Abstract
Microbial nucleic acids induce potent innate immune responses by stimulating the expression of type I interferons. Cyclic GMP-AMP synthase (cGAS) is a cytosolic dsDNA sensor mediating the innate immunity to microbial DNA. cGAS is activated by dsDNA and catalyzes the synthesis of a cyclic dinucleotide cGAMP with 2',5' and 3',5' phosphodiester linkages. cGAMP binds to the adaptor STING located on the endoplasmic reticulum membrane and mediates the recruitment and activation of the protein kinase TBK1 and transcription factor IRF3. Phosphorylated IRF3 translocates to the nucleus and initiates the transcription of the IFN-β gene. The crystal structures of cGAS and its complex with dsDNA, STING and its complex with various cyclic dinucleotides have been determined recently. Here we summarize the results from these structural studies and provide an overview about the mechanism of cGAS activation by dsDNA, the catalytic mechanism of cGAS, and the structural basis of STING activation by cGAMP.
Keywords: Innate immunity, pattern recognition receptors, nucleic acids, cyclic dinucleotides, type I interferons
1. Introduction
Nucleic acids, such as DNA, RNA, or nucleotides, from viral or bacterial pathogens induce potent immune responses in the infected cells [1–7]. The detection of microbial nucleic acids is a central strategy by which the host senses infection and initiates protective immune responses [6, 7]. A number of innate sensors for microbial DNA or RNA have been identified [3, 4, 8, 9]. For example, Toll-like receptor TLR9 recognizes CpG DNA, TLR3 recognizes dsRNA, and the RIG-I like receptors recognize dsRNA. Microbial DNA in the cytosol have long been known to induce potent innate immune responses by inducing the expression of type I interferons [10–12]. The search for cytosolic DNA sensors first lead to the discovery of STING (also known as MITA, ERIS, MPYS, and TMEM173), an adaptor protein located on the ER membrane, that mediates the signaling to cytosolic DNA and bacterial cyclic dinucleotides such as c-di-GMP and c-di-AMP [13–18] (Fig.1). Although STING serves as a direct sensor of cyclic dinucleotides, it is not a direct sensor for cytosolic DNA and exhibits very low affinity for dsDNA [7]. Another key cytosolic DNA sensor, AIM2, was also identified in this search as well [19–21]. AIM2 mediates the inflammatory response by activating caspase-1 through the AIM2 inflammasome and inducing the maturation of proinflammatory cytokines IL-1β and IL-18. Although the response mediated by the AIM2 inflammasome is important for host defense to microbial infection, the dominant response to cytosolic DNA is mediated by the transcriptional regulation of type I interferons [5].
Figure 1.
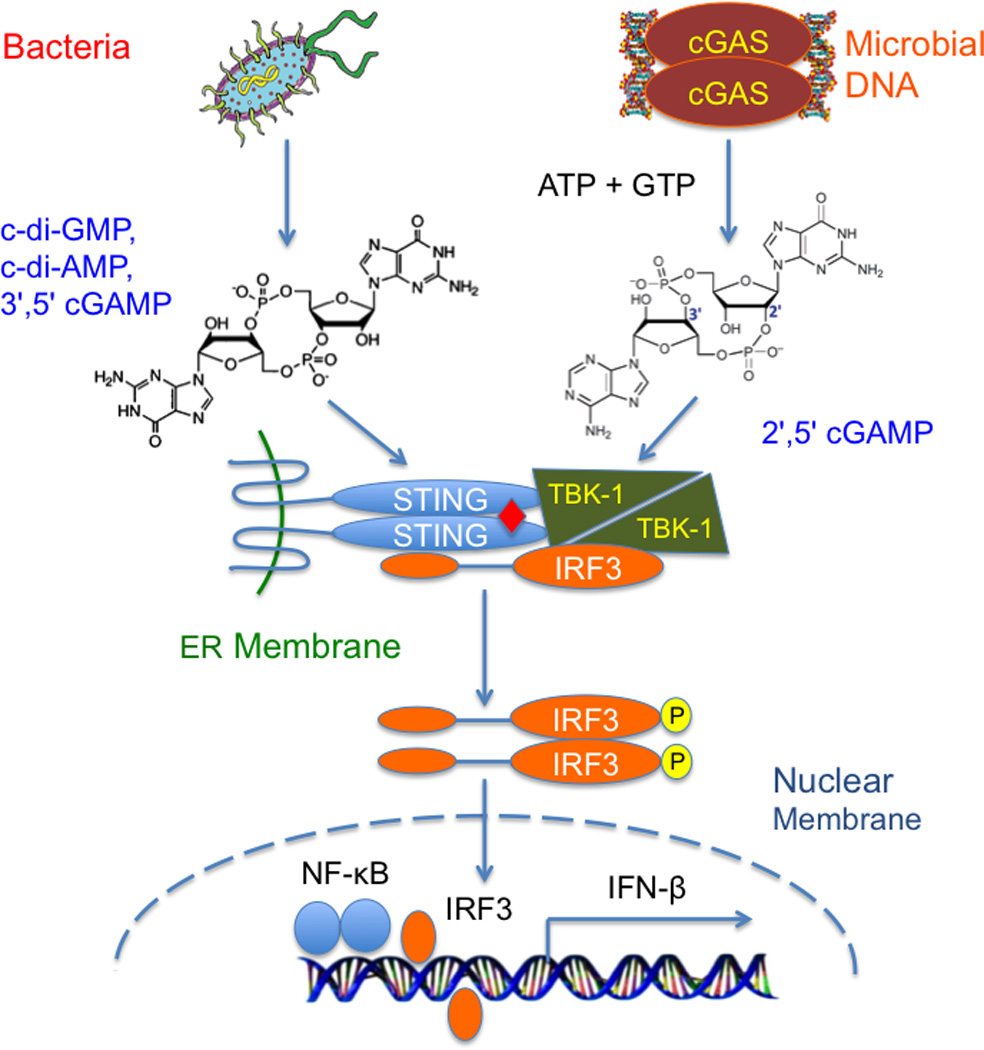
Innate immune sensing of bacterial cyclic dinucleotides and microbial DNA through the cGAS-STING pathway.
In a search for the cytosolic DNA sensor mediating the induction of type I interferons, Zhijian Chen and his colleagues used brute force biochemical approaches and identified the enzyme cyclic GMP-AMP synthase (cGAS) as the dsDNA sensor upstream of STING [22]. cGAS is activated by dsDNA and catalyzes the synthesis of a noncanonical cyclic dinucleotide 2',5' cGAMP (referred to as cGAMP hereafter) from ATP and GTP [23–26] (Fig. 1). cGAMP serves as a endogenous second messenger to stimulate the induction of type I interferons via STING. Ligand binding by STING induces the recruitment of the protein kinase TBK1 and transcription factor IRF3 to the signaling complex [27] (Fig. 1). Phosphorylation of IRF3 by TBK1 at the signaling complex promotes the oligomerization of IRF3 and its translocation into the nucleus, where it activates the transcription of the IFN-β gene together with NF-κB [27–29] (Fig. 1).
Recent structural studies of human, mouse, and porcine cGAS catalytic domains in isolation and in complex with dsDNA provided critical insights into the mechanism of cGAS activation by dsDNA and the catalytic mechanism of the enzyme [24, 30–34]. Although the conclusions from these studies are different from each other, the emerging picture, supported by extensive biochemical, biophysical, and in vivo characterization of cGAS mutants, is that cGAS is activated by dsDNA induced oligomerization [30, 32, 35]. In addition, the structures of STING in isolation and in complex with various cyclic dinucleotides including cGAMP have also been determined as well [23, 36–41]. In this review, we will give a brief overview of the mechanisms of cGAS activation by dsDNA, the structural basis of STING activation by cGAMP, and discuss several unresolved questions about the mechanism of cytosolic DNA sensing via the cGAS-STING pathway.
2. cGAS is activated by dsDNA and catalyzes the synthesis of cGAMP, a high affinity ligand for STING
The antiviral activity of cGAS (also known as C6orf150) was first described by Charles Rice’s group in a systematic screening of the antiviral activities of interferon inducible genes (ISGs) [42]. However, the mechanism underpinning the antiviral activity of cGAS was not established in this study. In a search of the cytosolic dsDNA sensor, Zhijian Chen’s group used classical biochemical fractioning technique and identified cGAMP as the type I interferon inducing molecule in cells stimulated with dsDNA [43]. Although the exact molecular structure of cGAMP was not determined in this study due to the technical limitations of mass spectrometry, the identification of cGAMP as the second messenger was a major breakthrough in understanding the mechanism of dsDNA sensing in innate immunity. To search for the enzyme that catalyzes the synthesis of cGAMP, the Chen group identified cGAS as the dsDNA sensor upstream of STING. cGAS is activated by dsDNA and catalyzes the synthesis of cGAMP from ATP and GTP [22]. The critical roles of cGAS in antiviral immunity and cytosolic DNA sensing were confirmed by recent studies of cGAS-deficient mice [44, 45].
cGAS is protein of around 500 amino acid residues. The catalytic domain of cGAS, which include residues 160 to 522 in human cGAS (hcGAS), is highly conserved in the sequences of cGAS from fish to human [7]. The catalytic domain of cGAS shows some sequence homology to 2'–5'-oligoadenylate synthase (OAS1). The N-terminal domain of cGAS is not highly conserved and rich of positively charged residues. Secondary structure prediction showed that the N-terminal domain of cGAS is flexible. Truncation of the N-terminal domain of cGAS does not abolish its catalytic activity as well as its ability to induce IFN-β gene expression in cells [22]. Both the catalytic domain and the N-terminal domain of cGAS have dsDNA binding activity [22]. DNA pull-down assays using a biotinylated 45 bp interferon stimulatory DNA (ISD) showed that full-length cGAS as well as its catalytic domain and N-terminal domain bind dsDNA, but not dsRNA [22]. DNA binding studies shows that the catalytic domain of cGAS binds the 45 bp ISD with an affinity of a few micromolars and the catalytic domain binds the ISD with an affinity of ~10 µM [30]. DNA binding studies by isothermal titration calorimetry (ITC) showed the catalytic domain of hcGAS binds the ISD at an affinity of ~20 µM [30]. In addition, DNA binding studies by fluorescence anisotropy showed that full-length cGAS binds dsDNA with an affinity of ~90 nM and associate with a ssDNA with an affinity of ~1.5 µM [46].
Both full-length cGAS and its the catalytic domain catalyzes the synthesis of cGAMP upon the stimulation by dsDNA [22]. The catalytic activity of cGAS is dependent on the length of dsDNA [30]. However, dsDNA of more than 20 bp in length are fully active compared to salmon sperm DNA, which contains a mixture of dsDNA with an average length of ~1 kb. In contrast, both ssRNA and dsRNA failed to activates cGAS [22]. cGAS is highly specific and only catalyzes the synthesis of cGAMP with ATP and GTP as substrates [22, 24, 25]. The Chen group initially proposed that cGAMP is a cyclic dinucleotide containing only 3',5' phosphodiester linkages [43]. Further studies at cellular and molecular level revealed that cGAMP synthesized by cGAS contains a 2',5' phosphodiester linkage between the 2'-OH of GMP and the 5' phosphate of AMP and a 3',5' phosphodiester linkage between the 3'-OH of AMP and the 5' phosphate of GMP [24–26]. The structure of cGAS product was further confirmed by extensive chromatographic and NMR analysis of chemically and enzymatically synthesized cGAMP and other cyclic dinulceotides [23–26].
Nucleotide binding studies by ITC demonstrated that human STING binds cGAMP at an affinity of ~60 nM, which is over 50 folds higher compared to its affinity for 3',5' cGAMP [30]. The significant difference between the binding affinities of STING for 3', 5' cGAMP and 2', 5' cGAMP was also obvious in binding studies by gel filtration chromatography [30]. Consistent with these results, Zhang et al. also showed that STING binds 2',5' cGAMP with much higher affinity (~5 nM) compared to 3',5' cGAMP (~ 1.0 µM) [23]. In addition, cell based studies showed cGAMP is more potent in stimulating the expression of IFN-β compared to 3',5' cGAMP [23, 30]. In contrast, the studies by Gao et al. showed similar binding affinities in the micromolar range for 2',5' and 3',5' cGAMP with various forms of mouse and human STING [41].
3. The structures of DNA free cGAS. Why it is not active?
The crystal structures of DNA free human, mouse, and porcine cGAS catalytic domains (residues 157 to 522 for hcGAS) have been determined recently [24, 30–34]. The overall structures of cGAS from different species are similar to each other [30]. The structure of cGAS catalytic domain also shows significant structural similarities to the structure of the dsRNA activated enzyme OAS1 (rmsd of 3.1 Å between hcGAS and OAS1). The cGAS catalytic domain exhibits a bi-lobed structure (Fig. 2A). The N-lobe of cGAS contains a canonical nucleotidyl transferase (NTase) fold. The C-lobe contains a tight five-helix bundle. The active site of cGAS is located on the edge of a deep groove between the N-lobe and the C-lobe (Fig. 2B). Mutations of any of the three catalytic residues, Glu225, Asp227, and Asp319 in hcGAS abolish the enzymatic activity cGAS [30]. Comparison of the structures of ligand free human cGAS and the structure of OAS1 shows that cGAS contains a unique zinc-binding loop (also known as zinc-thumb) between residues His390 and Cys404. Residues His390, Cys396, Cys397, and Cys404 in this loop coordinate with a Zn2+ ion in tetrahedral geometry [30]. The zinc-binding loop is conserved in mouse, human, and porcine cGAS [30, 31]. Although both mouse and human cGAS catalytic domains are monomeric in solution, they form similar dimers in the crystals via either crystallographic or non-crystallographic interactions [24, 30, 33] (Fig. 2B). The buried surface area at the hcGAS dimer interface is ~1330 Å2 [30], which is near the lower end of a typical protein-protein interface, explaining why dimerization of DNA free cGAS was not observed in solution. The two cGAS molecules in the dimer interact with each other mainly through hydrogen bonds involving residues from the zinc-binding loop [30] (Fig. 2B).
Figure 2.
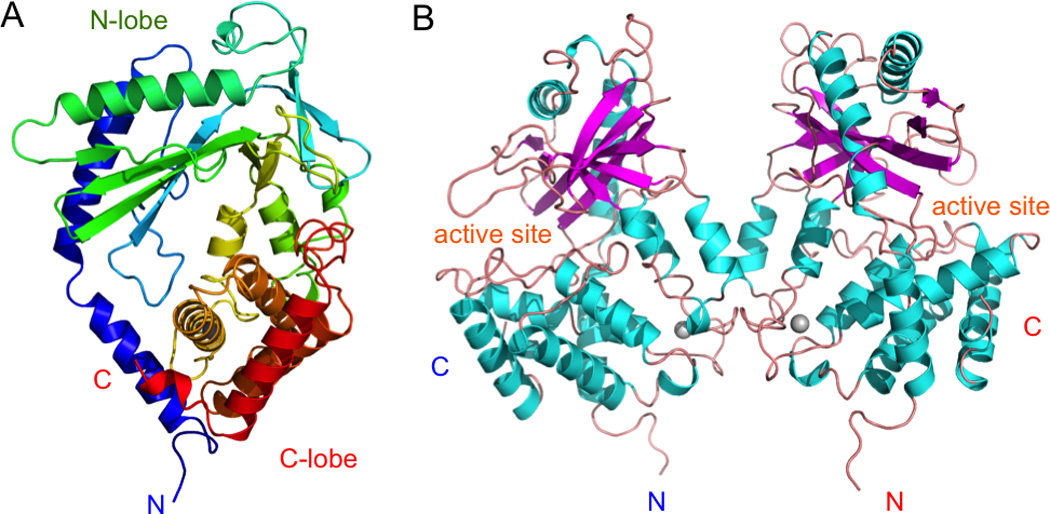
Structure of cyclic-GMP-AMP synthase (cGAS). (A). Structure of human cGAS catalytic domain colored rainbow. (B). Non-crystallographic symmetry related human cGAS dimer. The zinc ions are shown by the gray spheres.
Comparison of the DNA free structures of human, mouse and porcine cGAS showed that their C-lobe structures are highly conserved, while the NTase domains show considerable flexibility [30]. Other minor structural differences between these structures are likely due to the different crystal packing contacts. Since cGAS is not active without binding to dsDNA, none of these DNA free structures should represent the active conformation of cGAS. The reason that cGAS is not active without binding to dsDNA is likely due to the local destabilization of the NTase domain and the scrambled structure of the active site [30].
4. cGAS is activated by dsDNA induced oligomerization
A number of structures of cGAS catalytic domains bound to dsDNA have been determined recently [24, 30–32]. However, the mechanisms of cGAS activation by dsDNA proposed in these studies are different from each other. In the first two reports on the structures of mouse and porcine cGAS:dsDNA complexes [24, 31], the authors proposed that cGAS binds to dsDNA with a 1:1 stoichiometry and interacts with dsDNA through a single binding site in a similar manner as how OAS1 binds dsRNA [47]. In contrast, two other studies published recently showed that two cGAS molecules form a 2:2 complex with two dsDNA and each cGAS molecule interacts with two dsDNA through two binding sites [30, 32] (Fig. 3A). Both of these studies proposed that cGAS is activated by dsDNA induced oligomerization instead of simple 1:1 DNA binding [30, 32]. The first DNA binding site (site A) revealed by these two new papers is essentially the same as the DNA binding site described by Gao and Civril et al. [24, 31]. However, the second DNA binding site (site B) is new and not described previously. The buried surface area between mouse cGAS (mcGAS) and an 18 bp dsDNA at site A is about ~1,500 Å2 [30]. The buried surface area between mcGAS and the 18 bp dsDNA at site B is 800–900 Å2, which account for ~35% of the total buried surface area between mcGAS and the dsDNA [30]. Both of the DNA binding sites are rich of positively charged residues and show shape and charge complementarity to negatively charged dsDNA. cGAS associates with the dsDNA through extensive electrostatic interactions and hydrogen bonds. The dimerization of cGAS plays a critical role in the formation of the 2:2 cGAS:dsDNA complexes. Analysis of all the published structures to date showed that similar 2:2 complexes are formed in all the cGAS/dsDNA complex structures through either crystallographic or non-crystallographic interactions [30], so it is unlikely that the dsDNA induced dimerization of cGAS is an artifact of crystal packing contacts.
Figure 3.
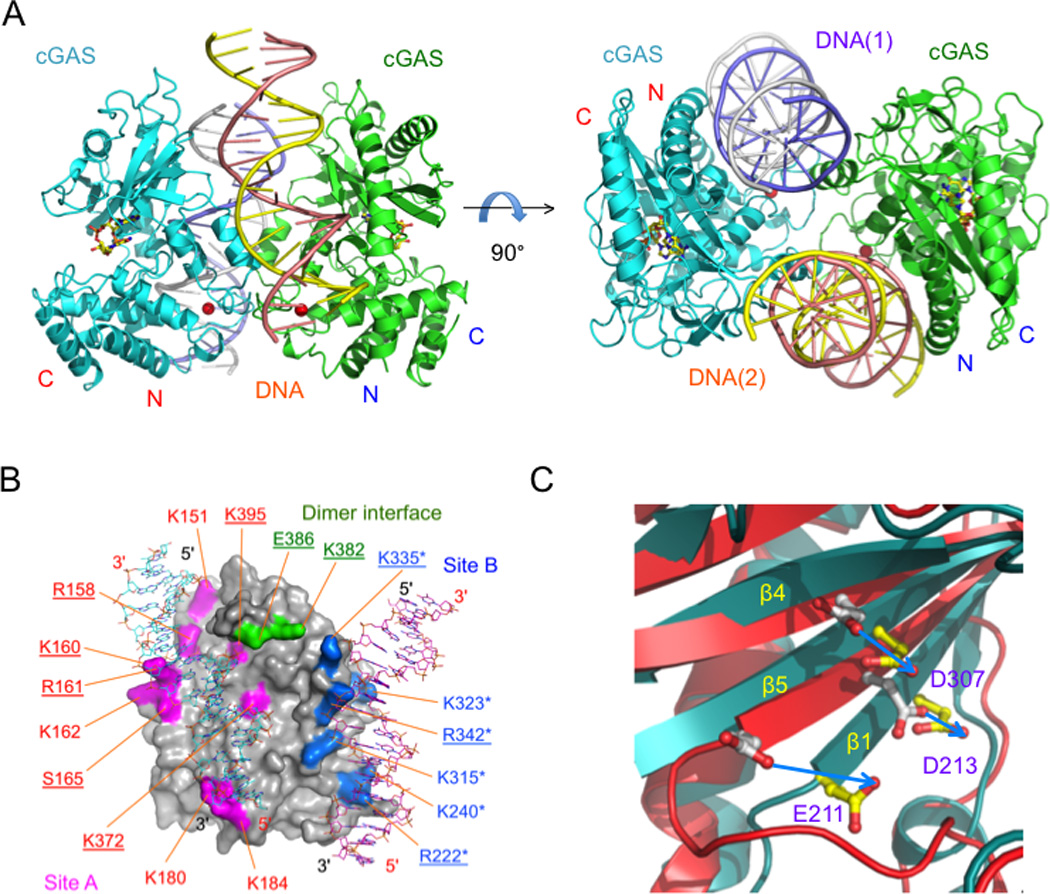
cGAS is activated by dsDNA induced oliogomerization. (A). Structure of mouse cGAS catalytic domain bound to an 18 bp dsDNA and cGAMP (ball-and-stick models). (B). The DNA two binding sites of cGAS. Mutations of residues that result over 50% loss of enzyme activity are underlined. Mutations of residues that dramatically reduced DNA binding are labeled with asterisks. (C). Conformational changes of mouse cGAS active sites upon dsDNA binding.
To confirm the dsDNA binding induced oligomerization of cGAS, Li et al. conducted small angle X-ray scattering (SAXS) studies of cGAS in isolation and its mixture with dsDNA and observed the formation of a higher order complex in solution [30]. Consistent with these observations, analytical ultracentrifugation (AUC) analysis of cGAS alone and its mixture with dsDNA also showed the formation of higher order complexes [30, 32]. In addition, mutations of key residues at either DNA binding site A or B disrupted the oligomerization of cGAS [30, 32]. Moreover, mutations at the cGAS dimer interface also abolished the formation of higher order complex [30]. To confirm that cGAS is activated by dsDNA induced oligomerization, Li et al. purified 18 mutants of mcGAS at the two DNA binding sites and the dimer interface and analyzed how these mutations affect the catalytic activity of cGAS [30]. These assays demonstrated that mutations of key residues at either site A, site B, or the cGAS dimer interface influence the activity of cGAS (Fig. 3B). These results clearly showed that dsDNA binding at site A, site B, and the dimerization of cGAS are needed for its activation. Consistent with results from these enzyme assays, IFN-β reporter assays with several mutants of full-length cGAS also showed that mutations at the two DNA binding sites and the dimer interface affect the activity of cGAS in cells [30]. Additional cell based assays showed that similar mutations of hcGAS affect the phosphorylation and oligomerization of IRF3 in cells [32]. In summary, these in vitro and in vivo studies of cGAS mutants confirmed that DNA binding at site A and site B as well as the dimerization are essential for cGAS activation.
A comparison of crystal structures of cGAS in isolation and in complex with dsDNA revealed that DNA binding induces significant conformational changes in the catalytic domain of cGAS [24, 30–32] (Fig. 3C). The three active site residues are realigned for catalysis when cGAS binds dsDNA (Fig. 3C). In addition, the activation loop (residues 197 to 206 in mcGAS) is restructured upon DNA binding. Residues in this restructured activation loop are likely involved in substrate binding. For example, it has been shown Ser199 in this loop interacts with the γ phosphate of ATP or GTP [24]. Crosslinking of the cGAS dimer by dsDNA plays a critical role in stabilizing the active conformation of cGAS. As reflected by the uniform distribution of temperature factors of the dsDNA bound cGAS structures, dsDNA binding reduces the structural flexibility of the NTase domain and stabilizes the active conformation of cGAS [30].
To further investigate how DNA binding activates cGAS, Li et al. purified a number of mutants of mouse cGAS catalytic domain and full-length mouse cGAS and conducted DNA binding studies [30]. Surprisingly, these studies demonstrated that mutation of each of the ten key residues at site A does not affect dsDNA binding by cGAS significantly [30]. In contrast, mutations of any of the six residues at site B dramatically reduced DNA binding [30] (Fig. 3B). It is hard to draw a direct correlation between DNA binding and the activity of these mutants. In addition, mutations at the cGAS dimer interface also have limited effect on DNA binding [30] (Fig. 3B). The explanation for these observations is that cGAS binds dsDNA cooperatively through the two binding sites and site B plays a more important role in the cooperative DNA binding [30]. It is likely that DNA binding at site A plays a major role in driving the conformational change that is needed for the activation of cGAS, while the crosslinking of the cGAS dimer by dsDNA stabilizes the active conformation of the enzyme [30].
5. The catalytic mechanism of cGAS
Based on extensive structural studies of cGAS bound to ATP, GTP, and various reaction products or intermediates and biochemical studies of cGAS catalyzed reaction with various kinds of substrates, two groups proposed similar catalytic mechanisms for cGAS [24, 25]. cGAS is highly specific when both ATP and GTP are used as substrates. In the first step of the cGAS catalyzed reaction, the triphosphate group of ATP binds to the active site (Fig. 4). The 2'-OH of GTP attack the α-phosphate of ATP, resulting in the formation of a linear intermediate pppGp(2',5')A (Fig. 4). This intermediate flips over in the active site to allow the triphosphate of GTP to associate with the active site (Fig. 4). Next, the 3'-OH group of AMP attacks the α-phosphate of GTP, resulting in the formation of the second phosphodiester bond with 3',5' linkage (Fig. 4). The formation of the 2',5' phosphodiester bond was demonstrated by the structure of mcGAS bound to pppGp(2',5')G [24], which is a major product of cGAS when GTP alone is used as a substrate. The structure of mouse cGAS bound to cGAMP revealed how the product interacts with cGAS immediately after the formation of the 3',5' phosphodiester bond [30] (Fig. 4). Another structure of mcGAS bound to cGAMP revealed that the product bound to cGAS in a different orientation [24]. This structure likely represents how cGAMP associates with cGAS before its release from the active site. Although it is not complete clear why cGAS is specific for ATP and GTP as substrate, the structure of porcine cGAS active site mutant associated with ATP and GTP revealed roughly how cGAS binds the two substrates [31]. However, the resolution of data (3.08 Å) does not allow the identity of ATP or GTP to be accurately determined. It is likely GTP associate with the active site Mg2+, while ATP is positioned to attack the α-phosphate of GTP with its 2'-OH group (Fig. 4). When GTP alone is used as a substrate, the predominant product is pppGp(2',5')G and a little cyclic product [pG(2',5')Gp(3',5')] [25]. In contrast, the product when ATP alone is used as substrate is pppAp(3',5')A and this product cannot be further cyclized by cGAS [25].
Figure 4.
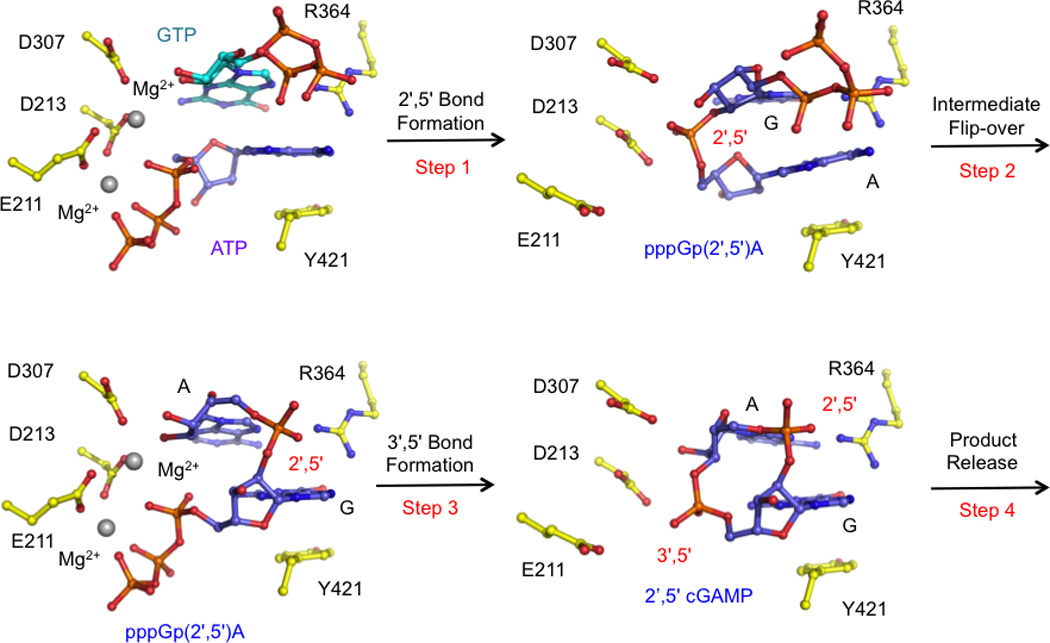
The catalytic mechanism of cGAS. These models are generated based on the crystal structures of porcine cGAS bound to ATP and GTP (PDB 4KB6), mouse cGAS bound to pppGp(2',5')G (PDB 4K98), and mouse cGAS bound to cGAMP (PDB 4LEZ).
6. The mechanism of STING activation by exogenous and endogenous ligands
The critical role of STING as a direct sensor of exogenous cyclic dinucleotide was first demonstrated by the landmark studies of the Vance group [14]. In this work, they expressed the cytosolic domain of STING and confirmed its interaction with c-di-GMP by in vitro binding studies [14]. The first glimpse into the mechanism of cyclic dinucleotide sensing by STING were demonstrated by the structural studies of human STING (hSTING) ligand binding domain in isolation and in complex with c-di-GMP from five labs [36–40]. These structures revealed that residues 155 to 174 of STING are part of the ligand binding domain instead of a transmembrane helix as predicted previously [13]. These structures showed that ligand free STING forms a dimer and adopt an open V-shaped conformation (Fig. 5). When STING binds to c-di-GMP, it can adopt either an open or a closed conformation and binds c-di-GMP at dimer interface. The STING/c-di-GMP complex structure determined by Huang et al. using the G230A/H232R variant of hSTING adopts a unique closed conformation [39]. It was suggested that this ligand induced conformational change was involved in the activation of STING by c-di-GMP [39].
Figure 5.
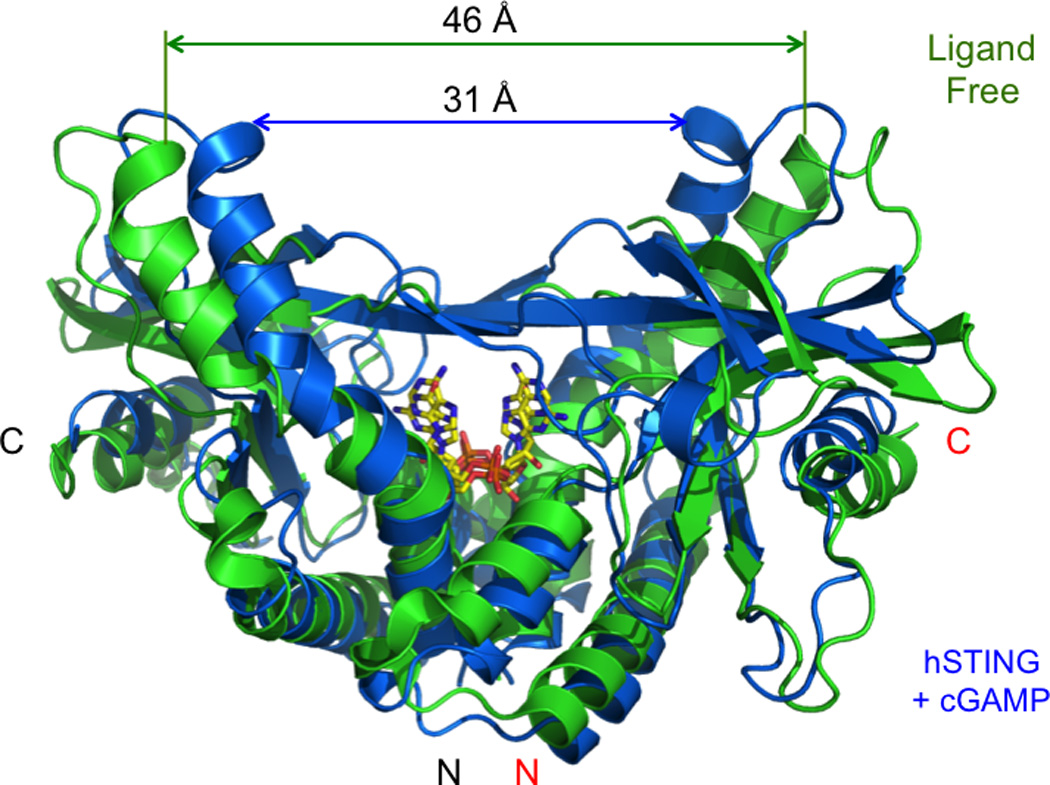
Conformational changes of human STING upon cGAMP binding. Ligand free STING is shown by the green ribbons (PDB 4EMU). STING bound to cGAMP is shown by the blue ribbons (P. Li, unpublished data). cGAMP in two different orientations are shown by the yellow stick models.
After the endogenous ligand of STING was identified, the structures of both human and mouse STING bound to cGAMP were determined [23, 41]. These structures revealed that both mouse and human STING adopt a closed conformation when they bind to cGAMP [23, 41] (Fig. 5). Although two different variant of hSTING, G230/H232 and G230/R232, were used in these structural studies, the structures of the complexes are essentially the same and are very similar to the closed structure of hSTING bound to c-di-GMP described by Huang et al [39]. In addition, similar structures were also observed for the complexes of mouse STING (mSTING) bound to either 3', 5' or 2', 5' cGAMP [41]. The long hairpin containing residues Arg232 and Arg238 in hSTING or Arg231 and Arg237 in mSTING seems play a critical role in forming the closed or active conformation of STING upon ligand binding. Residue Arg238 of hSTING or Arg237 of mSTING interacts with the phosphate groups of the c-di-GMP or cGAMP through electrostatic interactions [23, 39, 41]. Variants of hSTING at residues Gly230 and Arg232 in this loop have different binding properties to endogenous and exogenous ligands [41]. The replacement of Gly230 in hSTING by Ile229 in mSTING likely contributes to the different ligand binding and signaling activities of mSTING. It has been shown that the mutation of Arg231 to alanine of mSTING in this loop disrupts its response to c-di-GMP but does not affect the response to dsDNA [14]. It is likely that this mutation disrupts the ligand induced conformational change of the STING dimer induced by c-di-GMP but not that induced by cGAMP. Taken together, these structural studies of STING bound to both endogenous and exogenous cyclic dinucleotides demonstrated that the conformational changes of the STING dimer upon ligand binding (Fig. 5) is likely how these ligands activates the downstream signaling via STING. Consistent with this hypothesis, structures of two synthetic activators CMA and DMXAA bound to mSTING also showed similar ligand binding induced conformational changes [41, 48].
7. New developments and future directions
The major components of the cGAS-STING pathway have been identified in the last few years. The mechanism of how these molecules mediate the signaling of the cGAS-STING pathway is emerging. However, details of the cGAS-STING pathway are still not completely clear. For example, the mechanism of how full-length cGAS recognizes dsDNA is still not clear. The contribution of the N-terminal regions of cGAS to dsDNA sensing and cGAS activation remains to be established. The detailed mechanism of cGAS activation by dsDNA still needs further investigations. Although a plausible catalytic mechanism of cGAS has been proposed, the molecular details are not fully established. For example, it is still not quite clear how the enzyme catalyzes the synthesis of two different kinds of phosphodiester bonds with a single active site. The specificity of cGAS for ATP and GTP remains to be elucidated.
Although the identity of the cGAS product has been determined and its interactions with STING were studied extensively, the kinetic properties of endogenous and exogenous ligands binding by STING have not been studied. The significant differences in ligand binding and signaling by exogenous and endogenous cyclic dinucleotides are still not fully understood. Even though the structures cGAMP bound to either human or mouse STING have been determined, this does not mean that we know exactly how cGAMP interacts with STING and how ligand binding induces the conformational changes of the STING dimer. Since cGAMP is an asymmetrical dimer, the electron density maps observed in all of these structures are an average of cGAMP in two different orientations and do not allow an unambiguous modeling of the ligand at the resolution of around 2.0 Å [23, 41]. Further studies of cGAMP with STING by other techniques such as NMR should provide additional insight into these questions.
Another question that remains to be addressed is how ligand binding activates the signaling via STING. The current model suggests that ligand binding by STING facilitate the recruitment and activation TBK1 at the signaling complex. IRF3 is then recruited to the signaling complex and phosphorylated by TBK1 and translocates to the nucleus. The structures of TBK1 and IRF3 have been studied extensively, but the exact mechanism of their recruitment and activation via the cGAS-STING pathway remains to be established. It is also not clear why the over expression of STING can also activated TBK1 and IRF3.
Recently, a number of new molecules have been identified in the cGAS-STING pathway. For example, the protein kinase ULK1 seems negatively regulate the signaling via STING by phosphorylating the C-terminal tail of STING [49]. The nucleotide-binding, leucine-rich repeat-containing-protein NLRC3 also negatively regulates STING-dependent responses to dsDNA, c-di-GMP, and DNA viruses [50]. It was suggested that NLRC3 interacts directly with STING and TBK1. In addition, a recent study showed that Beclin-1 suppresses cGAMP synthesis by cGAS and inhibits IFN-β induction by dsDNA transfection and HSV infection [51]. It is likely these new proteins play critical roles in regulating the signaling of the cGAS-STING pathway. However, the mechanism about how these protein functions need to be confirmed by further in vitro and in vivo studies.
In our studies, we observed that suspension cultures of human THP-1 cells can be stimulated by cGAMP added to the culture medium and the intravenous injection of cGAMP in mice induces the production of IFN-β [30]. A recent study by Ablasser et al. shows that cGAMP can transfer from a producing cell to neighboring cells via the gap junction and stimulate the anti-viral responses in those cells directly [52]. The mechanism of how cells pick up cGAMP still needs further investigation.
As a novel endogenous second messenger, cGAMP likely plays other roles in addition to regulating the induction of type I interferons. It is evident cGAMP also mediates the activation of NF-κB via STING [30, 53]. However, the mechanism of NF-κB activation by cGAMP is still not clear. Moreover, our studies shows cGAMP regulates the induction of a wide spectrum of genes, including a number of cytokines, chemokines, and non-coding RNAs (P. Li unpublished data). Interestingly, cGAMP does not induce the synthesis or maturation of the inflammatory cytokine IL-1β in THP1 cells (P. Li unpublished data). It has been shown the cGAS-STING pathway is also critical for the restriction of RNA viruses in addition to DNA viruses [45, 54]. In contrast, the RIG-I like receptors only mediates the response to RNA viruses. However, the mechanism of how cGAS mediates the restriction of such a wide spectrum is still not fully understood.
Since cGAMP is an endogenous second messenger with a molecular mass of 675 daltons, it has the potential to be used directly as an adjuvant for vaccines, antiviral reagent, and antitumor reagent. Indeed, a recent study already demonstrated that cGAMP stimulates the adaptive responses mediate by both B-cells and T-cells [44]. The activity of cGAMP to induce IFN-β expression in cells and in mice suggest that it has the potential to replace IFN-β to treat infectious disease and cancer that are currently treated with IFN-β. These potentials of cGAMP need to be investigated in appropriate animal models and tested in clinical trials. These studies may lead to conceptually new approaches to treat viral diseases and cancer.
Acknowledgements
This study is supported by the National Institute of Health (Grant AI 087741 to P. Li) and the Welch Foundation (Grant A-1816 to P. Li). This work is dedicated to the memory of my dad Changrong Li (1938–2014, of P. Li).
Biography

Author Biography of Pingwei Li
Dr. Pingwei Li was born in 1967 in the suburb of Xi’an, China. He obtained his Ph.D. from Peking University in 1996, majoring in X-ray crystallography. During his graduate studies, he determined the crystal structure of Cu, Zn SOD from duck erythrocytes. He came to the United States in the spring of 1998 and did his first post-doc with Roland Strong in the Division of Basic Science at the Fred Hutchinson Cancer Research Center in Seattle. He determined the structures of the noncanonical MHC class I like molecules MICA, RAE-1β and their complexes with the natural killer cell and γδ T cell receptor NKG2D. He moved on to work in the Department of Molecular Biology at Princeton University as a staff scientist in the fall of 2001, studying the structures of the UBP family deubiquitinating enzymes HAUSP (USP7) and USP14 in the laboratory of Yigong Shi. He moved to the California Institute of Technology in the summer of 2003 and did another post-doc with Pamela Bjorkman. His research focuses on the structural studies of a monoclonal antibody (MW1) against polyglutamine. He cloned the VH and VL genes of the antibody, generated the Fv fragment by refolding, and determined the structure of MW1 Fv in isolation and in complex with a polyglutamine containing peptide GQ10G. He started his own lab in the Department of Biochemistry and Biophysics at Texas A&M University in the fall of 2005. His current research focuses on the structural basis of microbial nucleic acid sensing in innate immunity. His lab determined the structures of LGP2 and RIG-I C-terminal domains bound to dsRNA, the structure of STING bound to c-di-GMP and cGAMP, and the structure of TBK1. In his recent studies, his lab determined the structures of cGAS in isolation and in complex with dsDNA and elucidated the mechanism of cGAS activation by dsDNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nature immunology. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 2.Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Current opinion in immunology. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 4.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 5.Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Molecular cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annual review of immunology. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annual review of immunology. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 8.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends in immunology. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nature immunology. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 12.Hornung V, Latz E. Intracellular DNA recognition. Nature reviews Immunology. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, et al. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2012;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Molecular and cellular biology. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, et al. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, et al. Cyclic [G(2',5')pA(3',5')p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013;153:14. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2'–5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013 doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, Chen ZJ. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Science signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nature immunology. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 29.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:6. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell reports. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell reports. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato K, Ishii R, Goto E, Ishitani R, Tokunaga F, Nureki O. Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS. PloS one. 2013;8:e76983. doi: 10.1371/journal.pone.0076983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kranzusch PJ, Vance RE. cGAS Dimerization Entangles DNA Recognition. Immunity. 2013;39:992–994. doi: 10.1016/j.immuni.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature structural & molecular biology. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nature structural & molecular biology. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 38.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, et al. Structural Analysis of the STING Adaptor Protein Reveals a Hydrophobic Dimer Interface and Mode of Cyclic di-GMP Binding. Immunity. 2012 doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature structural & molecular biology. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 40.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, et al. Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA. Cell. 2013 doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoggins JW, Macduff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2013 doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. The EMBO journal. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, et al. NLRC3, a Member of the NLR Family of Proteins, Is a Negative Regulator of Innate Immune Signaling Induced by the DNA Sensor STING. Immunity. 2014;40:329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell host & microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013 doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi G, Brendel VP, Shu C, Li P, Palanathan S, Cheng Kao C. Single Nucleotide Polymorphisms of Human STING Can Affect Innate Immune Response to Cyclic Dinucleotides. PloS one. 2013;8:e77846. doi: 10.1371/journal.pone.0077846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]


