Abstract
Multiple circuitries ensure that cells respond correctly to the environmental cues within defined cellular programs. There is increasing evidence suggesting that cellular memory for these adaptive processes can be passed on through cell divisions and generations. However, the mechanisms by which this epigenetic information is transferred remain elusive largely because it requires that such memory survive through gross chromatin remodeling events during DNA replication, mitosis, meiosis and developmental reprogramming. Elucidating the processes by which epigenetic information survives and is transmitted is a central challenge in biology. Here we consider recent advances in understanding mechanisms of epigenetic inheritance with a focus on histone segregation at the replication fork and how an epigenetic memory may get passed through the paternal lineage.
Keywords: epigenetic, inheritance, replication, histone, polycomb, gamete
Beyond the Mendelian Rules of Inheritance
Through classical genetics and the advent of modern sequencing we have developed a comprehensive understanding of traditional modes of Mendelian inheritance, yet these advances cannot fully explain how organisms propagate vastly different phenotypes across generations independently of alterations in gene sequence, i.e. epigenetically. Conservatively, epigenetic inheritance (see glossary) requires that the transmitted phenotype be: 1) independent of changes in DNA sequence, 2) conveyed in the absence of the initial stimulus that caused the phenotype in the parental cell or organism (F0) and 3) propagated via a bona fide mechanism. Despite a rich and growing literature on epigenetic inheritance in a multitude of species, uncovering phenomena that satisfy all of these criteria has been a challenge, with the mechanism itself often being the most controversial (See Box 1 and [1] [2]). Here we review possible mechanisms of epigenetic inheritance with an emphasis on recent insights derived from the chromatin level. First, we consider transmission of epigenetic memories by examining the most fundamental constituent of conveying information in a dividing cell, the nucleosome, with emphasis on the replication fork. Second, we examine the complexities of inheritance across generations in multi-cellular organisms by highlighting exciting new discoveries involving chromatin dynamics that may convey epigenetic inheritance through the paternal lineage. Through these two fronts, we intend to shed light on possible mechanisms guiding the transmission of an epigenetic memory across multiple developmental stages.
Box 1. Transgenerational inheritance; considering caveats and alternative mechanisms.
Non-chromatin based mechanisms likely contribute to transgenerational inheritence. For example, some of these phenotypes might arise from cryptic genetic variation given that inbred strains, nearly identical clones or even neighboring cells in the same organism may possess marked genetic differences ([108]). Such genetic variation could be passed on to offspring or arise de novo (e.g., transposable elements, mutations) and account for differences. Unfortunately, these alternatives are seldom examined in transgenerational studies. Furthermore, establishing transgenerational inheritance in its purest sense is often confounded by maternal care, social transmission, or other variables that may propagate a phenotype without requirement for epigenetic memory per se. Indeed recent studies suggest that maternal care may play a significant role even in the transmission of phenotypes originating from the father [109].
Even if a phenotype is transmitted in a transgenerational epigenetic fashion, chromatin events may not always be responsible for their propagation. Transcriptional loops are one example [110]. As in somatic tissue, noncoding RNAs such as siRNA, piRNAs as well as miRNA contribute to inheritance and might function independently of changes at the level of chromatin (recently reviewed by [63]). In fact, a recent study showed that miRNAs are important for transmitting the experience of trauma to progeny through the paternal lineage [65]. Studying the importance of these varied contributions to transgenerational inheritance is important in understanding whether they are truly epigenetic.
Dismantling and Restoring Chromatin throughout DNA Replication
The post-replicative restoration of DNA methylation on the newly synthesized DNA via the maintenance DNA methyltransferase, DNMT1, is perhaps one of the better-understood examples of epigenetic inheritance (recently reviewed elsewhere [3]). In contrast, other epigenetic factors are thought to segregate onto replicated DNA to produce two phenotypically identical daughter cells at the end of mitosis. This is particularly true of histones—integral components of chromatin and the center of the following discussion.
Pioneering studies established that pre-existing, parental nucleosomes contribute to approximately half of the histones on nascent DNA, suggesting that parental histones likely contribute to shaping the epigenome of daughter cells [4]. The segregation of histones is cooperative and dispersive resulting in the equal and random distribution of histones, in clusters, onto both daughter DNA strands (reviewed by Annunziato [4]). Deposition is closely coupled to the replication machinery, as nucleosomes re-appear ~200-300 bases behind the replication fork on both leading and lagging strands [5, 6]. This relationship to the replication machinery is further apparent with Okazaki fragments in yeast that are nearly nucleosomal in size, with junctions clustering over nucleosomal dyads [7]. The exact molecular mechanism by which nucleosomal histones and their associated PTMs redistribute behind the replication fork is believed to involve epigenetic processes. A number of histone chaperones (see glossary) have been proposed to contribute to the segregation of histones, yet their respective modus operandi is quite distinctive.
Nucleosomal histones predominantly dissociate as two H2A-H2B histone dimers and a central (H3-H4)2 tetramer in vitro and in vivo at the replication fork [8-10]. Because H2A-H2B dimers are susceptible to internucleosomal exchange throughout interphase, the (H3-H4)2 tetrameric core of the nucleosome at the replication fork is the likely candidate for transmitting epigenetic information. Evidence suggests that parental (H3-H4)2 nucleosomal cores are immediately re-assembled behind the replication fork, followed by deposition of H2A-H2B dimers and linker histone H1 [4]. Pulse-chase analyses of isotope-labeled histones recently confirmed long-established biochemical data that the bulk of H3-H4 is transferred onto replicating DNA as intact (H3-H4)2 tetrameric units [9, 10]. This is in stark contrast to newly-synthesized histones which are brought onto replicating DNA as H3-H4 dimers. The Anti-silencing Factor 1 (ASF1) histone chaperone extensively binds the histone dimer hindering the formation of H3-H3’ contacts seen within (H3-H4)2 tetramers [11]. ASF1 associates with new cytoplasmic histones, which translocate into the nucleus as cargo on the importin-4 karyopherin [12, 13]. In the nucleus, ASF1 channels the replication-coupled H3.1/H3.2 and replication-independent H3.3 histone variants (see glossary) through different deposition pathways [14] (the deposition of various histone variants is reviewed elsewhere [15]). Dimers consisting of newly synthesized replication-coupled histone H3.1 are transferred from ASF1 to the Chromatin Assembly Factor 1 (CAF-1) chaperone [14, 16] to counteract the dilution of segregating parental histones. CAF-1 associates with the PCNA scaffold ring and is responsible for the de novo assembly of (H3-H4)2 tetrasome intermediates (nucleosomes lacking histones H2A-H2B) on replicated DNA (figure 1) [17]. Recent thermodynamic analyses established increasing binding affinities towards histones from ASF1, to CAF-1, and DNA, nicely illustrating the chain of successive handoffs [18, 19]. The same studies further imply the likely formation of tetramers on CAF-1, immediately prior to deposition. CAF-1 handles newly synthesized histone molecules that are largely unmodified save for H4 acetylation [20], and doubts remain as to whether CAF-1 deposits parental nucleosomal histones under normal conditions. Hence, once tetrameric cores are formed, they likely remain as such through subsequent rounds of replication and may no longer be channeled through CAF-1.
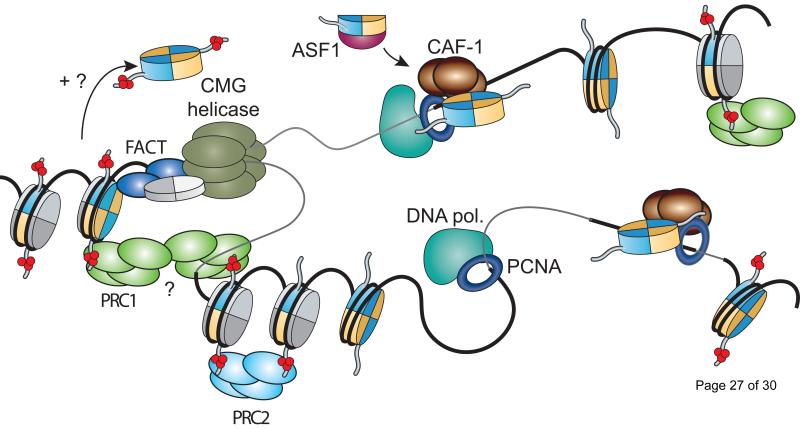
Figure 1.
Histone dynamics and inheritance of epigenetic information at the replication fork, as exemplified by the methylation of histone H3 on lysine 27. De novo nucleosome assembly proceeds through the nuclear import of histone H3-H4 dimers via the ASF1 histone chaperone. Differential thermodynamic affinities towards histones facilitate the transfer of these predominantly unmodified histones to the PCNA-bound CAF-1. The latter facilitates the formation and deposition of stable (H3.1-H4)2 tetramers to be completed by the addition of two juxtaposed H2A-H2B dimers. Nucleosomes encountering the replication fork are transiently bound and dissociated by the MCM2 subunit of the CMG replicative helicase as well as by the histone chaperone FACT. The exact mechanism by which these multi-PTM decorated histone tetramers segregate onto nascent DNA strands remains to be fully elucidated. Polycomb proteins persist at the replication fork through internucleosomal contacts. Only histone H3 N-terminal tails marked on lysine 27 are shown for simplicity. Histones H3, H4, and H2A-H2B are shown in blue, yellow and grey, respectively. Methyl marks are shown in red. Figure is adapted from [2].
Histone Chaperones and the Replicative Helicase
In addition to interacting with CAF-1, ASF1 has also been co-purified with other components of the replication machinery, such as the replicative clamp loader RFC [21], and the MCM subunits of the replicative helicase [22]. The latter led to the compelling suggestion that ASF1 disassembles and splits nucleosomal (H3-H4)2 tetramers to transfer epigenetic information in the form of two equivalent H3-H4 dimers onto both nascent DNA strands. This semi-conservative model is now however, countered in favor of a conservative segregation of histones as the (H3-H4)2 tetramers (notably H3.1-containing nucleosomes) largely remain intact through cell division [9, 10]. Moreover, it is uncertain whether ASF1 merely associates with inactive MCM subunits or an actual processive helicase (see below). Further mass spectrometry studies revealed that nucleosomes do not necessarily harbor symmetric epigenetic information on their two sister H3-H4 dimers [23]. While the central (H3-H4)2 tetramer is unlikely to be severed over the bulk of replicating chromatin (readers are directed to further views on H3-H4 segregation models [2, 9, 24]), the semi-conservative, partition-based model may still operate on a specific subset of nucleosomes given that a fifth of post-replicative H3.3 nucleosomal pools contain mixed parental and newly-synthesized species after two rounds of replication [10]. These nucleosomes cluster on active, tissue-specific enhancers [25], implying a unique and restricted route to partitioning at these important regulatory sites.
If ASF1 does not dissociate the bulk of nucleosomal histones, and CAF-1 handles newly-synthesized histones, how then are parental nucleosomes segregated at the replication fork? A breakthrough discovery in yeast revealed that a histone chaperone associates with nucleosomal histones while encountering the active replicative helicase [26], minimally composed of CDC45, MCM2-7, and the GINS subcomplex (CMG) [27, 28]. A relatively well-conserved acidic patch on the N-terminal extremity of MCM2 interacts with histone H3 [26, 29] and cooperates with the FACT histone chaperone to bind core nucleosomal histones at active CMG helicases [26]. Though predominantly viewed as an H2A-H2B histone chaperone, FACT is capable of binding the four canonical histones as well as core nucleosomal particles [30, 31]. Importantly, the material dissociated by the FACT-CMG interaction is composed of parental histones that had been released from chromatin rather than newly synthesized counterparts [26]. Supporting a role in the transmission of epigenetic information, cells expressing a mutant MCM2 deficient in histone binding exhibit a loss of subtelomeric heterochromatin [26]. Whether the interaction between FACT and CMG suffices to dissolve nucleosomes at the active helicase remains to be tested in vitro. During transcription, FACT acts by dissociating an H2A-H2B dimer to destabilize the core nucleosomal particle [30], a mechanism that might pertain during DNA replication as well. DNA helicases are capable of great disruption and it is likely that the piconewton forces generated through strand displacement further contribute towards the translocation of nucleosomal histones [32].
Acquiring and Restoring Epigenetic Information through Parental Histone Clusters
The aforementioned data argues for the maintenance of chromatin domains, rather than precise histone deposition, as guiding the propagation of epigenetic information at the replication fork such that the distinctive compositions of PTMs help dictate chromatin maturation through subsequent stages of the cell cycle. It is likely that putative epigenetic histone PTMs can be minimally transmitted in cis between neighboring nucleosomes, such that the deposition of parental nucleosomal clusters on both nascent DNA strands may be sufficient to ‘seed’ the propagation of epigenetic histone PTMs. Caveats do exist, at least in the case of rapidly dividing Drosophila embryos (see below) [33]. However, parental histones harboring key PTMs have been readily isolated and detected from active replication forks in cultured human cells adding validity to their epigenetic nature in somatic mammalian models [34]. Indeed evidence suggests that histone posttranslational modifications—notably those involved in gene silencing and chromatin compaction—do persist through multiple rounds of cell divisions in the absence of the causative signals (see Box 2). Although these parental histone clusters may help bookmark epigenetic signals, the restoration of original chromatin states occurs through the subsequent maturation of chromatin.
Box 2. Perseverance of histone PTMs.
Epigenetic forces regulating chromatin have long been documented. Phenotypic variegation is a classical example, as gene activity decreases when placed in proximity to heterochromatin (an effect known as Position Effect Variegation—or PEV) [111, 112]. In examining histones per se, truncations over the N-terminal tails of histone H4 abolishes silencing over the yeast silent mating cassettes [113]. The disruption of the Silent Information Regulator deacetylase machinery, however, creates mitotically-stable states where a majority of cells lose silencing over the same genetic loci, but a minority persist in a silent state [114]. This observed bistability argues towards histones and their posttranslational modifications having a direct epigenetic role.
An elegant in vivo system was recently developed in the laboratory of Gerald Crabtree that uncouples the initial signaling event leading to changes in chromatin and the inheritance of the ensuing chromatin state through subsequent cell divisions [115]. In this system, repressive HP1α or activating VP16 proteins was transiently recruited to a reporter gene. Their recruitment was dependent on and tightly controlled by the presence of small molecule ligands. Transient recruitment of HP1α led to the recruitment of endogenous HP1, the formation of large H3K9me3 domains that spread in cis, chromatin compaction, and silencing of the reporter gene. Importantly, the H3K9me3 domains persisted upon a number of cell divisions and in a clonal manner following abrogation of the initial stimulus. These proof of principle experiments highlight another key observation: parallel epigenetic pathways reinforce one another to reduce bistability through interlinked feedback mechanisms. Whereas short-lived HP1α tethering induced H3K9me3, a prolonged recruitment further instituted local DNA methylation, which in turn led to fewer stochastic epigenetic interconversions in dividing cells.
A prime example of how parental nucleosomal clusters may seed propagation of epigenetic histone PTMs involves the Polycomb Repressive Complex 2 (PRC2), a histone methyltransferase complex that catalyzes H3K27me3 and is involved in gene repression [35-37]. PRC2 spreads H3K27me3 over chromatin by simultaneously binding and catalyzing the mark [38]. The PRC2 subunit EED binds nucleosomes harboring H3K27me3 to stimulate further trimethylation of H3K27 within neighboring histones via the PRC2 catalytic subunit EZH2 [38], a mechanism of propagation reminiscent of the previously described spreading of H3K9 trimethylation [39] and yeast sirtuin-mediated histone deacetylation [40]. H3K9me3, another mark correlated with gene silencing, is bound by the Heterochromatin Binding Protein 1 (HP1), which in turn further recruits the H3K9 methyltransferase SUV3/9H1/2 [39].
The PTMs present on the inherited parental (H3-H4)2 tetramers are therefore expected to regulate the establishment of histone marks over maturing chromatin. As mentioned, the two H3-H4 dimers comprising (H3-H4)2 nucleosomal cores can harbor both symmetric and asymmetric PTMs. Indeed, a surprising proportion of such tetramers can not only harbor asymmetry between the two fastened H3-H4 dimers, but can also exhibit bivalent H3K4me3 and H3K27me3 marks within a single nucleosome [23]. PRC2 can spread H3K27me3, but only on nucleosomes containing asymmetric as opposed to symmetric H3K4me3 [23]. Therefore, the configuration of histone PTMs on inherited (H3-H4)2 tetramers carries consequences for the incorporation of subsequent histone modifications. Since histone PTMs are subject to stochastic fluctuations, the dosage of inherited parental histone marks is likely to influence the ensuing chromatin states.
Histone Marks: Beyond S-phase
Interestingly enough, the restoration of parental histone PTMs is not restricted to S-phase (figure 2). This is evident for both H3K27me2/3 and H3K9me2/3, two of the most commonly studied histone marks enforcing transcriptional silencing. Both PTMs are readily detected on nascent chromatin, even upon inhibition of de novo protein synthesis, highlighting the important contribution of parental histones in retaining these marks [34]. Flow cytometry and mass spectrometry based analyses illustrate a slight dilution of H3K9me2/3 through DNA replication, followed by active di- and tri-methylation by SUV39H1/2 after completion of S-phase [41-43]. Indeed, a limited pool of H3K9me2/3 is restored from mono- and di-methylated precursors throughout S-phase. This is in accordance with the postulation that monomethylation of H3K9 on newly-synthesized histone H3 by PRDM3, PRDM16, and SETDB1 serves as a substrate for di- and tri-methylation within heterochromatin [44-47]. However, steady-state H3K9me2/3 levels are only completely restored by the subsequent G1 phase through de novo methylation of unmodified residues [42]. Thus, pre-existing parental histones likely transmit H3K9me2/3 to maturing newly-synthesized counterparts in a post-replicative manner. These studies determined similar cell cycle kinetics for H3K27 methylation [42, 43]. It should be noted that a recent report failed to detect methylated histone H3 behind the replication fork in gastrulating Drosophila embryos [48]. Questions remain as to why histone marks were not detected or may even be actively erased in this system. Regardless of the cause, or how universal or unique this phenomenon may be across species, the study does highlight the importance for inheriting the histone-binding and modifying polycomb proteins.
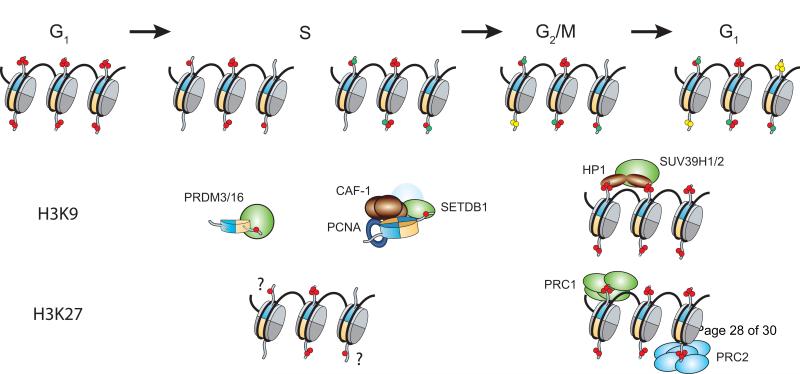
Figure 2.
Restoration of repressive H3K9 and H3K27 methylation marks throughout the cell cycle. Replicated nascent DNA strands receive an equal influx of newly synthesized and parental histones effectively diluting pre-existing posttranslational modifications. H3K9me1 increases in S-phase through the deposition of PRDM3/16 and SETDB1-modified histones. A subset of the monomethylated histones are perhaps converted into higher methylation states, however, steady states are achieved though the propagation of the marks onto unmodified histones from the end of replication until the subsequent G1 phase. Although H3K27me mirrors these cell cycle coupled kinetics, the enzyme responsible for H3K27 monomethylation has yet to be unequivocally identified. Red, green, and yellow dots represent parental methyl marks, and subsequent methylation events on modified and unmodified substrates, respectively.
Acquiring Cellular Memory through Histone-binding Proteins
Many chromatin-associated proteins modulate the transcriptional output of genes by converging on histones. Polycomb-group (PcG) and trithorax (Trx) proteins (see glossary) are key epigenetic regulators of transcription overseeing a large number of developmental genes. A linear mode of function was initially proposed for two core PcG protein complexes, Polycomb Repressive Complex 1 (PRC1) and PRC2: PRC1 recognizes and binds to H3K27me3 catalyzed by PRC2, ultimately enforcing gene silencing through PRC1-mediated chromatin compaction. While the model may hold under certain circumstances recent reports have revealed that it is far from universal [49-54] and the post-replicative recruitment of both repressive complexes is highly relevant. The importance of PcG proteins in replicating Drosophila embryos further underscores their role in the transmission of epigenetic information [33].
Studies using SV40-driven replication of PcG-bound chromatin templates in vitro revealed that Drosophila PRC1 remains associated with newly replicated daughter strands through its Posterior Sex Combs (Psc) subunit [55]. The same subunit also mediates chromatin compaction by PRC1 in vitro [56]. At first, this finding with PRC1 seems reminiscent of that with the SV40 replication machinery, which can traverse cross-linked histone octamers without histone eviction, leaving nucleosomes intact [57], however, the mechanism by which PRC1 remains associated with replicated DNA is quite different. Psc self-associates and is capable of bridging nucleosomal templates [58]. This has led to the proposal of a mechanism of epigenetic inheritance by which PRC1 complexes transiently associate with multiple contacts ahead and behind the replication fork (figure 1). This would allow PRC1 to remain associated with replicating via a looping event, suggesting an elegant mechanism of epigenetic inheritance.
PcG and Trx proteins were readily detected behind the fork on labeled replicated DNA in Drosophila systems [33, 55]. In human cells, the incorporation of thymidine analogues further confirmed the presence of both PRC1 and PRC2 protein complexes on replicated DNA. However, further SILAC analysis comparing biotin-dUTP labeled chromatin fragments after a 15 min or a 2 hour chase revealed an enrichment for both polycomb complex components on mature (2 hr) over nascent (15 min) chromatin [34]. The recruitment of these protein complexes over maturing chromatin in human cells therefore either reflects an important disparity between cultured human cells and gastrulating Drosophila embryos, or a second wave of recruitment following initial segregation of PcG components at the fork. In other words, two separate events may be at play: First, mammalian PcG protein complexes segregate at the fork as proposed in Drosophila models, thus bookmarking genomic targets for transcriptional silencing.
The process is vital since PRC2 mediated H3K27me3 can recruit PRC1 and the H2AK119 ubiquitin ligase activity of PRC1 can in turn stimulate the recruitment of PRC2 [50-52]. Additional PcG complexes may therefore be subsequently recruited in a post-replicative manner to their genomic targets as chromatin matures, through self-association or binding of the end marks. The data poses an exciting new epigenetic layer bestowing cellular identity that warrants further investigation. Whether human PRC1 and PRC2 complexes can self-associate to perpetuate themselves through multiple nucleosomal contacts at the replication fork, as is the case with Drosophila PRC1, remains to be determined. This is important since unlike Drosophila, mammalian models do not possess PRE and TRE (polycomb and trithorax responsive element) regulatory DNA elements to guide their binding onto chromatin.
Likewise, HP1 is immediately found on replicated DNA but in both mammals and flies [34, 48]. A model for the transmission of HP1 via CAF-1 association has been proposed, however the interaction is found on lingering PCNA molecules and is uncoupled from histone deposition [59, 60]. Yet, since HP1 dimerizes and is capable of bridging nucleosomes [61, 62], it is tempting to speculate that a pool of HP1 proteins may remain associated with replicating constitutive heterochromatin through a looping mechanism similar to the one proposed for Psc. A number of premises can be verified should the hypothesis hold true for these proteins, as looping and bridging of parental and daughter nucleosomal templates should be regulated in space and time and occur within a relatively short distance from the replicative helicase.
Several pathways clearly exist to recapitulate and maintain epigenetic information in somatic tissue. Recent publications now suggest that some of this memory may even persist through the germline to affect subsequent generations. Although still obscure, molecular insights into this form of Lamarkism are beginning to emerge.
Against all odds: the possibility and impossibility of transgenerational epigenetic inheritance
A central challenge to epigenetic inheritance is that a multitude of events occur in the germline, during gametogenesis and in the developing embryo, that all seem aimed at erasing the epigenetic memory of their parent-of-origin in order to give rise to a totipotent tabula rasa from which a complex multicellular organism is generated. Despite this, there are a growing number of recent reports suggesting a role for epigenetic mechanisms in the inheritance of stress-linked, metabolic, oncogenic and even complex behavioral phenotypes such as learning, memory, depression and drug response in filial generations, F2 and F3 [63-66]. Yet, even the best controlled studies using combinations of cross-fostering and in vitro fertilization still leave questions as to whether other modes of inheritance might be at play (e.g., cryptic genetic variation) as well as the mechanism by which this epigenetic information survives through development and across generations. Here we consider each step required for an epigenetic memory to survive with special consideration of mechanisms that might allow for its inheritance. In light of recent, more general reviews [1, 63, 64, 67], the emphasis will be on the latest insights into histone dynamics in sperm and evidence for the inheritance of epigenetic information, through the paternal lineage.
Epigenetic propagation through spermatogenesis
The first step in the inheritance of an epigenetic memory requires the transmission of the F0 experience to the gametes, a process that remains largely elusive. The subsequent passage of the epigenetic signal through gametogenesis faces tremendous challenges in light of the complex chromatin alterations observed during spermatogenesis (see glossary). During meiotic interphase S, transmission across the replication fork may involve mechanisms outlined in the first part of this review because canonical nucleosomes or slight variations remain largely intact and similar machinery has been shown to mediate both replication in mitosis and meiosis [68]. While some testis-specific histone variants may exist even in the early spermatogonia (e.g., H3T), many appear to be incorporated shortly before the second meiotic division [69]. This includes linker histones H1t, H1t2, HILS1 as well as core histones H2A.Lap1, H2AL1/2, H3T and TH2A/B [70]. Such histone exchange questions the notion that histones themselves may retain the epigenetic memory. In particular, it remains elusive whether these variants are modified in the same way as the canonical histones and whether such marks are influenced by canonical histones. Recent evidence suggest that this may indeed be possible in principle, as TH2B is retained post-fertilization [71].
Further complicating the survival of an epigenetic memory is the nucleosome instability required for the eviction of histones, incorporation of transition proteins and the subsequent packaging of mature sperm DNA with protamines (PRM1 and PRM2 in mammals—see glossary). However, consideration of each step required to make this transition reveals some clues. Initially, testis-specific histone variants are key players because their acidic C-terminal surface weakens DNA-protein interactions. In fact, if TH2B is depleted, the spermatogenic system compensates by increasing modifications in the histone core domains (e.g., crotonylation) to destabilize the nucleosome [71]. Also critical for histone eviction is histone hyperacetylation, following meiosis I [72]. Counterintuitively, this hyperacetylation ultimately leads to chromatin compaction likely through binding of the bromodomain protein BRDT, which in turn facilitates histone eviction through its chaperone-like action [73, 74]. Whether BRDT or other bromodomain proteins highly expressed in early spermatogenesis such as BRD2 [75] may bookmark certain loci is an intriguing, but untested hypothesis that would likely require their association with the small percentage of histones retained during spermatogenesis (see below). Following histone eviction, protamine replacement occurs, often orchestrated through intermediate transition proteins (TP1 and TP2) [70, 76]. A conceivable, yet untested hypothesis is that the protamines themselves carry certain epigenetic information at select loci given their post-translational modifications [77]. One mark, phosphorylation of serine within the arginine rich domains of PRM2, likely loosens the association of DNA with PRM2 surrounding those specific residues, perhaps representing such a mark but with unknown functional consequences [77, 78].
Perhaps the most promising mode of paternal epigenetic inheritance was revealed upon the discovery that histone-bearing nucleosomes are retained throughout spermatogenesis [79, 80] in approximately ~1% to 10% of the genome in mice and humans, respectively [81, 82]. It is important to note that the specific composition of these retained nucleosomes in mature sperm is unknown, but presumably comprises the four core histones which bear marks similar to that of canonical nucleosomes [77]. Despite some outstanding questions, these retained histones are prime candidates for epigenetic inheritance, as some evidence suggests their retention at CpG-rich sequences devoid of DNA methylation, including a large number of imprinted loci, as well as Polycomb and Trithorax targets [81-83]. However, these descriptions of histone retention are in contrast to recent reports suggesting that histones are depleted at promoters but are enriched over gene poor regions [84, 85] or alternatively have a relatively normal distribution throughout the genome [83]. The discrepancy between these results may be due to subtle variations in experimental procedure [86] thus requiring additional work to resolve the exact genomic location of sperm nucleosomes. Regardless of their precise localization, what is clear is that these histone are retained and modified in a manner consistent with their relevance in shaping certain phenotypes during embryogenesis. Indeed, a recent study showed that paternal diet could affect H3K27me3 status in sperm, suggesting that histones in the gametes may acquire and retain signatures of an epigenetic stimulus [87].
While there are multiple potential chromatin signatures of an epigenetic memory in each step of spermatogenesis, the transfer of this memory between each step remains largely unknown. If protamines indeed retain and transfer epigenetic information, CHD5 may represent one key regulator because it has been shown to be critical in orchestrating multiple steps in the histone-protamine transition including histone hyperacetylation, expression of histone variants and transition protein, as well as protamine deposition [88]. The exact CHD5 mechanism of action is unclear, but it may bridge epigenetic PTMs on histones with protamines, given the preference of its PHD domains for H3 lacking K4me3 and the recognition of H3K27me3 by its chromodomains [89, 90]. The poly(ADP-ribose) pathway is also a viable candidate as disruptions in this pathway lead to dramatic increases in histone retention in mature sperm [91]. Other chaperones may also be involved in this process as a diverse array of chaperones such as NASP and NAP1L likely associate with a variety of histone variants, protamines and transition proteins in sperm [92]. In fact, it has recently been proposed that the p75 subunit of Drosophila CAF-1 may be critical for protamine deposition [93].
Histone-mediated epigenetic transmission across the various stages of spermatogenesis likely involves a combination of replication dependent factors described above during meiosis I (e.g., ASF1, CAF-1 and PCNA) and other recently discovered chaperones during later periods of replication-independent nucleosome turnover. Because H3.3 is enriched in sperm, H3.3-specific chaperones such as HIRA or ATRX/Daxx are likely involved. In fact, HIRA may deposit histone H3.3, particularly to destabilized nucleosomes or in the presence of RNA Pol II at transcriptional start sites [94]. The converse also appears to be true, with HIRA mediating the deposition of H3.3 at polycomb-repressed loci [95]. These observations point to the interesting possibility that histone-specific chaperones as well as PRC2 may be critical mediators of maintaining epigenetic memory during periods of high histone turnover in spermatogenesis.
Survival of epigenetic memory in the zygote; transfer to F1
After successful retention of the paternal epigenetic memory through gametogenesis, such memory must then survive the dramatic reprogramming that occurs in the zygote. Prior to the first round of replication after fertilization, one of the initial reprogramming events is the decondensation of the sperm pronucleus and the replacement of protamines by maternal H3.3 via HIRA. This event is critical for heterochromatin establishment at loci important in maintaining chromatin segregation and genome stability (e.g., pericentromere; [96, 97]). If and how epigenetic information carried by the protamine survives this process are unknown. However, in contrast to protamines, histones retained in sperm may persist in the zygote as loci marked by H3K27me3 given that mouse sperm continue to show repression in the pre-implantation embryo [82, 83]. In humans, the H3K4me3 mark retained in sperm appears to correlate with early gene expression in the embryo. Therefore, H3K27me3 and H3K4me3 may be viable conduits for epigenetic inheritance, a notion that still requires direct testing. In light of evidence that certain chromatin states (e.g., bivalent domains) can mark loci for subsequent DNA methylation [98], an intriguing possibility is that CpG-associated histone modifications retained throughout spermatogenesis might serve as a signal for “re”-methylation of genes important for epigenetic memory.
Chromatin dynamics through the first mitotic division of the zygote and later transitions through implantation are less well understood. Using methods that largely detect bulk histone modifications, early studies suggested that there is parental asymmetry such that the maternal pronucleus shows enrichment for a number of histone modifications that were relatively low in the paternal pronucleus (e.g., H3K9me3). The finding of parental asymmetry, combined with evidence that PRC1 marks paternal chromatin during early pronuclear phases, followed by the establishment of H3K27me3 around the first replication of DNA, led to the suggestion that the paternal lineage is initially targeted for heterochromatin formation by maternal PRC1 and Suv39h [99, 100]. Thus, maternally mediated reprogramming poses a serious threat to any putative paternal epigenetic memory. Despite this large scale reprogramming, many paternal epigenetic programs are likely spared. In fact, a recent study provides a convincing case for the possibility that the small fraction of histones retained in sperm is transferred to the 2-cell stage embryo and mediate early embryonic transcription [91]. In this study, disruption of the poly(ADP-ribose) pathway (e.g., PARP and PARG) led to increased retention of histones at select loci in sperm with a resultant alteration in transcriptional profiles at histone-enriched loci. Interestingly, when these histone-enriched sperm fertilized an egg, the resulting embryo (2-cell) showed significant correlation with the histone rich regions and gene expression profiles observed in the sperm. In principle, this demonstrates that sperm histones can have a meaningful impact on early embryonic programming, and suggests that any histones retained during spermatogenesis may be substrates for transfer of an epigenetic memory even through the first mitosis.
With regard to propagation of active gene expression programs, another untested possibility is that bromodomain proteins may bookmark acetylated and thus previously active genes for later activation in the developing embryo. Indeed, recent studies give credibility to this idea by showing that bromodomain proteins such as BRD4 or p300 can bookmark active genes and facilitate the retention of pre-mitotic gene expression during and after mitosis in somatic cells [101, 102].
Epigenetic Inheritance during early gametogenesis
The last, critical step involves the faithful propagation of epigenetic memory through the germline in the developing F1 embryo, to be subsequently passed on to F2. It is well established that germline reprogramming of both DNA methylation and histone marks (e.g., H3K9me2) occurs following embryonic day 8 [67, 103]. Similar to the maternally-mediated reprogramming following fertilization, this leaves questions as to how a paternal engram survives. Elegant work in the germline is beginning to show that histone PTMs establish epigenetic programs that mark select loci for later transcriptional activation or repression. For example, in C. Elegans, MES-4, the homologue of the mammalian NSD family of H3K36me3 methyltransferases, is recruited to genes that were previously expressed in the maternal germline [104]. In mammals, the interplay of H3K27me3, H3K4me3 and DNA methylation is also beginning to emerge as targets for epigenetic programming throughout gametogenesis. This requires the movement of these epigenetic programs from ESC to primordial germ cells and on to self-renewing and committed adult germline stem cells, giving rise to spermatocytes. Recently, it was shown that certain epigenetic programs such as nucleosomal bivalency might be transferred from the early developing zygotic ESC to the germline. Once in the germline stem cells, these profiles exhibit a number of typical (e.g., co-occurrence of H3K27me3, H3K4me3 and DNA hypomethylation), as well as atypical profiles such as enrichment of locus-specific H3K9ac, H3K4me3 and DNA methylation [105]. Together, these confer specific transcriptional programs that may be transferred through spermatogenesis, thus representing a viable, yet untested platform from which to transfer epigenetic memory from fertilized embryo through gametogenesis [105-107].
Concluding Remarks
Despite such robust reprogramming throughout development, numerous mechanisms ranging from chromatin dynamics to non-chromatin based mechanisms may be capable of linking the seemingly impossible path from an experience at F0 to the transgenerational epigenetic inheritance of that phenotype. Many of these remain to be directly tested, but those reviewed above have emerged as leading candidates, such as those that may propagate repression in somatic cells through PRC1 and PRC2. In contrast, less is known about how heterochromatin formation might also be systematically thwarted by the concerted action of mechanisms aimed to retain active transcriptional profiles, such as those involving bromodomain-containing proteins, thereby facilitating epigenetic tagging of gene expression profiles important for epigenetic memory. What we can say for sure is that histones and other core components of chromatin clearly exert a surprising phenotypic influence over progeny at the cellular and organismal levels. Evidence to their epigenetic nature is fast progressing as some of these proteins are shown to survive the passage of replication forks and even perhaps an entire meiotic process. Such molecular ‘bookmarks’ are expected to facilitate a biological adaptation to changing environments but are also pertinent to disease as they regulate developmental programs. While many of these mechanisms are largely speculative and are not without caveats, they surely foster many exciting, future studies that should ultimately advance fundamental principles in basic biology and disease.
Highlights.
Histone modifications can persist over multiple cell divisions.
The acquisition of histone marks and full maturation of chromatin continues beyond S-phase.
Polycomb Group (PcG) proteins remain associated with replicated DNA.
Retained histones in sperm may be viable substrates for epigenetic inheritance.
Histones represent a mechanism for the survival of epigenetic information thru generations.
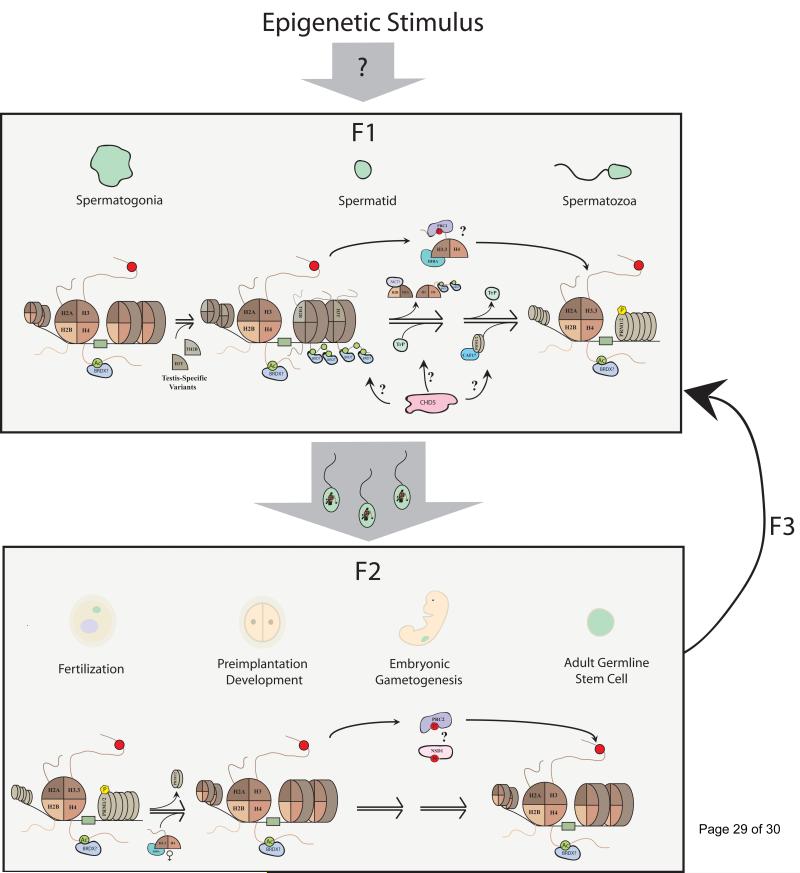
Figure 3. Survival of a Paternal Epigenetic Memory Across Generations.
Propagation of an epigenetic memory across generations in mammals requires its passage from the original epigenetic signaling event to the germline, into the embryo and through development. Potential routes of transmission are shown with the locus of epigenetic inheritance itself depicted by a green box and the forward facing nucleosome. (Top Panel) How the epigenetic stimulus itself arrives in the developing sperm is unclear. While little is known about how the epigenetic memory remains intact through the large-scale nucleosomal restructuring that occurs during the spermatogonia to sperm transition (e.g., testis-specific histone incorporation, transition proteins [Trp] and replacement with protamines [PRM1/2]), there are a number of possibilities. With their ability to mark loci through mitosis, it is possible that bromodomain proteins may mark certain loci through this transition. Perhaps the strongest evidence is for H3K27me3 either alone or together with H3K4me3 that are retained in the mature sperm and may be propagated post-fertilization. The mechanisms involved are largely speculative, but likely involve PRC2 bridging these transitions either alone or in concert with histone chaperones such as HIRA during the dynamic nucleosome turnover throughout spermatogenesis. (Bottom Panel) Faithful transmission to F2 then requires survival through maternally mediated reprogramming events such as the replacement of protamines with maternal H3. Recent evidence suggests that despite this reprogramming, certain marks such as H3K27me3 might be retained from sperm through the 2-cell stage. As in spermatogenesis, it remains unknown but plausible that bromodomains or other readers may similarly mark epigenetic loci. Gametogenesis also poses a threat to the epigenetic memory, however mounting evidence suggest that H3K27me3 and even H3K36me3 can be faithfully propagated throughout gametogenesis perhaps through the methyltransferase NSD1. From gametogenesis, the cycle would then repeat thus allowing propagation to F3 and on until this epigenetic memory is either diluted or otherwise erased.
Acknowledgments
We would like to thank Drs Lynne Vales, Wee-Wei Tee, and Philipp Voigt for recommendations and insightful discussions. Additional support was provided to J.M.S. by National Institutes of Health grant 1F32AA022842 and as a Simons Foundation Junior Fellow as well as by the National Institute of Health (NIH) (GM-64844 and R37-37120) and the Howard Hughes Medical Institute support to D.R.
Glossary
- Epigenetic Inheritance
The inheritance of a phenotype in a manner that is independent of the DNA sequence and that remains self-perpetuating in the absence of the initial stimulus that caused the phenotype in the parental cell or organism.
- Histone Variant
Core canonical and linker histones are encoded by a number of different histone genes, resulting in a number of non-synonymous substitutions and divergent domains. This variation adds complexity to the epigenetic landscape.
- Histone Chaperone
Proteins or protein complexes that specifically bind histones, thwarting non-specific interactions, and that promote their deposition or removal from DNA in an ATP-independent manner.
- PcG
Polycomb Group Proteins. A group of proteins involved in the regulation and transcriptional silencing of key developmental genes, including the Homeotic (or Hox) gene loci. Human PcG proteins assemble into Polycomb Repressive Complexes (PRCs), of which PRC2 catalyzes the methylation of H3K27 and PRC1 guides the ubiquitin ligation of H2AK119.
- Protamine
Low molecular weight proteins that tightly package DNA in late spermatids and mature sperm largely due to their arginine-rich DNA anchoring domains. Their precise function is unknown but might include protecting the paternal genome from DNA damage, facilitating formation of a small elongated sperm head for better motility and/or conveying epigenetic information.
- Spermatogenesis
The process of generating mature, haploid sperm (spermatozoa) from a diploid spermatogonium. This process initially requires mitosis to create spermatocytes, their subsequent meiotic divisions to create spermatids and finally maturation of spermatids to spermatozoa. During this chain of events, chromatin undergoes dynamic changes whereby canonical histones are largely replaced by protamines through a number of intermediate steps, including histone variant incorporation, nucleosomal destabilization, histone eviction and replacement with transition proteins prior to protamine deposition.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kar S, et al. An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics. 2012;7:994–1007. doi: 10.4161/epi.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annunziato AT. Assembling chromatin: The long and winding road. Biochim Biophys Acta. 2012;1819:196–210. doi: 10.1016/j.bbagrm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Herman TM, et al. Structure of chromatin at deoxyribonucleic acid replication forks: location of the first nucleosomes on newly synthesized simian virus 40 deoxyribonucleic acid. Biochemistry. 1981;20:621–630. doi: 10.1021/bi00506a027. [DOI] [PubMed] [Google Scholar]
- 6.Sogo JM, et al. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 7.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eickbush TH, Moudrianakis EN. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978;17:4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- 9.Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 10.Xu M, et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 11.English CM, et al. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campos EI, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez F, et al. Sequential establishment of marks on soluble histones H3 and H4. J Biol Chem. 2011;286:17714–17721. doi: 10.1074/jbc.M111.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagami H, et al. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 15.Filipescu D, et al. Developmental roles of histone H3 variants and their chaperones. Trends in genetics: TIG. 2013;29:630–640. doi: 10.1016/j.tig.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Tyler JK, et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 18.Liu WH, et al. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 2012;40:11229–11239. doi: 10.1093/nar/gks906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler DD, et al. Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 2012;40:10139–10149. doi: 10.1093/nar/gks812. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Verreault A, et al. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 21.Franco AA, et al. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 23.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacAlpine DM, Almouzni G. Chromatin and DNA replication. Cold Spring Harbor perspectives in biology. 2013;5:a010207. doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, et al. H3.3-H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet. 2013;9:e1003558. doi: 10.1371/journal.pgen.1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foltman M, et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell reports. 2013;3:892–904. doi: 10.1016/j.celrep.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 27.Moyer S, et al. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang Y, et al. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci U S A. 2012;109:6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishimi Y, et al. Binding of human minichromosome maintenance proteins with histone H3. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 30.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 31.Stuwe T, et al. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc Natl Acad Sci U S A. 2008;105:8884–8889. doi: 10.1073/pnas.0712293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranjith P, et al. Nucleosome hopping and sliding kinetics determined from dynamics of single chromatin fibers in Xenopus egg extracts. Proc Natl Acad Sci U S A. 2007;104:13649–13654. doi: 10.1073/pnas.0701459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petruk S, et al. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alabert C, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuzmichev A, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 37.Laible G, et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lachner M, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 40.Strahl-Bolsinger S, et al. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 41.McManus KJ, et al. Dynamic changes in histone H3 lysine 9 methylations: identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J Biol Chem. 2006;281:8888–8897. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- 42.Zee BM, et al. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol. 2012;32:2503–2514. doi: 10.1128/MCB.06673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu M, et al. A model for mitotic inheritance of histone lysine methylation. EMBO Rep. 2012;13:60–67. doi: 10.1038/embor.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 45.Loyola A, et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loyola A, et al. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 47.Pinheiro I, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150:948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 48.Petruk S, et al. Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication. Nature communications. 2013;4:2841. doi: 10.1038/ncomms3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper S, et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell reports. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 53.Morey L, et al. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell reports. 2013;3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 54.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Francis NJ, et al. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Francis NJ, et al. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 57.Vestner B, et al. Histone octamer dissociation is not required for in vitro replication of simian virus 40 minichromosomes. J Biol Chem. 2000;275:8190–8195. doi: 10.1074/jbc.275.11.8190. [DOI] [PubMed] [Google Scholar]
- 58.Lo S, et al. A bridging model for persistence of a polycomb group protein complex through DNA replication in vitro. Mol Cell. 2012;46:784–796. doi: 10.1016/j.molcel.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murzina N, et al. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 60.Quivy JP, et al. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cowieson NP, et al. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 62.Brasher SV, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim JP, Brunet A. Bridging the transgenerational gap with epigenetic memory. Trends in genetics: TIG. 2013;29:176–186. doi: 10.1016/j.tig.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bohacek J, Mansuy IM. Epigenetic inheritance of disease and disease risk. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:220–236. doi: 10.1038/npp.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gapp K, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature neuroscience. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature neuroscience. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature reviews. Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 68.Recht J, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trostle-Weige PK, et al. Isolation and characterization of TH3, a germ cell-specific variant of histone 3 in rat testis. The Journal of biological chemistry. 1984;259:8769–8776. [PubMed] [Google Scholar]
- 70.Rathke C, et al. Chromatin dynamics during spermiogenesis. Biochimica et biophysica acta. 2014;1839:155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 71.Montellier E, et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes & development. 2013;27:1680–1692. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Govin J, et al. Histone acetylation-mediated chromatin compaction during mouse spermatogenesis. Ernst Schering Research Foundation workshop. 2006;155:172. doi: 10.1007/3-540-37633-x_9. [DOI] [PubMed] [Google Scholar]
- 73.LeRoy G, et al. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Molecular cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dhar S, et al. Insights into role of bromodomain, testis-specific (Brdt) in acetylated histone H4-dependent chromatin remodeling in mammalian spermiogenesis. The Journal of biological chemistry. 2012;287:6387–6405. doi: 10.1074/jbc.M111.288167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shang E, et al. Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene expression patterns: GEP. 2004;4:513–519. doi: 10.1016/j.modgep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 76.Meistrich ML, et al. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- 77.Brunner AM, et al. Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics & chromatin. 2014;7:2. doi: 10.1186/1756-8935-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seligman J, et al. Tyrosine phosphorylation, thiol status, and protein tyrosine phosphatase in rat epididymal spermatozoa. Biology of reproduction. 2004;71:1009–1015. doi: 10.1095/biolreprod.104.028035. [DOI] [PubMed] [Google Scholar]
- 79.Gatewood JM, et al. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 80.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 81.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Erkek S, et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nature structural & molecular biology. 2013;20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 83.Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature structural & molecular biology. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 84.Carone BR, et al. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Developmental cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samans B, et al. Uniformity of nucleosome preservation pattern in Mammalian sperm and its connection to repetitive DNA elements. Developmental cell. 2014;30:23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 86.Saitou M. Paternal nucleosomes: are they retained in developmental promoters or gene deserts? Developmental cell. 2014;30:6–8. doi: 10.1016/j.devcel.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 87.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li W, et al. Chd5 orchestrates chromatin remodelling during sperm development. Nature communications. 2014;5:3812. doi: 10.1038/ncomms4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paul S, et al. Chd5 requires PHD-mediated histone 3 binding for tumor suppression. Cell reports. 2013;3:92–102. doi: 10.1016/j.celrep.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Egan CM, et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Developmental cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 91.Ihara M, et al. Paternal poly (adp-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS genetics. 2014;10:e1004317. doi: 10.1371/journal.pgen.1004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Govin J, et al. Proteomic strategy for the identification of critical actors in reorganization of the post-meiotic male genome. Molecular human reproduction. 2012;18:1–13. doi: 10.1093/molehr/gar063. [DOI] [PubMed] [Google Scholar]
- 93.Doyen CM, et al. Subunits of the histone chaperone CAF1 also mediate assembly of protamine-based chromatin. Cell reports. 2013;4:59–65. doi: 10.1016/j.celrep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 94.Ray-Gallet D, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Molecular cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 95.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santenard A, et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nature cell biology. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Loppin B, et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 98.Voigt P, et al. A double take on bivalent promoters. Genes & development. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Santos F, et al. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Developmental biology. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 100.Puschendorf M, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nature genetics. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 101.Zhao R, et al. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature cell biology. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Byun JS, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Messerschmidt DM, et al. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes & development. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS genetics. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hammoud SS, et al. Chromatin and Transcription Transitions of Mammalian Adult Germline Stem Cells and Spermatogenesis. Cell stem cell. 2014 doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 106.Ng JH, et al. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Developmental cell. 2013;24:324–333. doi: 10.1016/j.devcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 107.Lesch BJ, et al. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16061–16066. doi: 10.1073/pnas.1315204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dietz DM, et al. Paternal transmission of stress-induced pathologies. Biological psychiatry. 2011;70:408–414. doi: 10.1016/j.biopsych.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arico JK, et al. Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS genetics. 2011;7:e1001391. doi: 10.1371/journal.pgen.1001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dubinin NP, Sidorov BN. The position effect of the hairy gene. Biol. Zhur. 1935;4:555–568. [Google Scholar]
- 112.Panshin IB. New Evidence for the position effect hypothesis. Compt. Rend. Acad. Sci. U.R.S.S. N. S. 1935;4:85–88. [Google Scholar]
- 113.Kayne PS, et al. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 114.Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 115.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]