expression is inversely correlated with promoter DNA methylation9–13 and has been proposed as mediator of metabolic programming14. In male rodents, in utero exposure to a low protein diet is associated with LEP promoter hypomethylation in adipose tissue, changes in body composition and increased food intake15, 16. In humans, in utero famine exposure has been associated with LEP promoter hypermethylation in blood of adult men compared to their non-exposed siblings17. In humans and rodents, maternal over-nutrition produces similar adverse metabolic offspring phenotypes to under-nutrition14. Hence, in this study, we sought to investigate associations between maternal prepregnancy obesity and GDM and placental LEP DNA methylation in a birth cohort of healthy newborns.
2. MATERIALS AND METHODS
2.1 Study Population
Study participants are part of the Rhode Island Child Health Study (RICHS), which enrolls mother-infant dyads following delivery at Women and Infants Hospital of Rhode Island.18 Term infants born small for gestational age (SGA, <10th percentile), or large for gestational age (LGA, >90th percentile), based on birth weight percentiles19 are selected, and infants appropriate for gestational age (AGA, ≥10th and ≤90th percentile) matched on sex, gestational age (±3 days), and maternal age (±2 years) to SGA and LGA participants are enrolled20. Only singleton, viable infants without congenital or chromosomal abnormalities were recruited. Additional exclusion criteria include maternal age <18 years and life-threatening conditions. Post-recruitment infants were re-classified into birth weight groups using sex-specific growth charts.21 In this analysis, we examined the first 535 RICHS participants enrolled between September 2009 and October 2012 with placental LEP methylation information. A structured chart review served to collect information from inpatient medical record from delivery, and mothers completed an interviewer-administered questionnaire. Self-report of weight and height obtained during the interview served to calculate maternal prepregnancy body mass index (BMI). GDM status was obtained from medical charts. All subjects provided written informed consent. Protocols were approved by the Institutional Review Boards for Women and Infants Hospital of Rhode Island and Dartmouth College and carried out in accordance with the Declaration of Helsinki.
2.2 LEP DNA methylation analysis and genotyping
Placental samples were collected from all subjects within two hours following delivery. Twelve fragments of placental parenchyma, three from each quadrant, were obtained two centimeters (cm) from the umbilical cord and free of maternal decidua. Collected tissue was immediately placed in RNAlater solution (Life Technologies, Grand Island, NY, USA) and stored at 4°C. After at least 72 hours, tissue segments from each placental region were blotted dry, snap frozen in liquid nitrogen, homogenized by pulverization using a stainless steel cup and piston unit (Cellcrusher, Cork, Ireland) and stored at −80°C until needed. DNA was extracted from homogenized placental samples using the DNAeasy Blood & Tissue Kit (Qiagen, Inc, Valencia, CA, USA) and quantified using the ND 2000 spectrophotometer (Thermo Fisher Scientific Inc., Watham, MA, USA). DNA (500 ng) was sodium bisulfite-modified using the EZ DNA methylation Kit (Zymo Research, Irvine, CA, USA). For DNA methylation detection, bisulfite pyrosequencing was employed. Bisulfite PCR conditions, primer sequences (Integrated DNA Technologies, Inc, Coralville, IA) and pyrosequencing assays are detailed Supplementary Table 1. We measured DNA methylation at 23 CpGs in the LEP promoter using the PyroMark MD (Qiagen) and genotyped the SNP rs2167270 (+19 G>A) in the region. Genotype calls were made by comparing peak heights; triplicate wells were called independently and compared for quality control. All procedures were performed following manufacturer’s protocols.
Table 1.
Study population characteristics n % mean SD missing data
| n | % | mean | SD | missing data | |
|---|---|---|---|---|---|
| Infant characteristics | |||||
| Gestational age | 535 | 39.0 | 1.0 | ||
| Birth weight | 535 | 3486.6 | 696.6 | ||
| AGA | 281 | 52.5 | |||
| LGA | 140 | 26.2 | |||
| SGA | 114 | 21.3 | |||
| Sex | |||||
| Male | 270 | 50.5 | |||
| Female | 265 | 49.5 | |||
| Delivery Method | |||||
| C-Section | 273 | 51 | |||
| Vaginal | 262 | 49 | |||
| Genotype (rs2167270) | |||||
| G/G,G/A | 457 | 85.4 | |||
| A/A | 78 | 14.6 | |||
| Maternal characteristics | |||||
| Age | 535 | 29.6 | 5.6 | ||
| BMI (Kg/m2) | 529 | 26.8 | 7.1 | 6 | |
| Prepregnancy obesity | 6 | ||||
| No (BMI <30) | 394 | 74.5 | |||
| Yes (BMI ≥ 30) | 135 | 25.5 | |||
| Gestational diabetes mellitus | 56 | ||||
| No | 432 | 90.2 | |||
| Yes | 47 | 9.8 | |||
| Gestational weight gain | 9 | ||||
| Inadequate | 101 | 19.2 | |||
| Adequate | 135 | 25.7 | |||
| Excessive | 290 | 55.1 | |||
| Ethnicity | 3 | ||||
| Other | 138 | 25.9 | |||
| White | 394 | 74.1 | |||
| Tobacco use during pregnancy | 7 | ||||
| No | 502 | 95.1 | |||
| Yes | 26 | 4.9 | |||
| History diabetes type I | 59 | ||||
| No | 471 | 98.9 | |||
| Yes | 5 | 1.1 | |||
| History diabetes type II | 59 | ||||
| No | 472 | 99.2 | |||
| Yes | 4 | 0.8 | |||
| Pregnancy hypertension | 10 | ||||
| No | 490 | 93.3 | |||
| Yes | 35 | 6.7 |
SD, standard deviation; AGA, adequate for gestational age, LGA, large for gestational age, SGA, small for gestational age
2.3 Statistical analysis
Pairwise Pearson correlations were used to compare continuous LEP methylation between the 23 CpGs loci analyzed. Self-reported gestational weight gain (GWG) data was combined with prepregnancy BMI to construct a categorical variable following the Institute of Medicine cutoffs.22 Bivariate analyses were performed using Student’s t-test, one-way ANOVA or Pearson’s correlation, as appropriate. χ2 tests were used to assess frequency distributions. Multivariable analyses were completed using linear regression models, with continuous LEP methylation as the outcome and maternal and infant characteristic as predictor variables. A structural equation model (SEM) was used to assess mediation effects between predictors using Mplus, version 7.11 (Muthén & Muthén, Los Angeles, CA). A bootstrap method23 was used to estimate the mediational effect. All other analyses were conducted in R 3.0.1. The multivariable regression and SEM were adjusted for potential confounders: rs2167270 genotype, infant sex, maternal age, birth weight group. Confounders included in the models were significantly associated with methylation in the bivariate analysis and also associated with methylation at a P= 0.1 level in a fully adjusted multivariable linear model (data not shown) or are part of the RICHS study matching criteria (maternal age and birth weight group). All tests were two-sided and statistical significance was determined at P<0.05.
3. RESULTS
3.1 Study Population
The study population characteristics are summarized in Table 1. In accordance with the study design, all infants were born at term with overrepresentation of LGA and SGA and even distribution by sex. The majority of infants were born to Caucasian mothers (74.1%) that ranged between 18 and 40 years (yrs.) of age (mean=30 yrs.). The prevalence of maternal prepregnancy obesity and GDM in this study was 26% (n=135) and 10% (n=47), respectively. In addition, amongst study participants with medical chart diagnosis of GDM 61% were prepregnancy obese. There were no significant differences between the sample of participants analyzed in this study and the larger RICHS cohort in terms of maternal age, prepregnancy maternal obesity, GDM, infant sex or birth weight group.
3.2 Placental LEP DNA methylation
There was a high degree of inter-correlation of DNA methylation at each of the 23 CpGs (mean r = 0.7), thus we used the mean across the region. Mean LEP methylation was normally distributed and ranged from 9 to 45%. Genotypes frequencies at rs2167270 were in Hardy-Weinberg equilibrium, with 15% of the infants homozygous for the variant allele (A), 44% heterozygous and 41% and homozygous for the dominant allele (G).
3.3 Infant and maternal predictors of placental LEP DNA methylation
The results of the bivariate analyses between LEP methylation and maternal and infant characteristics are presented in Table 2. As previously reported10, placental LEP methylation extent was higher in infants with the A/A genotype and in males. Strikingly, we did not observe associations with infant birth weight. We observed higher methylation in placentas from infants born to prepregnancy obese mothers (P=0.03) and from those diagnosed with GDM (P=0.01). Subsequently, we constructed a multivariable linear regression model to predict LEP methylation adjusted for all significant covariates from the bivariate analyses and the study matching criteria (Table 3). Consistently, we observed associations between placental LEP methylation and infant sex and genotype. In addition, placentas from infants exposed to GDM had 1.89% higher methylation compared to those from the non-GDM group. In contrast, in the multivariable model, maternal prepregnancy obesity was no longer a significant predictor of LEP methylation (β=1.17, P=0.06). However, obesity was strongly associated with GDM (P<0.001, χ2 test). This attenuation pattern suggested that the initial association between obesity and placental LEP methylation may be mediated through GDM. We did not observe associations with GWG or any other maternal characteristic.
Table 2.
Bivariate analysis of infant and maternal variables and placental LEP methylation
| Infant | n | Mean LEP |
SD | P | Maternal | n | Mean LEP |
SD | P |
|---|---|---|---|---|---|---|---|---|---|
| Birtd weight group | 0.80 | Prepregnancy obesity | 0.03 | ||||||
| AGA | 281 | 24.1 | 6.0 | No | 394 | 23.6 | 5.9 | ||
| LGA | 140 | 23.9 | 5.8 | Yes | 135 | 24.9 | 6.2 | ||
| SGA | 114 | 23.7 | 6.3 | Gestational diabetes mellitus | 0.01 | ||||
| Sex | <0.0001 | No | 432 | 23.7 | 5.8 | ||||
| Female | 265 | 22.8 | 5.5 | Yes | 47 | 26.2 | 6.4 | ||
| Male | 270 | 25.1 | 6.3 | Ethnicity | 0.51 | ||||
| Genotype | 0.006 | Other | 138 | 24.3 | 6.5 | ||||
| Any G | 457 | 23.7 | 6.0 | White | 394 | 23.9 | 5.9 | ||
| A/A | 78 | 25.7 | 5.8 | Tobacco use during pregnancy | 0.51 | ||||
| No | 502 | 24.0 | 6.1 | ||||||
| Yes | 26 | 24.6 | 4.9 | ||||||
| Gestational weight gain | 0.46 | ||||||||
| Low | 101 | 24.2 | 6.7 | ||||||
| Adequate | 135 | 23.3 | 5.3 | ||||||
| High | 290 | 24.0 | 6.0 | ||||||
| Pregnancy hypertension | 0.76 | ||||||||
| No | 490 | 24.0 | 6.0 | ||||||
| Yes | 35 | 24.3 | 6.4 | ||||||
| History of type II diabetes | 0.40 | ||||||||
| No | 472 | 23.9 | 5.9 | ||||||
| Yes | 4 | 21.5 | 4.9 | ||||||
| History of type I diabetes | 0.95 | ||||||||
| No | 471 | 23.9 | 5.9 | ||||||
| Yes | 5 | 24.1 | 4.7 | ||||||
| r | P | ||||||||
| Maternal age | −0.05 | 0.26 | |||||||
SD, standard deviation; AGA, adequate for gestational age, LGA, large for gestational age, SGA, small for gestational age
Table 3.
Multivariable linear regression model† of placental LEP methylation predictors
| (n=473) | |||
|---|---|---|---|
| Estimate | Standard Error | P | |
| Prepregnancy obesity | |||
| No | Reference | ||
| Yes | 1.17 | 0.62 | 0.06 |
| Gestational diabetes mellitus | |||
| No | Reference | ||
| Yes | 1.89 | 0.92 | 0.04 |
| Maternal age | −0.07 | 0.05 | 0.15 |
| Infant sex | |||
| Female | Reference | ||
| Male | 2.28 | 0.53 | <0.0001 |
| Infant genotype (rs2167270) | |||
| G/G and G/A | Reference | ||
| A/A | 2.17 | 0.73 | 0.003 |
| Birth weight group | |||
| AGA | Reference | ||
| LGA | −0.70 | 0.61 | 0.25 |
| SGA | −0.25 | 0.70 | 0.73 |
Model is adjusted for all variables in the table
AGA, adequate for gestational age; LGA, large for gestational age; SGA, small for gestational age.
3.4 GDM mediates prepregnancy obesity effects on placental LEP methylation
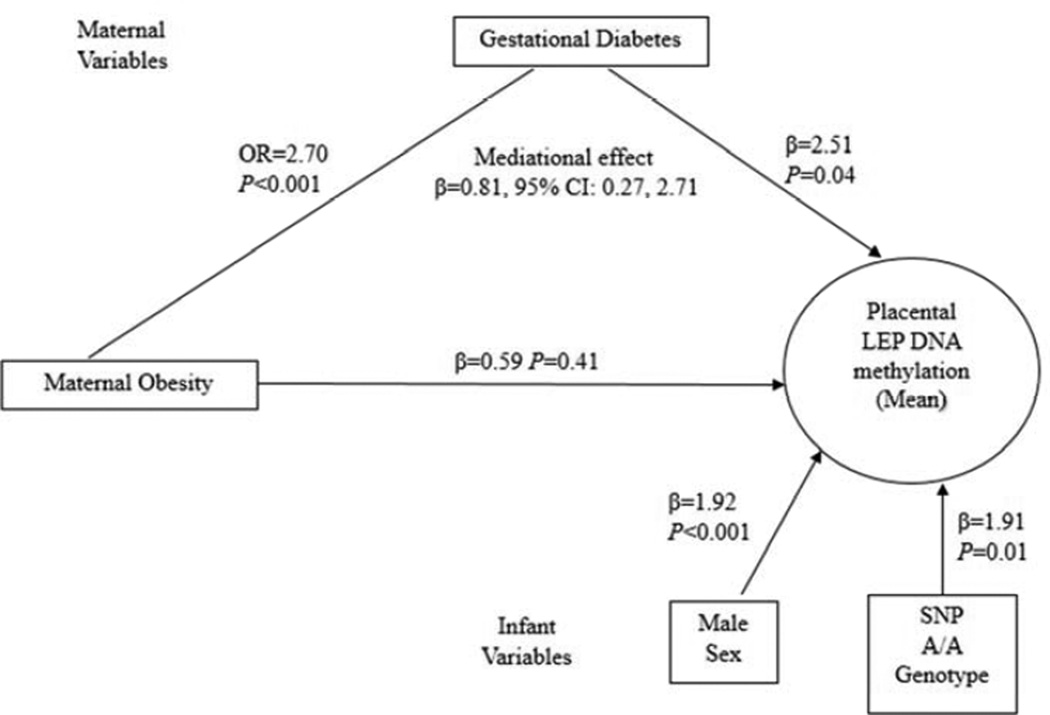
To build a formal path from maternal obesity to LEP methylation that accounts for the effects of GDM, we constructed a SEM (Figure 1) adjusted for the variables used in Table 3. This model has a good fit according to the fitness indices (e.g., Tucker Lewis Index=1). In this model, we found a significant association in the path from prepregnancy obesity to GDM (OR=2.7, P<0.001) and from GDM to increased placental LEP methylation (β=2.51, P=0.04), suggesting that placentas from infants born to a GDM mother have 2.5% higher methylation levels than those born to a non-GDM mother. Moreover, using a bootstrap method, we observed that GDM is a significant mediator between prepregnancy obesity and LEP methylation (β=0.81, 95% CI: 0.27, 2.71). In the SEM, the direct effect of prepregnancy obesity on LEP methylation is non-trivial (β=0.59, P=0.41) although is not significant, implying that GDM is likely a partial mediator. As expected infant sex and genotype also contributed independently to LEP methylation.
Figure 1.
Structural equation model relating maternal prepregnancy obesity to placental LEP DNA methylation through gestational diabetes in RICHS participants (n=473). Model is adjusted for variables in the figure, birth weight group and maternal age (continuous). Odds ratios (OR) are provided from logistic regression when the outcome (in this case gestational diabetes) was dichotomous, and the betas provided individual paths are standardized partial regression coefficients that can be interpreted similar to betas for linear regression.
4. DISCUSSION
This is the first study that has associated in utero exposure to preconception maternal obesity and GDM with placental LEP DNA methylation. Our findings suggest that methylation is higher in infants born to prepregnancy obese mothers and that this association is mediated by GDM. These findings are in line with other epidemiological studies that described associations between periconceptional parental obesity and cord blood DNA methylation patterns at imprinted24 and non-imprinted loci25, suggesting that metabolic exposures can influence offspring epigenetic signatures and possibly later-life disease risk.
The placenta is a substantial source of leptin.8 LEP mRNA is expressed on the maternal and fetal sides, while the leptin receptor (LEPR) is expressed predominantly on the maternal side, suggesting that placental and maternal serum leptin could regulate placental production of the hormone.8 In obese pregnancies, downregulation of LEPR mRNA in the syncytiotrophoblast without increased leptin protein levels26, suggests the existence of placental leptin resistance; analogous to hypothalamic resistance encountered in obesity.8 Furthermore, during pregnancy obese women exhibit overall higher serum leptin, but non-obese women display higher increases of leptin production per BMI unit27, suggesting differential placental leptin production. A recent study supports these findings, maternal obesity was associated with a lipotoxic placental environment characterized by widespread of changes in placental gene expression, including reduced LEP gene expression compared to placentas from non-obese controls.28 Hence, we can hypothesize that our observation of higher LEP promoter DNA methylation in placentas from obese pregnancies could result in lower placental leptin production due to resistance mechanisms in response to basal hyperleptinemia. This could serve as a placental adaptive response to control fetal growth in cases of positive energy balance such as prepregnancy obesity.8 However, we cannot directly investigate this hypothesis, and future research should address this issue.
We observed higher LEP methylation in GDM placentas compared to the non-GDM group. In contrast, a previous report9 did not find differences in placental LEP DNA methylation in infants from mothers with normal (n=25) compared to impaired glucose tolerance (IGT) (n=22). However, they observed a negative correlation between glucose levels (at 24–28 weeks) and LEP methylation on the fetal side of the placenta and, intriguingly, a positive correlation between glucose and maternal side LEP methylation. A more recent study29, measured LEP methylation in chorionic villus samples at birth from 100 newborns, and observed lower methylation in the GDM group (n=59), but these results did not withstand adjustment for confounders including BMI and infant sex. We have consistently shown10, 30 that sex is an important predictor of placental LEP methylation. Given the sexual dimorphism displayed by leptin8, the differential effect of LEP methylation on expression by sex10, and sexually dimorphic placental biology31, future research should take infant sex into account. With regards to placental LEP gene expression in GDM, the literature shows mixed results32; a number of studies report higher expression and others no differences in expression.26, 32 These differences might result from inter-study variability and low sample size. Interestingly, a study in tissue explants found significantly higher leptin release from placenta, amnion and choriodecidua obtained from normal pregnancies compared to GDM33, result consistent with our findings.
During normal gestation, placental leptin production induces a hyperleptinemic state compensated by hypothalamic leptin resistance, necessary to increase food intake.8 Additionally, around mid-gestation progressive insulin resistance occurs mediated in part by increased adiposity and placental hormones.34 In GDM, pregnancy-induced insulin resistance usually occurs over the chronic insulin resistance state frequently related to prepregnancy obesity.1 Moreover, maternal prepregnancy overweight status has been associated with overweight and abdominal obesity in offspring, with stronger associations when GDM is also present.35 Given the metabolic similarities within the endocrine milieu of obesity and GDM, it is possible that these states produce similar effects on placental LEP methylation. Our results support this hypothesis and given the temporality and comorbidity between these conditions, it is plausible that GDM acts as a partial mediator of the effects of maternal obesity on epigenetic control of placental LEP DNA methylation.
Previously, we observed an association between prepregnancy obesity and lower LEP DNA methylation in maternal blood, possibly reflecting hyperleptinemia and increased adiposity.30 Additionally, LEP methylation in maternal blood correlated to cord blood and consequently methylation was lower in cord blood of infants born to obese mothers. This study complements those findings and supports the known inter-tissue variability of DNA methylation.5 Interestingly, these findings show that maternal obesity can produce contrasting patterns of LEP promoter methylation between fetal tissues. As a key endocrine organ, placental methylation patterns could reflect adaptive responses to adverse metabolic intrauterine environment of prepregnancy obesity and GDM. Importantly, we have also demonstrated a link between increased placental LEP methylation and membership in a neurobehavioral profile characterized by lethargy and hypotonicity10, similar to the behavioral phenotypes of Lep deficient ob/ob mice. This association was only observed in male infants and could be partially explained by differential relation between LEP methylation and gene expression between sexes10. However, sex differences in offspring outcomes have been observed before in human and animal studies of developmental programming36 and could result from differential fetal and placental adaptations to the early-life environment. It remains to be determined whether placental LEP methylation could program obesity risk during childhood or later in life and help explain the relation between adverse nutritional in utero environments and offspring disease risk.37
Our study has several strengths, including a large sample of placentas from healthy infants and reliable measurements of LEP DNA methylation. However, this study is limited in our ability to define mechanisms behind these associations and we only studied one gene in two complex polygenic phenotypes. Also, in this study we used the average DNA methylation value across a region of 23 CpG sites. This could add a variance component that is not accounted in our analyses. Additionally, maternal BMI was derived from self-reported data that might lead to misclassification, although is likely under-reported and would bias our results to the null. GDM diagnoses were collected from medical chart records and we could not reliably obtain data on laboratory testing for this condition.
In summary, we established an association between prepregnancy obesity and placental LEP DNA methylation mediated by GDM in the largest study to date of healthy term infants. We confirmed previously observed associations between infant sex and genotype and placental LEP methylation10. These data suggest that placental epigenetic alteration of LEP may be one mechanism through which maternal phenotypes can program offspring health.
Supplementary Material
Acknowledgements
Thanks to Joyce Lee for her hard work in recruitment of subjects into this study at the Women and Infants Hospital's (RI). We also like to recognize the support of the staff of the Brown Center for the Study of Children at Risk for their efforts and the RICHS cohort participants for their collaboration.
This work was funded by the National Institute of Health (NIH) through the following grants: R01MH094609 (NIH-NIMH), R01ES022223 (NIH-NIEHS), P01 ES022832 (NIH-NIEHS), RD83544201 (US EPA), and P30CA23108 (NIH-NCI). Its contents are the responsibility of the authors and do not necessarily represent the official views of the funding institutions
Abbreviations
- LEP
leptin gene
- GDM
gestational diabetes mellitus
- AGA
adequate for gestational age
- LGA
large for gestational age
- SGA
small for gestational age
- GWG
gestational weight gain
- SEM
structural equation model
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Simmons D. Diabetes and obesity in pregnancy. Best Practice & Research Clinical Obstetrics & Gynaecology. 2011;25:25–36. doi: 10.1016/j.bpobgyn.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Gluckman PD, Hanson MA, Pinal C. The developmental origins of adult disease. Maternal & child nutrition. 2005;1:130–141. doi: 10.1111/j.1740-8709.2005.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zambrano E, Nathanielsz PW. Mechanisms by which maternal obesity programs offspring for obesity: evidence from animal studies. Nutrition reviews. 2013;71:S42–S54. doi: 10.1111/nure.12068. [DOI] [PubMed] [Google Scholar]
- 4.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 5.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 6.Ruchat S-M, Hivert M-F, Bouchard L. Epigenetic programming of obesity and diabetes by in utero exposure to gestational diabetes mellitus. Nutrition Reviews. 2013;71:S88–S94. doi: 10.1111/nure.12057. [DOI] [PubMed] [Google Scholar]
- 7.Burton GJ, Barker DJ, Moffett A, Thornburg K. The placenta and human developmental programming. Cambridge University Press; Number of pages. [Google Scholar]
- 8.Tessier D, Ferraro Z, Gruslin A. Role of leptin in pregnancy: Consequences of maternal obesity. Placenta. 2013;34:205–211. doi: 10.1016/j.placenta.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 9.Bouchard L, Thibault S, Guay SP, et al. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes care. 2010;33:2436–2441. doi: 10.2337/dc10-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesseur C, Armstrong DA, Murphy MA, et al. Sex-specific associations between placental leptin promoter DNA methylation and infant neurobehavior. Psychoneuroendocrinology. 2014;40:1–9. doi: 10.1016/j.psyneuen.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melzner I, Scott V, Dorsch K, et al. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. The Journal of biological chemistry. 2002;277:45420–45427. doi: 10.1074/jbc.M208511200. [DOI] [PubMed] [Google Scholar]
- 12.Noer A, Sorensen AL, Boquest AC, Collas P. Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Molecular biology of the cell. 2006;17:3543–3556. doi: 10.1091/mbc.E06-04-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchi M, Lisi S, Curcio M, et al. Human leptin tissue distribution, but not weight loss-dependent change in expression, is associated with methylation of its promoter. Epigenetics. 2011;6:1198–1206. doi: 10.4161/epi.6.10.16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vickers MH, Sloboda DM. Leptin as mediator of the effects of developmental programming. Best practice & research Clinical endocrinology & metabolism. 2012;26:677–687. doi: 10.1016/j.beem.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Seki Y, Williams L, Vuguin PM, Charron MJ. Minireview: Epigenetic programming of diabetes and obesity: animal models. Endocrinology. 2012;153:1031–1038. doi: 10.1210/en.2011-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jousse C, Parry L, Lambert-Langlais S, et al. Perinatal undernutrition affects the methylation and expression of the leptin gene in adults: implication for the understanding of metabolic syndrome. The FASEB Journal. 2011;25:3271–3278. doi: 10.1096/fj.11-181792. [DOI] [PubMed] [Google Scholar]
- 17.Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Human molecular genetics. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filiberto AC, Maccani MA, Koestler D, et al. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6:566–572. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenton T. A new growth chart for preterm babies: Babson and Benda's chart updated with recent data and a new format. BMC pediatrics. 2003;3:13. doi: 10.1186/1471-2431-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong DA, Lesseur C, Conradt E, Lester BM, Marsit CJ. Global and gene-specific DNA methylation across multiple tissues in early infancy: implications for children's health research. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014 doi: 10.1096/fj.13-238402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenton TR, Kim JH. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC pediatrics. 2013;13:59. doi: 10.1186/1471-2431-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasmussen KM, Abrams B, Bodnar LM, Butte NF, Catalano PM, Siega-Riz AM. Recommendations for weight gain during pregnancy in the context of the obesity epidemic. Obstetrics & Gynecology. 2010;116:1191–1195. doi: 10.1097/AOG.0b013e3181f60da7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrout PE, Bolger N. Mediation in experimental and nonexperimental studies: new procedures and recommendations. Psychological methods. 2002;7:422. [PubMed] [Google Scholar]
- 24.Soubry A, Murphy SK, Wang F, et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. International journal of obesity. 2013 doi: 10.1038/ijo.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gemma C, Sookoian S, Alvarinas J, et al. Maternal pregestational BMI is associated with methylation of the PPARGC1A promoter in newborns. Obesity. 2009;17:1032–1039. doi: 10.1038/oby.2008.605. [DOI] [PubMed] [Google Scholar]
- 26.Farley DM, Choi J, Dudley DJ, et al. Placental Amino Acid Transport and Placental Leptin Resistance in Pregnancies Complicated by Maternal Obesity. Placenta. 2010;31:718–724. doi: 10.1016/j.placenta.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Misra VK, Trudeau S. The influence of overweight and obesity on longitudinal trends in maternal serum leptin levels during pregnancy. Obesity. 2011;19:416–421. doi: 10.1038/oby.2010.172. [DOI] [PubMed] [Google Scholar]
- 28.Saben J, Lindsey F, Zhong Y, et al. Maternal obesity is associated with a lipotoxic placental environment. Placenta. 2014;35:171–177. doi: 10.1016/j.placenta.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Hajj N, Pliushch G, Schneider E, et al. Metabolic Programming of MEST DNA Methylation by Intrauterine Exposure to Gestational Diabetes Mellitus. Diabetes. 2013;62:1320–1328. doi: 10.2337/db12-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesseur C, Armstrong DA, Paquette AG, Koestler DC, Padbury JF, Marsit CJ. Tissue-specific Leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Molecular and cellular endocrinology. 2013;381:160–167. doi: 10.1016/j.mce.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clifton VL. Review: sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31:S33–S39. doi: 10.1016/j.placenta.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Kleiblova P, Dostalova I, Bartlova M, et al. Expression of adipokines and estrogen receptors in adipose tissue and placenta of patients with gestational diabetes mellitus. Molecular and cellular endocrinology. 2010;314:150–156. doi: 10.1016/j.mce.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Lappas M, Permezel M, Rice GE. Leptin and adiponectin stimulate the release of proinflammatory cytokines and prostaglandins from human placenta and maternal adipose tissue via nuclear factor-kappaB, peroxisomal proliferator-activated receptorgamma and extracellularly regulated kinase 1/2. Endocrinology. 2005;146:3334–3342. doi: 10.1210/en.2005-0406. [DOI] [PubMed] [Google Scholar]
- 34.Catalano PM. Obesity, insulin resistance, and pregnancy outcome. Reproduction. 2010;140:365–371. doi: 10.1530/REP-10-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pirkola J, Pouta A, Bloigu A, et al. Risks of overweight and abdominal obesity at age 16 years associated with prenatal exposures to maternal prepregnancy overweight and gestational diabetes mellitus. Diabetes care. 2010;33:1115–1121. doi: 10.2337/dc09-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aiken CE, Ozanne SE. Sex differences in developmental programming models. Reproduction. 2013;145:R1–R13. doi: 10.1530/REP-11-0489. [DOI] [PubMed] [Google Scholar]
- 37.Baptiste-Roberts K. Obesity During Pregnancy in Clinical Practice. Springer; 2014. Maternal Obesity and Implications for the Long-Term Health of the Offspring. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.