Abstract
Age is a major risk factor for cancer. Alterations in DNA methylation, histone modifications, chromatin structure and epigenetic regulatory mechanisms are prominent hallmarks of both the aging process and of cancer. Intriguingly – or possibly coincidentally - several chromatin features are common between aging and cancer. Here we ask whether, and if so how, aging-associated chromatin modifications contribute to tumor susceptibility and tumorigenesis.
At the intersection of aging, cancer and chromatin
The functional properties of genomes are shaped by the secondary features of chromatin [1, 2]. The hard-wired hereditary information contained in the primary genome sequence is modified by DNA methylation, histone modifications, higher order chromatin structure, non-coding RNAs and epigenetic regulatory mechanisms [1, 2]. These chromatin features determine and modulate gene expression programs and perform critical regulatory roles in numerous biological processes, particularly in response to environmental factors such as nutrition and chemical pollutants [3, 4].
Changes in chromatin features have been closely linked to cancer susceptibility and tumor growth [5, 6]. Many cancer cells and tissues exhibit global and/or local changes in histone modifications and chromatin structure. Chromatin alterations likely contribute substantially to tumorigenesis and numerous examples in which chromatin changes drive tumorigenesis have been described [5, 6]. Changes in chromatin features, brought about by environmental insult and intrinsic events, may influence multiple steps of tumorigenesis by stimulating cell proliferation, promoting genomic instability and facilitating cellular transformation [7].
Chromatin changes and epigenetic alterations are also one of the prominent hallmarks of aging cells and organisms [8]. The progressive loss of physical function over time, which characterizes the aging process, is often accompanied by epigenetic modifications. Exposure to environmental toxins and endogenous stress has been linked to aging, possibly via interference with chromatin regulatory mechanisms [9]. It has also been speculated that epigenetic alterations contribute to the age-related changes in biological functions as well as to the observed increased genomic instability in aging organisms and cells [7, 10].
Since advanced age is a primary risk factor for cancer, and because epigenetic alterations are hallmarks of both aging and cancer, an obvious question is whether aging-associated chromatin changes actively contribute to cancer susceptibility and growth and whether these changes are the basis for the strong age-dependence of cancer – or whether they are mere coincidental changes with no functional links [11]. In this review, we highlight several epigenetic features which are prominently affected both in mammalian aging and cancer, and we discuss their potential and mechanisms to drive age-related tumorigenesis.
DNA Methylation
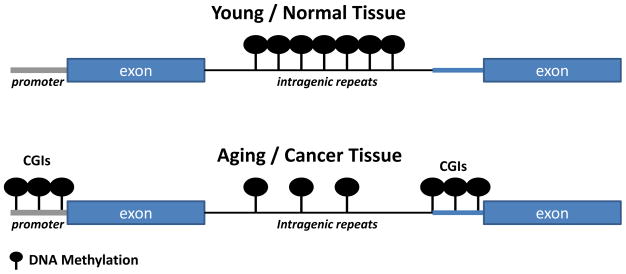
DNA methylation is one of the major genome regulatory mechanisms. Addition of a methyl group to the 5-carbon on cytosine (5mC) in cytosine-phosphate–guanine (CpGs) dinucleotides has been implicated in regulating gene expression by gene silencing and epigenetic memory in diverse biological processes [12]. Three DNA methyltransferases (DNMTs) mediate this modification: DNMT3a and DNMT3b function as de novo methyltransferases and DNMT1 acts as a maintenance methyltransferase [6, 8]. Changes in DNA methylation patterns have been documented in aging and in cancer (Table 1). Both processes are characterized by loss of methylation (DNA hypo-methylation) from CpGs located in repeat regions of the genome and concomitant increases in DNA methylation (DNA hyper-methylation) in CpGs located in the promoter regions of the genes known as CpG islands (CGIs) (Fig. 1; Table 1) [5, 9, 13, 14]. The same phenomena of DNA hypo-methylation and focal hyper-methylation is observed in senescence a state when cells cease to divide and which has been suggested to represent cellular aging and implicated as both a tumor protective and promoting mechanism [15].
Table 1.
Chromatin features in aging and cancer
| Aging | Cancer | |
|---|---|---|
| DNA Methylation | ||
| DNA hypo-methylation | ↑LINE and SINE regions 16–21 | ↑LINE and SINE regions13,19,24–26 |
| DNA hyper-methylation | ↑CGIs 20–21, 33, 35 | ↑ CGIs 5,14,34–36 |
| Histones modifications | ||
| H4K16ac | ↓ in vitro in aged cycling human fibroblasts52 | ↓various cancers55 |
| H4K20me3 | ↓ in vitro in aged cycling human fibroblasts52 ↑ in vivo in livers and kidneys of old rats78 ↑ Hutchinson-Gilford Progeria cells77 |
↓various cancers55, 74 ↓ rats model73 |
| H3K9ac | ↑ in vitro in aged cycling human fibroblasts52 | ↑ lung cancer81 ↓ breast cancer80 |
| H3K4me3 | ↑ in vitro in aged cycling human fibroblasts52 | ↓ breast cancer141 ↑ glioblastoma136, renal cell carcinomas137 |
| H3K9me3 | ↓ in vitro in aged cycling human fibroblasts52 | ↑ humans91 ↓ pancreatic carcinomas138 |
| H3K27me3 | ↓ Hutchinson-Gilford Progeria cells77 ↓ senescent cells99 |
↑ esophageal squamous cell carcinoma139 ↓ pediatric high-grade gliomas102 |
| LncRNA | ||
| HOTAIR | ND | ↑ breast114 |
| HOTTIP | ND | ↑ liver, leukemia 116, 117 |
| CTBP1-AS | ND | ↑ prostate118 |
| ANRASSF1 | ND | ↑ breast, prostate119 |
| XIST | ↓ senescence122 | ND |
| MALAT1 | ↓ senescence122 | ↑ various cancers140 |
| SAL-RNA1 | ↓ senescence122 | ND |
| ANRIL | ↓ senescence126 | ↑ prostate, leukemia 126–127 |
ND: not determined; Red: reduction of indicated feature; Green: increase of indicated feature
Figure 1. Changes in DNA methylation during aging and cancer.
CpGs located in the promoter regions of the genes known as CpG islands (CGIs) show increased propensity for methylation during aging and cancer. CpGs located in the repeat regions of the genome in contrast generally show loss of methylation with age and in cancer.
Loss of cytosine methylation from the repeat regions of the genome, such as in LINEs (long interspersed nucleotide elements) and SINEs (short interspersed nucleotide elements), which comprise ~50% of the genome, significantly contributes to a global decrease in DNA methylation observed during aging [16–21]. Hypo-methylation of these repeat regions is associated with the decline of organ functions during aging [17, 18]. Global DNA hypo-methylation of repeat-rich regions is also observed in numerous cancers [13, 19]. LINE-1 hypo-methylation is correlated with increased risk of developing cancer and poor survival of cancer patients as seen, for example, in head and neck squamous cell carcinoma, myeloma and bladder cancer [13, 22–26]. DNA hypo-methylation of repeat elements has also been proposed to contribute to cellular transformation by promoting chromosomal rearrangements and elevating mutation rates [13, 27–29]. Apart from hypo-methylation of repeat elements, loss of methylation from genic regions also causes aberrant gene expression and promotes tumorigenesis [13, 30].
Global DNA hypo-methylation associated with aging and senescence is, intriguingly, accompanied by local DNA hyper-methylation at CGIs[15, 20, 21]. CGI hyper-methylation in promoter regions generally aids in gene silencing by recruitment of enzymatic machinery to establish silent chromatin by proteins such as methyl-CpG binding domain (MBD) or may block binding of chromatin modifiers and transcription factors to methylated DNA [12, 31, 32]. CGIs methylation patterns are altered in a predictable fashion during aging and analysis of the methylation status of 353 defined CGIs is sufficient to accurately determine tissue age [33]. Hyper-methylation also appears to be relevant to cancer, since CGIs which are normally unmethylated are frequently hyper-methylated in cancer [5, 14]. Since CGI methylation has been observed in several tumor suppressor genes, it has been suggested that aberrant CGI methylation is a major contributor to neoplastic transformation via the stabilization of transcriptional repression, thereby leading to loss of function of tumor suppressor genes [11, 14]. CpG hyper-methylation of promoters also affects the expression of various non-coding RNAs that may have a role in malignant transformation [5, 34].
The phenomenon of CGI promoter hyper-methylation in aged tissues and cancer cells may suggest that methylation of CGIs pre-disposes aged cells to neoplastic transformation [11, 20]. For instance, similar patterns of CGI methylation of the estrogen receptor (ER) gene occur during aging in normal colon tissue and in colon cancer cells [35] and exogenous expression of ER suppresses colon cancer cell growth in vitro, suggesting that aging-associated repression of ER expression facilitates tumorigenesis [35]. In addition, promoters of genes which are targets of the repressive polycomb group of chromatin proteins are methylated during aging [36] as is the case for numerous cancers, where chromatin modifications by polycomb group proteins overlap with DNA hyper-methylated genes [37, 38]. Interestingly, some of these genes are involved in stem cell differentiation suggesting that age-dependent methylation of polycomb target genes may predispose the aging genome to neoplastic transformation by stabilizing stem cell-like features [36]. The link between aging, cancer and DNA hyper-methylation is also supported by the intriguing fact that genes whose mutations are associated with aggressive premature aging phenotypes, such as WRN in Werner Syndrome and LMNA, the causative agent of Hutchinson Gilford Progeria Syndrome, are frequently silenced by DNA methylation in a wide variety of cancers [39, 40]. Of note in this context is the fact, that some of these pre-mature aging disorders are characterized by increased tumor susceptibility [41].
The mechanisms underlying the opposing fate of DNA methylation, i.e DNA hypo-methylation of repeated sequences, and at the same time hyper-methylation of CGIs, observed during aging and cancer are not known [11, 13]. The DNA methylating activity of both maintenance methytransferase DNMT1 and de novo methyltransferases DNMT3a and DNMT3b are altered during aging and neoplastic transformation [42, 43]. Whereas the activities of DNMT1 and DNMT3a decrease, DNMT3b activity increases in cells approaching senescence [42, 43], suggesting that global DNA hypo-methylation during aging cells is due to reduced DNMT1 activity. DNMT1 has also been reported to be mislocalized in senescent cells possibly triggering hypo-methylation at specific genomic sites, particularly in lamin-associated domains [15]. CGI hyper-methylation during aging is assumed to be due to de novo activity of DNMT3b. In contrast to aging cells, which show reduced DNMT1 activity, several tumors show DNMT 1 over-expression and its activity has been linked to CGIs hyper-methylation [44], although DNMT1 over-expression is not always associated with CGI hyper-methylation [45]. DNMT3b and DNMT3a are also frequently over-expressed in tumors with DNMT3b generally at higher levels than DNMT3a [46]. It thus appears that, although aged cells and cancer cells share several common features of DNA methylation, no common mis-regulation of the DNA methylation machinery is apparent, suggesting that other, yet unknown, factors determine the methylation patterns of aging and cancer cells.
Histone modifications
Core histones throughout the genome are modified at numerous amino acid residues by a diverse set of chemical modifications, most prominently acetylation, methylation, ubiquitylation, SUMOylation and phosphorylation [47]. The addition or removal of these modifications creates binding sites for chromatin proteins and often alters the degree of compaction of chromatin [47, 48]. Aberrant patterns of histone modifications are hallmarks of both aging [8] and cancer [49] (Table 1).
Studies in yeasts and in senescent cells suggest that aging is accompanied by a reduction of the global histone levels thus affecting chromatin structure. H3, H4, H2A and H2B levels are decreased in replicatively old yeast budding cells [50, 51] and in senescent human fibroblasts generated either through replicative senescence or oncogene-induced senescence [52, 53]. General loss of core histones is thought to lead to nucleosome depletion and accompanying genome-wide increase in gene expression, elevated levels of DNA damage, retrotransposition, large-scale chromosome rearrangement, and translocation during yeast aging [50, 54].
One of the most prominently altered histone modification is H4K16 acetylation, which is reduced during the aging process [52] as well as in various types of tumors [55]. H4K16 acetylation has been implicated in higher-order chromatin organization [56, 57] and plays an important role in the DNA damage response, allowing for the possibility that aging-associated decrease in H4K16ac promotes genomic instability [58–60]. Although multiple, partially redundant, histone acetyltransferases promote acetylation of H4K16, Sirt1 appears to be the major modifying enzyme [61, 62]. Sirt1 is the focus of much attention in the aging field due to reports that its invertebrate homolog, Sir2, promotes longevity in yeast, worms and flies [63–65]. However, although over-expression of Sirt1 improves various aspects of health during aging and promotes survival in a mouse model of genomic instability, Sirt1 does not appear to increase longevity in mammals [66, 67]. Beyond its role in aging, Sirt1 also seems to play a role in cancer [68]. Its expression is up-regulated in a wide range of cancer types including leukemia, breast, prostate, lung, colon and gastric cancers [68, 69]. The reasons why Sirt1 may promote longevity in lower organisms, but promotes tumor growth in higher organisms are unknown.
Trimethylation of histone H4 on lysine 20 is also affected both in aging and cancer. H4K20me3 is highly enriched at pericentric heterochromatin, telomeres, imprinted genome regions and repetitive elements, suggesting that this modification is involved in transcriptional silencing [70, 71]. Levels of H4K20me3 are decreased in many cancers, including liver, breast, bladder and lung [72–74]. Loss of H4K20me3 in cancer may be due to reduced expression of the H4K20-specific methyltransferase Suv4–20h as observed in hepatocarcinoma in a rat model and in breast cancer cells [73, 74]. Interestingly, Suv4-20h has been implicated in maintenance of genome stability [75] and Suv4-20h double knock-out mice present with chromosomal aberrations due to less efficient DNA double-strand break repair [76]. H4K20me3 levels are also reduced in late-passage cycling human diploid fibroblasts (HDFs) [52]. Furthermore, exogenous expression of telomerase does not restore H4K20me3 levels, suggesting that replicative senescence and DNA damage responses are not responsible for elevation of trimethyl-H4K20 in HDFs [52]. In contrast, up-regulation of H4K20me3 has been observed in the premature aging disorder Hutchinson Gilford Progeria Syndrome [77] and in livers and kidneys of old rats [78]. Of note, in contrast to senescent fibroblasts derived by extensive passaging [52], H4K20me3 levels were found to be elevated in ras-induced senescent cells [79], suggesting differences in the senescence states depending on how senescence is induced [15].
Other aging-associated histone modifications include H3K9 acetylation, and trimethylation of H3K4, H3K9 and H3K27, and changes in these modifications have been observed in cancers, although their levels vary depending on tumor-type (Table 1). For example, breast cancer cells show low H3K9ac [80], whereas lung cancer cells show high H3K9ac levels [81]. Similarly, an increase in H3K9ac, the substrate for the histone deacetylase Sirt6 [82], which increases longevity in mammals, occurs in aged cycling human fibroblasts in vitro [52]. Mice deficient in SIRT6 exhibit accelerated aging [83], whereas transgenic mice overexpressing SIRT6 have a longer lifespan than control animals [84]. These findings are interesting in the light of the fact that much evidence points to a role of SIRT6 as a tumor suppressor, particularly via its role in DNA repair and genomic stability. Mouse embryonic fibroblasts from SIRT-6 deficient mice exhibit chromosomal aberrations including fragmented chromosomes and detached centromeres [83]. Along with H3K9ac levels, SIRT6 levels vary depending on the tumor type. SIRT6 is downregulated in several human cancers such as pancreatic cancer, colorectal cancer, and HCC [68], but up-regulated in prostate cancer [85]. It has been suggested that SIRT6 exerts its longevity function by reducing harmful levels of H3K9ac [86].
Aged cells are also characterized by increased levels of H3K4me3, a mark of active chromatin, and a decrease of H3K9me3, a mark of repressed chromatin [52]. In contrast H3K9me3 levels are elevated in oncogene-induced senescent cells and the mark accumulates in compacted heterochromatin foci [87, 88]. H3K9me3 has been associated with both positive and negative patient prognosis in several cancers depending on tumor type such as acute myeloid leukemia [89], salivary adenoid cystic carcinoma [90] and gastric adenocarcinoma [91]. Trimethylation of H3K4 also seems to play a role in cancer since the gene encoding one of the enzymes catalyzing the methylation of H3K4, the mixed lineage leukemia (MLL) protein lysine methyltransferase, is commonly translocated in leukemias leading to aberrant expression of developmental and hematopoietic genes [92]. A role of H3K4me3 in aging is supported by the fact that the deficiency in a member of the Ash-2 complex, which trimethylates H3K4, prolongs lifespan of Caenorhabditis elegans and conveys trans-generational longevity [93]. Based on observations in mouse cells, H3K4me3 also appears to be inter-dependent with the repressive mark H3K27me3, which is catalyzed by polycomb-group proteins, during development and cell differentiation [94–96]. Genome-wide distribution patterns of these two histones modifications differ in senescent cells compared to proliferating cells [97] and they exhibit bivalent regions enriched in H3K27me3 and H3K4me3 at lamin-associated domains and H3K27me3-depleted regions respectively referred to “mesas” and “canyons” between lamin-associated domains corresponding to genes and enhancers[97]. These “mesas” overlap with DNA hypomethylation regions observed in senescent [15] and cancer cells [98]. Loss of H3K27me3 and H3K4me3 “mesas” has also been reported in the premature aging disease Hutchinson-Gilford Progeria Syndrome [77, 97] and in senescent cells [99]. H3K27me3 is a target of EZH2, a methyltransferase and component of the polycomb repressive complex 2 (PRC2), whose expression is decreased in senescent cells [99]. Conversely, overexpression of EZH2 and H3K27me3 has been associated with poor survival in gastric, breast, prostate cancers and cutaneous melanoma [100, 101]. A reduction in H3K27me3 in pediatric high-grade gliomas harboring two mutations, K27M and G34R/V, in histone H3.3 has also been reported and this reduction of H3K27me3 has been associated with DNA hypo-methylation [102]. Indeed, in cancer cells, H3K27 seems to target loci for de novo DNA methylation [103]. However, the relationship between H3K27me3, DNA methylation and cancer is not clear since silencing of genes marked by trimethyl-H3K27 in the absence of DNA methylation has also been reported [104].
Most of the human studies on histone post-translational modifications in cancer and aged cells are correlative and it is unknown whether these modifications are causative or represent merely a consequence of these processes. In addition to the obvious local effects, changes in histone modification patterns may have on individual tumor-promoting or repressive genes, one of the most likely scenarios for how aging-associated chromatin modifications might contribute to tumorigenesis is by inducing changes to higher order chromatin structure which in turn may make chromatin susceptible to damage [105]. Since one of the major hallmarks of aging is the accumulation of DNA damage, it is possible that changes in higher order chromatin structure affect the DNA damage response in aged cells. In support, DNA-damage induced reorganization of chromatin due to redistribution of chromatin modifiers such as SIRT1 upon DNA damage has been reported. The resulting chromatin reorganization led to changes in gene expression patterns similar to those observed during aging [67].
Long non-coding RNAs
Non-coding RNAs have emerged as important regulatory molecules modulating gene expression [106, 107]. Using tiling arrays and deep sequencing to study gene expression at a genome-wide scale, it has become clear that the human genome is pervasively transcribed and transcription is not limited to protein coding regions [108–110]. In addition to different types of small non-coding RNAs [106], which function via interaction with RNA-binding proteins or mRNA, the human genome expresses a high number of non-coding transcripts larger than 200bp, known as long non-coding RNAs (lncRNAs), which play regulatory roles in various biological processes [107]. LncRNAs can affect the expression of genes in cis or trans via association with other RNAs [111, 112] or proteins such as transcription factors, RNA binding proteins or chromatin modifying enzymes [107].
Numerous lncRNAs have been linked to cancer [113], many of which associate with chromatin or chromatin-modifying enzymes (Table I). HOTAIR, which is transcribed from the HOX gene cluster, is over-expressed in a subset of breast cancers and its expression promotes metastasis and is correlated with poor prognosis [114]. HOTAIR binds to the repressive Polycomb complex 2 (PRC2) and the histone demethylase LSD1 complex to modulate target gene expression [114, 115]. Another lncRNA generated from the HOX gene cluster, HOTTIP, is over-expressed in hepato-cellular carcinoma where its expression is associated with poor patient survival [116]. Interestingly, HOTTIP acts by inducing gene expression by interacting with adaptor protein WDR5 of the MLL complex implicated in leukemia [117]. CTBP1-AS is an androgen responsive lncRNA, which is over-expressed in prostate cancer, where it exerts tumor suppressive effects by inhibiting the expression of its antisense gene CTBP1, a co-repressor for androgen receptor [118]. CTBP1-AS binds to the HDAC–mSin3A complex to decrease histone H3 acetylation and H3K4 methylation levels at the CTBP1 promoter [118]. The lncRNA ANRASSF1 is over-expressed in breast and prostate cells and it inhibits the tumor-suppressor gene RASSF1A [119] by forming an RNA/DNA hybrid that recruits the silencing complex PRC2 to the RASSF1A gene promoter [119]. The deletion of XIST, the prototypical lncRNA which mediates X chromosome inactivation and female dosage compensation by interacting with PRC2 [120], in the blood compartment of mice, results in genome instability and female-specific fully penetrant lethal blood cancer [121], suggesting a tumor protective role for XIST. It is generally not clear whether the observed changes in lncRNAs and the associated changes in gene expression are the drivers or consequence of cancer.
Early observations also point to the relevance of lncRNAs in aged cells and tissues. It has been noticed that young and senescent cells show differential expression of several lncRNAs [122]. The expression of some lncRNAs such as MALAT1, XIST and MIAT is lower in cells undergoing senescence and knockdown of MALAT1 and MIAT in early passage fibroblast induces senescence [122]. One lncRNA, SAL-RNA1, prevents senescence and its knockdown promotes phenotypic traits of senescence, including enlarged cell size and positive β-galactosidase staining, possibly via modulating p53 levels [122]. Further studies are required to probe whether lncRNAs play any direct functional roles in senescence and aging.
A possible model for the action of lncRNA in aging-associated tumorigenesis emerges from the analysis of the regulatory mechanisms of the INK4A-ARF-INK4B tumor suppressor gene cluster, which is deleted in several cancer types [123]. Expression of the protein coding genes p15INK4b, p16INK4a and ARF from this locus is increased during aging and the locus has been linked to aging-related diseases by genome-wide association studies (GWAS) [123, 124]. This locus also encodes the antisense lncRNA ANRIL, which negatively regulates the expression of protein coding genes from the INK4A-ARF-INK4B locus [125–127]. GWAS have identified ANRIL as a risk locus for cancer and other age-related diseases [128]. The levels of ANRIL are reduced in cells undergoing senescence [126] and ANRIL silences p15INK4b by heterochromatin formation [127]. ANRIL binds to polycomb repressive components CBX7 and SUZ12 and epigenetically represses the expression of p16INK4a and p15INK4b, respectively, by promoting H3K27 methylation [125, 126]. The fact that ANRIL is over-expressed in prostate cancer and leukemia and participates in epigenetic silencing of tumor suppressors suggests that it may be involved in cancer progression [126, 127].
A prominent feature of the aging process is the appearance of DNA damage [105]. Several lncRNAs are altered upon DNA damage, such as lncRNAp21 and PANDA [129]. Interestingly, lncRNA ANRIL is induced upon DNA damage, and elevated levels of ANRIL suppress the expression of tumor suppressors p15INK4b, p16INK4a and ARF upon DNA damage [130]. Another such DNA damage-inducible lncRNA is JADE, which upon DNA damage induces the expression of its neighboring gene Jade1, which acts as a cofactor in histone acetylation complex HBO1 (human acetylase binding to ORC1) [131]. Interestingly, lncRNA JADE is over-expressed in breast cancer, and its knockdown significantly reduces breast tumor growth in vivo [131]. Albeit preliminary, these observations suggest that lncRNAs function and expression is altered due to DNA damage during aging and contribute to subsequent tumor formation.
Models of chromatin-based aging-related cancer formation
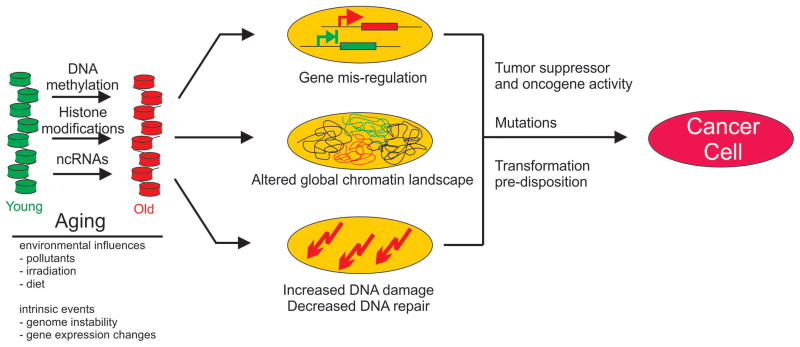
How might aging-related changes in chromatin contribute to tumor formation? Several models can be envisioned (Fig. 2). First, chromatin properties may directly interfere with regulation of individual, or groups of, genes involved in tumor suppression or promotion. While aging-associated changes to chromatin likely occur at a global scale, alterations of chromatin properties at particular sites as part of such global alterations of chromatin structures during aging may affect expression of key genes such as tumor suppressors or oncogenes. Similarly, lncRNAs may act in this model by affecting, either directly or indirectly, the expression of tumor suppressors or genes involved in cell proliferation or apoptosis (Fig. 2). Alternatively aging-associated changes in chromatin properties may increase susceptibility to oncogenic transformation by triggering aberrant differentiation of tissue stem cells or somatic cells allowing them to assume properties of cancer initiating cells (Fig. 2) [132]. Such global changes may not necessarily be sufficient to cause cancer, but they may set the stage for tumor formation by predisposing and facilitating transformation [133]. This notion is supported by reports of similarities in chromatin properties of tumor initiating cells and stem cells [134]. A further scenario, not mutually exclusive with others, relies on the fact that chromatin properties affect a cell’s response to DNA damage and aging-associated changes in chromatin features may thus impair DNA repair processes and in this way increase the frequency of mutations and elevate genomic instability, heightening the probability of oncogenic transformation [105, 135]. NITR [136–141].
Figure 2. Chromatin-based mechanism of aging-associated tumorigeneis.
Aging is accompanied by global and local changes in DNA methylation, histone modifications and ncRNA expression brought about by intrinsic and environmental factors. Aging-associated changes in chromatin may promote tumorigenesis by silencing tumor suppressors or activating oncogenes (top), altering the global chromatin landscape and chromatin organization (b), and leading to increased DNA damage (c).
Concluding remarks
Aging and tumorigenesis are two highly complex processes. It is not surprising to find alterations in chromatin properties in both, and even the occurrence of similar changes in both processes is expected. The question then arises whether common changes to chromatin features are merely coincidental or whether aging-associated chromatin features contribute to tumorigenesis. The jury is clearly still out. To understand the interplay of chromatin, aging and cancer it will be critical to extend the current studies beyond their correlative nature and specifically test the functional and causal role of aging-associated chromatin features in promoting tumor phenotype and delineating their mechanisms. A major challenge in these studies is the scarcity of experimental model systems. It is difficult to relate human cell culture systems to in vivo behavior. Similarly, animal models are only of limited relevance since both the aging and the tumorigenesis process in humans differs significantly from that in animal models. It will be important to develop suitable, well controlled experimental systems to directly test the impact of aging on cancer formation. This is a significant challenge. One interesting, albeit similarly imperfect, option is the study of naturally occurring human pre-mature aging disorders. Intriguingly, while some aging syndromes, such as Blooms Syndrome and Werner Syndrome, are associated with elevated tumor susceptibility, others, such as Cockayne Syndrome and Hutchinson Gilford Progeria Syndrome, are characterized by premature aging but no tumor formation. The similarities and differences between these diseases may provide insights into the link between aging and cancer.
Highlights.
Chromatin features are altered in aging and in cancer
Several changes to chromatin are common between aging and cancer
Several mechanisms by which aging-associated changes in chromatin may contribute to tumorigenesis can be envisioned
Acknowledgments
VS is supported by an NIH Khorana-Nirenberg Fellowship. Work in the Misteli laboratory is supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Graaf CA, van Steensel B. Chromatin organization: form to function. Current opinion in genetics & development. 2013;23:185–190. doi: 10.1016/j.gde.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T. The cell biology of genomes: bringing the double helix to life. Cell. 2013;152:1209–1212. doi: 10.1016/j.cell.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tollefsbol TO. Dietary epigenetics in cancer and aging. Cancer Treat Res. 2014;159:257–267. doi: 10.1007/978-3-642-38007-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldmann T, Schneider R. Targeting histone modifications--epigenetics in cancer. Current opinion in cell biology. 2013;25:184–189. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Cruickshanks HA, Adams PD. Chromatin: a molecular interface between cancer and aging. Current opinion in genetics & development. 2011;21:100–106. doi: 10.1016/j.gde.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huidobro C, et al. Aging epigenetics: causes and consequences. Mol Aspects Med. 2013;34:765–781. doi: 10.1016/j.mam.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Putiri EL, Robertson KD. Epigenetic mechanisms and genome stability. Clin Epigenetics. 2011;2:299–314. doi: 10.1007/s13148-010-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraga MF, et al. Cross-talk between aging and cancer: the epigenetic language. Annals of the New York Academy of Sciences. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- 12.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes & development. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nature reviews. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 15.Cruickshanks HA, et al. Senescent cells harbour features of the cancer epigenome. Nature cell biology. 2013;15:1495–1506. doi: 10.1038/ncb2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jintaridth P, Mutirangura A. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiological genomics. 2010;41:194–200. doi: 10.1152/physiolgenomics.00146.2009. [DOI] [PubMed] [Google Scholar]
- 18.Lange NE, et al. Alu and LINE-1 methylation and lung function in the normative ageing study. BMJ open. 2012;2 doi: 10.1136/bmjopen-2012-001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez J, et al. Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic acids research. 2008;36:770–784. doi: 10.1093/nar/gkm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen BC, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS genetics. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu ZZ, et al. Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence, and mortality in elderly individuals: the Normative Aging Study. Cancer causes & control: CCC. 2011;22:437–447. doi: 10.1007/s10552-010-9715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furniss CS, et al. Line region hypomethylation is associated with lifestyle and differs by human papillomavirus status in head and neck squamous cell carcinomas. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:966–971. doi: 10.1158/1055-9965.EPI-07-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baba Y, et al. LINE-1 Hypomethylation, DNA Copy Number Alterations, and CDK6 Amplification in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2014;20:1114–1124. doi: 10.1158/1078-0432.CCR-13-1645. [DOI] [PubMed] [Google Scholar]
- 25.Aoki Y, et al. Correction: Genomic vulnerability to LINE-1 hypomethylation is a potential determinant of the clinicogenetic features of multiple myeloma. Genome Med. 2013;5:88. doi: 10.1186/gm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilhelm CS, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16:1682–1689. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daskalos A, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. International journal of cancer. Journal international du cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 28.Chen RZ, et al. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 29.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 30.Sakatani T, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 31.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lujambio A, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Issa JP, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nature genetics. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 36.Teschendorff AE, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome research. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGarvey KM, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer research. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Easwaran H, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome research. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agrelo R, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agrelo R, et al. Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:3940–3947. doi: 10.1200/JCO.2005.11.650. [DOI] [PubMed] [Google Scholar]
- 41.Hoeijmakers JH. DNA damage, aging, and cancer. The New England journal of medicine. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 42.Casillas MA, Jr, et al. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Molecular and cellular biochemistry. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- 43.Lopatina N, et al. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. Journal of cellular biochemistry. 2002;84:324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- 44.Hermann A, et al. Biochemistry and biology of mammalian DNA methyltransferases. Cellular and molecular life sciences: CMLS. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eads CA, et al. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer research. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 46.Robertson KD, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic acids research. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Current opinion in genetics & development. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feser J, et al. Elevated histone expression promotes life span extension. Molecular cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Sullivan RJ, et al. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nature structural & molecular biology. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivanov A, et al. Lysosome-mediated processing of chromatin in senescence. The Journal of cell biology. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Z, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes & development. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraga MF, Esteller M. Towards the human cancer epigenome: a first draft of histone modifications. Cell Cycle. 2005;4:1377–1381. doi: 10.4161/cc.4.10.2113. [DOI] [PubMed] [Google Scholar]
- 56.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 57.Zhou BR, et al. Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome. Journal of molecular biology. 2012;421:30–37. doi: 10.1016/j.jmb.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma GG, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Molecular and cellular biology. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Molecular and cellular biology. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vempati RK, Haldar D. DNA damage in the presence of chemical genotoxic agents induce acetylation of H3K56 and H4K16 but not H3K9 in mammalian cells. Molecular biology reports. 2012;39:303–308. doi: 10.1007/s11033-011-0739-9. [DOI] [PubMed] [Google Scholar]
- 61.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 62.Pruitt K, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS genetics. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 64.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 66.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nature reviews. Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan H, et al. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther. 2013;6:1399–1416. doi: 10.2147/OTT.S37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calvanese V, et al. The role of epigenetics in aging and age-related diseases. Ageing research reviews. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 70.Schotta G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & development. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Regha K, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Molecular cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 73.Pogribny IP, et al. Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4-20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis. 2006;27:1180–1186. doi: 10.1093/carcin/bgi364. [DOI] [PubMed] [Google Scholar]
- 74.Tryndyak VP, et al. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther. 2006;5:65–70. doi: 10.4161/cbt.5.1.2288. [DOI] [PubMed] [Google Scholar]
- 75.Jorgensen S, et al. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic acids research. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & development. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarg B, et al. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. The Journal of biological chemistry. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 79.Chicas A, et al. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8971–8976. doi: 10.1073/pnas.1119836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elsheikh SE, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer research. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 81.Barlesi F, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:4358–4364. doi: 10.1200/JCO.2007.11.2599. [DOI] [PubMed] [Google Scholar]
- 82.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 84.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 85.Liu Y, et al. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein & cell. 2013 doi: 10.1007/s13238-013-3054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chandra T, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Molecular cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sadaie M, et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes & development. 2013;27:1800–1808. doi: 10.1101/gad.217281.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muller-Tidow C, et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116:3564–3571. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xia R, et al. High expression of H3K9me3 is a strong predictor of poor survival in patients with salivary adenoid cystic carcinoma. Archives of pathology & laboratory medicine. 2013;137:1761–1769. doi: 10.5858/arpa.2012-0704-OA. [DOI] [PubMed] [Google Scholar]
- 91.Park YS, et al. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Annals of surgical oncology. 2008;15:1968–1976. doi: 10.1245/s10434-008-9927-9. [DOI] [PubMed] [Google Scholar]
- 92.Aplan PD. Chromosomal translocations involving the MLL gene: molecular mechanisms. DNA repair. 2006;5:1265–1272. doi: 10.1016/j.dnarep.2006.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 95.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annual review of biochemistry. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 96.Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Current opinion in cell biology. 2012;24:374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shah PP, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes & development. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nature genetics. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & development. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bachmann IM, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 102.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 103.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 104.Kondo Y, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nature genetics. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 105.Burgess RC, et al. DNA damage, chromatin, and transcription: the trinity of aging. Current opinion in cell biology. 2012;24:724–730. doi: 10.1016/j.ceb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 109.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hangauer MJ, et al. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS genetics. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoon JH, et al. LincRNA-p21 suppresses target mRNA translation. Molecular cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.David R. RNA: a new layer of regulation. Nature reviews. Molecular cell biology. 2011;12:766. doi: 10.1038/nrm3225. [DOI] [PubMed] [Google Scholar]
- 113.Maass PG, et al. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014 doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 114.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quagliata L, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takayama K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beckedorff FC, et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS genetics. 2013;9:e1003705. doi: 10.1371/journal.pgen.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 121.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abdelmohsen K, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. 2013;12:890–900. doi: 10.1111/acel.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 124.Matheu A, et al. Anti-aging activity of the Ink4/Arf locus. Aging Cell. 2009;8:152–161. doi: 10.1111/j.1474-9726.2009.00458.x. [DOI] [PubMed] [Google Scholar]
- 125.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pasmant E, et al. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 129.Sharma V, Misteli T. Non-coding RNAs in DNA damage and repair. FEBS Lett. 2013;587:1832–1839. doi: 10.1016/j.febslet.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wan G, et al. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cellular signalling. 2013;25:1086–1095. doi: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wan G, et al. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013;32:2833–2847. doi: 10.1038/emboj.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Scaffidi P, Misteli T. In vitro generation of human cells with cancer stem cell properties. Nature cell biology. 2011;13:1051–1061. doi: 10.1038/ncb2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roukos V, et al. The cellular etiology of chromosome translocations. Current opinion in cell biology. 2013;25:357–364. doi: 10.1016/j.ceb.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scaffidi P, Misteli T. Cancer epigenetics: from disruption of differentiation programs to the emergence of cancer stem cells. Cold Spring Harbor symposia on quantitative biology. 2010;75:251–258. doi: 10.1101/sqb.2010.75.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Goodarzi AA, et al. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA repair. 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 136.Nagarajan RP, et al. Recurrent epimutations activate gene body promoters in primary glioblastoma. Genome research. 2014 doi: 10.1101/gr.164707.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ellinger J, et al. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. International journal of cancer. Journal international du cancer. 2010;127:2360–2366. doi: 10.1002/ijc.25250. [DOI] [PubMed] [Google Scholar]
- 138.Manuyakorn A, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.He LR, et al. Prognostic impact of H3K27me3 expression on locoregional progression after chemoradiotherapy in esophageal squamous cell carcinoma. BMC cancer. 2009;9:461. doi: 10.1186/1471-2407-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gibb EA, et al. Human cancer long non-coding RNA transcriptomes. PloS one. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Choe MK, et al. Functional elements demarcated by histone modifications in breast cancer cells. Biochemical and biophysical research communications. 2012;418:475–482. doi: 10.1016/j.bbrc.2012.01.042. [DOI] [PubMed] [Google Scholar]