Abstract
RAD51-associated protein 1 (RAD51AP1) is critical for homologous recombination (HR) by interacting with and stimulating the activities of the RAD51 and DMC1 recombinases. In human somatic cells, knockdown of RAD51AP1 results in increased sensitivity to DNA damaging agents and to impaired HR, but the formation of DNA damage-induced RAD51 foci is unaffected. Here, we generated a genetic model system, based on chicken DT40 cells, to assess the phenotype of fully inactivated RAD51AP1 in vertebrate cells. Targeted inactivation of both RAD51AP1 alleles has no effect on either viability or doubling-time in undamaged cells, but leads to increased levels of cytotoxicity after exposure to cisplatin or to ionizing radiation. Interestingly, ectopic expression of GgRAD51AP1, but not of HsRAD51AP1 is able to fully complement in cell survival assays. Notably, in RAD51AP1-deficient DT40 cells the resolution of DNA damage-induced RAD51 foci is greatly slowed down, while their formation is not impaired. We also identify, for the first time, an important role for RAD51AP1 in counteracting both spontaneous and DNA damage-induced replication stress. In human and in chicken cells, RAD51AP1 is required to maintain wild type speed of replication fork progression, and both RAD51AP1-depleted human cells and RAD51AP1-deficient DT40 cells respond to replication stress by a slow-down of replication fork elongation rates. However, increased firing of replication origins occurs in RAD51AP1−/− DT40 cells, likely to ensure the timely duplication of the entire genome. Taken together, our results may explain why RAD51AP1 commonly is overexpressed in tumor cells and tissues, and we speculate that the disruption of RAD51AP1 function could be a promising approach in targeted tumor therapy.
Keywords: Homologous recombination, DNA replication, RAD51 foci, RAD51AP1, DT40 cells
1. Introduction
Genome stability is dependent on the accurate repair of DNA double-strand breaks (DSBs). DSBs can arise from exogenous DNA damaging agents or, spontaneously, during DNA replication. One major mechanism by which DSBs are repaired is by homologous recombination DNA repair (HR). HR utilizes the sister chromatid as a template, and is considered a comparatively faithful DNA repair pathway that can be subdivided into three major stages: the pre-synaptic stage characterized by strand stabilization, the synaptic stage characterized by strand invasion and branch migration, and the post-synaptic stage which includes the formation of Holliday junctions and their resolution.
RAD51AP1 is a RAD51-associated protein that binds to both the RAD51 and DMC1 recombinases, and is critically important for wild-type levels of homologous recombination in mitotic and meiotic cells [1–3]. Knockdown of RAD51AP1 in human cells leads to increased levels of genomic instability and to decreased levels of homology-directed DNA repair [3,4]. Human cells with silenced RAD51AP1 are viable, but are sensitized to the cytotoxic effects of interstrand cross-linking agents and of ionizing radiation. However, human cells with depleted RAD51AP1 are able to form RAD51 DNA damage foci with normal kinetics [3,4]. Purified human RAD51AP1 binds avidly to DNA, and prefers D-loop structures over double-stranded DNA, and double-stranded DNA over single-stranded DNA [3,4]. Collectively, these results have shown that in mitotic human cells RAD51AP1 functions downstream of the pre-synaptic stage of HR and through its interaction with RAD51.
Here we generated RAD51AP1-deficient DT40 cells to assess the phenotypic consequences of the total loss of RAD51AP1 expression. In this model system, we are able to show, for the first time, that vertebrate cells are viable without any expression of RAD51AP1. Confirming our results for human cells with RAD51AP1 knockdown, chicken RAD51AP1 also is not essential for the formation of DNA damage-induced RAD51 foci. However, chicken RAD51AP1 is required for the timely resolution of RAD51 DNA damage foci, data that are difficult to obtain in human cells, because of the transient and likely incomplete depletion of the RAD51AP1 protein in RAD51AP1 knockdown experiments. We find that RAD51AP1-deficient DT40 cells are sensitized to the cytotoxic effects of cisplatin and of ionizing radiation, and that ectopic expression of GgRAD51AP1, but not of HsRAD51AP1, fully complements in these cell survival assays. Last, we show that RAD51AP1 is critical for overcoming replication stress in both human and in chicken cells, and we provide a model that may help explain why the human RAD51AP1 protein frequently is expressed at elevated levels in a variety of human cancer cells and tissues.
2. Materials and methods
2.1. Cell lines and reagents
Chicken DT40 cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, 10 μM beta-mercaptoethanol, 1% antibiotics and 1% L-glutamine and kept at 37 °C in a humidified incubator containing 5% CO2. Selection of individual clones derived from DT40 cells was performed in standard growth medium containing 1.7 mg/ml G418 and 0.5 μg/ml puromycin for gene targeting constructs, and 2 mg/ml hygromycin for lentiviral transduction to express the HsRAD51AP1 or GgRad51ap1 ORFs. U2OS cells were grown in Dulbecco’s modified Eagle’s Medium with 10% fetal calf serum at 37 °C in a humidified incubator containing 10% CO2. Forward transfections with siRNA were carried out using Lipofectamine RNAiMAX reagent (Invitrogen), essentially as recommended by the manufacturer. Down-regulation of RAD51AP1 was mediated by RAD51AP1-directed siRNAs with the target sense sequences 5′-CCTCATATCTCTAATTGCA-3′ (here: AP1_1) and 5′-TGAACAATCTCCGGAAAGA-3′ (here: AP1_2) and as previously described [3–5]. For negative control siRNA, the following target sequence was used: 5′-GATTCGAACGTGTCACGTC-3′, as previously described [6].
2.2. Growth-rate measurement and cell survival assays
For measuring the cell-doubling times of DT40 wild type and mutant cells, exponentially growing cells were cultured for 5 days and were counted twice a day using a Coulter Counter (Beckman Coulter). Cells were diluted appropriately in pre-heated medium to maintain cell density between 0.2 and 1.4 × 106 cells/ml and cell viability >95%. Each growth rate determined is the average of 3 independent cultures. Cells were treated with the indicated concentrations of cisplatin. Percent survival represents the number of colonies in the presence of the drug compared to the number of colonies in the absence of the drug.
2.3. Plasmids and lentiviral DNA
For the ectopic expression of the FLAG-tagged human and chicken RAD51AP1 ORFs, lentiviral vectors were constructed based on the Gateway System (Invitrogen), but modified by Dr. E. Campeau [7]. FLAG-HsRAD51AP1 was generated by sub-cloning a SalI to XhoI fragment containing the DNA sequence for the FLAG peptide 5′ to and in frame with the human RAD51AP1 ORF amplified from pOK24 [8] into pENTR1A (Invitrogen). To obtain pLenti CMV DEST #2 [7] encoding human FLAG-RAD51AP1, pLenti CMV DEST #2 was used and a standard LR recombination reaction was carried out, as described [7]. Similarly, FLAG-GgRAD51AP1 was generated by sub-cloning a SalI to EcoRI fragment containing the DNA sequence for the FLAG peptide 5′ to and in frame with the chicken RAD51AP1 ORF amplified from chicken EST cDNA clone 3GAL 54E24 into pENTR1A (Invitrogen), and the same strategy, as described above, was used to obtain pLenti CMV DEST #2 encoding chicken FLAG-RAD51AP1. Lentiviruses were produced in 293FT cells as described [7], aliquoted and stored at −80 °C. DT40 cells were transduced in the presence of 6 μg/ml polybrene in 6-well tissue culture plates at about 500,000 cells/ml growth medium without antibiotics. Two days after transduction, cells were spun out to replace the cell culture medium with fresh medium containing hygromycin. Cell cultures were expanded and single clones isolated that showed similar levels of expression for both HsRAD51AP1 and GgRAD51AP1, as determined by Western blot analysis.
2.4. Gene disruption constructs for generating the RAD51AP1 knockout in DT40 cells
For cloning of the chicken RAD51AP1 deletion construct, a BamHI-adapted forward and a BamHI-adapted reverse primers were used to amplify the puromycin and neomycin genes with β-actin promoters from pLoxPuro and pLoxNeo vectors (kindly obtained by Jean-Marie Buerstedde), respectively. To generate the 3.2 kb 5′ homology flank of the RAD51AP1 deletion construct a KpnI-adapted forward and a SalI-adapted reverse primer based on NCBI reference sequence: XM 417234.3 were used for amplification using DT40 genomic DNA as a template. To generate the 4.0 kb 3′ homology flank a NotI-adapted forward and a SacI-adapted reverse primer were used for DNA amplification. The homology arms were assembled into pBlueScript SK+ and the neomycin or puromycin resistance cassettes were cloned in as BamHI-digested fragments.
2.5. RT-PCR
Total RNA was isolated from DT40 wild type cells and derivatives using the SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions. For the generation of cDNA, a standard reverse transcription protocol was carried out using ImProm-II Reverse Transcriptase and oligo(dT)15primer (Promega). The following primers were used: forward 1 (F1; in exon 2) 5′-GGCGCGGATGGTGAGGAG-3′; reverse 3 (R3; in exon 7) 5′-GGAGTCTTCTTTTCTTTTCTAGTCTGCC; reverse 4 (R4; in exon 8) 5′-GGACATCATCTTTTGTGTCTGC-3′. β-Actin primers were used as control, as described previously [9].
2.6. Southern blotting and hybridization
Six μg of genomic DNA from DT40 wild type cells and derivatives was digested with PvuII, ethanol precipitated, re-suspended in TE buffer, and separated on a 0.8% agarose gel in 1 × TAE, at 35 V overnight. The DNA was transferred to a HybondXL membrane (Amersham) using a vacuum blotter, with 10 min hydrolysis in 0.125 N HCl and 1 h denaturation in denaturation buffer (0.5 M NaOH, 1.5 M NaCl). To neutralize, the membrane was rinsed in 2 × SSC for 30 min and stored wet until further use. Hybridizations were carried out at 60 °C in 5 × SSC, 5 × Denhardt’s and 0.5% SDS with 100 μg/ml sheared salmon sperm DNA. As a hybridization probe, a 32P-labeled 1.5 kb fragment with 100% homology to the 3′-end of the 5′-flank of the gene targeting vector was generated by standard random priming (Aligent Technologies) using 5 μl of α-32PdCTP (3000 Ci/mmol) and 1 μl Klenow enzyme (5 U/μl). The reaction was carried out at 37 °C for 60 min, stopped by the addition of 2 μl 0.5 M EDTA and purified over a G-25 column (GE Health-care Life Sciences). After hybridization, membranes were washed twice for 30 min in 2 × SSC with 0.1%SDS, once for 30 min in 1 × SSC with 0.1%SDS, and once for 50 min in 1 × SSC with 0.1%SDS at 60 °C. Phosphor screens were exposed for 48 h.
2.7. Mutagens and X-irradiation
X-ray experiments were performed using a 6.3 mA 160 kVp X-ray machine at dose rates of 0.5 Gy/min to 1 Gy/min. All exposures were carried out in 15 ml conicals at room temperature. Exponentially growing cells were exposed to the indicated concentrations of cisplatin in regular growth medium at 37 °C for 1 h.
2.8. Cell fractionation and indirect immunofluorescence
Fractionated extracts from DT40 cells overexpressing chicken or human FLAG-RAD51AP1 were prepared essentially as described in [10], and according to Dignam et al. [11]. Indirect immunofluorescence for RAD51 was carried out essentially as described previously [3], except that a Shandon Cytospin 4 (ThermoFisher Scientific) was used to prepare samples. Briefly, pre-wet cytospin filters (2%BSA in PBS) were used to spin 150,000 cells in 100 μl (1%BSA in PBS) onto cytospin slides to generate samples prior to fixation, permeabilization and immunocytohistochemistry.
2.9. DNA fiber assay
Exponentially growing cells were pulse-labeled with 25 μM CldU (Sigma) and 250 μM IdU (Sigma) for the times specified. Where indicated, the cells were exposed to 2 mM hydroxyurea (HU) as indicated. Labeled cells were harvested and DNA fiber spreads were prepared from 0.5 × 106 cells/ml as described previously [12]. Slides were incubated in 2.5 M HCl for 90 min and then washed several times in PBS, followed by incubation in blocking buffer (2% BSA, 0.1% Tween in PBS) for 1 h. Acid-treated fiber spreads were stained with monoclonal rat anti-BrdU antibody (Oxford Biotechnologies, 1:1000) to detect CldU, followed by monoclonal mouse anti-BrdU antibody (Becton Dickinson, 1:15,000) to detect IdU. Secondary antibodies were goat anti-rat AlexaFluor555 and goat anti-mouse AlexaFluor488 (both Invitrogen, 1:500). Primary antibodies were diluted in blocking buffer, incubated for 1.5 h with rat anti-BrdU antibody and over-night with mouse anti-BrdU antibody followed by extensive washes in PBS. Secondary antibodies were applied for 1.5 h, and slides were mounted in Immuno-Fluor mounting medium (MP Biomedicals). Fiber tracts were examined using an AxioVert 200 M fluorescence microscope (Zeiss). Pictures were taken from randomly selected fields with untangled fibers and analyzed using ImageJ software package. For structure analyses, the frequencies of the different classes of fiber tracks were classified as follows: red-green (elongating fork), red (stalled or terminated forks), green-red-red-green (first pulse origin) and green (second pulse origin). For fork speed analyses, the lengths of CldU and IdU tracks were measured and micrometer values were converted into kilobases. A conversion factor for the length of a labeled track of 1 μm = 2.59 kb was used [13]. A minimum of 100 individual fibers was analyzed for each experiment and the means of at least 3 independent experiments are presented.
2.10. Western blot analysis and antibodies
Western blot analysis was carried out according to our standard procedures [14]. The following primary antibodies were used: monoclonal mouse anti-FLAG (Sigma, M2; 1:3000), monoclonal mouse anti-α-Tubulin (Calbiochem, CP06; 1:3000), polyclonal rabbit anti-QM (Santa Cruz Biotechnology, C-17; 1:3000) and monoclonal rabbit anti-phosphoSer345-CHK1 (Cell Signaling, 133D3; 1:750). Our own polyclonal rabbit anti-human RAD51AP1 antibody was used, that has been described previously [15]. HRP-conjugated goat anti-rabbit or goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA; 1:10,000), IRDYE680-conjugated anti-mouse IgG or IRDYE800-conjugated anti-rabbit IgG (LiCor; 1:7500) were used as secondary antibodies.
2.11. BrdU incorporation
Cells were incubated at 37 °C in regular growth medium with 10 mg/ml BrdU on a shaking platform for 30 min. Cells were collected by centrifugation and fixed in 70% ethanol at −20 °C. Cells were spun down for 5 min and cell pellets (5 × 105 cells) were re-suspended in 1 ml 2 N HCl/0.5% Triton X-100 solution and incubated for 60 min on a vortex. Cells were spun down and flash-incubated in 2 ml borax solution before centrifugation. Cells were re-suspended in 0.5 ml 0.5% Tween-20/1% BSA in PBS and 10 μl of anti-BrdU-FITC antibody (BD 347583) was added directly to the cells and incubated for 60 min at room temperature. The supernatant was removed and cells were incubated in 1 ml PBS containing 20 μg/ml RNase and 30 μg/ml propidium iodide for at least 30 min before FACS analysis using a BD FACSCalibur flow cytometer with 488 nm excitation and 630 nm long pass filter.
2.12. Statistical analysis
The statistical analysis was performed using Prism (GraphPad Software Inc.) on the data from at least three independent experiments. Statistical significance was assessed by two-tailed unpaired Student’s t-test. P ≤ 0.05 was considered significant.
3. Results
3.1. Orthologs of RAD51AP1 are present in some invertebrates
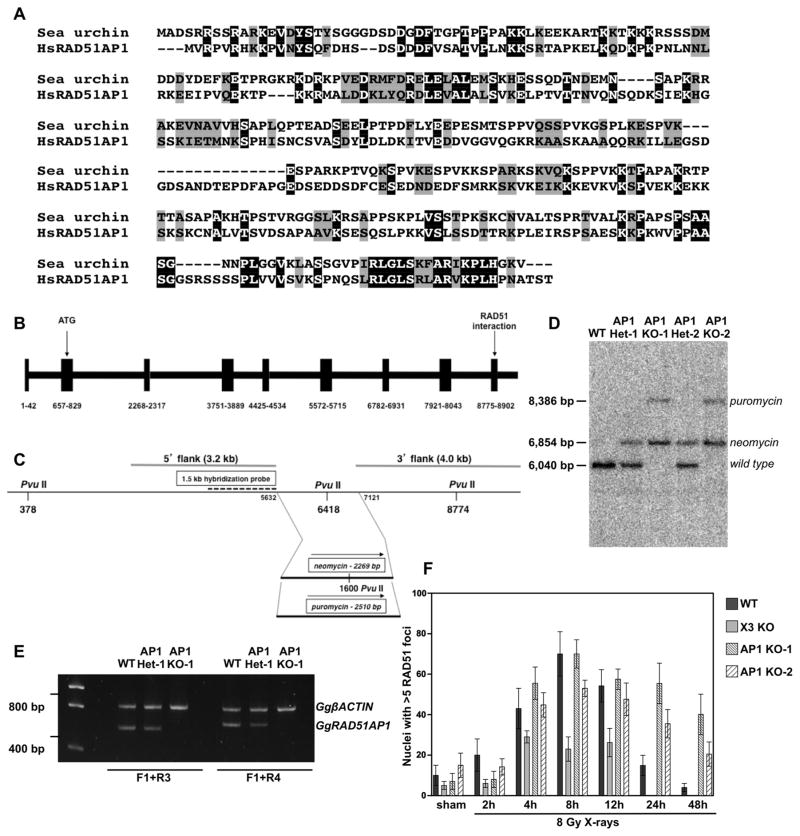
Previously, we suggested that RAD51AP1 likely was a vertebrate-specific gene, but had not excluded the possibility that invertebrates may possess unidentified functional equivalents to RAD51AP1 [3]. Meanwhile, however, sequence information for many more invertebrate genomes has become available. This led us to conduct further homology searches for orthologs of human RAD51AP1 to identify potential invertebrate models. Interestingly, we find that, although limited to metazoans, orthologs of human RAD51AP1 are present in some invertebrates, such as in the purple sea urchin (Strongylocentrotus purpuratus; Genbank XP 003729232) (Fig. 1A). RAD51AP1 also is present in the Pacific oyster (Crassostrea gigas; Genbank EKC29041.1), and in the adhering hairy plate (Trichoplax adhaerens; Genbank XP 002117334.1), the simplest animal known, lacking any recognizable organs ([16]; http://genome.jgi-psf.org/) (Supplemental Fig. 1A and B). In addition, RAD51AP1 orthologs are present in the Florida lancelet (Branchiostoma floridae), in the starlet sea anemone (Nematostella vectensis), in the polychaete worm (Capitella teleta), in the acorn worm (Balanoglossus claviegerus), and in molluscs like the California sea hare (Aplysia californica) and in the owl limpet (Lottia gigantea) (Supplemental Fig. 1C). However, RAD51AP1 orthologs do not appear to be present in any simple model organisms commonly used in the laboratory, such as Drosophila melanogaste, Caenorhabditis elegans or yeast.
Fig. 1.
RAD51AP1 is not limited to vertebrates, and a RAD51AP1 knockout in DT40 cells results in impaired resolution of RAD51 foci. (A) The presence of RAD51AP1 is not limited to vertebrates, although it does appear to be limited to multicellular animals (i.e. metazoans). A number of invertebrates do encode a RAD51AP1 ortholog, including the purple sea urchin (Strongylocentrotus purpuratus; Genbank XP 003729232.1). Shown is the ClustalW multiple sequence alignment of human and sea urchin RAD51AP1. Identical residues are shown in black and highly conserved residues are shown in gray (Note: The alignments of two additional invertebrates with human RAD51AP1 are in Supplementary Fig. S1). (B) Structure of the Gallus gallus (i.e. chicken) RAD51AP1 locus. Exons are in blocks; exons and introns are drawn to scale. (C) Diagram of GgRAD51AP1 knockout approach. The region deleted (3′ half of exon 6 and the entire exon 7, flanking the PvuII site at 6418 bp) was substituted by either a neomycin or a puromycin cassette transcribed from the chicken β-actin promoter. The 5′ and 3′ flanking regions used in the knockout constructs are also indicated, as are a hybridization probe and the PvuII restriction sites used for Southern blot analysis (see Section 2 and (D)). (D) Southern blot analysis using this hybridization probe and PvuII-cleaved genomic DNA isolated from wild type DT40 cells, two independently isolated GgRAD51AP1 heterozygotes (AP1 Het-1 and AP1 Het-2) and two homozygous knockouts (AP1 KO-1 and AP1 KO-2). (E) Semi-quantitative end-point RT-PCR independently confirming the RAD51AP1 heterozygotes and knockouts. (F) Time course of RAD51 foci formation in various DT40 derivatives after treatment with 8 Gy X-rays. Sham-irradiated control samples were treated identically except for the irradiation exposure and cells were fixed with together with the 8 Gy – 4 h time point. Data are the means from 3 independent experiments ± 1SEM.
3.2. Generation of RAD51AP1-deficient chicken DT40 B cell lines
Gene-specific knockdown of RAD51AP1 in human cells impairs their resistance to DNA damaging agents and increases their levels of genomic instability, as shown by us and by others [3,4,17]. However, the phenotypic consequences of the total loss of RAD51AP1 expression have remained elusive. In order to establish a genetic model system to study RAD51AP1-deficient cells, we disrupted the RAD51AP1 locus in the chicken DT40 B cell line. The chicken RAD51AP1 gene is located on chromosome 1, comprised of 9 exons (Fig. 1B), and its sequence information was obtained from the Gallus gallus-4.0 genome assembly (NCBI reference sequence: XM 417234.3). The start codon for chicken RAD51AP1 lies within exon 2, and the RAD51-interacting domain of chicken RAD51AP1, a peptide sequence that is virtually identical in amino acid sequence to the RAD51-interacting domain of human RAD51AP1, lies within exon 9 (Fig. 1B). This information was used to design a gene disruption construct, which deletes the 3′ half of exon 6 and the entire exon 7. Both alleles were disrupted by sequential transfection, and the gene disruption was confirmed by Southern blot analysis (Fig. 1C and D) and end-point RT-PCR (Fig. 1E). Currently, there is no antibody available that detects chicken RAD51AP1. The two RAD51AP1 heterozygous cell lines (obtained by two independent transfections), with the RAD51AP1 disruption of only one allele, are referred to as AP1 Het-1 and AP1 Het-2. The two independently isolated RAD51AP1 knockout cell lines (isolated from AP1 Het-1 and AP1 Het-2) have the RAD51AP1 disruptions of both alleles and are referred to as AP1 KO-1 and AP1 KO-2, respectively.
3.3. Targeted inactivation of RAD51AP1 in DT40 cells impairs the resolution of DNA damage-induced RAD51 foci
Even though siRNA knockdown of RAD51AP1 in human cells impairs homology-mediated DNA repair, functional loss of RAD51AP1 does not impair the formation of DNA damage-induced RAD51 foci, as we and others have shown [3,4,18]. However, these earlier studies were carried out under conditions of transient RAD51AP1 protein depletion, which made it difficult to assess the long-term consequences of RAD51AP1 loss on the resolution of RAD51 DNA damage foci. Here, we assessed both the formation and the resolution of RAD51 foci in DT40 wild type and in two independently isolated DT40 RAD51AP1 knockout cell lines (AP1 KO-1 and AP1 KO-2) by immunocytohistochemistry. For comparison purposes, we also assessed the previously described DT40 cell line that is a knockout for XRCC3 [19], a protein described to function in early stages of HR and required for wild type levels of RAD51 foci formation [19]. We find that the formation of RAD51 foci at 2–12 h post exposure to 8 Gy X-rays occurs at wild type levels in both AP1 KO-1 and AP1 KO-2 cells (Fig. 1F). These results show that chicken RAD51AP1 is not required for RAD51 foci formation in response to DNA damage. This is different than what we observed for the XRCC3 knockout DT40 cell line (Fig. 1F), which we find to be greatly impaired in RAD51 foci formation, as previously reported [19]. At 24 h and 48 h post exposure, however, the levels of RAD51 foci after exposure to 8 Gy X-rays remained much higher in AP1-KO1 and AP1 KO-2 cells than in wild type DT40 cells (Fig. 1F; Supplemental Fig. 1C), suggesting that RAD51AP1 is critical for the completion of HR in DT40 cells exposed to X-rays.
3.4. Ectopic expression of chicken RAD51AP1 but not of human RAD51AP1 rescues the increased cellular sensitivity of RAD51AP1 knockout DT40 cells to DNA damaging agents
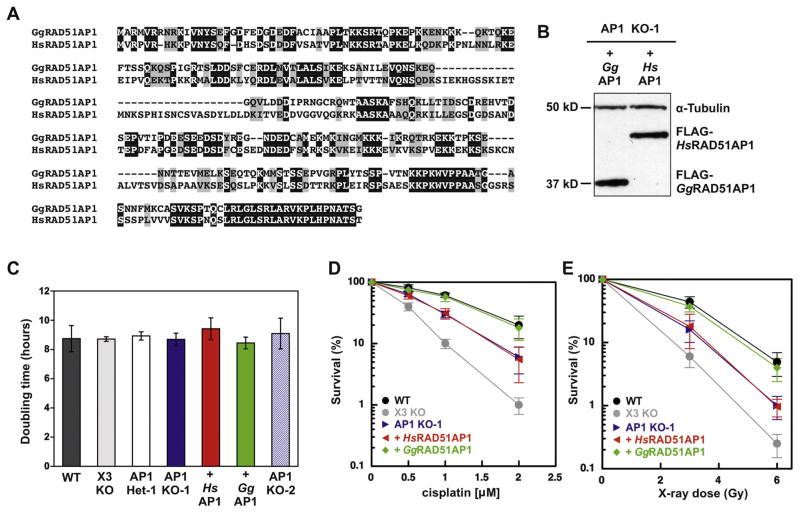
Next, we expressed FLAG-tagged versions of chicken RAD51AP1 or human RAD51AP1 in AP1 KO-1 cells. As depicted in Fig. 2A, both proteins share regions of homology, with 46% identical and 20% conserved residues based on the sequence of the smaller chicken RAD51AP1 protein. We produced lentivirus with the FLAG-tagged cDNAs for transduction and selected stably transduced clonal isolates that expressed similar levels of the ectopic proteins, as determined by Western blot analysis (Fig. 2B).
Fig. 2.
DT40 RAD51AP1 knockout cells exhibit enhanced sensitivity to DNA damaging agents, which is suppressed by ectopic expression of FLAG-tagged GgRAD51AP1, but not of FLAG-tagged HsRAD51AP1. (A) Sequence homology alignment between human and chicken RAD51AP1 using ClustalW analysis. Identical residues are shown in black and highly conserved residues are shown in gray. (B) Western blot analysis to show the levels of ectopically and stably expressed FLAG-tagged HsRAD51AP1 and GgRAD51AP1 in AP1 KO-1 cells. The signal for α-Tubulin is used as a loading control. (C) Cell doubling times of DT40 wild type cells and derivatives. Data are the means from at least three independent experiments ± 1 SEM. (D) and (E) Results from clonogenic cell survival assays to determine sensitivity of AP1 KO-1 cells and complemented AP1 KO-1 cells to cisplatin and X-rays, respectively. XRCC3 knockout DT40 cells were tested for comparison purposes. Data are the means from three independent experiments ± 1 SEM.
To examine the role of RAD51AP1 in cell viability, we determined the doubling times of all DT40 derivatives, including AP1 KO-1 cells stably expressing FLAG-tagged chicken or FLAG-tagged human RAD51AP1. We found that all DT40 cell lines proliferated with similar kinetics, and that AP1 KO-1 and AP1 KO-2 cells were not significantly different form DT40 wild type cells (Fig. 2C). Compared to wild type DT40 cells, AP1 KO-1 cells show increased cellular sensitivity to the cytotoxic effects of both cisplatin and X-ray exposure that, however, was somewhat less pronounced than the cytotoxicity observed for XRCC3-deficient DT40 (Fig. 2D and E). Interestingly, only ectopically expressed chicken, but not ectopically expressed human RAD51AP1 was able to complement the cellular sensitivity of AP1 KO-1 cells to cisplatin and to X-rays (Fig. 2D and E).
3.5. RAD51AP1 is required for the efficient recovery from spontaneously occurring replication stress
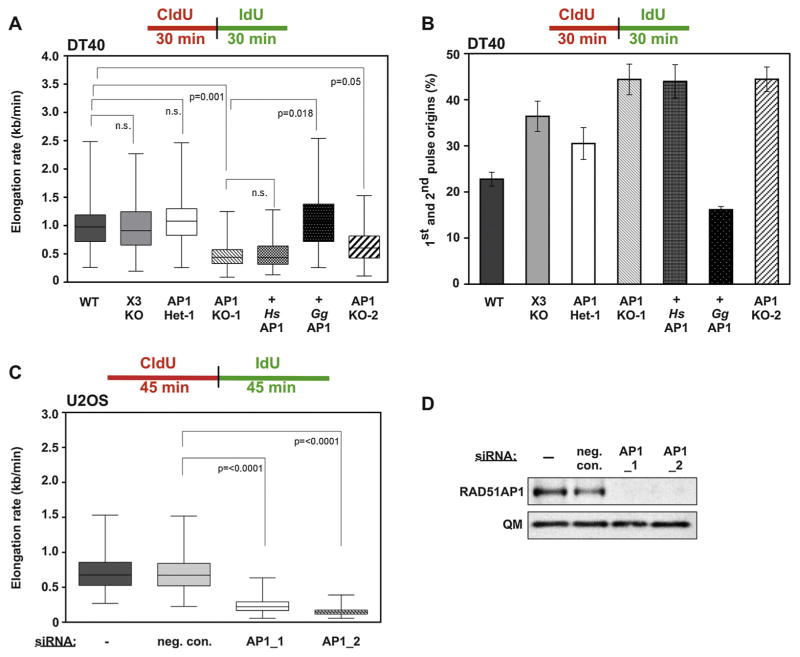
HR is essential to ensure faithful and complete DNA replication (for review see [20]). However, the effects of the functional loss of RAD51AP1 on DNA replication had not been determined. Here, we used our new RAD51AP1-deficient genetic model system in DT40 cells to assess the specific effects of RAD51AP1 loss on replication speed and origin firing using the DNA fiber technique. To do so, we labeled exponentially growing DT40 cells in two consecutive rounds for 30 min each with CldU first and then with IdU. Quantitative analysis of individual replication fibers revealed a two-fold and statistically significant decrease in elongation rates of DNA replication for the AP1 KO-1 and AP1 KO-2 cell lines compared to DT40 wild type cells (Fig. 3A, Supplemental Fig. 2A). Similarly, knockdown of human RAD51AP1 in U2OS cells by one of the 2 different siRNAs led to significantly decreased elongation rates of DNA replication compared to U2OS cells treated with negative control siRNA (Fig. 3C and D; Supplemental Fig. 2B). These results support the notion that both chicken and human RAD51AP1 are critically important for maintaining normal rates of DNA replication. However, in this assay neither heterozygous RAD51AP1 DT40 cells (i.e. AP1 Het-1) nor XRCC3-deficient DT40 cells are different from wild type DT40 cells (Fig. 3A). Importantly, ectopic expression of chicken RAD51AP1 but not of human RAD51AP1 fully reverted elongation rates back to wild type levels in AP1 KO-1 cells (Fig. 3A; Supplemental Fig. 2A), suggesting that human RAD51AP1 is unable to properly repair spontaneous replication-associated DNA damage in chicken AP1 KO-1 cells.
Fig. 3.
RAD51AP1 is required to maintain the progression of DNA replication forks and to suppress dormant origin firing. (A) RAD51AP1-deficient DT40 cells show greatly reduced DNA elongation rates under unperturbed conditions. Ectopic expression of GgRAD51AP1 (here: + GgAP1), but not of HsRAD51AP1 (here: + HsAP1) reverts elongation rates back to wild type levels. In box plots, boxes encompass the 25th–27th percentile, with error bars defining the minima and maxima. The black horizontal bars within the boxes indicate the means. (B) RAD51AP1-deficient DT40 cells show enhanced levels of first and second pulse replication origins under unperturbed conditions. Ectopic expression of GgRAD51AP1 (here: + GgAP1), but not of HsRAD51AP1 (here: + HsAP1) reverts origin firing close to the levels of wild type cells. (C) RAD51AP1-depleted U2OS cells show greatly reduced DNA elongation rates under unperturbed conditions. Knockdown of RAD51AP1 was obtained by transfection with one of the two different siRNAs targeting RAD51AP1. NCS: non-depleting control siRNA. In box plots, boxes encompass the 25th–27th percentile, with error bars defining the minima and maxima. The black horizontal bars within the boxes indicate the means. (D) Representative Western blots obtained for the experiments shown in (C).
Upon further analysis, we noticed closely spaced replication origins in AP1 KO-1 and AP1 KO-2 cells that were not present in DT40 wild type cells (Supplemental Fig. 3, upper panels). These observations led us to hypothesize that excessive origin firing may occur in RAD51AP1-deficient DT40 cells, as previously observed for deficiencies in other critical proteins of the HR pathway [21,22]. Indeed, when compared to wild type cells, the combined fraction of first and second pulse replication origins was significantly increased in AP1 KO-1 and AP1 KO-2 cells (Fig. 3B). Furthermore, compared to wild type cells, XRCC3-deficient DT40 cells also showed significantly increased levels of replication origins (Fig. 3B). Ectopic expression of human RAD51AP1 in AP1 KO-1 cells failed to complement and did not lead to reduced origin firing (Fig. 3B). However, compared to AP1 KO-1 cells, ectopic expression of chicken RAD51AP1 in AP1 KO-1 cells lead to reduced origin firing (Fig. 3B), further supporting the notion that ectopically expressed FLAG-tagged chicken RAD51AP1 is functional in AP1 KO-1 cells.
3.6. RAD51AP1 is required for efficient recovery from DNA replication stress induced by hydroxyurea
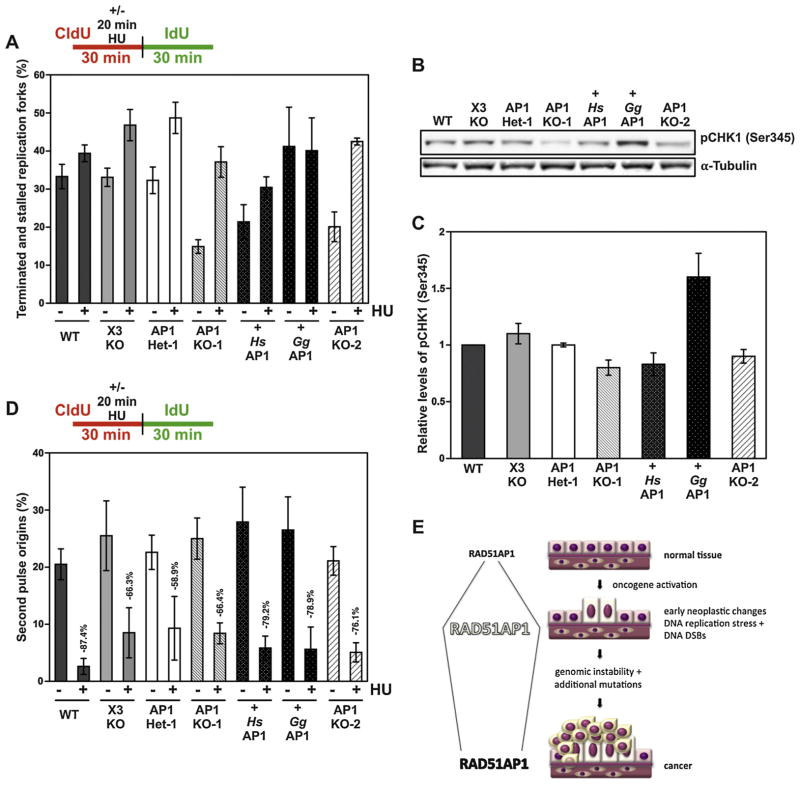
Next, we assessed the role of RAD51AP1 under conditions of stalled DNA replication. To do so, we labeled exponentially growing cells with CldU for 10 min first, incubated the cells 20 min with 2 mM HU in the presence of CldU, after which we replaced the medium with IdU-containing medium and incubated for 30 min before lysis. To assess the role of RAD51AP1 in repairing replication forks perturbed by exposure to HU, we determined the fraction of terminated/stalled forks in all DT40 derivatives and compared their levels with and without HU treatment. To determine the fraction of terminated/stalled forks, we measured the relative amounts of DNA fibers that were labeled in red only. Under spontaneous conditions (i.e. without HU treatment), the relative fraction of only red-labeled fibers is approximately the same in DT40 wild type, XRCC3-deficient and RAD51AP1 heterozygous DT40 cells (Fig. 4A, Table 1). Upon treatment with HU, these three cell lines, albeit to different extents, responded with increased levels of only red-labeled fibers, indicative of increased levels of fork stalling due to short-term HU exposure. Whereas a 20 min HU treatment of wild type DT40 cells only very moderately increased their levels of only red-labeled fibers (Fig. 4A, Table 1 and Supplemental Fig. 3), HU treatment elicited an approximately 1.5-fold increase in fork stalling in both XRCC3-deficient and RAD51AP1 heterozygous DT40 cells (Fig. 4A, Table 1). These results show that HR defective XRCC3-deficent DT40 cells are impaired in their ability to recover from stalled replication forks due to HU treatment. The somewhat surprising results for DT40 cells heterozygous for RAD51AP1 (i.e. AP1 Het-1) may point to a possible haploinsufficiency effect for RAD51AP1 detected by this assay.
Fig. 4.
RAD51AP1 is required to overcome HU-induced replication stress in DT40 cells. (A) Fraction of terminated and stalled replication forks (i.e. only red forks) with and without HU exposure, as depicted. (B) Representative Western blot to visualize the pCHK1(Ser345) for the analysis shown in (C). The signal for α-Tubulin serves as a loading control. (C) Quantification of pCHK1(Ser345) signals; data are the means of three independent experiments ± 1 SEM. (D) More second pulse replication origins are activated in HR-compromised RAD51AP1-deficient DT40 cells than in DT40 wild type cells. The fraction of second pulse origins (%) with and without HU exposure was assessed, as described in Material and methods. AP1 KO-1, AP1 KO-2 and X3 KO cells show increased firing of second pulse dormant replication origins compared to wild type cells. (E) Schematic to explain why elevated levels of RAD51AP1 protein may provide a selective advantage at early stages of neoplastic change during which time replication stress and, consequently, DNA double-strand breaks occur at higher than normal levels.
Table 1.
Percentage of stalled/terminated replication forks without and with HU treatment.
| Genotype 1* | Red-labeled fibers (%) unperturbed | Genotype 2* | Red-labeled fibers (%) unperturbed | P** |
|---|---|---|---|---|
| DT40 | ||||
| WT | 31.5 ± 3.2 | X3 KO | 33.1 ± 2.3 | n.s. |
| WT | 31.5 ± 3.2 | AP1 Het-1 | 32.2 ± 3.4 | n.s. |
| WT | 31.5 ± 3.2 | AP1 KO-1 | 14.9 ± 2.0 | 0.0044 |
| WT | 31.5 ± 3.2 | AP1 KO-2 | 20.1 ± 3.8 | 0.05 |
| WT | 31.5 ± 3.2 | + HsAP1 | 21.5 ± 4.5 | n.s. |
| WT | 31.5 ± 3.2 | + GgAP1 | 41.3 ± 10.3 | n.s. |
| AP1 KO-1 | 14.9 ± 2.0 | + HsAP1 | 21.5 ± 4.5 | n.s. |
| AP1 KO-1 | 14.9 ± 2.0 | + GgAP1 | 41.3 ± 10.3 | 0.0466 |
| AP1 KO-1 | 14.9 ± 2.0 | AP1 Het-1 | 32.2 ± 3.4 | 0.0047 |
| Genotype 1* | Red-labeled fibers (%) unperturbed | Genotype 2* | Red-labeled fibers (%) +20 min 2 mM HU | P** |
|---|---|---|---|---|
| DT40 | ||||
| WT | 31.5 ± 3.2 | WT | 39.4 ± 2.2 | n.s. |
| X3 KO | 33.1 ± 2.3 | X3 KO | 46.8 ± 4.1 | 0.0265 |
| AP1 Het-1 | 32.2 ± 3.4 | AP1 Het-1 | 48.7 ± 4.1 | 0.0270 |
| AP1 KO-1 | 14.9 ± 2.0 | AP1 KO-1 | 37.1 ± 4.0 | 0.0025 |
| AP1 KO-2 | 20.1 ± 3.8 | AP1 KO-2 | 42.5 ± 0.8 | 0.0049 |
| + HsAP1 | 21.5 ± 4.5 | + HsAP1 | 30.5 ± 2.8 | n.s. |
| + GgAP1 | 41.3 ± 10.3 | + GgAP1 | 40.1 ± 8.6 | n.s. |
Genotype 1 in comparison to genotype 2 to determine P-values. Data values are the mean ± 1 SEM.
All P-values were calculated by two-tailed Student’s t-test.
Under unperturbed conditions and compared to wild type cells, RAD51AP1-deficient AP1 KO-1 cells and AP1 KO-2 cells both showed significantly reduced fractions of only red-labeled fibers, as cells with slow elongation rates typically also show less terminating events (Fig. 4A, Table 1 and Supplemental Fig. 3). Furthermore, HU treatment of AP1 KO-1 or AP1 KO-2 cells increased the levels of only red-labeled fibers 2- to 3-fold (Fig. 4A, Table 1). Notably, ectopically expressed chicken RAD51AP1 (here: + GgAP1) fully rescues the inability of AP1 KO-1 cells to terminate/stall replication forks after spontaneous DNA damage (Fig. 4A, Table 1). Furthermore, AP1 KO-1 cells expressing chicken RAD51AP1 also are fully proficient in dealing with the HU-induced replication stress under the conditions tested here, as HU treatment did not lead to enhanced replication fork stalling in these cells (Fig. 4A, Table 1).
Although not significantly different from wild type cells, under unperturbed conditions AP1 KO-1 cells with ectopic human RAD51AP1 (here: + HsAP1) show a trend toward a smaller fraction of terminated/stalled replication forks (Fig. 4A, Table 1). After HU treatment, a small increase in replication fork stalling is observed for AP1 KO-1 cells with human RAD51AP1 (Fig. 4A, Table 1), further suggesting that, unlike AP1 KO-1 cells with ectopic chicken RAD51AP1, AP1 KO-1 cells with ectopic human RAD51AP1 are not fully able to overcome HU-induced replication stress under the conditions tested here. Taken together, these results show that RAD51AP1-deficient DT40 cells encounter increased levels of replication stress after short-term exposure to HU compared to DT40 wild type cells.
Cells that encounter increased levels of DNA replication stress typically activate the S-phase checkpoint via checkpoint kinase 1 (CHK1). Therefore, we tested for the levels of activated CHK1 in DT40 wild type cells and all derivatives by monitoring pCHK1(Ser345) signal intensities after HU treatment by Western blot analysis. One representative Western blot is shown in Fig. 4B, and the quantitative analysis from 3 independent experiments is shown in Fig. 4C. Treatment with 2 mM HU for 3 h elicited clear differences in the pCHK1(Ser345) signal intensities in all DT40 cell lines tested. In three independent experiments, AP1 KO-1 and AP1 KO-2 cells showed slightly impaired activation of CHK1 (as measured by pCHK1(Ser345) signals) compared to wild type cells. The combined analysis also shows, that ectopic expression of human RAD51AP1 in AP1 KO-1 cells had no major effect on the levels of activated pCHK1(Ser345) (Fig. 4C), in support of our findings that human RAD51AP1 cannot complement for the loss of chicken RAD51AP1 in chicken cells. However, compared to AP1 KO-1 cells, ectopic expression of chicken RAD51AP1 in AP1 KO-1 cells led to enhanced levels of pCHK1(Ser345) protein (Fig. 4C). These results suggest that, after HU treatment, RAD51AP1 activity may directly or indirectly be involved in facilitating intra-S phase checkpoint signaling. These results further suggest that chicken FLAG-RAD51AP1 may be expressed at somewhat higher levels in AP1 KO-1 cells than endogenous chicken RAD51AP1 in DT40 wild type cells.
CHK1 activation serves to suppress CDK activity to impair firing of dormant replication origins. Notably, increased origin activation has been described for cells with nonfunctional CHK1 [23–25]. Therefore, we assessed the levels of second pulse origins (i.e. green fibers only) following HU exposure in DT40 wild type cells and all derivatives. In line with our quantitative western blot analyses for activated CHK1, we find that second pulse origins are greatly diminished in DT40 wild type cells after HU treatment (Fig. 4D), supporting the notion that in DT40 wild type cells the S-phase checkpoint is functional. XRCC3-deficient and RAD51AP1-deficient DT40 cells are less proficient in suppressing firing of second pulse replication origins (Fig. 4D), suggesting that XRCC3 and wild type levels of RAD51AP1 are required to confer a fully functional S-phase checkpoint arrest. Compared to AP1 KO-1 cells, there is a trend toward an increased suppression of second pulse replication origins for the ectopic expression of both human and chicken RAD51AP1 in AP1 KO-1 cells. We speculate that fully functional RAD51AP1 contributes to the protection of genome integrity by transducing DNA damage signals in addition to engaging in the HR machinery. While both these functions appear to be fully transmitted by chicken RAD51AP1 in AP1 KO-1 cells (Figs. 2D and E, 3B and 4D), human RAD51AP1 in AP1 KO-1 retains some ability to transduce DNA damage signals for checkpoint arrest (Fig. 4D), but clearly is not fully functional both in DNA damage signal transduction (Fig. 3B) and in HR (Fig. 2D and E) in chicken cells.
4. Discussion
RAD51AP1 is a critical protein for DNA repair by homologous recombination (HR), and functions downstream of the assembly of DNA damage-induced RAD51 foci, as a late-acting accessory factor of the RAD51 recombinase [3,4]. Human cells with RAD51AP1 knockdown are moderately sensitized (~1.5- to 2.5-fold (D10 values; dose modification factor at 10% survival)) to the cytotoxic effects of inter-strand crosslinking agents and of ionizing radiation, and show enhanced levels of genome instability and DNA DSB repair defects [3,4,17,18]. Given the important biochemical role for RAD51AP1 in stimulating D-loop formation [3,4,15], the comparatively moderate phenotype determined for sensitivity to DNA damaging agents of human cells with RAD51AP1 knockdown had been somewhat surprising. In this regard, it was reasonable to assume that the low level RAD51AP1 protein remaining in cells after RAD51AP1 knockdown may contribute to masking more severe phenotypic effects. Unfortunately, human cells with inactivating mutations in RAD51AP1 or fully abrogated expression of RAD51AP1 protein have not yet been isolated.
Here, we generated a genetic model system to assess the phenotype of fully inactivated RAD51AP1 in vertebrate cells. We also expanded our previous studies aimed at detecting homologs of the RAD51AP1 gene across the animal kingdom in the hopes of identifying additional models in non-vertebrate cells, if they existed. We now find that RAD51AP1 is not exclusively vertebrate-specific, unlike we and others had suggested previously [3,4], at a time when overall fewer invertebrate genomes had been fully sequenced. However, RAD51AP1 appears to be limited to metazoan species and is not present in protozoans. As stated previously, RAD51AP1 appears to not be present in simple model organisms such as yeast [4], D. melanogaster and C. elegans, although RAD54, the second, late functioning accessory factor of RAD51, is. Therefore, the DT40 system currently is the best model system for characterizing RAD51AP1 deficiency at the cellular level.
We find that RAD51AP1 is not essential for viability in chicken DT40 cells. However, we show that RAD51AP1 absolutely is essential for the timely resolution of DNA damage-induced RAD51 foci in vertebrate cells. The impaired resolution of RAD51 foci in the absence of RAD51AP1 very much is in support of our previous biochemical studies in which we reported that RAD51AP1 stimulated D-loop formation and RAD51 recombinase activity in vitro [3,5,15]. In biochemical assays, we and others have shown that in the absence of RAD51AP1 the activity of RAD51 is comparatively low [3,5,15], leading to diminished levels of HR repair downstream of RAD51 filament formation.
Similar to human cells with RAD51AP1 knockdown [3,4,17], RAD51AP1 knockout DT40 cells are moderately sensitized to the cytotoxic effects of both inter-strand crosslinking agents and ionizing radiation (~1.5-fold (D10 values)). Importantly, species-specific differences exist between the human and chicken RAD51AP1 proteins. These findings are somewhat reminiscent for what has been reported on the species-specific interactions between RAD51 and RAD54 [26], and also on the species-specific stimulation of RAD54 ATPase activity by RAD51 [27]. Although we performed co-immunoprecipitation experiments to assess possible differences in complex formation between chicken RAD51 and ectopic human or chicken RAD51AP1, we did not observe any differences in these interactions (data not shown).
A role for RAD51AP1 in DNA replication and replication restart, which have been shown to rely on the HR pathway [22,28–30], had not been investigated. We show here that, in both human and chicken cells, RAD51AP1 modulates the progression of DNA replication forks. Under conditions of unperturbed DNA replication, we find that elongation speeds of individual replication forks are greatly reduced when RAD51AP1 is absent or depleted. Likely, the discrepancy between slower DNA replication rates of individual origins, but similar S-phase times (Supplemental Fig. 4), relates to the increased origin usage in RAD51AP1-deficient cells (discussed below). Ectopically expressed chicken RAD51AP1 is fully capable of reverting the elongation rates of AP1 KO-1 cells to wild type levels, supporting the notion that the observed phenotype for diminished speeds of DNA replication in AP1 KO-1 cells is solely due to RAD51AP1-deficiency. It is also interesting to note that XRCC3-deficiency does not lead to impaired rates of DNA replication, confirming observations by others [12,25]. In this context, it is worth mentioning that a recent study described a role for human RAD52 in replication-associated HR, which may substitute for XRCC3 [31].
The fraction of terminated replication forks, under conditions of spontaneous DNA replication, is much lower in RAD51AP1-deficient cells compared to wild type cells, as cells with slow elongation rates also typically show less terminating events [20,21]. RAD51AP1-deficient cells increase their usage of replication origins to ensure timely DNA replication, in line with previous reports that show that globally reduced replication fork speeds can elicit an increase in replication origin firing [32–34]. Taken together, these results show that RAD51AP1 is critical for maintaining DNA replication under unperturbed conditions. Even without exogenous DNA damage treatments, loss of RAD51AP1 function is expected to endanger genome integrity, resulting in chromosomal abnormalities, which has been observed by us in human cells [3].
Treatment with HU leads to a depletion of the dNTP-pools and to replication fork stalling. This effect can be more pronounced for cells with HR deficiencies [22,35], and this is the case for RAD51AP1-deficient cells, as we show here. While expression of ectopic chicken RAD51AP1 in AP1 KO-1 cells confers full resistance to HU exposure, this is not the case for human RAD51AP1 in chicken AP1 KO-1 cells. Since, after short-term HU treatment, any large-scale increase in new origin firing is prevented by the CHK1-mediated S-phase checkpoint [36], these results suggested that RAD51AP1-deficiency may lead to an aberrant CHK1-mediated DNA damage response. Indeed, we find mildly diminished activation of CHK1 and diminished suppression of second pulse dormant replication origins in AP1 KO-1 and AP1 KO-2 cells, and reversion to activated CHK1 by ectopically expressed chicken RAD51AP1. Human RAD51AP1 in AP1 KO-1 cells does not reverse CHK1 activation back to wild type levels, but confers some suppression on origin firing, suggesting that for mediating the DNA damage response the human protein is not totally dead in chicken cells.
Many cancer therapeutic agents lead to increased DNA replication stress. In addition, replication stress has emerged as a significant source of genome instability during early stages of carcinogenesis [37]. To improve both diagnosis of disease and targeted therapy, it is important to better understand the proteins involved in the replication stress response. RAD51AP1 clearly is one such candidate for which, for the first time, we have shown an important role in protecting against replication stress here. In this regard it is interesting to note that several primary human tumor types display elevated expression of RAD51AP1/RAD51AP1. For example, RAD51AP1 transcript and protein are elevated in the majority of cholangiocarcinomas [18], and a number of studies report on the up-regulation of RAD51AP1 in hepatocellular carcinoma [38], in acute myeloid leukemia [39], in aggressive mantle cell lymphoma [17], and in ovarian cancer [40]. RAD51AP1 also is part of the ‘cervical cancer proliferation cluster’ composed of 163 highly correlated transcripts [41], and is up-regulated in breast cancer cell lines [42]. We suggest that elevated RAD51AP1 may be critical during early steps of neoplasia during which, as a consequence of increased cell proliferation, replication stress occurs at higher than normal levels (see Fig. 4E). Overcoming this barrier to cell survival through elevated RAD51AP1 may allow for further genomic changes to occur that can drive the progression of precancerous lesions to cancer. Therefore, inhibiting RAD51AP1 during the initial stages of neoplasia may provide a useful approach in targeted therapy. However, dependent on genetic profile, RAD51AP1 may be down-regulated or may remain elevated in cancer tissues (see Fig. 4E). For example, RAD51AP1 showed significant association with BRCA1 status when the expression differences between BRCA1 and non-BRCA1 sporadic breast tumor was assessed [43].
Taken together, we describe a novel function for RAD51AP1 associated with maintaining DNA replication fork progression. We show that RAD51AP1-deficient DT40 and RAD51AP1-depleted human cells are impaired in fork velocities, and that RAD51AP1-deficient DT40 cells have problems overcoming stalled forks. We also provide some evidence that points to a role for RAD51AP1 in mediating CHK1 phosphorylation and speculate that fully functional RAD51AP1 may contribute to the protection of genome integrity by transducing DNA damage signals in addition to engaging in the HR machinery.
Supplementary Material
Acknowledgments
We are grateful to Shunichi Takeda for providing wild type and XRCC3-deficient DT40 cells, to Jean-Marie Buerstedde for pLoxNeo and pLoxPuro vectors, and to Christina Clover for chicken EST cDNA clone containing locus DN928517. This work was supported by grants from the National Institutes of Health [R01 CA120315 to D.S., and R01 ES021454 to C.W.].
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2014.09.007.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Contributor Information
David Schild, Email: dschild@lbl.gov.
Claudia Wiese, Email: claudia.wiese@colostate.edu, cwiese@lbl.gov.
References
- 1.Dunlop MH, Dray E, Zhao W, Tsai MS, Wiese C, Schild D, Sung P. RAD51-associated protein 1 (RAD51AP1) interacts with the meiotic recombinase DMC1 through a conserved motif. J Biol Chem. 2011;286:37328–37334. doi: 10.1074/jbc.M111.290015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dray E, Dunlop MH, Kauppi L, San Filippo J, Wiese C, Tsai MS, Begovic S, Schild D, Jasin M, Keeney S, et al. Molecular basis for enhancement of the meiotic DMC1 recombinase by RAD51 associated protein 1 (RAD51AP1) Proc Natl Acad Sci U S A. 2011;108:3560–3565. doi: 10.1073/pnas.1016454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 5.Dunlop MH, Dray E, Zhao W, San Filippo J, Tsai MS, Leung SG, Schild D, Wiese C, Sung P. Mechanistic insights into RAD51-associated protein 1 (RAD51AP1) action in homologous DNA repair. J Biol Chem. 2012;287:12343–12347. doi: 10.1074/jbc.C112.352161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner. PALB2, Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovalenko OV, Golub EI, Bray-Ward P, Ward DC, Radding CM. A novel nucleic acid-binding protein that interacts with human rad51 recombinase. Nucleic Acids Res. 1997;25:4946–4953. doi: 10.1093/nar/25.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovalenko OV, Wiese C, Schild D. RAD51AP2, a novel vertebrate- and meiotic-specific protein, shares a conserved RAD51-interacting C-terminal domain with RAD51AP1/PIR51. Nucleic Acids Res. 2006;34:5081–5092. doi: 10.1093/nar/gkl665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiese C, Galande S. Elimination of reducing agent facilitates quantitative detection of p53 antigen. Biotechniques. 2001;30:960–963. doi: 10.2144/01305bm07. [DOI] [PubMed] [Google Scholar]
- 11.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell. 2003;11:1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 13.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiese C, Hinz JM, Tebbs RS, Nham PB, Urbin SS, Collins DW, Thompson LH, Schild D. Disparate requirements for the Walker A and B ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res. 2006;34:2833–2843. doi: 10.1093/nar/gkl366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai MS, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 17.Henson SE, Tsai SC, Malone CS, Soghomonian SV, Ouyang Y, Wall R, Marahrens Y, Teitell MA. Pir51, a Rad51-interacting protein with high expression in aggressive lymphoma, controls mitomycin C sensitivity and prevents chromosomal breaks. Mutat Res. 2006;601:113–124. doi: 10.1016/j.mrfmmm.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Obama K, Satoh S, Hamamoto R, Sakai Y, Nakamura Y, Furukawa Y. Enhanced expression of RAD51 associating protein-1 is involved in the growth of intra-hepatic cholangiocarcinoma cells. Clin Cancer Res. 2008;14:1333–1339. doi: 10.1158/1078-0432.CCR-07-1381. [DOI] [PubMed] [Google Scholar]
- 19.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carr AM, Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol. 2013;425:4733–4744. doi: 10.1016/j.jmb.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 21.Nikkila J, Parplys AC, Pylkas K, Bose M, Huo Y, Borgmann K, Rapakko K, Nieminen P, Xia B, Pospiech H, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun. 2013;4:2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daboussi F, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M, Lopez BS. A homologous recombination defect affects replication-fork progression in mammalian cells. J Cell Sci. 2008;121:162–166. doi: 10.1242/jcs.010330. [DOI] [PubMed] [Google Scholar]
- 23.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golub EI, Kovalenko OV, Gupta RC, Ward DC, Radding CM. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J Biol Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 28.Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol Cell Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 31.Wray J, Liu J, Nickoloff JA, Shen Z. Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Res. 2008;68:2699–2707. doi: 10.1158/0008-5472.CAN-07-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 35.Parplys AC, Petermann E, Petersen C, Dikomey E, Borgmann K. DNA damage by X-rays and their impact on replication processes. Radiother Oncol. 2012;102:466–471. doi: 10.1016/j.radonc.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 38.Song H, Xia SL, Liao C, Li YL, Wang YF, Li TP, Zhao MJ. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J Gastroenterol. 2004;10:509–513. doi: 10.3748/wjg.v10.i4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoch C, Kern W, Kohlmann A, Hiddemann W, Schnittger S, Haferlach T. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosom Cancer. 2005;43:227–238. doi: 10.1002/gcc.20193. [DOI] [PubMed] [Google Scholar]
- 40.Miles GD, Seiler M, Rodriguez L, Rajagopal G, Bhanot G. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res Notes. 2012;5:164. doi: 10.1186/1756-0500-5-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosty C, Sheffer M, Tsafrir D, Stransky N, Tsafrir I, Peter M, de Cremoux P, de La Rochefordiere A, Salmon R, Dorval T, et al. Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene. 2005;24:7094–7104. doi: 10.1038/sj.onc.1208854. [DOI] [PubMed] [Google Scholar]
- 42.Wilson CA, Cajulis EE, Green JL, Olsen TM, Chung YA, Damore MA, Dering J, Calzone FJ, Slamon DJ. HER-2 overexpression differentially alters transforming growth factor-beta responses in luminal versus mesenchymal human breast cancer cells. Breast Cancer Res. 2005;7:R1058–R1079. doi: 10.1186/bcr1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1 deficient breast tumors. Cancer Res. 2007;67:9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.