Abstract
Drugs that target DNA topoisomerase II isoforms and alkylate DNA represent two mechanistically distinct and clinically important classes of anticancer drugs. Guided by molecular modeling and docking a series of etoposide analog epipodophyllotoxin-N-mustard hybrid compounds were designed, synthesized and biologically characterized. These hybrids were designed to alkylate nucleophilic protein residues on topoisomerase II and thus produce inactive covalent adducts and to also alkylate DNA. The most potent hybrid had a mean GI50 in the NCI-60 cell screen 17-fold lower than etoposide. Using a variety of in vitro and cell-based assays all of the hybrids tested were shown to target topoisomerase II. A COMPARE analysis indicated that the hybrids had NCI 60-cell growth inhibition profiles matching both etoposide and the N-mustard compounds from which they were derived. These results supported the conclusion that the hybrids displayed characteristics that were consistent with having targeted both topoisomerase II and DNA.
Keywords: epipodophyllotoxin, topoisomerase II, etoposide, hybrid, structure-based design, anticancer, DNA, alkylator, COMPARE, K562 cells, molecular modeling, docking, nitrogen mustard
1. Introduction1
Hybrid drug molecules that contain two distinct pharmacophores and that can simultaneously act at two pharmacological targets have the potential to exceed the potency, spectrum of activity, and efficacy of either of the constituent moieties from which they were derived. Several recent reviews have described the various approaches taken in designing hybrid drugs.1–5 In the anticancer drug development area these have included such molecules as an epipodophyllotoxin-lexitropsin hybrids; enediyne-lexitropsin hybrids;5 an epipodophyllotoxin-amsacrine hybrid;6 various N-mustard- and cisplatin-steroid hybrids;4 epipodophyllotoxin-camptothecin hybrids;7 and chlorambucil-distamycin A hybrids.3
The X-ray structure of two molecules of etoposide complexed in a ternary complex with topoisomerase IIβ (PDB ID: 3QX3) has recently been determined.8,9 In this structure the glycosidic moiety of etoposide occupies a spacious binding pocket in the protein. The bound epipodophyllotoxin moiety of etoposide, unlike planar DNA intercalators that stack with two bases, stacks only with a single +5 guanine base in a DNA deformed structure.9 Etoposide is an effective anticancer agent10 and interfacial topoisomerase II inhibitor11,12 which stabilizes a covalent topoisomerase II-DNA cleavable complex intermediate10–14 resulting in single- and double-strand DNA breaks that are cytotoxic. Compounds in which the glycosidic moiety of etoposide is modified (e.g. teniposide) or even replaced retain excellent activity.15–18 In previous studies we described the synthesis and biological and topoisomerase IIα inhibitory activities of photoaffinity etoposide probes with a substitution on the glycosidic moiety18 or with the glycoside moiety replaced with a substituted phenyl group.17 Here, guided by molecular modeling and docking into the etoposide binding site in the X-ray structure of topoisomerase IIβ, we designed and synthesized a series of epipodophyllotoxin-N-mustard hybrid compounds. These hybrids were designed to contain a bifunctional N-mustard moiety that could alkylate protein residues on topoisomerase II or alkylate DNA bases when DNA and topoisomerase II are in a ternary complex with inhibitor. These N-mustard hybrids could also alkylate uncomplexed DNA.19 It was hypothesized that binding of an epipodophyllotoxin-N-mustard hybrid in the cleaved DNA-topoisomerase II complex would result in an N-mustard-derived covalent adduct being produced, and that this covalent adduct would irreversibly inactivate topoisomerase II. Hence, the hybrid molecules would have an advantage over etoposide which is bound in a thermodynamic equilibrium within an established complex between topoisomerase II and DNA and is free to dissociate. In addition, since these hybrid compounds may exert their antitumor effects by two independent mechanisms (targeting topoisomerase II and alkylating DNA), cancer cells should be much less able to develop resistance. Due to the known clinical utility of the N-mustards19 and of etoposide10, a series of epipodophyllotoxin-N-mustard hybrids based on chlorambucil, melphalan and bendamustine moieties were designed, synthesized and tested for their biological activity in vitro and in cell-based assays.
Several of the newly synthesized hybrids incorporated a piperazine linker to extend the chain and to improve pharmacological properties. The most potent of the series had a National Cancer Institute (http://dtp.nci.nih.gov) mean NCI-60 cell line screen GI50 that was 17-fold more potent than etoposide and was 40-fold more potent than the most potent N-mustard melphalan. Using a variety of assays, all of the hybrids showed strong evidence for targeting topoisomerase II. In addition, the hybrids showed evidence consistent with the ability to function as alkylating agents as expected.
2. Chemistry
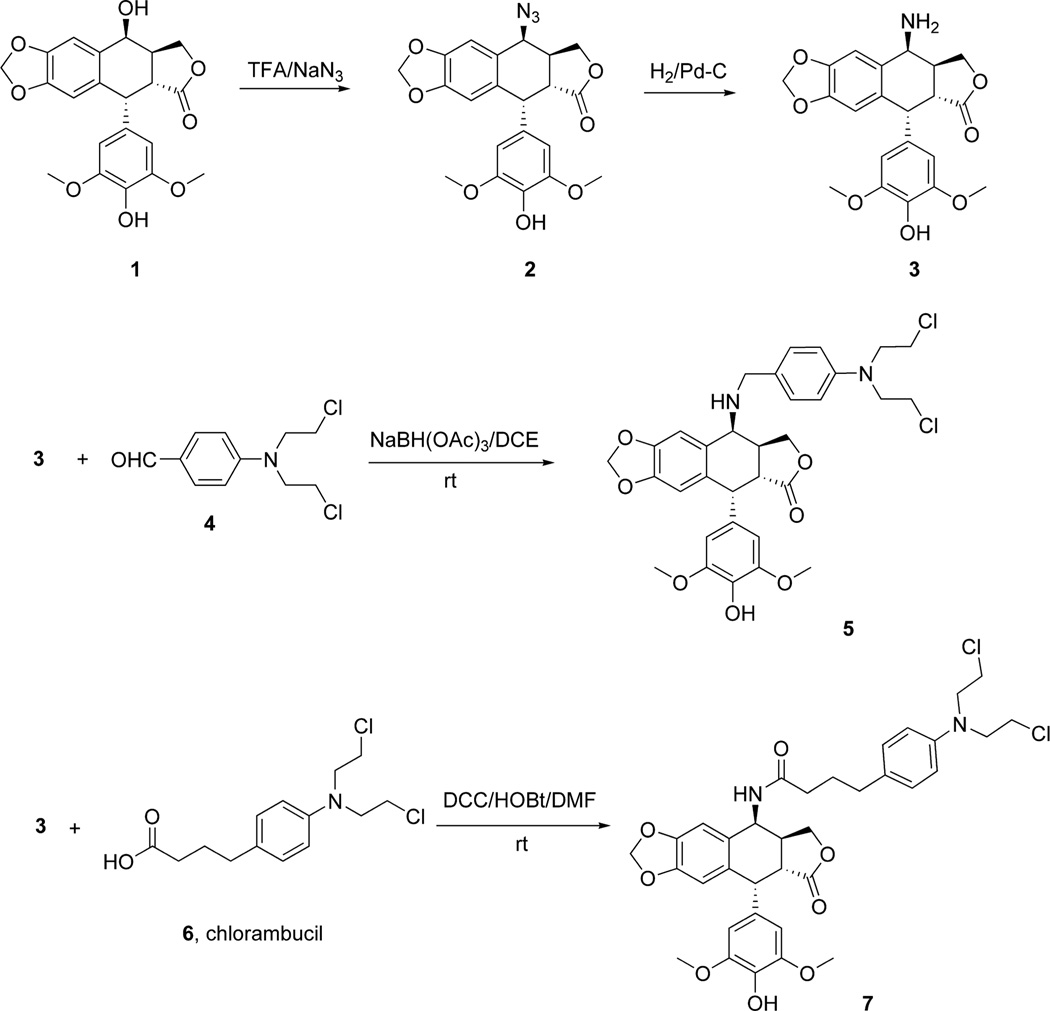
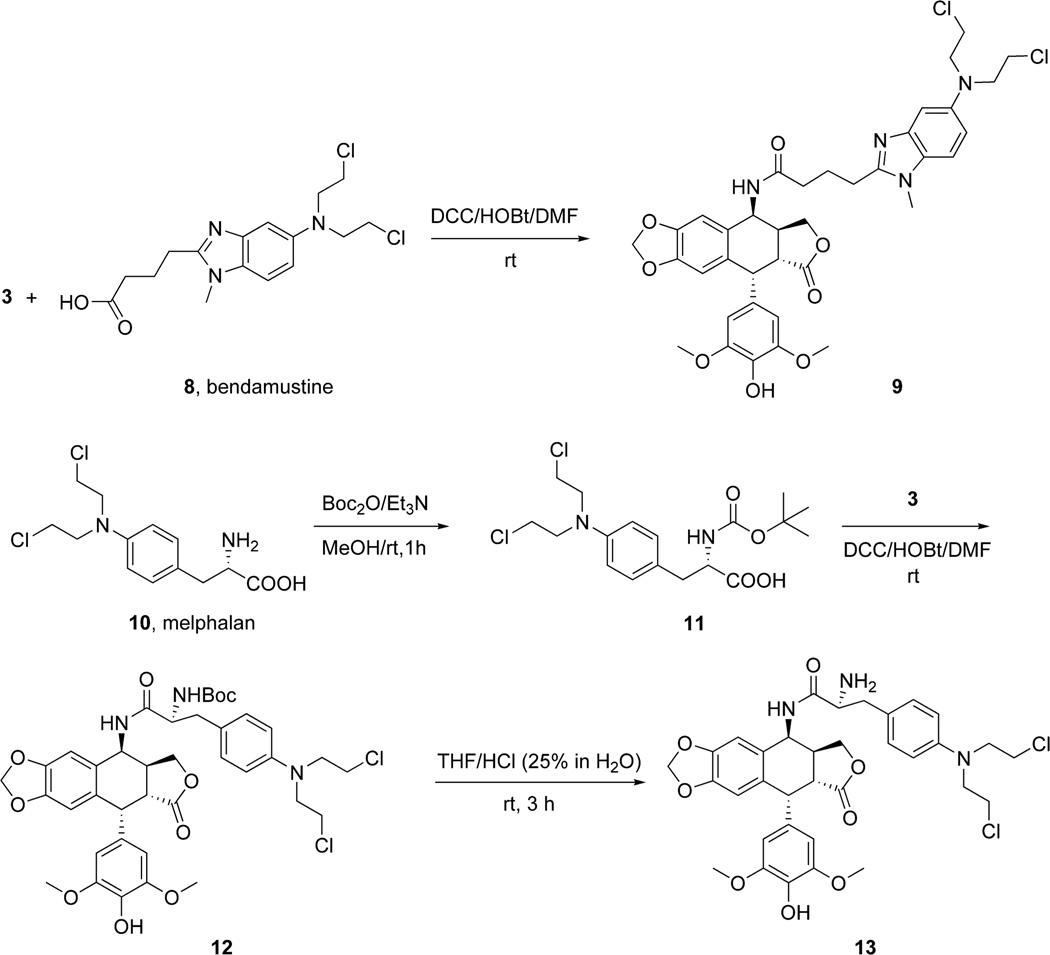
Two different strategies were used for the synthesis of the hybrid molecules containing both the epipodophyllotoxin and the N-mustard anticancer drug motifs. In the first approach the N-mustard compounds were directly linked to the epipodophyllotoxin as depicted in Schemes 1 and 2. In the second approach the N-mustard compounds were linked to the epipodophyllotoxin analog via a piperazine linker as shown in Schemes 3 and 4. In the first step (Scheme 1), the 4β-OH group in 1 (4'-O-demethylepipodophyllotoxin) (AvaChem, San Antonio, TX) was efficiently converted to the 4β-azide (compound 2) via nucleophilic substitution with NaN3 and TFA at room temperature as previously reported.20,21 The corresponding 4β-amino compound 3 ((10R,11R,15S,16S)-16-amino-10-(4-hydroxy-3,5-dimethoxyphenyl)-4,6,13-trioxatetracyclo[7.7. 0.03,7.011,15]hexadeca-1,3(7),8-trien-12-one) was prepared by reducing the azide 2 with catalytic hydrogenation.20,21 The advanced intermediate 3 was then subjected to a reductive amination reaction with 4-(bis(2-chloroethyl)amino)benzaldehyde 4 to furnish 5. Similarly the DCC coupling reactions of 3 with chlorambucil 6 and bendamustine 8 (OChem, Des Plaines, Il) gave compounds 7 and 9, respectively (Schemes 1 and 2). The epipodophyllotoxin-melphalan hybrid was synthesized in three steps. Melphalan 10 (OChem, Des Plaines, Il) was protected with the Boc group using Boc2O (di-tert-butyl dicarbonate) to give 11 ((2R)-3-{4-[bis(2-chloroethyl)amino]phenyl}-2-{[(tert-butoxy)carbonyl]amino}propanoic acid) which was prepared using a described procedure.22 Compound 11 on DCC coupling with 3 yielded 12 (Scheme 2). Finally deprotection with 25% (v/v) HCl gave the desired compound 13.
Scheme 1.
Synthesis of the epipodophyllotoxin advanced intermediate 3 and the N-mustard hybrid 5 and the chlorambucil-derived 7.
Scheme 2.
Synthesis of the epipodophyllotoxin-N-mustard bendamustine-derived hybrid 9 and the melphalan-derived 13.
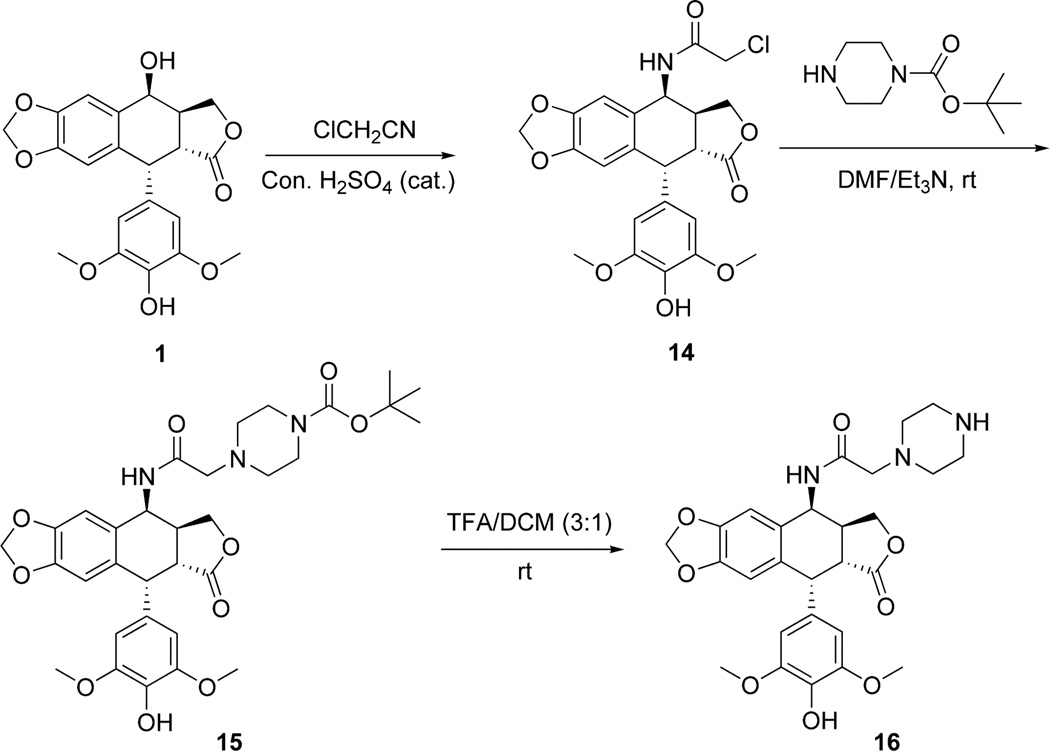
Scheme 3.
Synthesis of the epipodophyllotoxin piperazine-linked advanced intermediates 15 and 16.
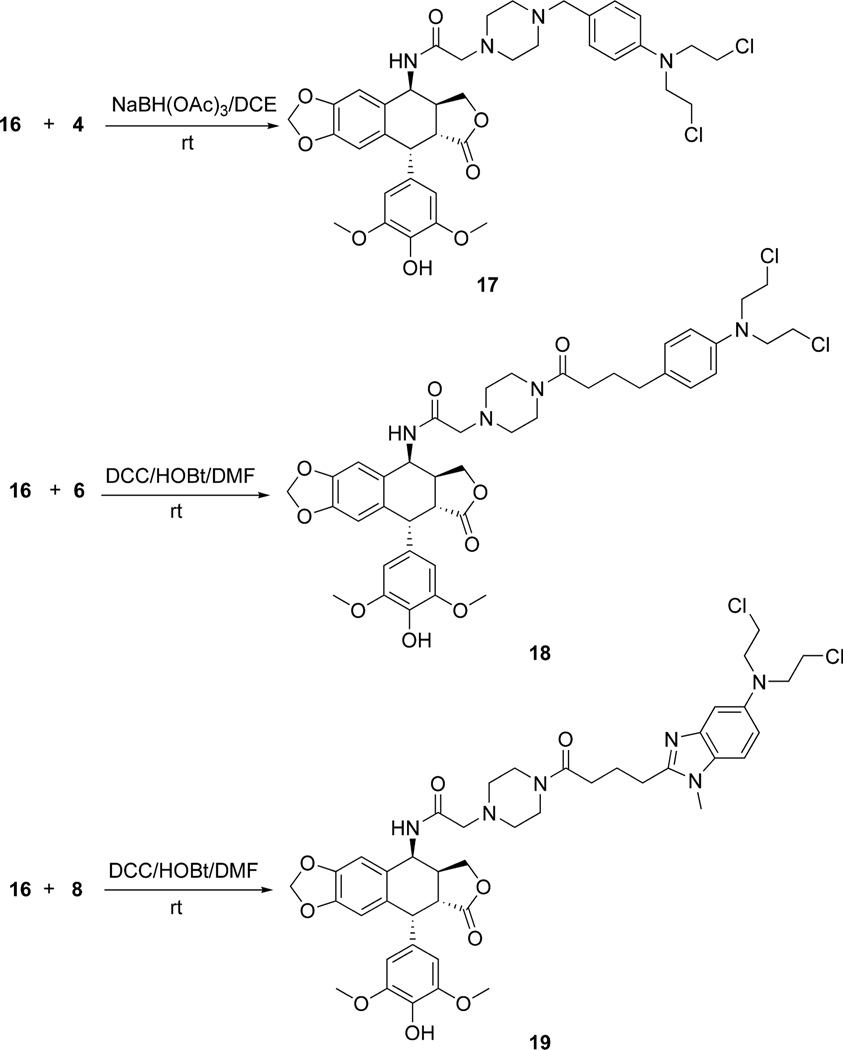
Scheme 4.
Synthesis of the epipodophyllotoxin-N-mustard piperazine-linked hybrids 17, 18 and 19
The Ritter reaction of chloroacetonitrile with 4'-O-demethylepipodophyllotoxin 1 was used23 to make the previously described24 4-chloroacetamido-4-deoxy-4'-demethylepipodophyllotoxin intermediate 14 in excellent yield (Scheme 3). In the next step 14 was subjected to nucleophilic substitution with 1-Boc-piperazine to give the intermediate 15, which on deprotection with TFA furnished the deprotected piperazine 16. The reductive amination reaction of 16 with 4 resulted in formation of 17 in good yield (Scheme 4). Similarly, DCC coupling of 16 with chlorambucil 6 and bendamustine 8 gave 18 and 19, respectively, in moderate to good yield (Scheme 4).
3. Results and discussion
3.1 Cell growth inhibitory effects of the hybrids and piperazine intermediates on human leukemia K562 cells and K/VP.5 cells with a decreased levels of topoisomerase IIα and topoisomerase IIβ
The cell growth inhibitory effects of the synthesized hybrids and the piperazine advanced intermediates on human leukemia K562 cells and K/VP.5 cells25,26 were compared to etoposide and the N-mustards chlorambucil, melphalan and bendamustine (Table 1). Most of the hybrids displayed sub-micromolar IC50 values comparable to that of etoposide. All of the hybrids were much more growth inhibitory than the three N-mustards tested. Cancer cells can acquire resistance to topoisomerase II poisons by lowering their level and/or activity of topoisomerase II.27 We have previously used28,29 a clonal K562 cell line selected for resistance to etoposide as a screen to determine the ability of compounds to act as topoisomerase II poisons. These K/VP.5 cells were determined to be 26-fold resistant to etoposide and to contain reduced levels of both topoisomerase IIα (~6-fold) and topoisomerase IIβ (~3-fold).25,26 In addition, these cells are cross-resistant to other known topoisomerase II poisons, but are not cross-resistant to camptothecin and other non-topoisomerase IIα targeted drugs.25 Decreased cellular topoisomerase II translates to fewer DNA strand breaks and reduced cytotoxicity. As shown in Table 1, while etoposide and the piperazine advanced intermediates 15 and 16 were highly cross-resistant (10- to 17-fold), the hybrids generally displayed low cross resistance (1.3- to 4.8-fold), or even collateral sensitivity in the case of 13. These results are consistent with most of the hybrids acting, at least in part, as topoisomerase II poisons in cells. However, because the cross-resistance of the hybrids is low compared to etoposide and the piperazine intermediates 15 and 16 that are essentially etoposide analogs, these results suggest that the hybrids are acting through an additional cytotoxic alkylation mechanism. In contrast, the N-mustards chlorambucil, melphalan and bendamustine all displayed collateral sensitivity towards the K/VP.5 cell line consistent with reports that DNA topoisomerase II contributes to enhanced alkylating agent adduct repair.30,31 Reduction of DNA topoisomerase II in K/VP.5 cells would result in compromised DNA repair and increased sensitivity.
Table 1.
Comparison of the cell growth inhibitory effects of the epipodophyllotoxin-N-mustard hybrids 5, 7, 9, 13, 17, 18, and 19, with the epipodophyllotoxin piperazine advanced intermediates 15 and 16 and some N-mustards and etoposide.
| Compound | K562 IC50 (µM) |
K/VP.5a IC50 (µM) |
RRb | mean NCI-60 cell GI50 (µM) |
|---|---|---|---|---|
| 5 | 0.46 | 0.77 | 1.7 | 0.98 |
| 7 | 0.72 | 1.8 | 2.5 | 1.3 |
| 9 | 2.0 | 2.5 | 1.3 | 4.2 |
| 13 | 1.5 | 0.66 | 0.4 | 6.9 |
| 15 | 0.21 | 3.4 | 16 | NDc |
| 16 | 0.83 | 8.4 | 10 | ND |
| 17 | 0.27 | 0.85 | 3.1 | 0.71 |
| 18 | 0.37 | 4.3 | 12 | ND |
| 19 | 5.4 | 26 | 4.8 | NDd |
| 6, Chlorambucil | 13 | 10 | 0.7 | 60 |
| 8, Bendamustine | >110 | 34 | < 0.3 |
70 |
| 10, Melphalan | 12 | 5.3 | 0.5 | 29 |
| Etoposide | 0.29 | 4.9 | 17 | 12 |
The etoposide resistant K/VP.5 cells that were derived from K562 cells have decreased levels of topoisomerase IIα and topoisomerase IIβ.25,26
RR is relative resistance (K/VP.5 IC50 ÷ K562 IC50).
ND is not determined.
1-dose NCI-60 data for this compound in given in Supplemental Figure 6.
3.2 Evaluation of cell growth inhibitory effects of the hybrids in the NCI-60 human tumor cell line screen
Compounds 5, 7, 9, 13, 17 and 19 were tested by the National Cancer Institute (http://dtp.nci.nih.gov) to identify which tumor cell types were most sensitive to these compounds and to be able to carry out NCI COMPARE analyses (http://dtp.nci.nih.gov/compare) to determine what other anticancer and cytotoxic drugs the hybrids shared tumor cell growth inhibition profiles with, and to identify putative mechanisms by which the hybrids exerted their activity (see below Figure 9 results and discussion of hierarchical cluster analysis). The NCI-60 cell line GI50 5-dose testing results for compounds 5, 7, 9, 13 and 17 are given in Table 1 and Supplemental Figures 1, 2, 3, 4 and 5, respectively (pages S5, S9, S13, S17 and S21, respectively). The 1-dose (10 µM) testing results for 19 are given in Supplemental Figure 6 (page S22). All of the hybrids tested had lower mean GI50 values compared to etoposide. It can also be seen from these results that the leukemia cell lines were as a group the most sensitive to the hybrids tested, with GI50 values generally in the sub-micromolar range. The hybrids also generally displayed very good growth inhibitory effects towards non-small cell lung cancer lines and renal cancer lines. Of the hybrids subjected to 5-dose testing, 17 was the most growth inhibitory with a mean GI50 over all cell lines tested of 0.71 µM, and 13 was the least growth inhibitory with an mean GI50 of 6.9 µM (Table 1). The hybrids were the least growth inhibitory towards colon and ovarian cancers with GI50 values in the low micromolar range. The most potent hybrid (compound 17) tested had a mean NCI-60 cell line GI50 value that was 17-fold more potent than etoposide and 41-fold more potent than the most potent N-mustard melphalan (Table 1).
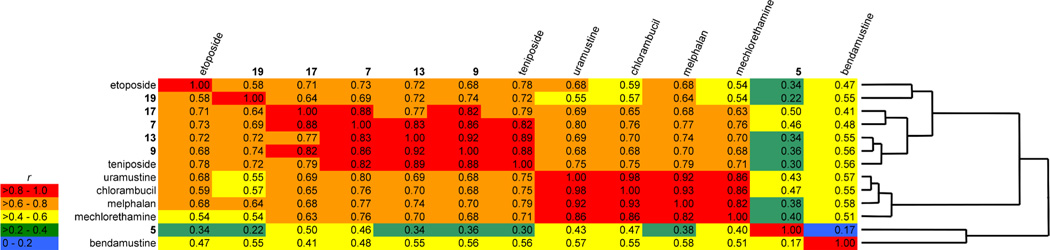
Figure 9.
Cross correlation and hierarchical cluster analysis obtained from a COMPARE analysis of NCI 60-cell log(GI50) data for the epipodophyllotoxin-N-mustard hybrid compounds, etoposide, teniposide and the N-mustard compounds bendamustine, uramustine, chlorambucil, melphalan and mechlorethamine. The values in the table are the Pearson correlation coefficients r. The individual cells are colored according to the range of r in which they fall.
3.3 The hybrids inhibited topoisomerase IIα decatenation activity, acted as a topoisomerase IIα poisons and induced DNA double-strand breaks in cells
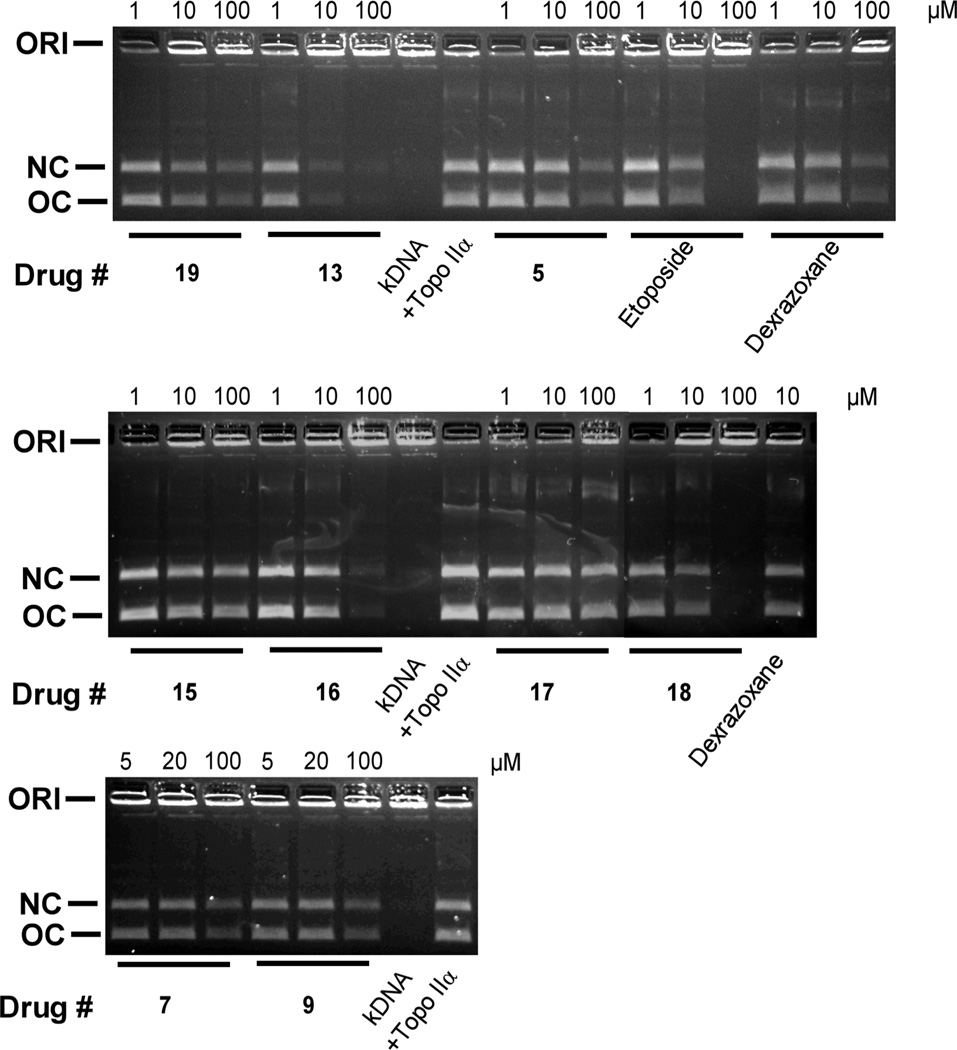
The torsional stress that occurs in DNA during replication and transcription can be relieved by topoisomerase II. In addition, normal chromosomal separation of daughter DNA strands during mitosis is facilitated by topoisomerase II. Topoisomerase II alters DNA topology by catalyzing the passing of an intact DNA double helix through a transient double-strand break made in a second helix.10,13,14 As shown in Figure 1, all of the hybrids inhibited the decatenation of concatenated kDNA by topoisomerase IIα. Dexrazoxane (ICRF-187), a known inhibitor of topoisomerase IIα decatenation,32 and etoposide were included as positive controls. The decatenation assay measures the ability of compounds to inhibit the catalytic activity only, and is not a measure of “interfacial” inhibitory or “poisoning” activity which can induce a stabilized topoisomerase II-DNA covalent complex.11,33 Based on the reduction in the amount of open circular (OC) DNA produced, compound 13, which inhibited the decatenation activity in the low micromolar range (compare 1 and 10 µM), was the most inhibitory, and even exceeded that of etoposide and dexrazoxane. The piperazine advanced intermediate 16 and the hybrid 18 were about as inhibitory as etoposide, while the rest of the hybrids were less inhibitory (compare at 10 and 100 µM).
Figure 1.
Effect of the hybrids, the piperazine advanced intermediates 15 and 16, and the etoposide and dexrazoxane positive controls on the inhibition of the topoisomerase IIα-mediated decatenation activity of kDNA. These fluorescent images of the ethidium bromide-containing gels show that in the absence of added drug (lanes marked +Topo IIα) topoisomerase IIα decatenated kDNA to its open circular (OC) and nicked circular (NC) forms. Topoisomerase IIα was present in the reaction mixture for all lanes but lanes marked kDNA. ORI is the gel origin. Concentration dependent inhibition of decatenation of topoisomerase IIα was shown for all agents tested. The results were typical of experiments carried out on 2 different days.
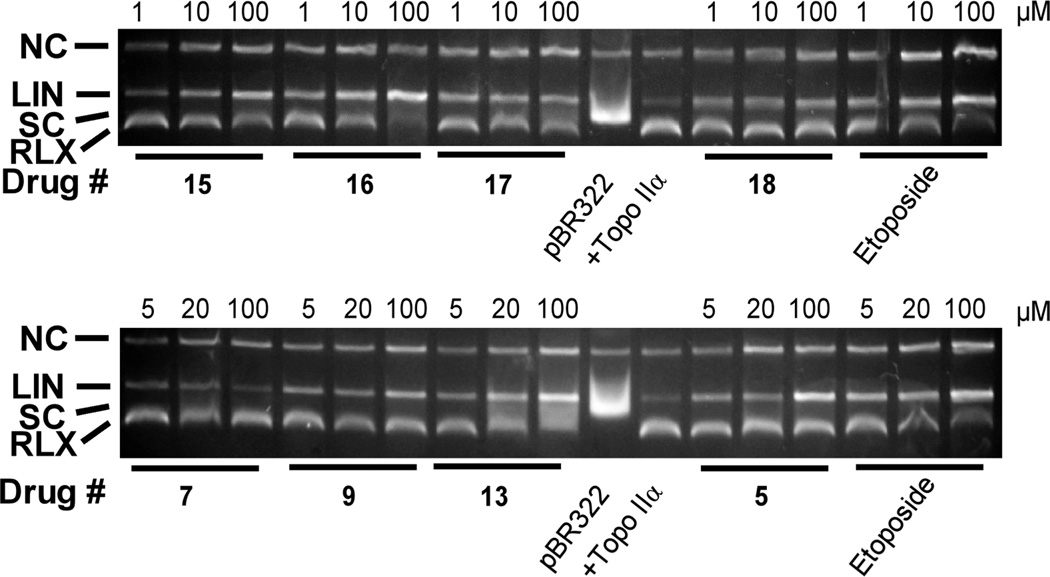
The topoisomerase II interfacial inhibitors/poisons are cytotoxic through their ability to stabilize a covalent topoisomerase II-DNA cleavable complex intermediate which can lead to frank double-strand DNA breaks.10,13,14 Thus, DNA cleavage assay experiments28,29,34 were carried out to see whether the hybrids stabilized the cleavable complex. Etoposide was used as a positive control. As shown in Figure 2, the addition of etoposide to the reaction mixture containing topoisomerase IIα and supercoiled pBR322 DNA induced formation of linear pBR322 DNA. Migration of linear DNA (LIN) was identified by treating closed circular pBR322 with the restriction enzyme HindIII to generate authentic linearized PBR322 (not shown). Figure 2 shows that all of the hybrids tested induced concentration dependent formation of linear DNA consistent with activity as topoisomerase IIα poisons. Based on the amount of linear DNA produced, hybrid 5 and the piperazine advanced intermediate 16 were more potent than etoposide, while the other hybrids exhibited similar or were less active compared to etoposide.
Figure 2.
Concentration dependent effect of the hybrids, the piperazine advanced intermediates 15 and 16, and etoposide on the topoisomerase IIα-mediated relaxation and cleavage of supercoiled pBR322 plasmid DNA to produce linear DNA. These fluorescent images of ethidium bromide-stained gels show that topoisomerase IIα (+Topo IIα) converted supercoiled (SC) pBR322 DNA to relaxed (RLX) DNA. In this assay the relaxed DNA runs slightly ahead of the supercoiled DNA because the gel was run in the presence of ethidium bromide. Topoisomerase IIα was present in the reaction mixture in all but the lanes marked pBR322. The etoposide positive controls produced significant amounts of linear DNA (LIN). A small amount of nicked circular (NC), which may arise from strand breakage during isolation, is normally present in pBR322 DNA. The results were typical of experiments carried out on 2 different days.
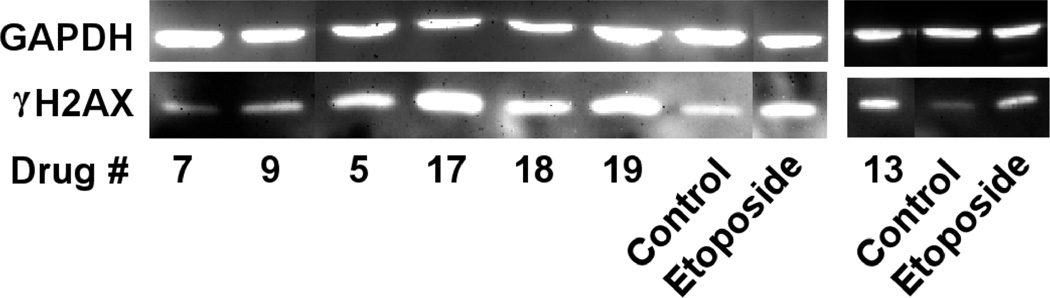
Phosphorylated H2AX (γH2AX), which is a variant of an H2A core histone, rapidly localizes at the site of double-strand DNA breaks upon treatment of cells with drugs or ionizing radiation.35 The thousands of γH2AX molecules that are localized at the site of DNA double-strand breaks are thought to amplify the DNA damage signal and are a widely accepted marker of double-strand breaks.35 Thus, in order to determine if the hybrids could induce double-strand breaks in intact K562 cells, the level of γH2AX protein was determined by Western blotting with etoposide as the positive control (all agents used at 20 µM).34 Experiments carried out as we previously described,29 and shown in Figure 3, indicate that the compounds 5, 17, 18, 19 and 13 increased levels of γH2AX in K562 cells while 7 and 9, whose topoisomerase cleavage activity was low (Figure 2), were relatively inactive. Compounds 17, 19 and 13 were even more potent than the etoposide positive control. These results suggest that the hybrids acted as topoisomerase II poisons in a cellular context to produce damaging DNA double-strand breaks.
Figure 3.
The hybrids 7, 9, 5, 17, 18, 19, 13 and the etoposide positive control induced double-strand DNA breaks in K562 cells as indicated by formation of γH2AX. K562 cells were treated with 20 µM of the drugs indicated for 4 h in growth medium, lysed and subjected to SDS-PAGE electrophoresis and Western blotting. The blots were probed with antibodies to γH2AX and with GAPDH (glyceraldehyde 3-phosphate dehydrogenase) as a loading control and a chemiluminescent-inducing horseradish peroxidase-conjugated secondary antibody. The results were typical of experiments carried out on 2 different days.
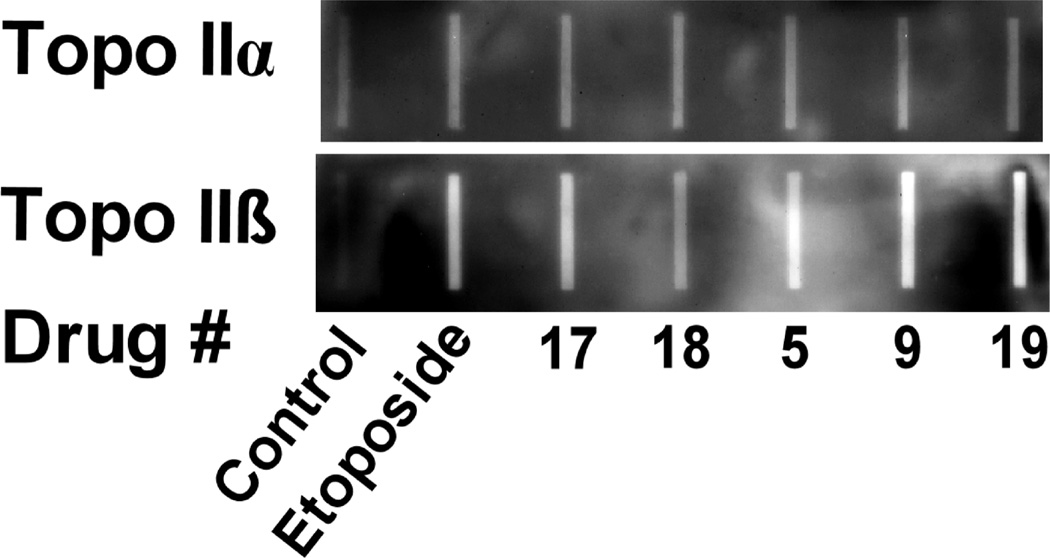
A cellular ICE (immunodetection of complexes of enzyme-to-DNA) assay was also used to determine if the hybrids could produce topoisomerase II-covalent complexes in K562 cells. Experiments were carried out as we previously described29 and are shown in Figure 4. At a concentration of 30 µM, all of the hybrids tested and the etoposide positive control increased the amount of topoisomerase IIα covalently bound to DNA compared to the untreated control in (Figure 4, upper image). The hybrids were about as potent as etoposide in producing topoisomerase IIα-covalent complexes. Likewise the hybrids and etoposide were about equipotent in producing increases in the amount of the topoisomerase IIβ-covalent complexes (Figure 4, lower image).
Figure 4.
Chemiluminescent images of a Western slot blot determination of cellular covalent topoisomerase IIα-DNA (upper image) and topoisomerase IIβ-DNA cleavage complexes (lower image) produced in K562 cells determined using an ICE (immunodetection of complexes of enzyme-to-DNA) assay. In these experiments K562 cells were either treated with DMSO vehicle or with 30 µM of the etoposide positive control or the drugs indicated for 1 h.
3.4 Comparison of the DNA cross-linking activity of the hybrids to that of chlorambucil and cisplatin
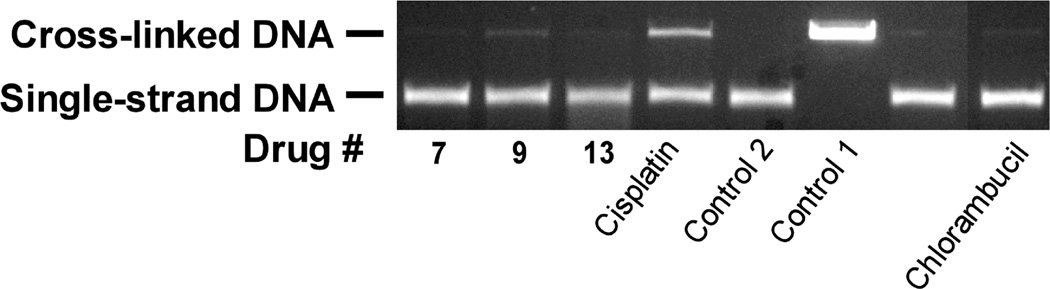
The hybrids contain an N-mustard moiety that is potentially capable of damaging DNA through the production of DNA cross links. Thus, a DNA cross-link assay was performed as we previously described36,37 using 10 µM of the agents indicated. Chlorambucil and cisplatin were used as positive controls (Figure 5). Of the hybrids tested only compound 9 produced a comparable amount of cross-linked DNA as did chlorambucil, and both of these agents produced much less cross-linked DNA compared to cisplatin. Detectable DNA cross-linking in the presence of 9 suggests that this mechanism may contribute, in part, to cellular growth inhibition and cytotoxicity.
Figure 5.
The DNA cross-linking effects of the hybrids, cisplatin and chlorambucil on linearized pBR322 DNA. This fluorescent image of the ethidium bromide-stained gel shows that the bifunctional alkylating agent chlorambucil control and cisplatin cross-linked DNA while the hybrids only slightly increased cross linking. All agents were tested at a concentration of 10 µM. Control 1 was not heated and thus consisted of only double stranded DNA. Control 2 and the other drug-treated samples were heated to 90 °C for 5 min to cause strand separation and then rapidly cooled. The strands that were cross-linked by drug treatment recombined to form double-strand DNA, whereas those that were not cross-linked, did not recombine. The amount of DNA in each well was the same. The band intensity of the single stranded DNA is less than that of double strand DNA because of reduced binding of ethidium bromide.
3.5 Cell cycle analysis and two-color flow cytometry
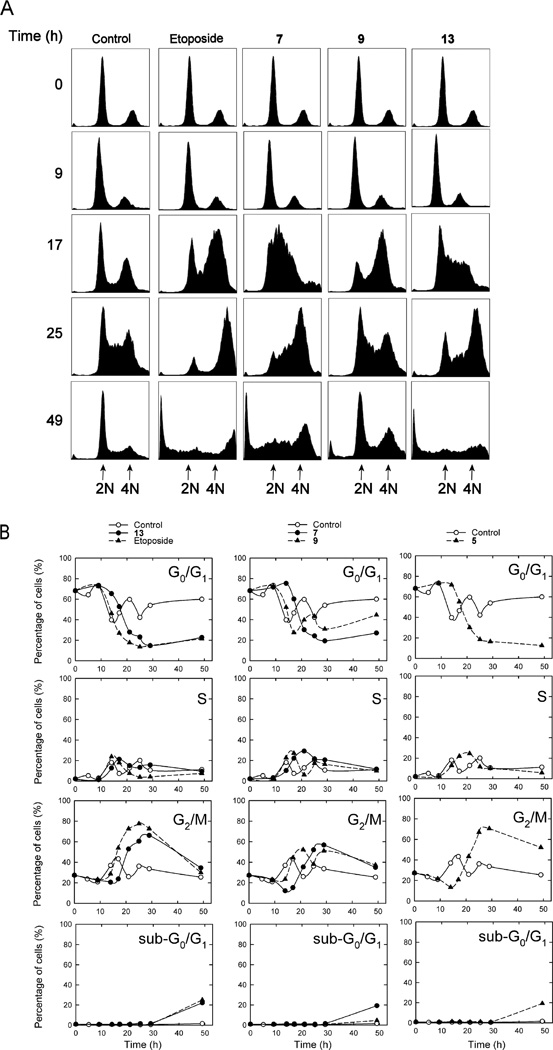
Cell cycle analysis was carried out on synchronized CHO cells as we previously described28,37 on the hybrids indicated (Figure 6) in order to determine the possible mechanism(s) by which these compounds might be acting. Drugs that act on topoisomerase IIα typically cause a block in G2/M, while N-mustard type DNA cross-linking drugs such as melphalan typically cause a cell cycle delay from G1.38 CHO cells (normal doubling time of 12 h) that were synchronized to G0/G1 through serum starvation were treated with 20 µM of the hybrid indicated, or etoposide (20 µM) for comparison, in order to determine whether these compounds induced a G2/M cell cycle block and/or a delay from G0/G1. CHO cells were chosen for these experiments as they are easily and effectively synchronized by serum starvation. Subsequent serum repletion resulted in the control (DMSO vehicle) cells advancing to G2/M by 17 h (Figure 6B). The control cells (Figure 6B, all panels) then went through several complete cell cycles as evidenced by the 12 h periodicity for peaks in the various cell cycle stages. Of the compounds tested 5, 7 and 13 displayed a significant percentage of sub-G0/G1 cells comparable to that observed for etoposide, which was indicative of apoptosis. The percentage of sub-G0/G1 cell was small in control- and 9-treated cells (< 2%). Treatment with etoposide caused a large block in G2/M by 25 h, which subsequently decreased at later times as the sub-G1 population, indicative of apoptosis, increased to about 25%. It can be seen from Figure 6B that the etoposide-treated and the control cells exited G0/G1 at almost identical rates as shown by the overlap of the their respective curves between 9 and 14 h. Importantly, compounds 5, 7 and 13 all displayed a delayed exit of about 5 h from G0/G1 compared to etoposide and the vehicle-treated control. This delayed progression from G0/G1 may be indicative of a mechanism typical of a DNA cross-linking agent such as melphalan.38 However, the large G2/M blocks observed with these compounds were also typical of a agent such as etoposide that targets topoisomerase II. Thus, these results are consistent with some of the hybrids acting both as DNA cross-linking agents and topoisomerase II poisons. However, it should also be noted that other agents that don't target topoisomerase II can also cause G2/M blocks.
Figure 6.
Cell cycle effects of hybrid treatment of synchronized CHO cells. CHO cells that had been synchronized in G0/G1 through serum starvation were repleted with serum and were treated with the DMSO vehicle or with 20 µM of the hybrid indicated directly after repletion and allowed to grow for the times indicated, after which they were subjected to cell cycle analysis of their propidium iodide-stained DNA. In (A) the cell counts are displayed on the vertical axis and the DNA content is plotted on the horizontal axis. In (B) the percentage of the cells in the sub-G0/G1, G0/G1, S and G2/M phases is plotted as a function of time for each of the hybrids indicated. The solid lines are a least-squares calculated spline fit to the data. As shown in the top plots a high percentage of the serum starved cells were initially present in G0/G1. After serum repletion the percentage of control cells in each phase varied periodically as the cells progressed through several cell cycles.
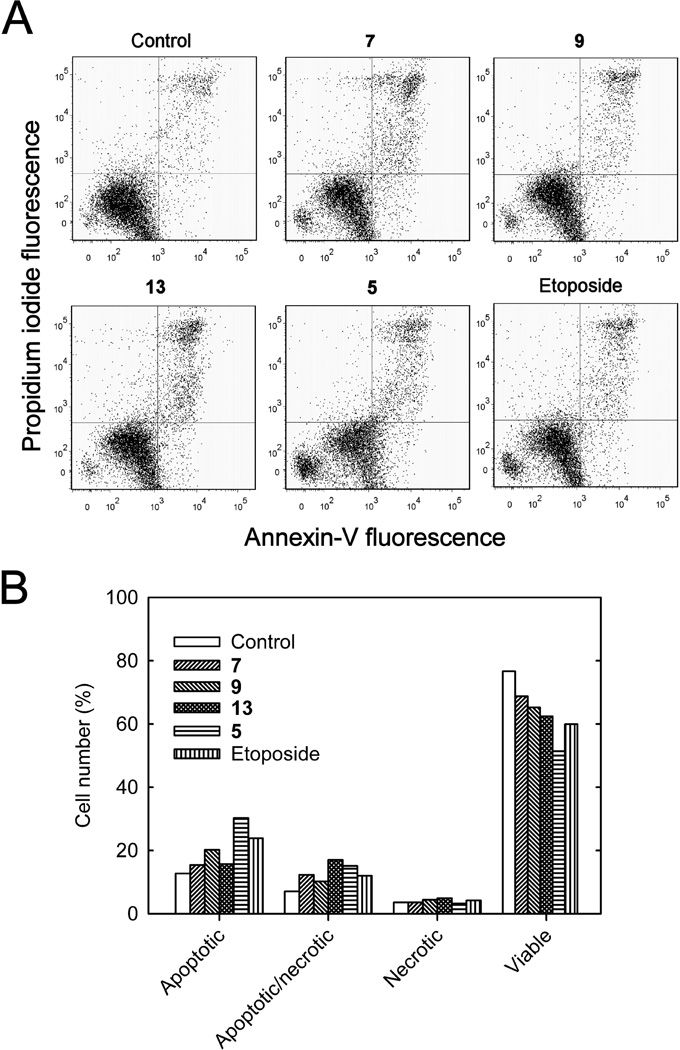
Early in the apoptotic process translocation of the phospholipid phosphatidylserine occurs from the inner to the external plasma membrane of cells. Fluorescent annexin V-FITC, which strongly binds phosphatidylserine on cells in this early stage of apoptosis,39 was used to identify apoptotic and apoptotic/necrotic cells by two-color flow cytometry as we previously described.37 Thus, annexin V-FITC stained cells in both the lower and upper right quadrants of Figure 7A were considered to be apoptotic. When used in combination with propidium iodide, which binds DNA, a measure of cells that are necrotic may be obtained since necrotic cell membranes are permeable to propidium iodide. Thus, cells in the lower right quadrant were apoptotic-only and those in the upper right quadrant were apoptotic/necrotic. As shown in Figures 7A and 7B, all treatments of K562 cells with hybrids 5, 7, 9, 13 or with etoposide (24 h, 10 µM) reduced the proportion of viable cells and increased the proportion of both apoptotic and apoptotic/necrotic cells.
Figure 7.
Treatment of K562 cells with the hybrids indicated or the etoposide positive control induced apoptosis as determined by annexin V-FITC/propidium iodide two-color fluorescence flow cytometry. (A) Two-color flow cytometry scatter plots of untreated control K562 cells, and 10 µM of the hybrid or the etoposide positive control treated for 24 h. Annexin V-FITC binds to the exterior surface of cells that are in the early stages of apoptosis and membrane-impermeant propidium iodide only binds the DNA of necrotic cells that have leaky membranes. Thus, the lower left quadrant contained viable cells; the upper left quadrant contained cells that were necrotic-only; the lower right quadrant contains cells that were apoptotic-only but were not necrotic; and the upper right quadrant contained cells that were both apoptotic and necrotic. (B) Changes in relative number of K562 cells that were classified as necrotic, apoptotic/necrotic, apoptotic or viable 24 h after no treatment, or after treatment with 10 µM of the hybrid or the etoposide positive control as indicated.
3.6 Docking of 7 into the etoposide binding site of etoposide complexed to cleaved DNA and topoisomerase IIβ
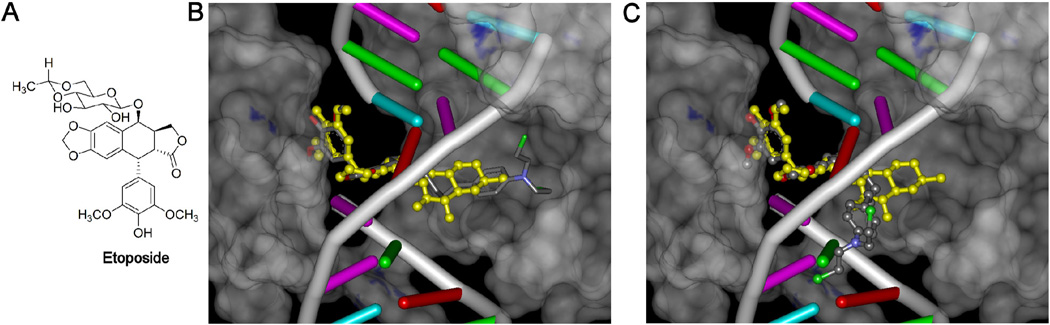
It is well known that the glycosidic moiety of etoposide (Figure 8A) can undergo substitution such as that found in teniposide16 or be substituted or replaced as in the two photoaffinity etoposide analogs we synthesized and biologically caracterized,17,18 and still retain good topoisomerase II-targeted activity. The glycosidic moiety may even be replaced with a variety of other moieties such as the polyamines found in F1451240 and in TOP-53 or a p-nitrophenyl moiety found in GL-311.16 It has been shown in the PDB ID: 3QX3 X-ray structure of two molecules of etoposide complexed to cleaved DNA and topoisomerase IIβ8,9 that the glycosidic moiety of etoposide occupies a spacious binding pocket in the protein with few protein interactions. This result explains why etoposide analogs are relatively insensitive to substituents that replace or substitute on the glycosidic moiety.
Figure 8.
Docking of the epipodophyllotoxin-chlorambucil hybrid 7 into the etoposide binding site of the PDB ID: 3QX3 X-ray structure of etoposide complexed to cleaved DNA and topoisomerase IIβ yields two equally high scoring poses. (A) Structure of etoposide. (B) Docking pose of hybrid 7 into the X-ray structure in which the N-mustard moiety extends into the spacious protein cavity is shown in a stick representation in atom colors. (C) Docking pose of hybrid 7 into the X-ray structure that more closely interacts with DNA is shown in a ball-and-stick representation in atom colors. The X-ray structure of etoposide bound to the 3QX3 structure is shown in a yellow ball-and-stick structure in both (B) and (C). For clarity DNA is shown in a ladder representation and the protein is shown as a gray semi-transparent surface. The poses shown in (B) and (C) are the nearly energetically equivalent highest scoring poses of 7 docked into the etoposide binding site of the PDB ID: 3QX3 X-ray structure. The H-atoms of the bound etoposide or the poses of 7 in (B) and (C) displayed are not shown for clarity. The etoposide was removed from the structure and 7 was docked into its place with the genetic algorithm docking program GOLD.
In order to determine if the hybrids synthesized could also fit into this spacious binding pocket, docking was carried out into the PDB ID: 3QX3 X-ray structure of etoposide complexed to cleaved DNA and topoisomerase IIβ in which etoposide had been removed. The results of the docking of 7 are shown in Figure 8B and 8C. The X-ray-determined pose of etoposide is shown in each panel in a yellow ball-and-stick representation. Docking of 7 resulted in two poses (Figure 8B and 8C) that gave nearly equal GOLD fitness scores. In one pose (shown in stick representation in atom colors Figure 8B) the N-mustard moiety is fully extended into the protein pocket normally occupied by the glycosidic moiety. In the other pose (shown ball-and-stick representation in atom colors in Figure 8C) the N-mustard moiety is in close proximity to the DNA. The polycyclic and pendant phenol moieties of both of these poses show a high degree of overlap with that of the X-ray structure of the bound etoposide (yellow ball-and-stick representation). The bifunctional N-mustard anticancer drugs such as chlorambucil, melphalan and bendamustine, which are capable of alkylating DNA bases or nucleophilic protein residues, act through a highly reactive aziridinium ion mechanism19 produced by loss of a chloride from one of the N-mustard arms. The two docking poses of 7 shown in Figure 8 indicate that alkylation of either nucleophilic amino acids on topoisomerase IIβ (Figure 8B) or on nucleophilic oxygen or nitrogen groups on DNA (Figure 8C) in the ternary complex could result in a covalent adduct with either protein or with DNA. In the case of alkylation of topoisomerase II, this adduct would be expected to result in irreversible inactivation of the enzyme. In contrast, the etoposide in the ternary etoposide-topoisomerase II-DNA complex is free to dissociate from the complex. If alkylation of DNA occurs it would be expected to result in permanent double- or single-strand DNA damage that would be difficult for the cell to repair.
There is a high degree of homology (~78%) in the catalytic domains of topoisomerase IIα and topoisomerase IIβ.41 While the X-ray structure of an etoposide-topoisomerase IIα-DNA complex has not yet been reported, the topoisomerase IIα-DNA complex with cleaved DNA has been reported.41 It was previously suggested8,9 that one of the two key glycosidic-interacting residues, Gln778, in the topoisomerase IIβ structure, which is replaced by methionine (Met762) in topoisomerase IIα, may be useful in developing isoform-specific anticancer drugs. An additional difference in the glycosidic binding pocket in the two topoisomerase II isoforms is the replacement of Ala816 in topoisomerase IIβ by the more nucleophilic Ser800 in topoisomerase IIα.9 If this Ser800 group in topoisomerase IIα were to form a covalent bond with the N-mustard hybrids, this would result in a desirable preferential selectivity of the hybrids for topoisomerase IIα over topoisomerase IIβ. It was shown in a recent study of 19 topoisomerase II inhibitors, which included etoposide and teniposide, that most had little or no preference for inhibition of topoisomerase IIα over topoisomerase IIβ.42 Additionally, it has been proposed that topoisomerase IIβ may be responsible for mediating chromosome translocations in therapy-related leukemias.43 Thus, a topoisomerase IIα specific agent may be less leukemogenic and hence more efficacious than the relatively non-selective topoisomerase II-targeted anticancer agents currently in use.
3.7 Cross correlation and hierarchical cluster analysis obtained from a COMPARE analysis of NCI 60-cell data
An NCI COMPARE analysis (http://dtp.nci.nih.gov/compare) of the NCI GI50 endpoint 60-cell line data was carried out on the epipodophyllotoxin-N-mustard hybrid compounds, and for comparison etoposide and teniposide, and the N-mustard compounds bendamustine, uramustine, chlorambucil, melphalan and mechlorethamine. These results were further subjected to hierarchical cluster analysis44 which identifies which groups of compounds had a similar tumor cell growth inhibition profile in the NCI 60-cell line data. COMPARE analysis on the NCI 60-cell data has been used to identify compounds that act through common mechanisms45 and is based on the assumption that compounds with a common mechanism of action will show a similar profile of log(GI50) values as identified by significant cross-correlation coefficients r. The results of the COMPARE analysis are shown in Figure 9. As might be expected all the N-mustards (except bendamustine) correlated very well with each other with r values in the 0.82 to 0.98 range and also clustered together very well. The second most highly clustered group consisted of the hybrid compounds 7, 9, 13, 17 and teniposide, all of which contain an epipodophyllotoxin moiety and had r values in the range of 0.77 to 0.92. The next most highly clustered group consisted of compound 19 and etoposide. These latter two results show that the hybrid compounds which contain an epipodophyllotoxin moiety all share an NCI GI50 endpoint 60-cell profile, and presumably a common mechanism of action, with the epipodophyllotoxins etoposide and teniposide. The hybrids and the N-mustards also clustered together. Thus, the data in Figure 9 was also examined to see how well the hybrid compounds correlated with the N-mustard compounds from which they were derived. Compound 7, which was derived from (Scheme 1) and contains the chlorambucil moiety, correlated well (r 0.76, p 10−11) and highly significantly with chlorambucil. Compound 13, which contains the melphalan moiety, correlated very well and highly significantly (r 0.74, p 10−10) with melphalan. Compound 9, which was derived from (Scheme 2) and contains the bendamustine moiety, correlated well and very significantly (r 0.56, p 7 × 10−6) with melphalan. What these latter set of correlations show is that in the NCI COMPARE analysis the hybrid compounds that contain an N-mustard moiety correlate well with the corresponding N-mustards from which they were derived, suggesting they may have a common mechanism of action. These results are consistent with the low cross-resistance to the etoposide-resistant K/VP.5 cell line (Table 1) and the delayed cell cycle progression results the hybrids displayed in Figure 6.
4. Conclusions
Etoposide and the N-mustards from which our hybrids are derived are thought to exert their anticancer activity by distinctively different mechanisms. Etoposide acts as a topoisomerase IIα interfacial inhibitor/poison10–13 that results in stabilization of cleaved DNA and frank double strand breaks that lead to cell death. The N-mustards are thought to act through the formation of cross-links with DNA that results in DNA damage that is difficult to repair and also leads to cell death. The hybrids were designed with the view that combining epipodophyllotoxin and N-mustard moieties in the same molecule would produce a potent anticancer drug that combined the anticancer activities of both of these classes of agent. Seven N-mustard-epipodophyllotoxin hybrid compounds (and the two piperazine advanced intermediates, 15 and 16) were synthesized and tested for cell growth inhibitory activity with K562 cells (Table 1). Six of the hybrids were tested in the NCI-60 cell line assay.
All of the hybrid compounds tested showed strong evidence that they targeted topoisomerase II. This was evidenced by their strong activities in the kDNA decatenation assay (Figure 1), the DNA cleavage assay (Figure 2) and the cellular ICE assay (Figure 4). Additionally, the results of the cellular γH2AX assays (Figure 3) and the cell cycle analyses which displayed G2/M blocks were also characteristic of topoisomerase II-targeted drugs. A DNA cross-link assay showed that 9 had N-mustard type activity as it formed DNA cross-links (Figure 5) about equal to that of chlorambucil. Also, the hybrids caused a delayed progression from synchronized CHO cells compared to that observed for etoposide (Figure 6) consistent with the hybrids possessing native N-mustard type activity. The hybrids tested (Figure 7) were comparable to etoposide in inducing apoptosis in K562 cells.
The COMPARE analysis showed that the hybrid compounds clustered well (Figure 9) with teniposide and etoposide with which they share the epipodophyllotoxin moiety. The NCI 60-cell growth inhibition profiles of the hybrids were also highly significantly correlated with the N-mustard compounds from which they were derived. These and our other results strongly suggested that the hybrids inhibited tumor cell growth both by inhibiting topoisomerase II and by alkylating DNA.
The molecular modeling and docking results of 7 (Figure 8) into the etoposide binding site of the PDB ID: 3QX3 X-ray structure of etoposide complexed to cleaved DNA and topoisomerase IIβ showed two nearly energetically equivalent poses (Figure 8B and 8C). In Figure 8B the N-mustard moiety of 7 was directed into the protein cavity. In Figure 8C an energetically equivalent pose 7 was found positioned such that it could possibly interact with the DNA. Alkylation of nucleophilic protein residues on topoisomerase II by the hybrids would be expected to permanently inactivate the enzyme and result in cell kill. Likewise, alkylation of nucleophilic oxygen or nitrogen bases on DNA would likewise be expected to result in cell kill. Given that these hybrids have two moieties through which they can independently, and in tandem, exert their anticancer activity, they would be expected to have a broader range of therapeutic activities than either one of their constituent moieties. Because the hybrids can act through two independent cell killing mechanisms, emergence of resistance should be delayed/reduced. In conclusion, a series of hybrid epipodophyllotoxin N-mustard analogs have been designed, synthesized and biologically characterized that were more potent than the prototype epipodophyllotoxin etoposide or their N-mustard analogs. Subsequent studies on the hybrids will focus on the molecular mechanisms by which the hybrids are cytotoxic, their selectivity for targeting topoisomerase IIα over topoisomerase IIβ, their pharmacokinetics and pharmacodynamics, and the characterization of the putative covalent adducts with topoisomerase II and DNA.
5. Experimental
5.1 Chemistry
1H and 13C NMR spectra were recorded on a Bruker Avance NMR spectrometer operating at 300 MHz or 500 MHz, as indicated, for 1H NMR and 75 MHz (or 125 MHz as indicated) for 13C NMR, respectively, in CDCl3 or DMSO-d6. The chemical shifts are expressed as 5 units with Si(CH3)4 as the internal standard. Melting points were uncorrected. The high resolution mass spectra were run on a Bruker microTOF Focus mass spectrometer (Fremont, CA) using electron spray ionization. TLC was performed on plastic-backed plates bearing 200 µm (thickness) silica gel 60 F254 (Silicycle, Quebec City, Canada). Where applicable compounds were visualized by quenching of fluorescence by UV light (254 nm). Unless otherwise specified chemicals were from Aldrich (Oakville, Canada) and were used without further purification.
The following general procedure for DCC coupling was used. To a solution of the appropriate amine (1 mmol) in anhydrous DMF under an inert atmosphere the appropriate acid (1 mmol), DCC (1 mmol) and HOBt (1-hydroxybenzotriazole) (1 mmol) were added. The reaction mixture was stirred at room temperature overnight. The medium was then taken up with CH2Cl2 and water. The organic phase was extracted with water (3 × 50 mL), then dried (MgSO4) and filtered. This product was further purified by silica gel chromatography using CH2Cl2/methanol as the eluant to give the pure compounds in good to moderate yields.
5.1.1 (10R,11R,15S,16S)-16-[({4-[bis(2-chloroethyl)amino]phenyl}methyl)amino]-10-(4-hydroxy-3,5-dimethoxyphenyl)-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-12-one(5)
3 (0.1 g, 0.25 mmol) and the benzaldehyde 4 (0.062 g, 0.25 mmol) were mixed in DCE (5 mL) and then treated with sodium triacetoxyborohydride (0.064 g, 0.3 mmol). The mixture was stirred at room temperature under a N2 atmosphere for 3 h. After completion of the reaction, the mixture was quenched by adding aqueous saturated NaHCO3, and the product was extracted with EtOAc. The EtOAc extract was dried (MgSO4) and the solvent was evaporated to give the crude free base 5 which was purified by column chromatography using hexane:EtOAc (80:20) as an eluant (0.1 g, 72%). mp: 168–172 °C (decomposed); 1H NMR (CDCl3, 300 MHz): δ 7.24–7.28 (m, 2H), 6.75 (m, 2H), 6.57 (s, 1H), 6.48 (s, 1H), 6.29 (s, 2H), 5.92 (s, 2H), 5.35 (bs, 1H), 4.53 (d, J = 6.5, 1H), 4.35 (m, 2H), 3.97 (m, 1H), 3.60–3.82 (m, 16H), 3.35 (m, 1H), 2.76 (m, 1H); 13C NMR (CDCl3, 75 MHz): δ 175.5, 147.6, 147.2, 146.3, 145.5, 133.9, 132.7, 131.6, 131.2, 129.8, 128.8, 112.2, 110.2, 108.3, 108.0, 101.3, 68.6, 56.4, 55.4, 54.0, 53.5, 43.6, 41.5, 40.4, 38.6; HRMS (ESI) for C32H35Cl2N2O7 m/z (M+H)+: calcd 629.1816, obsd 629.1800. The synthesis of 5 by a different method has been reported previously.46
5.1.2 4-{4-[bis(2-chloroethyl)amino]phenyl}-N-[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxy phenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]butana mide (7)
The DCC coupling reaction of 3 with chlorambucil 6 gave 7 (78%); mp: 138-142 °C (decomposed); 1H NMR (CDCl3, 300 MHz): δ 7.09 (d, J = 8.52 Hz, 2H), 6.74 (s, 1H), 6.74 (s, 1H), 6.67 (d, J = 8.58 Hz, 2H), 6.53 (s, 1H), 6.30 (s, 2H), 6.00 (d, J =2.61 Hz, 2H), 5.58 (d, J =7.05,1H), 5.43 (s, 1H), 5.23 (m, 1H), 4.59 (d, J = 4.8 Hz, 1H), 4.43 (m, 1H), 3.83-3.61 (m, 15H), 2.96 (m, 1H), 2.82 (dd, J = 6.12, 4.89 Hz, 1H), 2.60 (t, J = 7.38, 7.29, 2H), 2.25 (t, J = 7.62, 7.53, 2H), 1.97 (m, 2H); 13C NMR (CDCl3, 75 MHz): δ 174.4, 172.9, 148.4,147.6, 146.5, 134.1, 132.6, 130.2, 129.8, 128.9, 113.7, 110.1, 108.9, 107.8, 101.6, 69.1, 56.4, 54.3, 48.0, 43.5, 41.8, 39.8, 37.2, 35.6, 34.2, 27.2; HRMS (ESI) for C35H38Cl2N2NaO8 m/z (M+Na)+: calcd 707.1897, obsd 707.1890.
5.1.3 4-{5-[bis(2-chloroethyl)amino]-1-methyl-1H-1,3-benzodiazol-2-yl}-N-[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]butanamide (9)
Similarly the DCC coupling reaction of 3 with bendamustine 8 gave 9 (65%); mp: 265–268 °C (decomposed); 1H NMR (DMSO-d6, 300 MHz): δ 8.31 (d, J = 10.62 Hz, 1H), 8.24 (s, 1H), 7.34 (d, J = 8.73 Hz, 1H), 6.91 (d, J = 1.77 Hz, 1H), 6.81 (s, 1H), 6.77 (d, J = 2.1 Hz, 1H), 6.52 (s, 1H), 6.24 (s, 2H), 6.01 (d, J = 10.14 Hz, 2H), 5.20 (m, 1H), 4.49 (d, J = 4.95 Hz,1H), 4.29 (t, J = 7.62, 4.35 Hz, 1H), 3.58–3.82 (m, 18H), 3.18 (m, 1H), 2.95 (m, 1H), 2.85 (m, 2H), 2.27 (m, 2H), 2.02 (m, 2H); 13C NMR (CDCl3, 75 MHz): δ 174.6, 171.6, 151.8, 147.1, 146.8, 145.5, 134.4, 132.1, 130.1, 124.7, 113.2, 112.3, 109.3, 108.8, 108.2, 101.2, 94.9, 68.3, 55.9, 52.4, 46.6, 42.7, 41.1, 40.8, 38.0, 36.5, 24.3, 21.9; HRMS (ESI) for C37H41Cl2N4O8 m/z (M+H)+: calcd 739.2296, obsd 739.2317.
5.1.4 tert-butyl N-[(1R)-2-{4-[bis(2-chloroethyl)amino]phenyl}-1-{[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]carbamoyl}ethyl]carbamate (12)
The DCC coupling of 11 with 3 yielded 12 (66%); 1H NMR (CDCl3, 300 MHz): δ 7.14 (d, J = 8.46, 2H), 6.67 (d, J = 8.58, 2H), 6.49 (s, 1H), 6.31 (s, 1H), 6.28 (s, 1H), 6.19 (d, J = 7.32 Hz, 1H), 5.97 (d, J = 2.61 Hz, 2H), 5.44 (s, 1H), 5.08 (m, 1H), 4.98 (m, 1H), 4.53 (d, J = 4.8 Hz,1H), 4.36 (t, J = 8.6,7.5 Hz,1H), 4.18 (t, J = 8.4, 7.1 Hz, 1H), 3.82-3.63 (m, 14H), 3.06-2.85 (m, 3H), 2.80 (m, 1H), 1.42 (s, 9H). This intermediate was used in the next step without further purification.
5.1.5 (2R)-2-amino-3-{4-[bis(2-chloroethyl)amino]phenyl}-N-[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]propanamide (13)
12 (175 mg, 0.22 mmol) was dissolved in 3 mL of THF. To this solution 2.0 mL of hydrochloric acid (25 %, v/v) was added and the mixture was stirred for 20 h at room temperature. The solvent was removed under reduced pressure. The oil remaining was dissolved in methanol and the product was precipitated with diethyl ether, filtered and dried, to afford 123 mg (90%) of a colorless solid 13; mp: 282–285 °C; 1H NMR (DMSO-d6, 300 MHz): δ 8.45 (d, J = 9.8 Hz,1H), 8.33 (s, 3H), 7.10 (d, J = 8.43 Hz, 2H), 6.72 (d, J = 8.56, 2H), 6.50 (s, 1H), 6.31 (s, 1H), 6.28 (s, 1H), 6.19 (s, 2H), 5.98 (d, J = 2.60 Hz, 2H), 5.05 (m, 1H), 4.99 (m, 1H), 4.47 (d, J = 4.86 Hz,1H), 4.28 (t, J = 8.4,7.2 Hz, 1H), 4.18 (t, J = 8.2, 6.9 Hz, 1H), 3.82-3.63 (m, 14H), 3.09 (m, 1H), 3.02-2.89 (m, 3H); 13C NMR (DMSO-d6, 125 MHz): δ 174.3, 167.8, 147.1, 146.3, 145.5, 139.7, 134.6, 132.4, 130.5, 130.0, 129.0, 122.7, 111.4, 108.2, 55.8, 52.0, 47.3, 42.8, 40.0, 39.0, 36.6, 35.8; HRMS (ESI) for C34H38Cl2N3O8 m/z (M+H)+: calcd 686.2030, obsd 686.2038.
5.1.6 tert-butyl 4-({[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]carbamoyl}methyl)piperazine-1-carboxylate (15)
To a stirred solution of 14 (0.1 g, 0.21 mmol) and 1-Boc-piperazine (0.043 g, 0.23 mmol) in DMF (3 mL) triethylamine (0.042 g, 0.42 mmol) was added at room temperature. The reaction mixture was stirred overnight; it was then quenched with water and then extracted with CH2Cl2 (3 × 20 mL). The combined organic extract was washed with brine and dried over Na2SO4. Concentration under reduced pressure gave a crude product, which on column chromatography gave the title compound as a colorless solid (0.12 g, 92%); mp: 233 °C; 1H NMR (CDCl3, 300 MHz): δ 7.20 (d, J = 7.29 Hz, 1H), 6.69 (s, 1H), 6.56 (s, 1H), 6.32 (s, 2H), 6.01 (d, J = 1.2 Hz, 2H), 5.44 (s, 1H), 5.22 (m, 1H), 4.63 (d, J = 4.74 Hz,1H), 4.46 (t, J = 7.77, 8.79 Hz, 1H), 3.80 (m, 7H), 3.40 (m, 4H), 3.19–2.96 (m, 3H), 2.80 (dd, J = 4.89,4.95 Hz,1H), 2.49 (m, 4H), 1.47 (s, 9H); 13C NMR (CDCl3, 75 MHz): δ 174.2, 170.1, 154.6, 148.4, 147.7, 146.5, 134.2, 132.5, 130.1, 128.8, 110.3, 108.3, 107.9, 101.7, 80.1, 69.0, 61.3, 56.4, 53.3, 47.8, 43.6, 41.9, 37.2, 28.3; HRMS (ESI) C32H40N3O10 for m/z (M+H)+: calcd 626.2708, obsd 626.2682.
5.1.7 N-[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]-2-(piperazin-1-yl)acetamide (16)
15 (0.1 g, 0.16 mmol) was dissolved in CH2Cl2 (1 mL) to which TFA (3 mL) was added at room temperature. The reaction was stirred overnight and was then quenched with a saturated solution of NaHCO3. The usual work up procedure and purification by column chromatography gave the desired compound 16 (0.075 g, 90%); mp: 178–182 °C; 1H NMR (CDCl3, 300 MHz): δ 8.28 (bs, 1H), 8.02 (d, J = 8.22 Hz, 1H), 6.76 (s, 1H), 6.54 (s, 1H), 6.24 (s, 2H), 6.0 (s, 2H), 5.18 (m, 1H), 4.48 (d, J = 4.98 Hz, 1H), 4.32 (t, J = 7.98, 7.74 Hz, 1H), 3.72 (t, J = 10.44, 8.79 Hz, 1H), 3.63 (m, 6H), 3.30 (m, 5H), 3.09-2.86 (m, 3H), 2.65 (m, 4H); 13C NMR (CDCl3, 75 MHz): δ 174.3, 170.5, 148.4, 147.7, 146.6, 134.2, 132.5, 130.1, 128.9, 110.3, 108.4, 107.9, 101.6, 61.8, 56.4, 54.5, 47.7, 45.6, 43.6, 41.9, 37.2. HRMS (ESI) C27H32N3O8 for m/z (M+H)+: calcd 526.2184, obsd 526.2165. The synthesis of 16, isolated as the dihydrochloride salt, by a different method has been reported previously.47
5.1.8 2-[4-({4-[bis(2-chloroethyl)amino]phenyl}methyl)piperazin-1-yl]-N-[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7), 8-trien-10-yl]acetamide (17)
16 (0.1 g, 0.19 mmol) and the benzaldehyde 4 (0.056 g, 0.23 mmol) were mixed in 1,2-dichloroethane (5 mL) and then treated with sodium triacetoxyborohydride (0.081 g, 0.38 mmol). The mixture was stirred at room temperature under a N2 atmosphere for 3 h. After completion of reaction, it was quenched by adding aqueous saturated NaHCO3, and the product was extracted with EtOAc. The EtOAc extract was dried (MgSO4) and the solvent was evaporated to give the crude free base 17 which was purified by column chromatography using CH2Cl2:methanol (95:5) as eluant (0.115 g, 80%); mp: 155–158 °C; 1H NMR (CDCl3, 300 MHz): δ 7.18 (d, J = 8.31 Hz, 2H), 6.70 (s, 1H), 6.66 (d, J = 8.52 Hz, 2H), 6.56 (s, 1H), 6.32 (s, 2H), 6.02 (d, J = 2.25 Hz, 2H), 5.4 (bs, 1H), 5.21 (m, 1H), 4.67 (d, J = 5.7 Hz, 1H) 4.46 (t, J = 10.8, 8.88 Hz,1H), 3.83-3.58 (m, 15H), 3.40 (m, 2H), 3.10 (d, J = 6.3 Hz, 2H), 2.95 (m, 1H), 2.85 (dd, J = 4.77,3.33 Hz, 1H), 2.55 (m, 4H), 2.39 (m, 4H); 13C NMR (CDCl3, 75 MHz): δ 174.3, 170.6, 148.4, 147.6, 146.5, 145.3, 134.2, 132.5, 130.6, 130.1, 129.1, 111.8, 110.2, 108.6, 107.9, 101.6, 69.0, 62.0, 61.3, 56.4, 53.6, 52.5, 47.5, 43.6, 42.0, 40.4, 37.2; HRMS (ESI) C38H45Cl2N4O8 for m/z (M)+: calcd 755.2609, obsd 755.2589.
5.1.9 2-[4-(4-{4-[bis(2-chloroethyl)amino]phenyl}butanoyl)piperazin-1-yl]-N-[(10S,11S,15R,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]acetamide (18)
The DCC coupling of 16 with 6 yielded 18 (76%); mp: 159–163 °C (decomposed); 1H NMR (CDCl3, 300 MHz): δ 7.10 (d, J = 8.4 Hz, 2H), 6.70 (s, 1H), 6.68 (d, J = 8.58 Hz, 2H), 6.56 (s, 1H), 6.31 (s, 2H), 6.00 (s, 2H), 5.44 (bs, 1H), 5.24 (m, 1H), 4.64 (d, J = 4.8 Hz, 1H), 4.42 (m, 1H), 3.84-3.64 (m, 15H), 3.55-3.18 (m, 6H), 3.08-2.89 (m, 3H), 2.72-2.52 (m, 5H), 2.38 (m, 2H), 1.95 (m, 2H); 13C NMR (CDCl3, 75 MHz): δ 174.3, 171.4, 148.4, 147.7, 146.5, 144.4, 134.2, 132.8, 130.6, 129.6, 128.7, 112.2, 110.3, 108.4, 107.9, 101.6, 68.8, 56.5, 53.6, 47.9, 43.6, 42.0, 40.5, 37.1, 34.1, 32.3, 26.7; HRMS (ESI) C41H49Cl2N4O9 for m/z (M)+: calcd 811.2871, obsd 811.2857.
5.1.10 2-[4-(4-{5-[bis(2-chloroethyl)amino]-1-methyl-1H-1,3-benzodiazol-2-yl}butanoyl)piperazin-1-yl]-N-[(10S,11S,16R)-16-(4-hydroxy-3,5-dimethoxyphenyl)-14-oxo-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-10-yl]acetamide (19)
The DCC coupling of 16 with 8 yielded 19 (52%); mp: 198–202 °C (decomposed); 1H NMR (CDCl3, 300 MHz): δ 7.25 (d, J = 8.91 Hz, 1H), 7.13 (d, J = 7.26 Hz, 1H), 7.08 (d, J = 1.86 Hz, 1H), 6.85 (dd, J = 1.44, 1.35 Hz, 1H), 6.68 (s, 1H), 6.56 (s, 1H), 6.31 (s, 2H), 6.01 (d, J = 2.91 Hz, 2H), 5.21 (m, 1H), 4.63 (d, J = 4.59 Hz,1H), 4.43 (t, J = 7.83, 8.52 Hz, 1H), 3.83-3.40 (m, 22H), 3.20-2.94 (m, 5H), 2.86 (d, J = 4.8, 3.6 Hz, 1H), 2.55-2.40 (m, 6H), 2.25 (m, 2H); 13C NMR (CDCl3, 75 MHz): δ 174.2, 170.6, 169.9, 153.8, 148.5, 147.7, 146.5, 143.6, 134.2, 132.6, 130.1, 128.8, 128.2, 111.4, 110.4, 110.3, 108.3, 107.9, 101.7, 68.9, 61.2, 56.4, 54.5, 53.5, 47.8, 45.0, 43.6, 41.9, 41.2, 40.7, 37.2, 31.6, 30.1, 26.0, 22.4; HRMS (ESI) C43H51Cl2N6O9 for m/z (M)+: calcd 865.3089, obsd 865.3090.
5.2 Materials, cell culture and growth inhibition assays
Unless specified, reagents were obtained from Sigma-Aldrich (Oakville, Canada). The assay conditions and the expression, extraction and purification of recombinant full-length human topoisomerase IIα were described previously.48 Human leukemia K562 cells, obtained from the American Type Culture Collection and K/VP.5 cells (a 26-fold etoposide-resistant K562-derived sub-line with decreased levels of topoisomerase IIα and topoisomerase IIβ mRNA and protein)25,26 were maintained as suspension cultures in αMEM (Minimal Essential Medium Alpha) (Invitrogen, Burlington, Canada) containing 10% fetal calf serum. The spectrophotometric 96-well plate cell growth inhibition MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H–tetrazolium) CellTiter 96 AQueous One Solution Cell Proliferation assay (Promega, Madison, WI), which measures the ability of the cells to enzymatically reduce MTS after drug treatment, has been described.29,49 The drugs were dissolved in DMSO and the final concentration of DMSO did not exceed an amount (typically 0.4 % or less) that had any detectable effect on cell growth. The cells were incubated with the drugs for 72 h and then assayed with MTS. The IC50 values for cell growth inhibition were measured by fitting the absorbance-drug concentration data to a four-parameter logistic equation as described.28,29 Typically 9 different drug concentrations determined in duplicate and measured on two different days were used to characterize the growth inhibition curves. The catenated kDNA and the primary anti-topoisomerase IIα and topoisomerase IIβ antibodies were from TopoGEN (Port Orange, FL) and Santa Cruz (Dallas, TX), respectively, and the secondary horseradish peroxidase-conjugated antibody was from Cell Signaling Technology (Danvers, MA).
5.3 Topoisomerase IIα kDNA decatenation, pBR322 DNA relaxation and cleavage assays
A gel assay as previously described28,29 was used to determine if the hybrids inhibited the catalytic decatenation activity of topoisomerase IIα. kDNA, which consists of highly catenated networks of circular DNA, is decatenated by topoisomerase IIα in an ATP-dependent reaction to yield individual minicircles of DNA. Topoisomerase II-cleaved DNA covalent complexes produced by anticancer drugs may be trapped by rapidly denaturing the complexed enzyme with sodium dodecyl sulfate (SDS).28,34 The drug-induced cleavage of double-strand closed circular plasmid pBR322 DNA to form linear DNA at 37 °C was followed by separating the SDS-treated reaction products by ethidium bromide gel electrophoresis, essentially as described, except that all components of the assay mixture were assembled and mixed on ice prior to addition of the drug.28,29,34
5.4 γH2AX assay for DNA double-strand breaks in drug-treated K562 cells
The γH2AX assay was carried out essentially as described.29,37 K562 cells in growth medium (1.5 mL in a 24-well plate, 4 × 105 cells/mL) were incubated with drug or with DMSO as a control for 4 h at 37 °C. Cell lysates (60 µg protein) were subjected to SDS-polyacrylamide gel electrophoresis on a 14% gel. Separated proteins were transferred to polyvinylidene fluoride (PVDF) membranes and then treated overnight with rabbit anti-γH2AX primary antibody diluted 1:2000 (Upstate, Charlottesville, VA). This was followed by incubation for 1 h with peroxidase-conjugated goat-anti-rabbit secondary antibody (Cell Signaling Technology) diluted 1:2000. After incubation with luminol/enhancer/peroxide solution (Bio-Rad, Mississauga, Canada), chemiluminescence of the γH2AX band was imaged on a Cell Biosciences (Santa Clara, CA) FluorChem FC2 imaging system equipped with a charge-coupled-device camera.
5.5 Cellular assays for the detection of covalent DNA-topoisomerase IIα and DNA-topoisomerase IIβ protein complexes
The cellular ICE (immunodetection of complexes of enzyme-to-DNA), assays for topoisomerase IIα or topoisomerase IIβ covalently bound to DNA were carried out as described.28,29 The ICE assay used for the detection of covalent complexes of topoisomerase IIα and topoisomerase IIβ bound to DNA was a modification of the original cesium chloride ultracentrifugation gradient assay used to isolate DNA.50 The modification of this assay instead employed the selective precipitation of genomic DNA using DNAzol (Invitrogen).
5.6 DNA cross-link assay
The hybrids and the positive controls were tested for their ability to cross-link linearized pBR322 DNA as previously described.36,37 In this assay double-stranded linear DNA was heated which caused the strands to separate. Upon rapid cooling the strands that were cross-linked by drug treatment recombined to form double-stranded DNA, whereas those that were not cross-linked, did not recombine. Briefly, the pBR322 DNA (1.2 µg,) was linearized at a single restriction site with HindIII restriction enzyme (20 units, Invitrogen) in 20 µL of buffer (50 mM Tris/1 mM MgCl2/5 mM NaCl, pH 8.0) and purified by a Wizard SV Gel and PCR Clean-up System (Promega). The 10 µL reaction mixture, which contained linear DNA (70 ng), was incubated with 10 µM of the agents for 2 h at 37 °C. The bifunctional DNA cross-linking agents chlorambucil and cisplatin were used as a positive controls. The mixture was then heated at 90 °C for 5 min and was then immediately placed in an ice-water bath for 5 min. The double-stranded DNA was then separated by electrophoresis from single stranded DNA on an ethidium bromide agarose gel (1.1%, w/v). The DNA in the gel was imaged by its fluorescence on a Alpha Innotech Fluorochem 8900 imaging system.
5.7 Cell cycle synchronization, cell cycle analysis and annexin V flow cytometry
The cell cycle synchronization experiments were carried out as previously described.28,37 For the synchronization experiments CHO cells were grown to confluence in α-MEM supplemented with 10% fetal calf serum. Following serum starvation with α-MEM-0% fetal calf serum for 48 h, the cells were seeded at 2 x l05 cells/mL, repleted with α-MEM-10% fetal calf serum. Directly after repletion they were continuously treated with 20 µM of the hybrid in 35-mm diameter dishes for different periods of time. Cells were fixed in 75% ethanol and stained with a solution containing 20 µg/mL propidium iodide, 100 µg/mL RNase A in 0.1% (v/v) Triton X-100 at room temperature for 30 min. Flow cytometry was carried out on a BD FACSCanto II flow cytometry system (BD Biosciences, Mississauga ON, Canada) and analyzed with FlowJo software (Tree Star, Ashland OR) for the proportion of cells in sub-G0/G1, G0/G1, S, and G2/M phases of the cell cycle.
The fraction of apoptotic cells induced by treatment of K562 cells with the hybrids and the etoposide positive control were quantified by two-color flow cytometry by simultaneously measuring integrated green (Annexin V-FITC) fluorescence, and integrated red (propidium iodide) fluorescence as we previously described.37 The Annexin V-FITC binding to phosphatidylserine present on the outer cell membrane was determined using an Apoptosis Detection Kit (BD Biosciences, Mississauga, Canada). Briefly, K562 cells in suspension were untreated or treated with the compounds indicated at 37 °C for 24 h. The cells were collected by centrifugation at 1000 g for 3 min and the pooled cells were washed with the manufacturer-supplied binding buffer. Approximately 2.5 × 105 cells were resuspended in 500 µL of manufacturer-supplied binding buffer, and mixed with 5 µL of Annexin V-FITC and 5 µL of propidium iodide at a final concentration of 1 µg/mL. After 15 min of incubation in the dark, the cells were analyzed using flow cytometry.
5.8 Molecular modeling and docking of the hybrids into an X-ray structure of etoposide complexed to cleaved DNA and topoisomerase IIβ
The molecular modeling and the docking were carried out as described29 using the genetic algorithm docking program GOLD version 3.2 (CCDC Software, Cambridge, UK) with default GOLD parameters and atom types and with 50 starting runs as described.29 The hybrids were docked into one of the etoposide binding sites of the X-ray structure (www.rcsb.org/pdb; PDB ID: 3QX3)8 which contains two molecules of etoposide complexed to cleaved DNA and topoisomerase IIβ. The structure was prepared by removing the etoposide molecules, the water molecules and Mg2+ to avoid potential interference with the docking. The binding site (4 Å) was defined using etoposide in the PDB ID: 3QX3 structure. Etoposide docked back into its protein-DNA structure with a heavy atom rmsd distance of 0.48 Å, compared to the X-ray structure.8 Values of 2.0 Å or less in the extensive GOLD test set are considered to be good.51 The graphics were prepared with DS Visualizer 2.5 (Accelrys, San Diego, CA).
5.9 COMPARE and hierarchical clustering analysis of hybrid compounds
An NCI COMPARE analysis of the NCI GI50 endpoint 60-cell line data was carried out online (http://dtp.nci.nih.gov/compare) using both public data and private data (Supplemental Figures 1 – 6) for specific hybrid compounds tested by the NCI (data in Supporting Information) to obtain the Pearson correlation coefficients r. As a log(GI50) value was not determined for 19 percentage growth values from a single point (10 µM) NCI 60-cell result were used in the analysis. The p values were calculated in SigmaPlot (Systat Software, San Jose, CA). The hierarchical clustering analysis was carried out online (http://www.wessa.net/) using Ward's method.44
Supplementary Material
Acknowledgments
Supported by grants from the Canadian Institutes of Health Research, the Canada Research Chairs Program, and a Canada Research Chair in Drug Development to Brian Hasinoff and NIH grant CA090787 to Jack Yalowich. The authors declare no competing financial interest. The funding source(s) had no involvement in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations
Boc2O, di-tert-butyl dicarbonate; CHO, Chinese hamster ovary; Et3N, triethylamine; EtOAc, ethyl acetate; HOBt (1-hydroxybenzotriazole); ICE, immunodetection of complexes of enzyme-to-DNA; K562 cells, human erythroleukemic cells; kDNA, catenated DNA; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; NaBH(OAc)3, sodium triacetoxyborohydride; Pd-C, palladium on carbon; αMEM, Minimal Essential Medium Alpha.
Supplementary data
Supplementary data associated with article, which includes National Cancer Institute (http://dtp.nci.nih.gov) NCI 60-cell line panel testing results for compounds 5, 7, 9, 13, 17 and 19 and NMR data, can be found in the online version, at http://xx.
References and notes
- 1.Nepali K, Sharma S, Sharma M, Bedi PM, Dhar KL. Eur. J. Med. Chem. 2014;77:422–487. doi: 10.1016/j.ejmech.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Decker M. Curr. Med. Chem. 2011;18:1464–1475. doi: 10.2174/092986711795328355. [DOI] [PubMed] [Google Scholar]
- 3.Baraldi PG, Preti D, Fruttarolo F, Tabrizi MA, Romagnoli R. Bioorg. Med. Chem. 2007;15:17–35. doi: 10.1016/j.bmc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Fortin S, Berube G. Expert Opin. Drug Discov. 2013;8:1029–1047. doi: 10.1517/17460441.2013.798296. [DOI] [PubMed] [Google Scholar]
- 5.Tietze LF, Bell HP, Chandrasekhar S. Angew. Chem., Int. Ed. Engl. 2003;42:3996–4028. doi: 10.1002/anie.200200553. [DOI] [PubMed] [Google Scholar]
- 6.Arimondo P, Boukarim C, Bailly C, Dauzonne D, Monneret C. Anticancer Drug Des. 2000;15:413–421. [PubMed] [Google Scholar]
- 7.Denny WA, Baguley BC. Curr. Top. Med. Chem. 2003;3:339–353. doi: 10.2174/1568026033452555. [DOI] [PubMed] [Google Scholar]
- 8.Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 9.Wu CC, Li YC, Wang YR, Li TK, Chan NL. Nucleic Acids Res. 2013;41:10630–10640. doi: 10.1093/nar/gkt828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitiss JL. Nat. Rev. Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pommier Y, Marchand C. Nat. Rev. Drug Discov. 2012;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pommier Y. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pommier Y, Leo E, Zhang H, Marchand C. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deweese JE, Osheroff N. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordaliza M, Garcia PA, del Corral JM, Castro MA, Gomez-Zurita MA. Toxicon. 2004;44:441–459. doi: 10.1016/j.toxicon.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 16.You Y. Curr. Pharm. Des. 2005;11:1695–1717. doi: 10.2174/1381612053764724. [DOI] [PubMed] [Google Scholar]
- 17.Chee G-L, Yalowich JC, Bodner A, Wu X, Hasinoff BB. Bioorg. Med. Chem. 2010;18:830–838. doi: 10.1016/j.bmc.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasinoff BB, Chee G-L, Day BW, Avor KS, Barnabé N, Thampatty P, Yalowich JC. Bioorg. Med. Chem. 2001;9:1765–1771. doi: 10.1016/s0968-0896(01)00102-x. [DOI] [PubMed] [Google Scholar]
- 19.Kundu GC, Schullek JR, Wilson IB. Pharmacol., Biochem. Behav. 1994;49:621–624. doi: 10.1016/0091-3057(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 20.Zhou XM, Wang ZQ, Chang JY, Chen HX, Cheng YC, Lee KH. J. Med. Chem. 1991;34:3346–3350. doi: 10.1021/jm00116a001. [DOI] [PubMed] [Google Scholar]
- 21.Zhao M, Zhang Y, Yang Z, Cao B, Zheng Y, Chen H. Chin. J. Med. Chem. 2009;19:85–88. [Google Scholar]
- 22.Scutaru AM, Wenzel M, Gust R. Eur. J. Med. Chem. 2011;46:1604–1615. doi: 10.1016/j.ejmech.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Hansen HF, Jensen RB, Willumsen AM, Norskov-Lauritsen N, Ebbesen P, Nielsen PE, Buchardt O. Acta Chem. Scand. 1993;47:1190–2000. doi: 10.3891/acta.chem.scand.47-1190. [DOI] [PubMed] [Google Scholar]
- 24.Wang ZG, Yin SF, Ma WY, Li BS, Zhang CN. Acta Pharm. Sin. 1993;28:422–427. [PubMed] [Google Scholar]
- 25.Ritke MK, Roberts D, Allan WP, Raymond J, Bergoltz VV, Yalowich JC. Br. J. Cancer. 1994;69:687–697. doi: 10.1038/bjc.1994.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritke MK, Yalowich JC. Biochem. Pharmacol. 1993;46:2007–2020. doi: 10.1016/0006-2952(93)90643-b. [DOI] [PubMed] [Google Scholar]
- 27.Fattman C, Allan WP, Hasinoff BB, Yalowich JC. Biochem. Pharmacol. 1996;52:635–642. doi: 10.1016/0006-2952(96)00338-3. [DOI] [PubMed] [Google Scholar]
- 28.Yalowich JC, Wu X, Zhang R, Kanagasabai R, Hornbaker M, Hasinoff BB. Biochem. Pharmacol. 2012;84:52–58. doi: 10.1016/j.bcp.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasinoff BB, Wu X, Nitiss JL, Kanagasabai R, Yalowich JC. Biochem. Pharmacol. 2012;84:1617–1626. doi: 10.1016/j.bcp.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan KB, Mattern MR, Boyce RA, Schein PS. Biochem. Pharmacol. 1988;37:4411–4413. doi: 10.1016/0006-2952(88)90626-0. [DOI] [PubMed] [Google Scholar]
- 31.Tan KB, Mattern MR, Boyce RA, Schein PS. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7668–7671. doi: 10.1073/pnas.84.21.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasinoff BB, Kuschak TI, Yalowich JC, Creighton AM. Biochem. Pharmacol. 1995;50:953–958. doi: 10.1016/0006-2952(95)00218-o. [DOI] [PubMed] [Google Scholar]
- 33.Xiao Z, Loughlin F, George GN, Howlett GJ, Wedd AG. J. Am. Chem. Soc. 2004;126:3081–3090. doi: 10.1021/ja0390350. [DOI] [PubMed] [Google Scholar]
- 34.Burden DA, Froelich-Ammon SJ, Osheroff N. Methods Mol. Biol. 2001;95:283–289. doi: 10.1385/1-59259-057-8:283. [DOI] [PubMed] [Google Scholar]
- 35.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Biochem. Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 36.Hasinoff BB, Wu X, Yang Y. J. Inorg. Biochem. 2004;98:616–624. doi: 10.1016/j.jinorgbio.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Hasinoff BB, Wu X, Yalowich JC, Goodfellow V, Laufer RS, Adedayo O, Dmitrienko GI. Anticancer Drugs. 2006;17:825–837. doi: 10.1097/01.cad.0000224442.78211.27. [DOI] [PubMed] [Google Scholar]
- 38.Fernberg JO, Lewensohn R, Skog S. Eur. J. Haematol. 1991;47:161–167. doi: 10.1111/j.1600-0609.1991.tb01549.x. [DOI] [PubMed] [Google Scholar]
- 39.Kang YJ, Zhou ZX, Wang GW, Buridi A, Klein JB. J. Biol. Chem. 2000;275:13690–13698. doi: 10.1074/jbc.275.18.13690. [DOI] [PubMed] [Google Scholar]
- 40.Gentry AC, Pitts SL, Jablonsky MJ, Bailly C, Graves DE, Osheroff N. Biochemistry. 2011;50:3240–3249. doi: 10.1021/bi200094z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wendorff TJ, Schmidt BH, Heslop P, Austin CA, Berger JM. J. Mol. Biol. 2012;424:109–124. doi: 10.1016/j.jmb.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro AB, Austin CA. Anal. Biochem. 2014;448:23–29. doi: 10.1016/j.ab.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 43.Cowell IG, Sondka Z, Smith K, Lee KC, Manville CM, Sidorczuk-Lesthuruge M, Rance HA, Padget K, Jackson GH, Adachi N, Austin CA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8989–8994. doi: 10.1073/pnas.1204406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wessa P. [(accessed July 15, 2014)];Hierarchical Clustering (v1.0.3) in Free Statistics Software (v1.1.23-r7), Office for Research Development and Education. http://www.wessa.net/rwasp_hierarchicalclustering.wasp.
- 45.Mertins SD, Myers TG, Holbeck SL, Medina-Perez W, Wang E, Kohlhagen G, Pommier Y, Bates SE. Mol. Cancer Ther. 2004;3:849–860. [PubMed] [Google Scholar]
- 46.Wang Z, Ma W, Zhang C. 1993 CN1068330A. [Google Scholar]
- 47.Imbert T, Guminski Y, Barret JM, Kruczynski A. 2005 FR2869035A1. [Google Scholar]
- 48.Hasinoff BB, Wu X, Krokhin OV, Ens W, Standing KG, Nitiss JL, Sivaram T, Giorgianni A, Yang S, Jiang Y, Yalowich JC. Mol. Pharmacol. 2005;67:937–947. doi: 10.1124/mol.104.004416. [DOI] [PubMed] [Google Scholar]
- 49.Zhang R, Wu X, Guziec LJ, Guziec F, Jr, Chee G-L, Yalowich JC, Hasinoff BB. Bioorg. Med. Chem. 2010;18:3974–3984. doi: 10.1016/j.bmc.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian D, Furbee CS, Muller MT. Methods Mol. Biol. 2001;95:137–147. doi: 10.1385/1-59259-057-8:137. [DOI] [PubMed] [Google Scholar]
- 51.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.