Abstract
Advances in molecular biology and bioinformatics have resulted in the identification of a number of potential biomarkers that could be relevant in the management of patients with non-small cell lung cancer (NSCLC). Although there is an increasing amount of literature related to these biomarkers, major issues need to be resolved including validity and reproducibility of results. Additionally, in order to interpret the existing literature accurately a clear distinction must be made between the prognostic and predictive value of biomarkers. The practical applicability of biomarker discovery for patients with lung cancer includes the identification of patients with early-stage NSCLC who are most likely to benefit from adjuvant therapy. Information gleaned from biomarkers has the potential to help in evaluating the role of targeted therapies including immunotherapy in the neoadjuvant and adjuvant setting. The role of gene signatures and the use of newer platforms such as RNA, methylation and protein signatures is being explored in patients with early stage NSCLC. This review focuses on the applications of biomarker discovery in patients with early stage NSCLC
Introduction
Lung cancer is the leading cause of cancer related mortality in the United States and worldwide. Non-small-cell lung cancer (NSCLC) is the most common form of lung cancer. Early-stage NSCLC (ES-NSCLC; stages I and II) accounts for approximately 18% of cases.1 Most of these patients are treated with curative intent and often require multimodality therapy.2,3 Despite these aggressive measures, the survival associated with ES-NSCLC is less than optimal with 5-year overall survival (OS) ranging from 50% for stage IA disease to 15% for stage IIIA NSCLC.4
ES-NSCLC has assumed particular significance in recent years for two main reasons. Firstly, the incidence of ES-NSCLC is expected to rise due to the use of computed tomography screening of high-risk patients which has demonstrated a survival benefit.5 Secondly, the outcomes of ES-NSCLC may potentially benefit from an improved understanding of the molecular and immunologic basis of NSCLC which has already led to improved outcomes in advanced NSCLC.
Several clinical trials have demonstrated improved survival with post-operative chemotherapy in selected patients who undergo complete surgical resection.6,7 Available evidence supports the use of adjuvant chemotherapy for stage II and stage IIIA, but not for stage IA NSCLC.8
There are several shortfalls to the current approach of selecting patients for adjuvant therapy based on surgical stage alone. Given the marginal benefits and potential toxicities associated with chemotherapy, perhaps the greatest challenge lies in the identification of patients at greatest risk of recurrence. One approach to identifying high-risk patients focuses on the biology of ES-NSCLC in an effort to predict the risk of recurrence and potential for response to treatment by using biomarkers.
A biomarker is a “characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention”. A prognostic biomarker is a factor that is associated with an outcome that is independent of treatment, whereas a predictive biomarker interacts with treatment to influence outcome.9 There is good clinical evidence for a limited number of biomarkers that are used in clinical practice. Examples include the use of hormone receptor status in breast cancer. These biomarkers are prognostic of improved survival independent of cancer treatment and also predict the benefit of hormonal therapy with drugs such as tamoxifen.10 Despite a concerted effort, there is a lack of biomarkers with potential application in the management of ES-NSCLC.
The search for a prognostic and predictive biomarker has to take into consideration two key points: the strength of evidence to support its use and the depth of information provided by a biomarker that adds to what is already known about the disease based on clinical parameters. Although a plethora of potential prognostic biomarkers have been proposed in the past couple of decades, very few have been validated. In this review we have focused on a very small number of these biomarkers, including immune markers and molecular signatures relevant to ES-NSCLC because of the potential promise associated with them.
Prognostic biomarkers
P53
The tumor suppressor gene, p53 is frequently altered in NSCLC. 11 Although it is a well-established poor prognostic factor in many tumors12,13, in ES-NSCLC its prognostic role is controversial. A subgroup analysis of CALGB 9633, a phase III trial that randomized patients with stage IB NSCLC to observation or adjuvant chemotherapy, showed that p53 expression by immunohistochemistry (IHC) was detectable in 47% of tumors, and correlated with shorter disease-free survival (DFS) (Hazard ratio (HR) 1.95, P=0.003) and OS (HR 2.30, P=0.0005) in multivariate analyses.14 A meta-analysis of pooled patient data from 43 studies which included patients with ES-NSCLC who underwent potentially curative resection showed that p53 mutation or overexpression was an indicator of poor prognosis, especially in patients with adenocarcinoma (ADC). Compared to patients with no alterations, patients with ADC and p53 overexpression or mutations had a 21.8% (P=0.0000039) and 48% (P=0.000031) reduction in 5-year OS respectively.15
KRAS
RAS belongs to the family of small GTPase proteins. Rodenhuis et al first reported an association between KRAS mutations and NSCLC. They studied 39 NSCLC samples for the presence of NRAS, KRAS and HRAS mutations or amplifications and concluded that mutational KRAS activation may be an important early event in the pathogenesis of ADC of the lung.16 They also showed that KRAS mutations were present in more than 30% of ADCs and was more frequent in smokers.17 Studies in ES-NSCLC report that KRAS mutations, especially at codon 12, are associated with worse PFS and OS.18,19 Slebos et al. were the first to show that differences in PFS and OS in patients with ES-NSCLC with and without KRAS mutations were significant (P=0.038 and P=0.001).20 The prognostic significance of KRAS in NSCLC was evaluated in a combined analysis of 8 studies with a total of 881 patients. KRAS mutations were detected in 25% cases and involved codons 12, 13 and 61 of the KRAS gene. For the KRAS mutant group, the relative risk (RR) for mortality was 2.35 (95%CI 1.61–3.22), compared to patients with wild type KRAS. However, these studies were heterogeneous and there were no adjustments for other clinical variables.21 In recent studies, the relevance of different amino acid substitutions in KRAS has been analyzed. Pre-clinical and retrospective data point out the importance of specific KRAS mutations on the prognosis of NSCLC, such as G12C or G12V in contrast to other substitutions.22 Despite the data presented above, the prognostic significance of KRAS remains controversial. In an analysis involving 300 patients with ES-NSCLC with tumors harboring KRAS mutations enrolled in four adjuvant trials, the presence or absence of mutations in KRAS codon 12 did not confer a survival disadvantage in the observation arms of these trials.23
Prognostic and Predictive biomarkers
EGFR
The epidermal growth factor receptor (EGFR) is a member of the tyrosine kinase cell surface receptor family. Mutations in the gene encoding this protein results in constitutive activation and amplification of intracellular signals which lead to proliferation, invasion and migration of cancer cells.24 The overall implications of the presence of EGFR mutations or amplification in ES-NSLC are not well defined. Rusch et al. detected EGFR overexpression by IHC in 74 (71%) of 96 ES-NSCLC tumor samples. However, there was no association with OS.25 In a separate study 53 ES-NSCLC tumor samples (79% ADC) were analyzed for the presence of EGFR mutations in exon 19 and 21 by polymerase chain reaction (PCR), and 32% samples were found to be harbor mutations. Presence of an EGFR mutation was identified as a favorable prognostic factor, with 5- year OS of 92% for EGFR-mutated vs. 57% for EGFR wild-type tumors (P=0.037).26 The same group reported a retrospective analysis of 180 patients with either KRAS codon 12 mutation or EGFR mutation (exons 18-21). This study showed that presence of an EGFR mutation was associated with longer OS (P=0.048). However there was no impact on progression-free survival (PFS). 27 Liu et al. examined 130 ES-ADC samples for EGFR mutations in exon 19 and 21 by nested PCR, and detected mutations in 44.3% samples. Presence of an EGFR mutation did not have an impact on median PFS (36.6 months for EGFR-mutated vs. 25.7 months for EGFR wild-type tumors; P=0.56).28 A large retrospective study reported the outcome of 1118 patients with resected ES-NSCLC of whom 20% had an EGFR mutation. The presence of an EGFR mutation correlated with longer OS (HR 0.51; P < 0.001). A subgroup analysis was conducted in from a different dataset of 286 resected ES-NSCLC ADCs harboring EGFR mutations to determine the effect of adjuvant EGFR tyrosine kinase inhibitor (TKI) therapy. Among 286 patients receiving adjuvant TKI Cox regression analysis demonstrated a significant improvement in PFS (HR 0.43; P=0.001) but no significant differences in overall survival.29
Although, as illustrated above multiple retrospective analyses demonstrate improved survival in patients with completely resected EGFR-mutated ES-NSCLC, definitive conclusions can only be drawn by conducting large prospective clinical trials in this patient population.
Her2
Her2, a receptor tyrosine kinase and a member of the EGFR family is overexpressed in 20% of advanced NSCLC and mutated in less than 2%.30,31 In ES-NSCLC, studies suggest that overexpression of Her2 mRNA or protein is associated with an unfavorable prognosis.32,33 In a retrospective study 239 tumor samples of patients with ES-NSCLC were evaluated for Her2 overexpression, which was defined as an IHC score of 2+/3+ (scoring based on staining intensity and number of cells stained). Her2 overexpression was detected in 18% of tumors. The relapse rate for Her2-positive versus Her2-negative tumors was 60% versus 33% respectively (P=0.03) in the absence of adjuvant treatment.34 A different definition of Her2 positivity by IHC was used by Xia et al who considered Her2 overexpression as any positive staining. In a study involving only stage I and IIA NSCLC, 74% of ADCs and 54% of squamous cell carcinoma (SCC) were found to overexpress Her2. The 5 year OS was 65% versus 86% in Her2-positive versus Her2-negative tumors respectively (P=0.014).35 Despite differences in the criteria used to define Her2 overexpression, both studies attribute negative prognostic value to Her2 overexpression in ES-NSCLC. Although the presence of a Her2 mutation can predict for response to Her2-directed therapy in the metastatic disease setting, its predictive value remains unclear in ES-NSCLC.36
ERCC1
The excision repair cross-complementation group 1 (ERCC1) protein plays a rate-limiting role in the nucleotide excision repair (NER) pathway that recognizes and removes cisplatin-induced DNA adducts.37 In ES-NSCLC, tumor ERCC1 expression appears to have both prognostic and predictive value. In one of the first studies evaluating ERCC1, 51 resected samples of NSCLC were analyzed for ERCC1 RNA expression by PCR. Overexpression was associated with improved survival (HR 0.337, P=0.018), suggesting the value of ERCC1 expression as a prognostic biomarker.38 Subsequent studies focused on the role of this gene as a predictive marker, wherein low ERCC1 expression levels are predictive of benefit from cisplatin-based adjuvant chemotherapy. ERCC1 expression by IHC was studied in patients with stage I-III NSCLC enrolled in the International Adjuvant Lung Cancer Trial (IALT). Adjuvant chemotherapy prolonged survival among patients with ERCC1-negative tumors (HR 0.65, P=0.002), but not among patients with ERCC1-positive tumors (HR 1.14, P=0.40). However, subjects with ERCC1-positive tumors had better survival than subjects with ERCC1-negative tumors in the observation arm (HR 0.66, P=0.009). This and other studies have demonstrated the prognostic and predictive value of ERCC1 in ES-NSCLC.39,40
Despite these interesting results, utilization of ERCC1 as a predictive marker is limited by the lack of reproducibility of results with the antibodies used in the IHC assay. Friboulet et al repeated ERCC1 IHC staining on the original testing set of 589 samples from the IALT study, and attempted to unsuccessfully validate the results utilizing 494 samples collected from two independent phase 3 trials, JBR10 and CALGB 9633. There was a lack of correlation between the 16 different antibodies used, and an inability of these antibodies to identify various ERCC1 isoform specificities.41
RRM1
RRM1 is the regulatory protein subunit of ribonucleotide reductase and is involved in DNA repair, carcinogenesis and cancer progression. The mechanistic relationship between RRM1 levels and sensitivity of tumors cells to gemcitabine has been demonstrated in preclinical studies.42 In ES-NSCLC, several studies have suggested a prognostic role for RRM1. In a study of 126 patients with ES-NSCLC RRM1 expression was evaluated by PCR analysis. This study demonstrated better median OS in patients with higher levels of RRM1 (52 months vs. 24 months, P=0.013).43 Similarly, RRM1 expression by automated quantitative protein analysis (AQUA) was studied in 187 patients with stage I NSCLC. In this study, the median OS was 120 months vs. 60.2 months in the group with high RRM1 expression compared to the low expression group (P=0.01).40 To evaluate the predictive potential of RRM1 expression in the context of treatment with gemcitabine a phase II study was conducted in the ES-NSCLC population with the choice of adjuvant therapy determined by RRM1 protein expression. Eighty five patients with stage I NSCLC were enrolled and treatment was customized based on expression of ERCC1 and RRM1. Among patients with RRM1-expressing tumors the PFS at 2 years was 83% in patients receiving gemcitabine compared to 71% in patients in the observation arm. No statistical tests were performed to evaluate the significance of these observations. Hence, further studies are needed to determine the predictive value of RRM1 expression in ES-NSCLC.44
BRCA
The BRCA gene is a tumor suppressor involved in the homologous recombination repair pathway (HRRP), a mechanism for DNA repair. Methylation or mutations which decrease BRCA expression or activity have been shown to cause impairment in the HRRP. The value of BRCA1 and BRCA2 germ line mutations and the risk of developing breast and ovarian cancer are well established.45
BRCA protein level expression in ES-NSCLC was studied by IHC in 98 tumor samples, 50 of them ES-NSCLC. In this particular group, 50% of the cases showed low levels of BRCA1 or BRCA2 protein.46 Marist et al. described the methylation status of BRCA genes and its relationship to prognosis in 158 ES-NSCLC patients. Only 3.8% of the tumors were methylated (only ADC and large-cell carcinomas). No methylation in BRCA2 was detected and the methylation of BRCA1 was associated with worse OS (HR 3.1, 95%CI 1.2-7.9).47 Rosell et al examined 9 genes involved in the NER pathway, including BRCA1 in 126 ES-NSCLC patients, none of whom had received adjuvant chemotherapy. Among the genes studied, only high BRCA1 mRNA expression correlated significantly with worse OS (HR 1.98, P=0.02).48 In the adjuvant setting, the predictive value of BRCA1 was studied in 83 patients with ES-NSCLC (stage II to IIIA) and treatment was tailored according to BRCA1 mRNA levels. With a median follow-up of 41.6 months, the median time to progression (TTP) for the all patients was 22.9 months. Based on BRCA1 levels median TTP was 20.3, 56.5 and 51.9 months for the low, intermediate and high level groups respectively (P=0.31) and the differences were not statistically significant.49 Despite its promise as a potential predictive biomarker of platinum efficacy, there is a lack of evidence to define BRCA as a clinically relevant biomarker in ES-NSCLC.
Table 1 summarizes the biomarkers evaluated in ES-NSCLC
Table 1. Summary of biomarkers evaluated in ES-NSCLC.
| Gene | Protein function | Author Year | Method | Number of Patients | Comments |
|---|---|---|---|---|---|
| TP53 | Involved in growth arrest and apoptosis | *Mitsudomi14 2000 | IHC | 2579 | Prognostic for survival |
| Gene Sequencing | 650 | ||||
| KRAS | Transduction protein with GTPase activity | *Shepherd22 2013 | PCR | 1543 | Not of significant prognostic or predictive value |
| EGFR | Tyrosine kinase cell surface receptor | Liu27 2014 | PCR | 131 | Not significantly prognostic |
| ERBB2 | Tyrosine kinase cell surface receptor | Xia33 2012 | IHC | 172 | Prognostic for survival |
| ERCC1 | Enzyme involved in DNA damage repair | Friboulet39 2013 | IHC | 589 | Not predictive for response to chemotherapy |
| RRM1 | Enzyme essential for the production of deoxyribonucleotides | Zheng38 2007 | AQUA | 187 | Prognostic for survival |
| BRCA | Participate in the repair of DSB | Rosell45 2007 | PCR | 126 | Prognostic for survival |
Abbreviations: PCR, polymerase, IHC, immunohistochemistry, AQUA, absolute quantification, DSB, double strand break
Immune system and ES-NSCLC
Immunotherapeutic approaches are beginning to play an increasingly important role in the treatment of solid tumors. There have been important advances in this field leading to the approval of ipilimumab in melanoma 50, sipuleucel- T vaccine in prostate cancer 51 and reports of clinical activity of monoclonal antibodies (mAb) against programmed cell death-1 (PD-1) and its ligand (PDL-1) in solid tumors.52.
Past trials of immunotherapy in lung cancer have demonstrated limited activity. Immune mechanisms proposed to explain the lack of success in lung cancer include under-expression of classic major histocompatibility complex (MHC) molecules, overexpression of soluble factors such as PGE2 and TGF-β1 and overexpression of specific enzymes such as COX-2 and Ido-1+, help in circumventing successful immune control.53 More recently, checkpoint inhibition with anti-PD1 and anti-PDL1 therapies has yielded remarkable results in heavily pretreated patients with NSCLC and provided an impetus to further explore the role of immunotherapy in the treatment of lung cancer.
Tumor infiltrating lymphocytes as prognostic biomarkers
Previous studies have demonstrated the prognostic significance of tumor infiltrating immune cells and the location and density of these infiltrates in ES-NSCLC.54 In a retrospective study of 1290 patients with lung cancer a correlation was shown between tumor infiltrating lymphocytes (TILs), and prognosis in a subgroup of patients with stage I SCC. Presence of TILs was associated with a significant survival advantage over tumors without TILs (P= 0.03).55
Among studies that evaluated specific T cell subsets and their localization in ES-NSCLC, Wakayashi et al studied 178 ES-NSCLC and showed that the location of CD8(+) T cells (nests compared with stromal areas) had an effect on prognosis. The actuarial 5-year OS was 47% versus 60% (P=0.04).56 Similarly, the proportion of TILs [CD4(+),CD8(+),CD20(+)] in areas enriched for tumor epithelial cells compared to tumor stroma was examined in 335 cases of ES-NSCLC. It was observed that the presence of larger numbers of stromal CD4(+) and CD8(+) T-cells was an independent prognostic factors for longer disease specific survival (DSS) compared with epithelial infiltration with HRs of 2.6 (P<0.001) and 1.98 (P< 0.043) respectively.57 These observations merit further study to determine the significance of T-cell infiltration of the stroma.
Regulatory T cells (Tregs or Foxp3-positive T cells), are a specific subset of the T cell repertoire which have been demonstrated to be involved in decreasing the antitumor response.58 It has been shown that recurrence-free survival (RFS) of patients with tumors containing ≥ 3 Tregs in 10 high-power fields (hpf) was significantly worse than that of patients with tumors containing < 3 Tregs/hpf, especially in stage I and II disease (HR 5.38, P < 0.016).59 A recent study has shown the prognostic significance of the ratio of Tregs to CD3 compared to Tregs alone in 956 cases of stage I NSCLC. A low Tregs to CD3 ratio was found to be associated with a longer RFS (5-year RFS: 85% vs. 77%; P=0.004).60 Of note, the better-than-expected 5-year survival observed in both groups is probably a result of selection bias since a large fraction of patients enrolled on this study had favorable prognostic factors including the absence lymphatic and vascular invasion.
Checkpoint molecules as prognostic and predictive immune biomarkers
A relatively new area of research in cancer treatment focuses on the role of regulatory signals through molecules like CTLA-4 and PDL-1. CTLA-4 is a well-known inhibitory molecule of T cell activation expressed on Tregs. Its expression occurs in the context of the interaction between T-cell receptors and antigen presentation. The expression of CTLA-4 on Tregs can provide one explanation for the prognostic significance of Tregs infiltration in ES-NSCLC as previously described.59,61 CTLA-4 overexpression has been found to be more common in ADC compared to SCC and appears to be an independent favorable prognostic factor (5-year OS of patients with CTLA-4-overexpressing tumors: 64.8% versus 45.9%; P=0.078).62
PD-1 is a checkpoint receptor that has immunosuppressive functions similar to CTLA-4.63 However a few key differences between these immune checkpoints include the following: 1) PD-1 is activated during the effector stages of T cell activation, 2) The interaction of PD-1 with its ligand PD-L1 occurs primarily in peripheral tissues instead of lymph nodes and 3) PD-1 is expressed in tumor tissue tissues as well as hematopoietic cells.64 A phase I study by Topalian et al found that 50% of lung tumors with PD-1 expression (defined as ≥ 5% expression) responded to anti-PD-1 monoclonal antibody compared to 0% of PD-1 negative tumors.52 While the study included patients with advanced disease, these results are encouraging and potentially applicable to ES-NSCLC in the future.65 It should be noted, however that although associations have been demonstrated between expression of and response to monoclonal antibodies targeting PD-1 and PD-L1, recent observations also show that expression of PD-1 and PD-L1 is not a pre-requisite for the generation of a therapeutic response.66,67 This important point needs to be borne in mind as studies are designed in the future incorporating anti-PD-1/PD-L1 therapies for patients with ES-NSCLC.
Studies performed thus far to evaluate the prognostic value of PDL-1 expression in ES-NSCLC have generated discordant results. In one of the largest validation studies for PD-L1 expression (any level of expression in tumor cells above the negative control was considered a positive result) Velcheti et al, demonstrated PD-L1 protein expression in two separate cohorts (enrolled in two different countries) of NSCLC that included 88% and 84% cases with stage I – III disease respectively. Expression of PDL-1was detected in 36% and 25% cases in the two cohorts and was associated with better OS independent of others factors including histology in each of the cohorts (P= 0.036 and 0.027).68 Of note this study did not analyze the effects of treatment administered. Therefore, although it provides valuable hypothesis-generating data this study precludes the classification of PD-L1 expression as an established prognostic factor in ES-NSCLC.
Gene Signatures as biomarkers
A gene signature is a biomarker in which the expression of multiple genes, proteins, or microRNA (miRNA) is measured and combined into a group with possibly superior prognostic or predictive potential as compared to its individual components. Data from gene signatures may be presented in the form of risk scores as is the case with the Oncotype DX assay.69 Signatures may also be presented as categorical subgroups such as activated B-cell-like (ABC) or germinal center B-cell like (GCB) diffuse large B-cell lymphoma.70
In ES-NSCLC, several studies have attempted to identify gene expression signatures with prognostic or predictive potential by using diverse platforms and genes. Details of some gene signatures evaluated in ES-NSCLC are presented in Table 2 to illustrate the diversity of genes chosen, types of platforms and primary goals used to capture prognostic and/or predictive information. One of the challenges in developing gene signatures is to make them simple and reproducible. Chen et al identified a prognostic RT-PCR-based five-gene signature (including DUSP6, MMD, STAT1, ERBB3, and LCK) using risk scores based on microarray analyses of 125 ES-NSCLC tumor specimens. A high-risk score was associated with increased risk of death [HR for death 1.92, P=0.03)].71 Lau et al developed another RT-PCR-based signature utilizing a three gene cluster (STX1A, HIF1A, and CCR7) with prognostic significance. Based on analysis of 147 patients they were able to stratify stage I ES-NSCLC into two prognostic groups. When poor risk scores were compared to good risk scores the HR for survival was 5.9 (P=0.0019).72
Table 2. Examples of studies evaluating different types of signatures in ES-NSCLC.
| Signature | Characteristics | Independent validation | N of patients and Stage | Author Year | Comments |
|---|---|---|---|---|---|
| Immune genes signature (meta-analysis) | Different miRNA platforms | Yes | 1625* I to III | Suzuki54 2011 | Prognostic for PFS |
| Angiogenic signature | Lung tissue miRNA | No | 20I to III | Donnem79 2012 | Prognostic for PFS |
| DNA Methylation Signature | 450000 CpG sites analyzed | Yes | 237 stage I | Sandoval76 2014 | Prognostic for PFS |
| MicroRNA Signature | 440 miRNA | Yes | 251 I to IIIA | Landi78 2010 | Prognostic for OS |
| Proteomic Signature | Mass spectrometry 2630 signals | Yes | 116 I to IIIA | Yanagisawa82 2007 | Prognostic for OS |
Abbreviations: N, number, total number of patients from different studies; miRNA micro RNA; PFS, progression-free survival; OS, overall survival
These and other early studies of gene expression signatures using RT-PCR evaluated only a limited number of genes as opposed to larger microarray platforms that have tested hundreds of genes with a goal of developing a test that could be easily applied in the clinical setting. These studies are limited by the heterogeneous stages of disease in the study population, the lack of information on standard clinical predictive factors, and perhaps most importantly, the lack of blinded validation in independent cohorts in most cases. A review of 16 studies published on gene expression profiling in NSCLC from 2002 to 2009 found no evidence of the clinical utility of theses signatures beyond that obtained from currently used clinical parameters.73 More recently, Kratz et al reported a 14-gene prognostic signature using quantitative PCR in 361 patients with non-squamous ES-NSCLC and validated their results in a large cohort of stage I non-squamous NSCLC. In the validation cohort, the 5-year OS was 71% in low-risk, 58% in intermediate-risk, and 49% in high-risk patients (P=0.0003). Multivariate analysis indicated that the 14-gene signature provided information which added to that obtained from standard staging.74
Attempts have been made to develop prognostic gene signatures in ES-NSCLC based on methylation of DNA bases or acetylation of histones. A study was conducted to determine the methylation status of 7 genes in 51 stage I NSCLC patients and 116 matched controls. In multivariate analysis methylation of p16 and CDH13 was associated with a shorter time to recurrence (odds ratio for early recurrence 25.25; P=0.006).75 In a separate study in stage I NSCLC the methylation status of tumors was investigated in 237 cases and validated in a separate cohort of 143 cases. The analysis revealed a 5 gene methylation signature (HIST1H4F, PCDHGB6, NPBWR1, ALX1, HOXA9) that could identify patients at high risk of recurrence (HR 3.24; P =0 .001).76
In recent years, the prognostic value of miRNAs, which are small, noncoding, single stranded RNAs that are involved in regulation of gene expression, has been explored in ES-NSCLC.77 Raponi et al profiled global miRNA expression of 54 SCC samples and found that high miR-146b expression was associated with a median OS of 26 months versus 95 months, (HR= 2.7, P< 0.0035) in the low miR-146b expression group. Landi et al studied 165 ADC and 125 SCC using a miRNA platform and found that 5 miRNAs of the let-7 family strongly differentiated ADC from SCC (P<0.0001) and a group of 5 miRNA signatures correlated with OS, independent of stage and age (P<0.017).78
A number of studies have also been published using different platforms for gene enrichment. This has been done to study clusters in specific pathways, such as immune response or angiogenesis.54,79 Donnem et al used this approach to analyze a set of 128 miRNA in a cohort of ES-NSCLC patients. They found a significant correlation between a signature of miRNAs and angiogenic marker fibroblastic growth factor 2 (FGF2) (r=0.34, P <0.001). There was no significant correlation with VEGF-A and HIF-1α.79 Although miRNAs shows promise as a prognostic marker, most of the miRNAs are non- overlapping in different studies, most likely due to a selection bias and design issues.
Far fewer studies have been published on predictive signatures in lung cancer. Zhu et al performed a study with a 15-gene expression signature in 133 frozen tumor samples in the JBR.10 adjuvant clinical trial. The signature separated patients without adjuvant treatment into 2 groups, high-risk and low-risk. The high risk group had a mortality HR of 15.02 (P =0 .001) in both stage I and II compared to the low risk group. Among 71 patients receiving chemotherapy, the high risk group was found to have a mortality HR of 0.33 (P=0.005) versus a HR of 3.67 (P=0.013) in the low risk group.80 This signature was found to be both prognostic for survival of untreated patients and predictive for survival after adjuvant chemotherapy.
To summarize, several studies have described gene-expression signatures that are associated with outcome in ES-NSCLC. However, most of these studies have one or more caveats; non-prospective validation, inability to provide additional information to that gleaned from standard prognostic factors, and lack of strong data to alter clinical treatment decisions. Table 3 illustrates key aspects related to the evaluation of prognostic signatures in ES-NSCLC with an emphasis on gene signatures.81
Table 3. Key points in the evaluation of biomarker studies in ES-NSCLC.
| Objectives of the trial | Is the objective of the trial to identify prognostic markers, predictive markers or both? |
| Type of study | Is it a biomarker discovery or validation study or both? |
| Population | What subgroup of ES-NSCLC does the study include? For example, stage IA or stage IB, which histology squamous vs. non-squamous. Other subgroups EGFR mutated etc. |
| Specimen | Is adequate technical information provided regarding tissue processing and storage so that technical bias can be minimized in future validation studies? Is the study performed on easily available samples or on fresh-frozen specimens? |
| Assay | Is the assay clinically applicable? Is the technique widely available? Was it performed on blinded clinical data? |
| Validation | Is the biomarker validated in an independent cohort of patients with similar characteristics? |
| Benefit over standard risk-factors | Does the biomarker offer significant benefit or information over standard risk factors? Was independent statistical validation performed? |
| Benefit over standard risk-factors | Does the biomarker offer significant benefit or information over standard risk factors? Was independent statistical validation performed? |
Abbreviations: ES-NSCLC, early stage non-small cell lung cancer
Conclusions
In the present review we have described several potential biomarkers that have been evaluated in patients with ES-NSCLC (Figure 1). Significant challenges remain in the incorporation of information derived from these biomarkers into the management of patients with ES-NSCLC. A clear distinction has to be made between prognostic and predictive factors especially in relation to clinical trials designed to evaluate potential biomarkers or when a retrospective analysis is conducted to evaluate a putative biomarker. Figure 2 envisions a future scenario illustrating the use of biomarkers in clinical practice.
Figure 1.
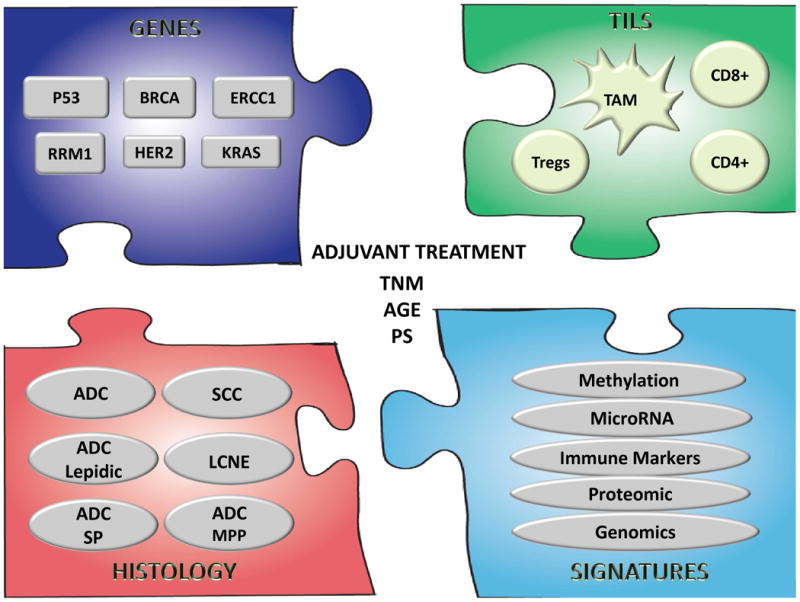
Potential biomarkers in ES-NSCLC: A) single genes; B) tumor infiltrating lymphocytes (TILs) including macrophages (TAM), T lymphocytes CD4(+), CD8(+), and T regulatory cell (Tregs); C) histology subtypes: ADC (adenocarcinoma), SCC (squamous cell carcinoma), BRC ( bronchoalveolar carcinoma), LCNE (large cell neuroendocrine tumor), SP (solid predominant) and MPP (micropapillary); D) signatures or group of genes, proteins, methylation patterns, microRNAs etc. In the center, prognostic factors used in clinic, staging (TNM), performance status and age.
Figure 2.
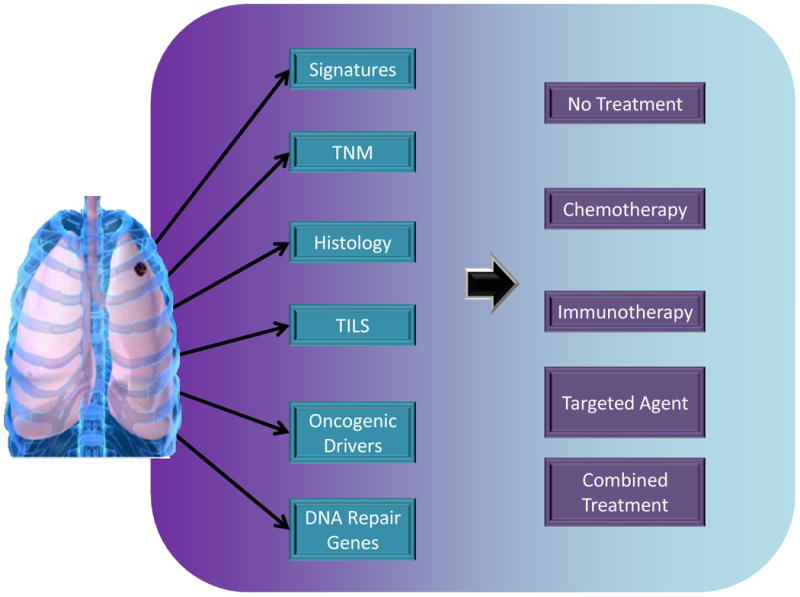
Potential future scenarios for using biomarkers in early stage non-small cell lung cancer. Besides the stage (TNM) and histologic subtype, molecular information from single genes (e.g., oncogenic drivers, DNA repair genes) or groups of genes (signatures) will help in identifying patients who could derive benefit from adjuvant treatment. Immune biomarkers (TILs) could provide prognostic or predictive information relevant to emerging immune therapies.
Many important issues need to be settled before biomarker-derived information and gene signatures can find widespread acceptance for patients with ES-NSCLC. One important issue is the validity and reproducibility of data. Other aspects that are no less important include reliability in trial implementation and tissue sampling. Uniformity in obtaining and handling biopsy samples is of critical importance. Despite these challenges, utilizing technological advances in bioinformatics and genomics has the potential to result in the discovery of newer molecular markers that might have prognostic and predictive value for patients with ES-NSCLC.
Finally, just as in the case of advanced lung cancer where biomarkers have an established role in predicting response to specific tyrosine kinase inhibitor therapies, a similar paradigm needs to be explored in ES-NSCLC to identify patients who could derive benefit from neoadjuvant and adjuvant therapy. Immune system-related biomarkers have the potential to represent the next step forward in the effort to develop prognostic and predictive markers for patients with ES-NSCLC.
Acknowledgments
Financial Support: Intramural Program, National Cancer Institute, National Institutes of Health
References
- 1.SEER Cancer Statistics Review 1975-2011. National Cancer Institute; [Google Scholar]
- 2.Fruh M, Rolland E, Pignon JP, et al. Pooled analysis of the effect of age on adjuvant cisplatin-based chemotherapy for completely resected non-small-cell lung cancer. J Clin Oncol. 2008;26:3573–81. doi: 10.1200/JCO.2008.16.2727. [DOI] [PubMed] [Google Scholar]
- 3.Arriagada R, Auperin A, Burdett S, et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet. 2010;375:1267–77. doi: 10.1016/S0140-6736(10)60059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review. National Cancer Institute; Bethesda, MD: 2012. [Google Scholar]
- 5.Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–60. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 7.Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol. 2006;7:719–27. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 8.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–9. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 9.Buyse M, Sargent DJ, Grothey A, et al. Biomarkers and surrogate end points--the challenge of statistical validation. Nat Rev Clin Oncol. 2010;7:309–17. doi: 10.1038/nrclinonc.2010.43. [DOI] [PubMed] [Google Scholar]
- 10.Group EBCTC. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 11.Mogi A, Kuwano H. TP53 mutations in nonsmall cell lung cancer. J Biomed Biotechnol. 2011;2011:583929. doi: 10.1155/2011/583929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oscier DG, Gardiner AC, Mould SJ, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–84. [PubMed] [Google Scholar]
- 13.Abudu A, Mangham DC, Reynolds GM, et al. Overexpression of p53 protein in primary Ewing's sarcoma of bone: relationship to tumour stage, response and prognosis. Br J Cancer. 1999;79:1185–9. doi: 10.1038/sj.bjc.6690190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graziano SL, Gu L, Wang X, et al. Prognostic significance of mucin and p53 expression in stage IB non-small cell lung cancer: a laboratory companion study to CALGB 9633. J Thorac Oncol. 2010;5:810–7. doi: 10.1097/jto.0b013e3181d89f95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsudomi T, Hamajima N, Ogawa M, et al. Prognostic significance of p53 alterations in patients with non-small cell lung cancer: a meta-analysis. Clin Cancer Res. 2000;6:4055–63. [PubMed] [Google Scholar]
- 16.Rodenhuis S, van de Wetering ML, Mooi WJ, et al. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–35. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 17.Rodenhuis S, Slebos RJ, Boot AJ, et al. Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res. 1988;48:5738–41. [PubMed] [Google Scholar]
- 18.Rosell R, Li S, Skacel Z, et al. Prognostic impact of mutated K-ras gene in surgically resected non-small cell lung cancer patients. Oncogene. 1993;8:2407–12. [PubMed] [Google Scholar]
- 19.Vega F, Iniesta P, Caldes T, et al. Association of K-ras codon 12 transversions with short survival in non-small cell lung cancer. Int J Oncol. 1996;9:1307–11. doi: 10.3892/ijo.9.6.1307. [DOI] [PubMed] [Google Scholar]
- 20.Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–5. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 21.Huncharek M, Muscat J, Geschwind JF. K-ras oncogene mutation as a prognostic marker in non-small cell lung cancer: a combined analysis of 881 cases. Carcinogenesis. 1999;20:1507–10. doi: 10.1093/carcin/20.8.1507. [DOI] [PubMed] [Google Scholar]
- 22.Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J Clin Oncol. 2013;31:1112–21. doi: 10.1200/JCO.2012.43.0454. [DOI] [PubMed] [Google Scholar]
- 23.Shepherd FA, Domerg C, Hainaut P, et al. Pooled Analysis of the Prognostic and Predictive Effects of KRAS Mutation Status and KRAS Mutation Subtype in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol. 2013 doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rusch V, Klimstra D, Venkatraman E, et al. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin Cancer Res. 1997;3:515–22. [PubMed] [Google Scholar]
- 26.Sonobe M, Nakagawa M, Takenaka K, et al. Influence of epidermal growth factor receptor (EGFR) gene mutations on the expression of EGFR, phosphoryl-Akt, and phosphoryl-MAPK, and on the prognosis of patients with non-small cell lung cancer. J Surg Oncol. 2007;95:63–9. doi: 10.1002/jso.20547. [DOI] [PubMed] [Google Scholar]
- 27.Sonobe M, Kobayashi M, Ishikawa M, et al. Impact of KRAS and EGFR gene mutations on recurrence and survival in patients with surgically resected lung adenocarcinomas. Ann Surg Oncol. 2012;19(Suppl 3):S347–54. doi: 10.1245/s10434-011-1799-8. [DOI] [PubMed] [Google Scholar]
- 28.Liu WS, Zhao LJ, Pang QS, et al. Prognostic value of epidermal growth factor receptor mutations in resected lung adenocarcinomas. Med Oncol. 2014;31:771. doi: 10.1007/s12032-013-0771-9. [DOI] [PubMed] [Google Scholar]
- 29.D'Angelo SP, Janjigian YY, Ahye N, et al. Distinct clinical course of EGFR-mutant resected lung cancers: results of testing of 1118 surgical specimens and effects of adjuvant gefitinib and erlotinib. J Thorac Oncol. 2012;7:1815–22. doi: 10.1097/JTO.0b013e31826bb7b2. [DOI] [PubMed] [Google Scholar]
- 30.Heinmoller P, Gross C, Beyser K, et al. HER2 status in non-small cell lung cancer: results from patient screening for enrollment to a phase II study of herceptin. Clin Cancer Res. 2003;9:5238–43. [PubMed] [Google Scholar]
- 31.Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–6. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 32.Brabender J, Danenberg KD, Metzger R, et al. Epidermal growth factor receptor and HER2-neu mRNA expression in non-small cell lung cancer Is correlated with survival. Clin Cancer Res. 2001;7:1850–5. [PubMed] [Google Scholar]
- 33.Takenaka M, Hanagiri T, Shinohara S, et al. The prognostic significance of HER2 overexpression in non-small cell lung cancer. Anticancer Res. 2011;31:4631–6. [PubMed] [Google Scholar]
- 34.Korrapati V, Gaffney M, Larsson LG, et al. Effect of HER2/neu expression on survival in non-small-cell lung cancer. Clin Lung Cancer. 2001;2:216–9. doi: 10.3816/clc.2001.n.006. [DOI] [PubMed] [Google Scholar]
- 35.Xia Q, Zhu Z, Wang J, et al. Expression and association of HER2 with prognosis in early-stage (T1-T2N0M0) non-small cell lung cancer. Tumour Biol. 2012 doi: 10.1007/s13277-012-0429-9. [DOI] [PubMed] [Google Scholar]
- 36.Mazieres J, Peters S, Lepage B, et al. Lung Cancer That Harbors a HER2 Mutation: Epidemiologic Characteristics and Therapeutic Perspectives. J Clin Oncol. 2013 doi: 10.1200/JCO.2012.45.6095. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Elledge SJ, Peterson CA, et al. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci U S A. 1994;91:5012–6. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon GR, Sharma S, Cantor A, et al. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127:978–83. doi: 10.1378/chest.127.3.978. [DOI] [PubMed] [Google Scholar]
- 39.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–91. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 40.Zheng Z, Chen T, Li X, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–8. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 41.Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med. 2013;368:1101–10. doi: 10.1056/NEJMoa1214271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidson JD, Ma L, Flagella M, et al. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64:3761–6. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 43.Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–85. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Bepler G, Zinner RG, Moon J, et al. A phase 2 cooperative group adjuvant trial using a biomarker-based decision algorithm in patients with stage I non-small cell lung cancer (SWOG-0720, NCT00792701) Cancer. 2014 doi: 10.1002/cncr.28714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med. 2008;359:2143–53. doi: 10.1056/NEJMra0802968. [DOI] [PubMed] [Google Scholar]
- 46.Lee MN, Tseng RC, Hsu HS, et al. Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer. Clin Cancer Res. 2007;13:832–8. doi: 10.1158/1078-0432.CCR-05-2694. [DOI] [PubMed] [Google Scholar]
- 47.Marsit CJ, Liu M, Nelson HH, et al. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23:1000–4. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- 48.Rosell R, Skrzypski M, Jassem E, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One. 2007;2:e1129. doi: 10.1371/journal.pone.0001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez JM, Cobo Manuel, Arrabal Rafael, et al. Pilot SCAT trial: Spanish customized adjuvant chemotherapy (CT) based on BRCA1 mRNA expression levels (l) in resected stage II-IIIA non-small cell lung cancer (NSCLC) patients (p) Journal of clinical oncology. 2012;30 [Google Scholar]
- 50.Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–22. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 52.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jadus MR, Natividad J, Mai A, et al. Lung cancer: a classic example of tumor escape and progression while providing opportunities for immunological intervention. Clin Dev Immunol. 2012;2012:160724. doi: 10.1155/2012/160724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki K, Kachala SS, Kadota K, et al. Prognostic immune markers in non-small cell lung cancer. Clin Cancer Res. 2011;17:5247–56. doi: 10.1158/1078-0432.CCR-10-2805. [DOI] [PubMed] [Google Scholar]
- 55.Ruffini E, Asioli S, Filosso PL, et al. Clinical significance of tumor-infiltrating lymphocytes in lung neoplasms. Ann Thorac Surg. 2009;87:365–71. doi: 10.1016/j.athoracsur.2008.10.067. discussion 371-2. [DOI] [PubMed] [Google Scholar]
- 56.Wakabayashi O, Yamazaki K, Oizumi S, et al. CD4+ T cells in cancer stroma, not CD8+ T cells in cancer cell nests, are associated with favorable prognosis in human non-small cell lung cancers. Cancer Sci. 2003;94:1003–9. doi: 10.1111/j.1349-7006.2003.tb01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Shibli KI, Donnem T, Al-Saad S, et al. Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin Cancer Res. 2008;14:5220–7. doi: 10.1158/1078-0432.CCR-08-0133. [DOI] [PubMed] [Google Scholar]
- 58.deLeeuw RJ, Kost SE, Kakal JA, et al. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res. 2012;18:3022–9. doi: 10.1158/1078-0432.CCR-11-3216. [DOI] [PubMed] [Google Scholar]
- 59.Shimizu K, Nakata M, Hirami Y, et al. Tumor-infiltrating Foxp3+ regulatory T cells are correlated with cyclooxygenase-2 expression and are associated with recurrence in resected non-small cell lung cancer. J Thorac Oncol. 2010;5:585–90. doi: 10.1097/JTO.0b013e3181d60fd7. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki K, Kadota K, Sima CS, et al. Clinical impact of immune microenvironment in stage I lung adenocarcinoma: tumor interleukin-12 receptor beta2 (IL-12Rbeta2), IL-7R, and stromal FoxP3/CD3 ratio are independent predictors of recurrence. J Clin Oncol. 2013;31:490–8. doi: 10.1200/JCO.2012.45.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melero I, Hervas-Stubbs S, Glennie M, et al. Immunostimulatory monoclonal antibodies for cancer therapy. Nat Rev Cancer. 2007;7:95–106. doi: 10.1038/nrc2051. [DOI] [PubMed] [Google Scholar]
- 62.Salvi S, Fontana V, Boccardo S, et al. Evaluation of CTLA-4 expression and relevance as a novel prognostic factor in patients with non-small cell lung cancer. Cancer Immunol Immunother. 2012;61:1463–72. doi: 10.1007/s00262-012-1211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–34. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res. 2013;19:5300–9. doi: 10.1158/1078-0432.CCR-13-0143. [DOI] [PubMed] [Google Scholar]
- 65.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Antonia SJ, Grosso F, Horak CE, et al. Association of tumor PD-L1 expression and immune biomarkers with clinical activity in patients with non-small cell lung cancertreated with nivolumab. 2013 [Google Scholar]
- 67.Gettinger S, Shepherd F, Antonia SJ, et al. First-line nivolumab (anti-PD-1; BMS-936558, ONO-4538) monotherapy in advanced NSCLC: Safety, efficacy, and correlation of outcomes with PD-L1 status. J Clin Oncol. 2014;32:5s–32. [Google Scholar]
- 68.Velcheti V, Schalper KA, Carvajal DE, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94:107–16. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 70.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 71.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 72.Lau SK, Boutros PC, Pintilie M, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–9. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 73.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102:464–74. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379:823–32. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–28. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 76.Sandoval J, Mendez-Gonzalez J, Nadal E, et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol. 2013;31:4140–7. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 77.Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012;18:268–74. doi: 10.1097/PPO.0b013e318258b743. [DOI] [PubMed] [Google Scholar]
- 78.Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Donnem T, Fenton CG, Lonvik K, et al. MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer. PLoS One. 2012;7:e29671. doi: 10.1371/journal.pone.0029671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–24. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subramanian J, Simon R. What should physicians look for in evaluating prognostic gene-expression signatures? Nat Rev Clin Oncol. 2010;7:327–34. doi: 10.1038/nrclinonc.2010.60. [DOI] [PubMed] [Google Scholar]
- 82.Yanagisawa K, Tomida S, Shimada Y, et al. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst. 2007;99:858–67. doi: 10.1093/jnci/djk197. [DOI] [PubMed] [Google Scholar]


