Abstract
Short tandem repeat (STR) variation has been proposed as a major explanatory factor in the heritability of complex traits in humans and model organisms. However, we still struggle to incorporate STR variation into genotype-phenotype maps. Here, we review the promise of STRs in contributing to complex trait heritability, and highlight the challenges that STRs pose due to their repetitive nature. We argue that STR variants are more likely than single nucleotide variants to have epistatic interactions, reiterate the need for targeted assays to accurately genotype STRs, and call for more appropriate statistical methods in detecting STR-phenotype associations. Lastly, we suggest that somatic STR variation within individuals may serve as a read-out of disease susceptibility, and is thus potentially a valuable covariate for future association studies.
Keywords: short tandem repeats, microsatellites, heritability, epistasis, sequencing technologies
The ‘missing heritability’ of complex diseases and STR variation
Complex diseases such as diabetes, various cancers, cardiovascular disease, and neurological disorders cluster in families, and are thus considered to have a genetic component [1–3] (Glossary). The identification of these genetic factors has proven challenging; although genome-wide association (GWA) studies have identified many genetic variants that are associated with complex diseases, these generally confer less disease risk than expected from empirical estimates of heritability. This discrepancy, termed the ‘missing heritability’, has been attributed to many factors [1–6]. A trivial explanation is that shared environments among relatives may artificially inflate estimates of heritability. However, missing heritability may also be due to variants in the human genome that are currently inaccessible at a population scale [1,2]. One such class of variation is short tandem repeat (STR) unit number variation. Some have previously suggested that adding STR variation to existing genetic models would considerably increase the proportion of heritability explained by genetic factors in human disease [7,8]. Three percent of the human genome consists of STRs [9] and 6% of human coding regions are estimated to contain STR variation [10,11]. Recently, the first catalog of genome-wide population-scale human STR variation has appeared [12], opening up new possibilities for understanding the contribution of STRs to human genetic diseases. This catalog, and similar data sources [13], have appeared decades after initial calls for the assessment of the role of STRs in phenotypic variation [14], lagging behind surveys of other genomic elements. Much of the initial interest in STRs was generated by the discovery of phenomena such as genetic anticipation, which are mediated by the unique features of STRs [15]. As we discuss, new and forthcoming data sources will help to realize the long-deferred promise of STRs for explaining heritability.
STRs consist of short (2–10 bp) DNA sequences (units) that are repeated head-to-tail multiple times. This structure causes frequent errors in recombination and replication that add or subtract units, leading to STR mutation rates that are 10-fold to 104-fold higher than those of non-repetitive loci [16,17]. Due to technical barriers, STR variation has until very recently remained inaccessible to genome-wide assessment.
STRs are often conserved (even if their unit number or even sequence changes), especially in coding sequences [18–21]. In both humans and the yeast Saccharomyces cerevisiae, promoter regions are known to be dramatically enriched for STRs [22,23]. In coding regions, STRs tend to occur in genes with roles in transcriptional regulation, DNA binding, protein-protein binding, and developmental processes [16,21, 22]. These consistent functional enrichments across vastly diverged lineages suggest important functional roles for STRs.
Indeed, analysis of STR variation in the Drosophila Genetic Reference Panel identified dozens of associations between STR variants and quantitative phenotypes in recombinant inbred fly lines [13]. Moreover, accumulating evidence from exhaustive genetic studies shows that STR variation has dramatic, often background-dependent phenotypic effects in model organisms [25–29]. Together, these findings suggest that STR variation has the potential to dramatically revise the heritability estimates attributable to genetic factors.
The high STR mutation rate also leads to substantial somatic variation of STR loci within individuals. In fact, this somatic variation, also called microsatellite instability (MSI), has been used for decades as a biomarker for different classes of cancer [30]. Recent studies demonstrate that organisms exposed to various environmental stresses and perturbations show increased genome instability, including MSI [31–34]. MSI may be useful as a biomarker for cellular stress states that may predispose to disease.
The broad interest in STR variation has led to the development of techniques for high-throughput genotyping of STRs [35,36] and an explosion of analysis tools for extracting STR variation from existing sequence data [37–39]. However, the precision of these methods remains limited, due to a combination of low effective coverage of STRs and the lack of robust models for distinguishing technical error from somatic variation. Attempts to use STR variation for GWA in a fashion equivalent to SNV variation may be underpowered and confounded by the unique characteristics of this class of variants. In this review, we discuss the latest advances in these fields, and lay out a set of priorities for the future study of STRs.
STR variation is associated with human genetic diseases
Within coding regions, STR mutations are generally in-frame additions and subtractions of repeat units, resulting in proteins with variable, low-complexity amino acid runs [21]. These mutations can result in phenotypic effects and lead to genetic disorders; several neurological diseases (spinocerebellar ataxias, Huntington’s disease, spinobulbar muscular atrophy, dentatorubral-pallidoluysian atrophy, intellectual disability, etc.) are a consequence of dramatically expanded STR alleles [7,40,41]. Many of these disease-associated STR expansions behave as dominant gain-of-function mutations [7]. However, even comparatively modest coding STR variation may confer disease risk or behavioral phenotypes, according to a variety of single-marker association studies [42–45]; for instance, variants in separate coding STRs in RUNX2 are associated with defects in bone mineralization and higher incidence of fractures [46,47]; STR variation in this gene in dogs is also associated with craniofacial phenotypes [48]. Noncoding STR variation in regulatory sequences can affect transcription, RNA stability, and chromatin organization. For instance, certain STR variants alter CFTR expression and thus cystic fibrosis status [16]. We take these studies as evidence that STR variation, even in the absence of large expansions, may contribute significantly to the heritability of human traits and genetic diseases.
The severity of the STR expansion-associated diseases may suggest that natural selection should eliminate STRs in functional regions, but several recent studies across many organisms indicate that variable STRs are globally maintained [19,20,24,49,50]. For example, the pre-expansion polyQ-encoding STR in the human gene SCA2 is under positive selection, suggesting that this variable STR is actively maintained in spite of the pathogenic expansions that occasionally occur and cause spinocerebellar ataxia [51]. Considering both the evidence of positive selection on STRs and the functional enrichments of STR-containing genes, several authors have proposed that functional STRs are maintained because they confer ‘evolvability’, or the capacity for fast adaptation [21,22,52–54]. This suggestion is intriguing, in part because many STR mutations are dominant, and, when beneficial, can quickly sweep to fixation. Although we do not further discuss these evolutionary considerations here, they underscore the phenotypic potential of STR variation.
STR variation has dramatic background-dependent effects on phenotype
To date, the functional consequences of unit number variation in selected STRs have been studied in plants, fungi, flies, voles, dogs, and fish [25,27,28,55–57], among other organisms. In S. cerevisiae, STR unit number in the FLO1 gene accurately predicts the phenotype of cell-cell and cell-substrate adhesion (flocculation); flocculation provides protection against various stresses [57,58]. STR variation in yeast promoters has been shown to alter gene expression [22]. In Drosophila melanogaster, Neurospora crassa, and Arabidopsis thaliana, natural coding STR variation in circadian clock genes alters diurnal rhythmicity and developmental timing [25–27,59]. Some have proposed that the large phenotypic responses to selection observed in the Canidae are a consequence of elevated STR mutation rates relative to other mammalian clades [48,53]. We can state unambiguously that naturally variable STRs underlie dramatic phenotypic variation in model organisms.
Beyond the observable fact that variable STRs affect phenotype, we can make specific predictions about the components of phenotypic variation that they affect. Both theoretical expectations and empirical data indicate that STR variants are likely to participate in epistatic interactions, and probably more so than most SNVs. One plausible hypothesis is that STRs act as mutational modifiers of other loci, as may be expected intuitively from their elevated mutation rate (Box 1, Figure I).
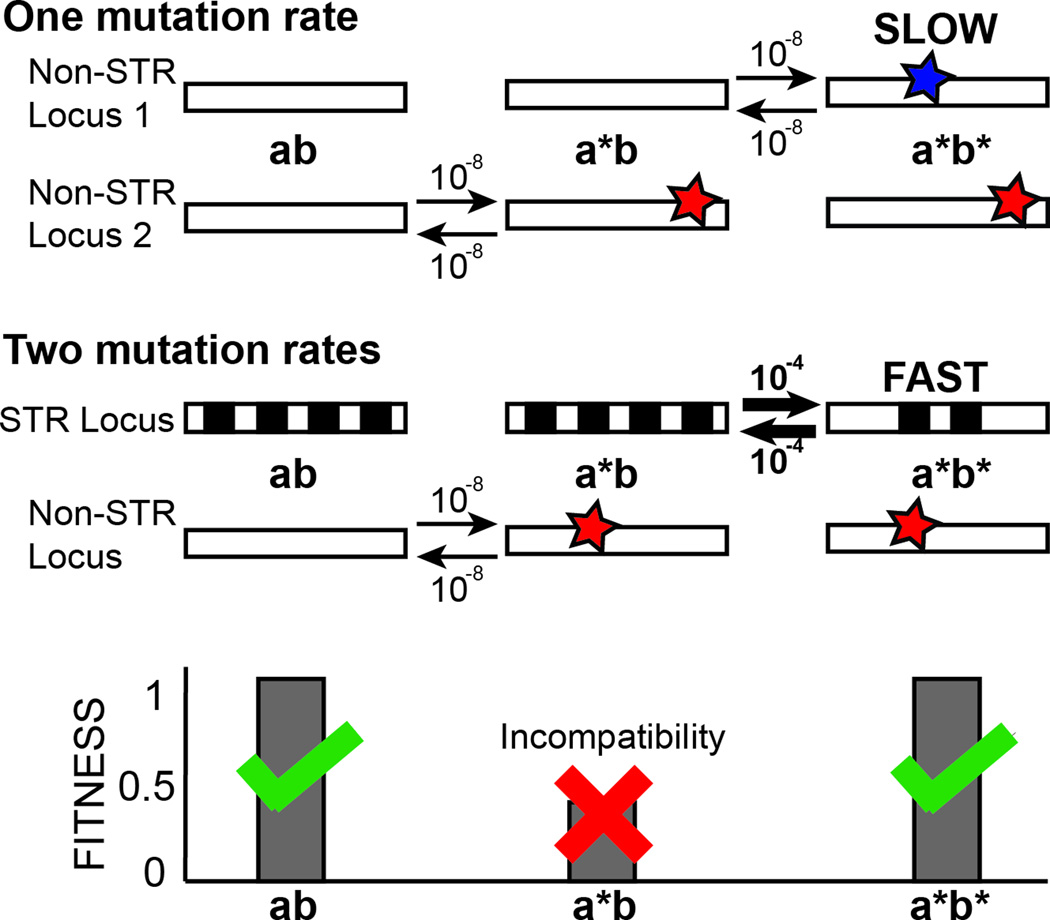
BOX 1: Modifier mutations leading to epistasis are expected in STRs.
We have previously proposed that STRs might be more susceptible to genetic interactions [25], as we will briefly explicate here. Consider a simple two-locus haploid model under panmixis, in which loci A and B each start with a single allele (ab) and have the same probability p per generation of mutating to a second allele (a* or b*), with p also as the probability per generation of reverting mutations (Figure I). Let us further assume that A and B are in sign epistasis [99] (that is, a*b and/or ab* have fitness less than ab and a*b*). To escape the unfavorable a*b genotype, the organism may either revert to ab or mutate forward to a*b*. When the A and B loci have equal mutation rates, we expect that the reversion of a single mutant is just as likely as a second mutation, and consequently that a*b* individuals will appear only relatively rarely and slowly. However, consider a similar model, in which locus B has an elevated mutation rate pb > pa. In this case, the a*b genotype has a higher probability of a second, modifying mutation to a*b* than of a reversion to ab. Moreover, flux along the other mutational path (ab → ab* → a*b*) will be increased. In sum, a*b* genotypes will arise at higher rates, and will attain their equilibrium frequency much more rapidly, if either A or B has an elevated mutation rate [100] (p.131). This scenario can lead quickly to an equilibrium population in which incompatible epistatic alleles are frequent, even though recombinants have lower fitness. Relaxing the assumption of no population structure will further speed this process. Consequently, we would expect STRs and other loci with high mutation rates to be more likely to modify other alleles than loci with lower mutation rates, as long as we assume that all loci are equally capable of genetic interactions. This process may be referred to as ‘coadaptation’. For a rigorous model of the evolution of hybrid incompatibility, see Orr [101].
Figure I.
A locus with higher mutation rates allows genetic modification of unfavorable genotypes at interacting loci. Top, a model of evolution under epistasis with only one slow mutation rate. Middle, a model of evolution under epistasis with a slow and a fast mutation rate. Boxes represent loci, stars represent SNV-type mutations, black and white checkering indicates an STR locus (a/b, a*/b, and a*/b* signify different genotypes). Arrows with numbers represent possible mutations and their respective rates. Bottom, fitness of each genotype under both models. We expect that the model with two mutation rates will occupy the fully derived state (a*/b*) more quickly.
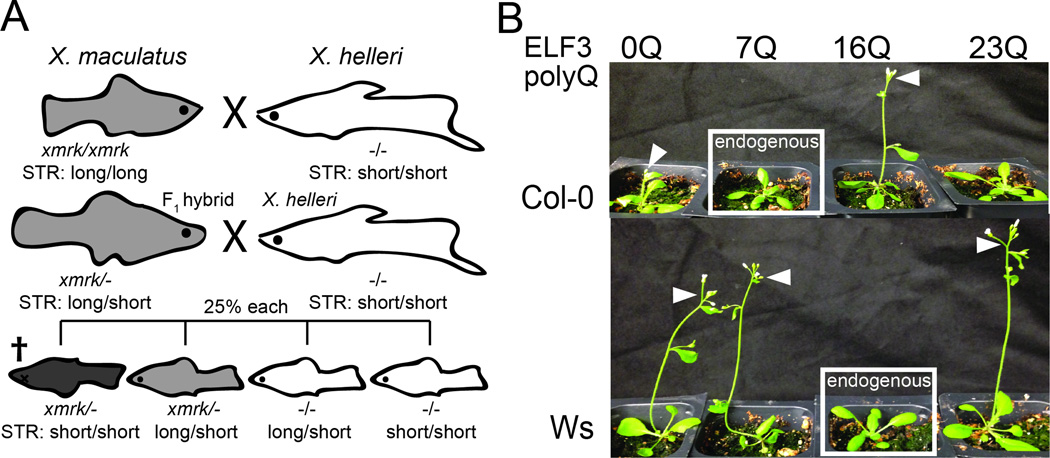
This expectation is borne out in the handful of studies reporting exhaustive genetic analysis of STRs. For instance, in the Xiphophorus genus of fish, a genetic incompatibility has recently been attributed to the interaction between the xmrk oncogene and an STR in the promoter of the tumor suppressor cdkn2a/b [29,60]. If the xmrk gene product is not properly regulated by cdkn2a/b, fish develop fatal melanomas, a two-locus Bateson-Dobzhansky-Muller incompatibility described in classic genetic experiments (Figure 1A) [61–63]. Expansions in the cdkn2a/b promoter STR are associated with the presence of a functional copy of the xmrk oncogene across species, and are thought to functionally repress the activity of the xmrk gene product through increased dosage of the tumor suppressor [29].
Figure 1. Genetic and transgenic analysis reveals STR-mediated incompatibilities.
A, the Gordon-Kosswig-Anders cross shows a genetic incompatibility between two fish species in the Xiphophorus genus. Modified from Meierjohann and Schartl [63]. F1 hybrids back-crossed to their X. helleri parent yield a 3:1 ratio of viability, where the inviables result from co-segregation of the functional xmrk gene and a short STR allele in the cdkn2a/b promoter. Shading indicates melanism conferred by xmrk. B, genetic background is epistatic to effects of ELF3 STR variation in A. thaliana. Expression-matched transgenic plants with various alleles of the ELF3 STR in the Columbia (Col-0) and Wassilewskija (Ws) backgrounds, showing endogenous, exogenous, and synthetic (“0”) alleles in each background [25]. White boxes indicate transgenic plants carrying the ELF3 STR endogenous to their respective background; white arrowheads indicate early-flowering ELF3 STR genotypes (elf3 mutants and poorly-functioning ELF3 STR alleles confer early flowering).
Similarly, we have shown that natural variation in the polyQ-encoding ELF3 STR significantly affects all ELF3-dependent phenotypes in the plant A. thaliana, with ELF3 STR length and phenotype showing a strikingly nonlinear relationship (Figure 1B)[25]. Some naturally occurring ELF3 STR variants phenocopy elf3-loss-function mutants in a common reference background (Figure 1B), suggesting background-specific modifiers. Indeed, when we compare the phenotypic effects of each ELF3 STR variant between two divergent backgrounds, Columbia (Col-0) and Wassilewskija (Ws), we find dramatic differences. The endogenous STR alleles from these two strains (Col-0 7 units, Ws 16 units) show mutual incompatibility when exchanged between backgrounds. The ELF3 protein is thought to function as an “adaptor protein” or physical bridge in diverse protein complexes [64,65]. We speculated that background-specific polymorphisms in these interacting proteins underlie the ELF3 STR-dependent background effect.
Also in A. thaliana, a variable STR in the promoter of the CONSTANS gene has been linked to phenotypic variation in the onset of flowering [28]. CONSTANS encodes a major regulatory protein that promotes flowering. Transgenic experiments demonstrate that this regulatory STR variation affects CONSTANS expression and hence onset of flowering. However, the effects of this STR variation depend on the presence of a functional allele of FRIGIDA, a negative regulator of flowering that is highly polymorphic across A. thaliana populations.
A dramatic example of incompatibility can be found in an intronic repeat in the IIL1 gene in A. thaliana, which was found to be dramatically expanded in one strain [55]. The expansion delayed flowering under high temperatures, but when crossed into the reference genetic background, a strongly interacting locus modifies this phenotype.
In the Drosophila genus, coding STR variation in the per gene co-evolves with other variants [59,66]. Transgenic flies expressing chimeric per genes with a D. melanogaster STR domain fused to a D. pseudoobscura flanking region (and vice versa) have arrhythmic circadian clocks, indicating the modifying effect of flanking variation in generating an STR-based genetic incompatibility. Among STRs subjected to exhaustive genetic study, to our knowledge, only the yeast FLO1 coding STR has no known modifiers due to variation in genetic background [57].
In addition to these exhaustive genetic studies, there are several other observations that support the role of the genetic background in controlling the phenotypic effects of STRs. For instance, experiments in Caenorhabditis elegans and human cells indicate that the phenotypic effects of proteins with expanded polyQ tracts are modulated by genetic background [67], or by variants in interacting proteins [68]. In humans, genetic association studies indicate the existence of genetic modifiers of polyQ expansion disorders for both Huntington’s disease [69] and spinocerebellar ataxias [70]. Taken together, these experimental and observational data support our argument that functional STRs are likely to be enriched for variants in epistasis with other loci.
STRs with background-dependent phenotypic effects tend to either encode polyQ tracts or reside in promoter regions. There are good reasons to expect that these STR classes might be enriched in DNA/protein-protein interactions that could underlie epistasis. PolyQ tracts, specifically, often bind DNA surfaces [71], and an analysis of human protein interactome data found that polyQ-containing proteins engage in more physical interactions with other proteins than those without polyQs [72]. Similarly, noncoding STRs in regulatory regions may compensate for mutations in trans-acting factors, as observed for the STRs in the cdkn2a/b promoter in Xiphophorus [29] and in the CONSTANS promoter in A. thaliana [28]. We suggest that polymorphisms in protein interaction partners or in transcriptional regulators are plausible explanations for the observed background effects. In summary, we expect that STR variation is likely to contribute a substantial epistatic component to heritability, which has important implications for their use in explaining phenotypic variation.
Analytical tools and genotyping methods continue to struggle with STR-specific challenges
To fulfill the promise of STR variation for explaining heritability, we need accurate, genome-wide assessment of STR variation in populations of humans and other organisms. The scientific community has tackled this problem in a flurry of recent studies describing methods for genotyping STRs genome-wide (Table 1). Specifically, in the last two years, several analytical tools have been developed to call STR genotypes from whole-genome-sequencing data [37–39]. These tools attempt to address the two major challenges for genotyping STRs: poor mappability due to low sequence complexity and high technical error rate due to amplification stutter.
Table 1.
Technologies for assessing STR variation by high-throughput sequencing.
| Name | Data Source | Analysis strategy |
Accepted coveragea |
Reported accuracy |
Reported efficiency |
Limitations | Ref. |
|---|---|---|---|---|---|---|---|
| lobSTR | Human, whole-genomeb,c | Align to modified reference | 1 read | 88%–95% | 0.2% of reads are informative | Depends on depth of sequencing and length of reads | [38] |
| RepeatSeq | Human, whole-genomeb,d | Align to reference, locally realigned | 2 reads | 92% | Not reported | Depends on depth of sequencing and length of reads | [37] |
| STRViper | A. thaliana whole-genomeb,d | Compare insert size to reference | 10 reads | 74% | Not reported | Cannot call STR unit number genotypes | [39] |
| Array Capture | Human, array captureb | RepeatSeq | 2 reads | 88%–92% | 2.2% informative reads | Low enrichment for STR-spanning reads | [35] |
| SureSelect RNA probe capture | Human, target enrichment, Roche 454 | Locally align flanking regions | 4 reads | 88%–95% | 27% informative reads | Expensive probe design, captured only 60% of targeted STRs | [36] |
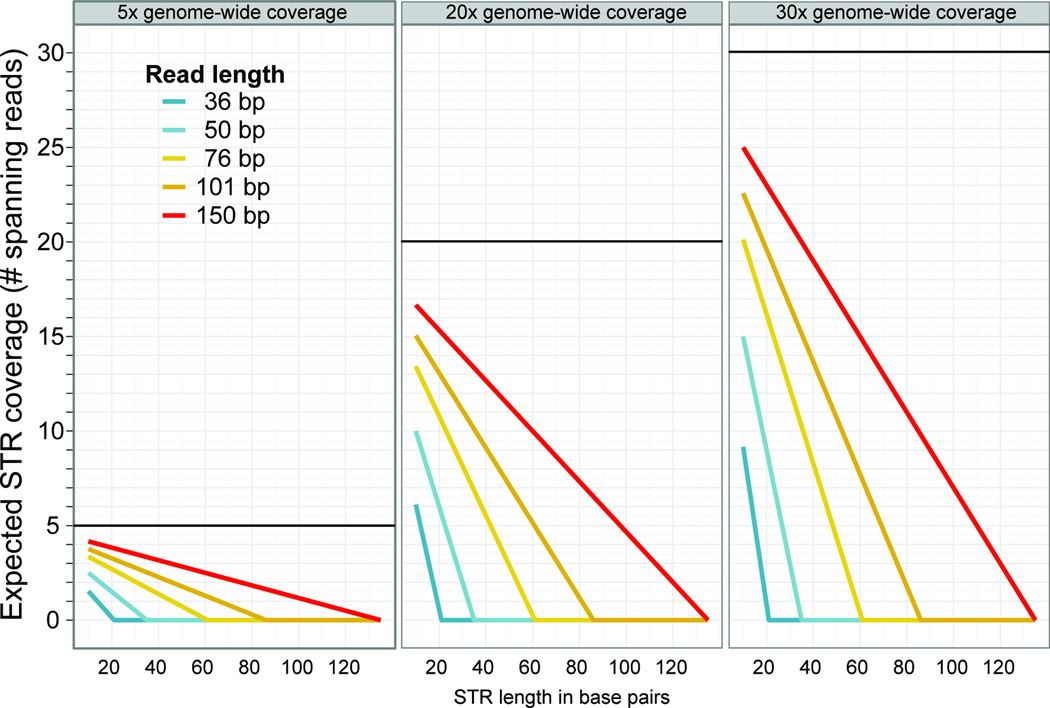
To accurately map an STR sequence read and retrieve its unit number genotype, the sequence read must span the STR of interest and include some unique flanking sequence. This requirement limits the length of STRs that can be accurately genotyped and decreases effective STR coverage compared to average whole-genome-sequencing coverage (Figure 2). For this reason, much of the existing sequencing data, which consists largely of short reads (36 bp, 50 bp, or 76 bp) with only modest genome coverage (5–20×) is not suitable for accurate, genome-wide calls of STR genotypes; only a fraction of STRs, mostly short ones, can be assessed with some confidence (Figure 2).
Figure 2. Effective reduction in STR coverage in whole-genome sequencing.
Expected coverage of STRs for various sequencing depths and read lengths. We assumed 8 bp of flanking sequence on either side (per requirement for LobSTR software [38]). Black bars indicate nominal sequencing coverage for each scenario. 4–5× coverage (left panel) is typical for genomes in the human 1000 Genomes Project [95]; 15–20– coverage is typical for genomes in the A. thaliana 1001 Genomes Project [97,98].
Moreover, these analytical tools estimate technical error based on STR genotypes from sequenced homozygous or haploid genomes, ignoring somatic alleles within individuals (which are expected for STRs even in primary tissues, occurring at rates 104–105 times higher than SNV somatic mutations) [73–76]. Probabilistic error models have been formulated to quantify variation arising from technical sources [37,38], but in the face of somatic STR variation, these models presumably require substantial read coverage to call germ-line STR genotypes with confidence. However, because of the low effective coverage of STR loci (Figure 2), STR genotype calls are based on as few as one to two STR-spanning reads [37,38] (Table 1). Calls based on so few reads may not be accurate even for homozygous germline alleles. Calling heterozygous STR genotypes remains difficult with the modest coverage of most available whole-genome-sequencing data, such as found in the 1000 Genomes Project [12], which becomes even more challenging when potential somatic mutations contribute to a heterogeneous sample population. To illustrate this challenge, consider a heterozygous ~30 bp-STR locus and whole-genome sequencing with 101 bp-reads at 5× coverage – this scenario is likely to yield just three STR-spanning reads (Figure 2). These three reads may represent one, two, or three different alleles, representing any mixture of two different germ-line alleles, somatic alleles, or technical error, making an accurate call difficult. Consequently, an increase in the sequencing depth of available data may be required before these tools reach their full potential.
Others have attempted to genotype STRs using whole-genome-sequencing data from paired-end reads (50bp) of size-selected genomic fragments [39], similar to strategies used to detect large insertions or deletions [77–80]. This approach is limited by the resolution of gel electrophoresis in the size selection of DNA fragments. Consequently, this method cannot determine STR unit number genotypes, but rather reports whether an STR is variable across samples. The authors argue that this approach is the most accurate for population-level detection of STR variability [81], but it is not informative for discerning the relationship between STR unit number genotype and phenotype.
Although these analysis tools represent important and useful advances, their limitations illustrate that ‘dustbin-diving’ of whole-genome-sequencing data may not suffice for accurate population-scale genotyping of STRs genome-wide. Alternative approaches that enrich for STR-spanning sequencing reads are needed. Indeed, two such approaches have been recently published. Both use targeted capture of STRs to enrich for STR-spanning reads combined with high-throughput sequencing compatible with midsize-reads (101 bp, 500 bp) [35,36]. Targeted STR capture requires the design of STR-specific probes (or rather probes specific to their unique flanking sequences) and involves additional sequencing, but these approaches can dramatically increase the number of informative reads, therefore providing substantial STR coverage for accurate genotyping calls (Table 1). For example, the SureSelect-RNA-probe capture method reports 27% informative STR-spanning reads compared to the 0.2 % informative reads found in whole-genome-sequencing data (Table 1). This increase in informative reads is a major advantage over whole-genome resequencing because STRs represent only a small fraction of the genome overall [35,36]. Although targeted capture combined with high-throughput sequencing appears to be a cost-effective alternative for accurate STR genotyping compared to whole-genome sequencing, distinguishing heterozygous alleles, somatic variants, and technical error remains a challenge. We suggest that recent innovations in single-molecule targeted capture [82] should be useful in distinguishing these categories and in further increasing enrichment of informative, STR-spanning reads.
Lack of statistical models for detecting STR-phenotype associations in GWA
Assuming that we obtain accurate, population-scale genotype data for STRs, we may not yet have statistical tools appropriate for detecting STR associations with phenotype [8]. In diploid organisms, a biallelic SNV is typically analyzed by modeling phenotype as a function of the number of non-reference alleles at that locus (0, 1, or 2) in each individual. A null hypothesis of no monotonic relationship between phenotype and the allele count is then formulated and tested [83]. This framework cannot accommodate more than two alleles, which we would expect for many STRs. Simply using tagged SNVs linked to STRs to perform GWA is unfeasible, because linkage disequilibrium decays very quickly between SNVs and STRs across human populations [12].
To address these complications, a previous study attempted GWA between STR genotypes and human disease phenotypes by comparing relative frequencies of various alleles in pooled DNA from cases and controls [84]. By pooling samples, this approach eases the analysis of multiallelic loci, but it loses information by ignoring specific individuals.
In a more recent study, the authors used logistic regression and the analysis of variance to detect associations between STR alleles and quantitative phenotypes in an inbred Drosophila mapping population [13]. Given that significant associations were detected, such approaches may be sufficiently powerful in recombinant inbred lines. However, their strategy relied on homozygosity, and considered multiallelic STRs in a pairwise fashion, so these straightforward methods will lose power with outbred populations and multiallelic STRs.
The central confounder of these studies is that most STRs of appreciable variability (and thus, interest) are multiallelic, as a simple consequence of the STR mutational mechanism [17]. This multiallelic feature could be accommodated by treating STR alleles categorically, but this choice entails a corresponding reduction in power, because many alleles are rare.
Some studies have reported linear associations between STR unit number and quantitative phenotypes [27,57], suggesting that using simple tests of linear correlation between these variables may be a powerful option. However, this linearity (or even monotonicity) of the relationship between STR unit number genotype and phenotype is a poorly-supported assumption [25]. Nonetheless, STR unit number is a numerical variable, and it would be preferable to gain power from treating it as such. For instance, more similar STR unit number genotypes might be associated with more similar phenotypes, but this intuition may be difficult to generalize.
Lastly, both intuition (Box 1) and the studies discussed above lead us to expect that relatively many phenotypically relevant variable STRs will show epistasis with other loci. This epistasis will reduce power in tests of association between STRs and phenotype [85], given the inadequacy of the current paradigm of quantitative genetics in detecting and modeling the effects of epistasis [85,86]. At present, targeted and exhaustive genetic studies (as described above) are the only effective method for understanding the effects of epistasis.
In total, these obstacles present a daunting challenge for the integration of STR genotypes into the current genotype-phenotype maps. Overall, we call for a reappraisal of statistical methodologies for use in GWA with STR variation to account for these various STR-specific confounders.
Somatic STR variation may be a sensitive marker for increased disease susceptibility
It has been appreciated for some time that the high STR mutation rate leads to somatic variation within individuals in addition to germ-line variation between individuals [71]. This somatic STR variation is particularly noticeable in tumor tissues, but is also measurable in primary tissues [73,87]. While these findings immediately led to systems of classification for tumor types and clones [76,88,89], the investigation of somatic STR variation (or MSI) may also inform us about general phenotypic states and disease susceptibility.
Patients with various complex diseases tend to carry a greater load of rare germ-line variants than unaffected control groups [6]. It is widely assumed that these rare variants contribute in some fashion to these disorders [90]; however, an alternative interpretation holds that they are signs of stochastic genome instability, which when increased leads to higher susceptibility to complex diseases. [6]. Increased genome instability will increase somatic variation, which may then serve as a read-out of disease susceptibility [6]. This alternative interpretation has some support from empirical data. For instance, perturbation of the molecular chaperone Hsp90, which stabilizes diverse DNA repair proteins, leads to increased somatic STR mutation rates in human cells; in various model organisms Hsp90 perturbation increases transposon mobility and intrachromosomal homologous recombination [31–34]. Hsp90 perturbation also increases the penetrance of many genetic variants in flies, plants, fish, worms and yeast, suggesting that increased genome instability and increased phenotypic heritability are associated [34]. If this association also applies to disease phenotypes, increased genome instability may predict higher disease susceptibility.
Consequently, although somatic MSI may not be the cause of disease phenotypes, it may serve as a biomarker for individuals who are more vulnerable to environmental and genetic perturbations leading to disease. Again, this strategy hinges on the development of cost-effective technologies for screening panels of STRs for somatic mutations across many humans, which will require new strategies to distinguish technical error from somatic STR variation.
Another possibility is that somatic variation is itself phenotypically relevant, or even plays a role in developmental processes. It is known that STRs are enriched in genes with neuronal function [91]; some have even proposed that such somatic mutation is a component of normal neuronal development in humans [92]. If this is the case, then a greater appreciation of somatic variation will be necessary to understand canonical developmental processes. Collectively, STR variation within (in addition to between) individuals has great potential as a read-out for disease susceptibility, and perhaps also as a cause of phenotypic variation itself.
Concluding remarks
The study of STRs and other under-ascertained genomic elements has the potential to reshape our model of the heritability of complex diseases and traits, both in terms of the overall proportion of heritability explained, and in terms of the components of heritability themselves (Outstanding Questions). Experimental studies in model organisms have taught us that the phenotypic effects of genome-wide STR variation are both dramatic and impossible to understand without taking epistasis into account. In the future, our understanding will be improved by 1) accurate STR population-scale and somatic genotyping, 2) more appropriate statistical methods for analyzing STR-phenotype associations, and 3) a broader description of epistasis between STR variation and other loci in determining phenotype.
OUTSTANDING QUESTIONS.
In light of wide-spread epistasis, what statistical and experimental tools can quantify the effect of STR variation on phenotype?
Can inexpensive, accurate tools be developed for germ-line and somatic STR genotyping?
Will somatic STR variation be effective as a readout for disease susceptibility?
HIGHLIGHTS.
Short tandem repeat (STRs) variation is understudied but may majorly contribute to heritability.
STR variation likely engages in epistatic interactions with other loci.
Association of STR genotype with phenotype poses unique challenges.
Assessment of somatic STR variation may hold promise as a covariate in future GWA studies
Acknowledgements
This work was supported by grants from the National Human Genome Research Institute Interdisciplinary Training in Genomic Sciences (2T32HG35-16 to MOP, T32 HG00035 to KDC) and the National Institute of Health New Innovator Award (DP2OD008371 to CQ). We are grateful to members of the Queitsch and Shendure laboratories for helpful discussions, and to Wen Huang and David Mittelman for responding to email inquiries.
GLOSSARY
- Short tandem repeat (STR)
a repetitive nucleotide sequence that consists of many copies of a short sequence in tandem (ex. CAGCAGCAGCAG). STRs are frequently called microsatellites.
- Single nucleotide variant (SNV)
Variant that consists of a change at a single nucleotide position. Common SNVs are sometimes called single nucleotide polymorphisms (SNPs).
- Heritability
The fraction of variation in a phenotype across a population that can be attributed to genetic differences.
- Epistasis
Non-reciprocal interactions of non-allelic gene variants, due for instance to functional interdependence between gene products in a protein complex or metabolic pathway.
- Genome-wide association (GWA)
A set of methods by which each of a large number of genetic variants genome-wide is tested for statistical associations with a phenotype. Often referred to in the context of genome-wide association studies (GWAS).
- Complex disease, complex traits
Complex diseases or traits are phenotypic characters thought to be affected by multiple genetic and environmental factors.
- Somatic variation
Genetic variation across somatic cells or tissues of an organism, which are generally not inherited by offspring (which inherits instead germ-line variation). Generally arises from mutations in specific cell lineages after early development.
- Microsatellite instability (MSI)
Somatic variation of STRs (microsatellites) associated with phenotypic changes such as cancer, often due to mutations in DNA repair genes.
- Bateson-Dobzhansky-Muller incompatibility
Hybrid incompatibilities observed when crossing two close species or divergent strains of a species with one another. Caused by the co-segregation of non-parental allele combinations, resulting in a dysfunctional genetic interaction (negative epistasis).
- Genetic anticipation
A mode of disease inheritance characterized by progressively earlier ages of disease onset as generations progress. Generally caused by the gradual expansion of STRs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuk O, et al. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom JS, et al. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494:234–237. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heng HHQ. Missing heritability and stochastic genome alterations. Nat. Rev. Genet. 2010;11:813. doi: 10.1038/nrg2809-c3. [DOI] [PubMed] [Google Scholar]
- 7.Fondon JW, et al. Simple sequence repeats: genetic modulators of brain function and behavior. Trends Neurosci. 2008;31:328–334. doi: 10.1016/j.tins.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Hannan AJ. Tandem repeat polymorphisms: modulators of disease susceptibility and candidates for “missing heritability”. Trends Genet. 2010;26:59–65. doi: 10.1016/j.tig.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian S, et al. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Dushlaine CT, et al. Tandem repeat copy-number variation in protein-coding regions of human genes. Genome Biol. 2005;6:R69. doi: 10.1186/gb-2005-6-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mularoni L, et al. Mutation patterns of amino acid tandem repeats in the human proteome. Genome Biol. 2006;7:R33. doi: 10.1186/gb-2006-7-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willems TF, et al. The Landscape of Human STR Variation. bioRxiv. 2014 doi: 10.1101/gr.177774.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay TFC, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashi Y, et al. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland GR, et al. Hereditary unstable DNA: a new explanation for some old genetic questions? Lancet. 1991;338:289–292. doi: 10.1016/0140-6736(91)90426-p. [DOI] [PubMed] [Google Scholar]
- 16.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol. Carcinog. 2009;48:379–388. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legendre M, et al. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Z, et al. Genome-wide analysis of tandem repeats in plants and green algae. G3. 2014;4:67–78. doi: 10.1534/g3.113.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaper E, et al. Deep Conservation of Human Protein Tandem Repeats within the Eukaryotes. Mol. Biol. Evol. 2014;31:1132–1148. doi: 10.1093/molbev/msu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, et al. Insight into role of selection in the evolution of polyglutamine tracts in humans. PLoS One. 2012;7:e41167. doi: 10.1371/journal.pone.0041167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gemayel R, et al. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 22.Vinces MD, et al. Unstable tandem repeats in promoters confer transcriptional evolvability. Science (80-.) 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawaya S, et al. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS One. 2013;8:e54710. doi: 10.1371/journal.pone.0054710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persi E, Horn D. Systematic analysis of compositional order of proteins reveals new characteristics of biological functions and a universal correlate of macroevolution. PLoS Comput. Biol. 2013;9:e1003346. doi: 10.1371/journal.pcbi.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Undurraga SF, et al. Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc. Natl. Acad. Sci. 2012 doi: 10.1073/pnas.1211021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michael TP, et al. Simple sequence repeats provide a substrate for phenotypic variation in the Neurospora crassa circadian clock. PLoS One. 2007;2:e795. doi: 10.1371/journal.pone.0000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawyer LA. Natural Variation in a Drosophila Clock Gene and Temperature Compensation. Science (80-.) 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- 28.Rosas U, et al. Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS. Nat. Commun. 2014;5:3651. doi: 10.1038/ncomms4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scarpino SV, et al. Evolution of a genetic incompatibility in the genus Xiphophorus. Mol. Biol. Evol. 2013;30:2302–2310. doi: 10.1093/molbev/mst127. [DOI] [PubMed] [Google Scholar]
- 30.Boland CR, et al. A National Cancer Institute Workshop on Microsatellite Instability for Cancer Detection and Familial Predisposition: Development of International Criteria for the Determination of Microsatellite Instability in Colorectal Cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 31.Kovalchuk I, et al. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature. 2003;423:760–762. doi: 10.1038/nature01683. [DOI] [PubMed] [Google Scholar]
- 32.Specchia V, et al. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 33.Mittelman D, et al. Hsp90 modulates CAG repeat instability in human cells. Cell Stress Chaperones. 2010;15:753–759. doi: 10.1007/s12192-010-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Queitsch C, et al. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet. 2012;8:e1003041. doi: 10.1371/journal.pgen.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilmatre A, et al. Rapid multiplexed genotyping of simple tandem repeats using capture and high-throughput sequencing. Hum. Mutat. 2013;34:1304–1311. doi: 10.1002/humu.22359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duitama J, et al. Large-scale analysis of tandem repeat variability in the human genome. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Highnam G, et al. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 2013;41:e32. doi: 10.1093/nar/gks981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gymrek M, et al. lobSTR: A short tandem repeat profiler for personal genomes. Genome Res. 2012;22:1154–1162. doi: 10.1101/gr.135780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao MD, et al. Inferring short tandem repeat variation from paired-end short reads. Nucleic Acids Res. 2014;42:e16. doi: 10.1093/nar/gkt1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 41.Poeta L, et al. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am. J. Hum. Genet. 2013;92:114–125. doi: 10.1016/j.ajhg.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caspi A, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. IL1 receptor antagonist gene IL1-RN variable number of tandem repeats polymorphism and cancer risk: a literature review and meta-analysis. PLoS One. 2012;7:e46017. doi: 10.1371/journal.pone.0046017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisenegger C, et al. DAT1 polymorphism determines L-DOPA effects on learning about others’ prosociality. PLoS One. 2013;8:e67820. doi: 10.1371/journal.pone.0067820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inanir A, et al. Association of IL-4 gene VNTR variant with deep venous thrombosis in Behçet’s disease and its effect on ocular involvement. Mol. Vis. 2013;19:675–683. [PMC free article] [PubMed] [Google Scholar]
- 46.Morrison NA, et al. Glutamine repeat variants in human RUNX2 associated with decreased femoral neck BMD, broadband ultrasound attenuation and target gene transactivation. PLoS One. 2012;7:e42617. doi: 10.1371/journal.pone.0042617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrison NA, et al. Polyalanine repeat polymorphism in RUNX2 is associated with site-specific fracture in post-menopausal females. PLoS One. 2013;8:e72740. doi: 10.1371/journal.pone.0072740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fondon JW, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mularoni L, et al. Highly constrained proteins contain an unexpectedly large number of amino acid tandem repeats. Genomics. 2007;89:316–325. doi: 10.1016/j.ygeno.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Mularoni L, et al. Natural selection drives the accumulation of amino acid tandem repeats in human proteins. Genome Res. 2010;20:745–754. doi: 10.1101/gr.101261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu F, et al. Positive selection of a pre-expansion CAG repeat of the human SCA2 gene. PLoS Genet. 2005;1:e41. doi: 10.1371/journal.pgen.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gemayel R, et al. Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes (Basel) 2012;3:461–480. doi: 10.3390/genes3030461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laidlaw J, et al. Elevated Basal Slippage Mutation Rates among the Canidae. J. Hered. 2007;98:452–460. doi: 10.1093/jhered/esm017. [DOI] [PubMed] [Google Scholar]
- 54.King DG. Indirect selection of implicit mutation protocols. Ann. N. Y. Acad. Sci. 2012;1267:45–52. doi: 10.1111/j.1749-6632.2012.06615.x. [DOI] [PubMed] [Google Scholar]
- 55.Sureshkumar S, et al. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science. 2009;323:1060–1063. doi: 10.1126/science.1164014. [DOI] [PubMed] [Google Scholar]
- 56.Hammock EAD, Young LJ. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- 57.Smukalla S, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verstrepen KJ, et al. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peixoto aa, et al. Molecular coevolution within a Drosophila clock gene. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4475–4480. doi: 10.1073/pnas.95.8.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butler AP, et al. Regulation of CDKN2A/B and Retinoblastoma genes in Xiphophorus melanoma. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2007;145:145–155. doi: 10.1016/j.cbpc.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 61.Gordon M. The genetics of a viviparous top-minnow platypoecilus; the inheritance of two kinds of melanophores. Genetics. 1927;12:253–283. doi: 10.1093/genetics/12.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kosswig C. Über Kreuzungen zwischen den Teleostiern Xiphophorus helleri und Platypoecilus maculatus. Z. Indukt. Abstammungs-Vererbungsl. 1928;47:150–158. [Google Scholar]
- 63.Meierjohann S, Schartl M. From Mendelian to molecular genetics: the Xiphophorus melanoma model. Trends Genet. 2006;22:654–661. doi: 10.1016/j.tig.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 64.Yu J-W, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell. 2008;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nusinow Da, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011 doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peixoto A, et al. Molecular evolution of a repetitive region within the per gene of Drosophila. Mol. Biol. Evol. 1993;10:127–139. doi: 10.1093/oxfordjournals.molbev.a039993. [DOI] [PubMed] [Google Scholar]
- 67.Gidalevitz T, et al. Natural genetic variation determines susceptibility to aggregation or toxicity in a C. elegans model for polyglutamine disease. BMC Biol. 2013;11:100. doi: 10.1186/1741-7007-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Metzger S, et al. Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum. Mol. Genet. 2008;17:1137–1146. doi: 10.1093/hmg/ddn003. [DOI] [PubMed] [Google Scholar]
- 69.Rubinsztein DC, et al. Genotypes at the GluR6 kainate receptor locus are associated with variation in the age of onset of Huntington disease. Proc. Natl. Acad. Sci. 1997;94:3872–3876. doi: 10.1073/pnas.94.8.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zühlke C, et al. Spinocerebellar ataxia type 17: report of a family with reduced penetrance of an unstable Gln49 TBP allele, haplotype analysis supporting a founder effect for unstable alleles and comparative analysis of SCA17 genotypes. BMC Med. Genet. 2005;6:27. doi: 10.1186/1471-2350-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Escher D, et al. Conservation of Glutamine-Rich Transactivation Function between Yeast and Humans. Mol. Cell. Biol. 2000;20:2774–2782. doi: 10.1128/mcb.20.8.2774-2782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaefer MH, et al. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012;40:4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slagboom PE, et al. Somatic mutations and cellular aging: two-dimensional DNA typing of rat fibroblast clones. Mutat. Res. 1991;256:311–321. doi: 10.1016/0921-8734(91)90022-4. [DOI] [PubMed] [Google Scholar]
- 74.Golubov A, et al. Microsatellite instability in Arabidopsis increases with plant development. Plant Physiol. 2010;154:1415–1427. doi: 10.1104/pp.110.162933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J-M, et al. A novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst. Biol. 2010;4:29. doi: 10.1186/1752-0509-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chapal-Ilani N, et al. Comparing algorithms that reconstruct cell lineage trees utilizing information on microsatellite mutations. PLoS Comput. Biol. 2013;9:e1003297. doi: 10.1371/journal.pcbi.1003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hajirasouliha I, et al. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics. 2010;26:1277–1283. doi: 10.1093/bioinformatics/btq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi J, Zhao F. inGAP-sv: a novel scheme to identify and visualize structural variation from paired end mapping data. Nucleic Acids Res. 2011;39:W567–W575. doi: 10.1093/nar/gkr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grimm D, et al. Accurate indel prediction using paired-end short reads. BMC Genomics. 2013;14:132. doi: 10.1186/1471-2164-14-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao MD, et al. Sequencing technologies and tools for short tandem repeat variation detection. Brief. Bioinform. 2014 doi: 10.1093/bib/bbu001. [DOI] [PubMed] [Google Scholar]
- 82.Hiatt JB, et al. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hayes B. Overview of Statistical Methods for Genome-Wide Association Studies (GWAS) Methods Mol. Biol. 2013;1019:149–169. doi: 10.1007/978-1-62703-447-0_6. [DOI] [PubMed] [Google Scholar]
- 84.Oka A, et al. Localization of a non-melanoma skin cancer susceptibility region within the major histocompatibility complex by association analysis using microsatellite markers. Tissue Antigens. 2003;61:203–210. doi: 10.1034/j.1399-0039.2003.00007.x. [DOI] [PubMed] [Google Scholar]
- 85.Mackay TFC. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 2014;15:22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nelson RM, et al. A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet. 2013;29:669–676. doi: 10.1016/j.tig.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 87.Armour JAL, et al. Analysis of somatic mutations at human minisatellite loci in tumors and cell lines. Genomics. 1989;4:328–334. doi: 10.1016/0888-7543(89)90338-8. [DOI] [PubMed] [Google Scholar]
- 88.Lee HS, et al. Microsatellite instability in synchronous gastric carcinomas. Int. J. Cancer. 2001;91:619–624. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1105>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 89.Kim T-M, et al. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Girirajan S, et al. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolton KA, et al. STaRRRT: a table of short tandem repeats in regulatory regions of the human genome. BMC Genomics. 2013;14:795. doi: 10.1186/1471-2164-14-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nithianantharajah J, Hannan AJ. Dynamic mutations as digital genetic modulators of brain development, function and dysfunction. Bioessays. 2007;29:525–535. doi: 10.1002/bies.20589. [DOI] [PubMed] [Google Scholar]
- 93.Green RE, et al. A draft sequence of the Neandertal genome. Science (80-.) 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 99.Weinreich DM, et al. Perspective: sign epistasis and genetic costraint on evolutionary trajectories. Evolution (N. Y) 2005;59:1165–1174. [PubMed] [Google Scholar]
- 100.Felsenstein J. Theoretical Evolutionary Genetics. 2013 http://evolution.gs.washington.edu/pgbook/pgbook.pdf. [Google Scholar]
- 101.Orr HA. The Population Genetics of Speciation: The Evolution of Hybrid Incompatibilities. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]