Abstract
NLRs are innate immune sensors that monitor the sanctity of the cytosolic compartment. Tenthorey et al. reveal a novel ligand-sensing interface within regions of the oligomerization domain (NOD/NACHT) of the NAIPs, rather than within the leucine-rich repeats (LRR), as was anticipated.
The immune system must appropriately distinguish friend from foe. To achieve this, TLRs (toll-like receptors) survey the extracellular/vacuolar space, while NLRs (NOD-like receptors) survey the cytosolic space. NLRs are defined by the presence of a NOD/NACHT (nucleotide oligomerization binding domain) domain that is comprised of several subdomains, including a nucleotide binding domain (NBD) and several discrete helical domains (HD1, WHD, and HD2). NLRs also typically contain an N-terminal signaling domain (CARD, PYD, or BIR) and C-terminal LRR (leucine-reach repeat) domain [1]. Certain NLRs oligomerize to form inflammasomes that activate caspase-1, resulting in secretion of IL-1β/IL-18 and initiation of pyroptosis, an inflammatory form of cell death. Although various activating ligands for NLRs have been characterized, the molecular basis for ligand recognition for most NLRs remains obscure. Assembly of the inflammasome is often paralleled to that of the apoptosome, which forms upon oligomerization of APAF-1, an NLR relative. The crystal structure of the apoptosome revealed a disk-shaped homoheptamer, where the WD40 repeats (functionally parallel to the LRR domain) bind the APAF-1 ligand, cytochrome c. The analogous LRR domain of plant NLRs (NB-LRRs) confers ligand specificity [2] as does the LRR of TLRs. Hence, it was expected that the LRR domain would confer NLR specificity in mammals. However, recent work by Tenthorey et al reveal that ligand specificity in a subset of NLRs does not map to the LRR domain, but rather to several NBD-associated domains within the NACHT [3].
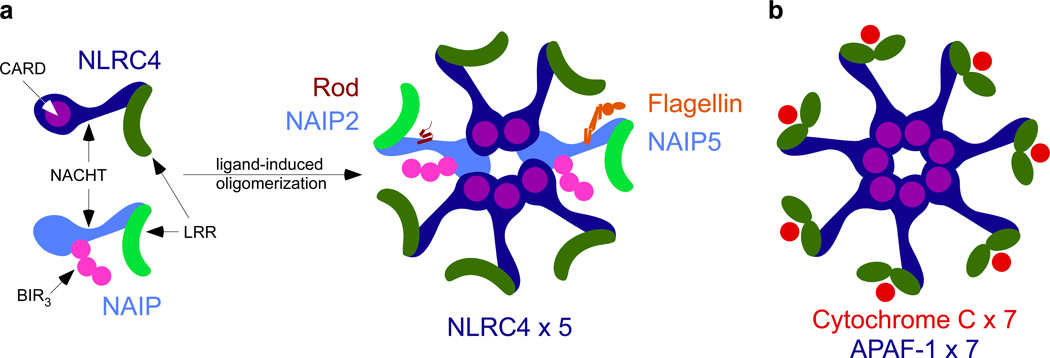
NAIPs (neuronal apoptosis inhibitor proteins) are NLR family members with a NOD/NACHT, LRR, and 3 BIR domains (baculovirus inhibitor of apoptosis protein repeat) (Figure 1A). NAIPs are relatively unique amongst mammalian NLRs in that they require another NLR, NLRC4, in order to activate caspase-1. Some plant NLRs have been shown to require “helper NB-LRRs” to fully potentiate immune signaling, analogous to the relationship between NAIPs and NLRC4 [4]. Upon ligand binding, a trimeric complex including the ligand, NAIP, and NLRC4 forms. While mice are known to express NAIP1, NAIP2, NAIP5, and NAIP6, only one human NAIP has been identified. NAIP5 and 6 detect bacterial flagellin, NAIP2 detects the type III secretion system rod protein (including PrgJ), and mouse NAIP1 and human NAIP (hNAIP) detect the T3SS needle protein [5]. The NAIP ligands (rod, needle, and flagellin) enter the host cytosol aberrantly via the T3SS, while the bacteria are actively secreting virulence factors to alter host functions. Despite a high degree of sequence identity between different NAIPs, they exhibit remarkable specificity for distinct ligands. To rigorously map the ligand-recognition domain in NAIP proteins, Tenthorey et al. generated 43 NAIP2/5 and NAIP2/6 chimeras. They transfected these constructs, NLRC4, and 6myc-tagged ligand (PrgJ/FlaA) into HEK293T cells and monitored oligomerization capability via immunoblot; 31 of the 43 were determined to be competent. To verify signaling ability, caspase-1 and pro-IL-1β were transfected alongside the inflammasome structural components, and monitored for the cleaved p17 fragment of IL-1β. The authors found they could switch ligand specificity by swapping the α-helices and a short distal region associated with the NBD of NAIPs. Once bound to ligand, it is known that NAIPs are licensed to oligomerize with NLRC4. However, Tenthorey et al. reveal that NAIPs will bind to their cognate ligand but will not oligomerize on their own. Only when NLRC4 was transfected alongside the tagged NAIPs was oligomerization detectable by immunoblot in an average 5:2 ratio of NLRC4:NAIP. Surprisingly, the NAIPs within the NLRC4 inflammasome need not be the same, as NAIP5-flagellin and NAIP2-rod complexes can form a single inflammasome hub with NLRC4 [3].
Figure 1. Structural and functional similarities between the APAF-1 apoptosome and the NAIP/NLRC4 inflammasome.
(A) Comparison of NAIP and NLRC4 NLR proteins. (B)The apoptosome is an oligomer of APAF-1, which consists of 3 distinct domains, an N-terminal caspase activation and recruitment domain (CARD, in purple), an expanded nucleotide binding domain (NBD, in blue), and WD40 repeats (green) at the C-terminus. The WD40 repeats bind cytochrome c (red) and oligomerize to form a homo-heptamer, with each APAF-1 bound to one cytochrome c. The NAIP/NLRC4 inflammasome can exist as a mixed oligomer. In a proposed heptamer formation, paralleling the apoptosome, different NAIPs (light blue) can bind their cognate ligand (rod-NAIP2, flagellin-NAIP5), recruiting NLRC4 (dark blue). This relieves autoinhbition by the LRR (green) and could twist the baculovirus inhibitor of apoptosis protein repeat (BIR, in pink) for potential interactions with NLRC4.
The findings surrounding the NAIP LRR domain in this study are striking, as they indicate that of the two functions of the LRR, autoinhibition and ligand binding, the latter is dispensable in NAIPs. There is ample evidence showing autoinhibition of both plant and mammalian NLRs by their LRR, for example, NLRC4ΔLRR constitutively oligomerizes [6]. Recent structural studies have also revealed that the LRR domain obscures the NBD when NLRC4 is in an inactive state [7]. In light of Tenthorey et al.’s findings, ligand binding to the NBD-associated helical domains, not the LRR, must transmit a signal via conformational change to expose the NACHT, permitting oligomerization. This is a reasonable hypothesis to explain alleviation of autoinhibition within the NAIP; how this conformational change is accomplished within the NLRC4 monomer remains unknown. Perhaps when the NAIP binds its ligand, the three BIR domains are exposed, and these interact with an unidentified ligand sensing region of NLRC4 to relieve autoinhibition. Interestingly, the ligand specificity domain of NAIPs aligns with a phosphorylation site (serine 533) on NLRC4. However, the function of this phosphorylation event remains to be fully elucidated [8, 9].
The potential for NAIP/NLRC4 inflammasomes composed of heterogeneous NAIPs is novel and provocative. Halff et al. previously proposed that a singular NAIP5-flagellin complex could result in an 11–12 NAIP/NLRC4 oligomer [10]. Although the data in Halff et al. could not rule out multiple NAIP5 monomers, they were indicative of an excess of NLRC4, which is consistent with the average 5:2 NAIP:NLCR4 ratio observed in Tenthorey et al. Both studies employed inflammasome reconstitution rather than native proteins, however, so whether their proposed compositions of the NAIP/NLRC4 inflammasome occur in vivo remains to be determined. Assuming a heptameric inflammasome hub, Tenthorey et al.’s results suggest that one NAIP5-flagellin and one NAIP2-rod would have the potential to recruit and activate 5 NLRC4 monomers to form a functional inflammasome hub (Figure 1B). This feature is suggestive of a signal amplification mechanism, wherein detection of distinct pathogen-associated molecules would trigger inflammasome signaling in an additive manner. This could be particularly important for rod and needle detection, as these proteins are produced in vanishingly small quantities. In contrast, 7 molecules of cytochrome c are required to unlock 7 APAF-1 monomers to oligomerize into the apoptosome (Figure 1B). Thus, the unique characteristics of the NAIP/NLRC4 oligomer could make it one of the most sensitive and rapid inflammasomes characterized so far. It remains to be seen whether other NLRs will follow the APAF-1 paradigm, or the NAIP model.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wen H, Miao EA, Ting JPY. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. 2013;39:432–441. doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonardi V, Cherkis K, Nishimura MT, Dangl JL. A new eye on NLR proteins: focused on clarity or diffused by complexity? Current Opinion in Immunology. 2012;24:41–50. doi: 10.1016/j.coi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular Basis for Specific Recognition of Bacterial Ligands by NAIP/NLRC4 Inflammasomes. Molecular Cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proceedings of the National Academy of Sciences. 2011;108:16463–16468. doi: 10.1073/pnas.1113726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Yang J, Shi J, Gong Y-N, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. Available at: http://www.nature.com/doifinder/10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 6.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, et al. Crystal Structure of NLRC4 Reveals Its Autoinhibition Mechanism. Science. 2013 doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 8.Qu Y, Misaghi S, Izrael-Tomasevic A, Newton K, Gilmour LL, Lamkanfi M, Louie S, Kayagaki N, Liu J, Kömüves L, et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 2012 doi: 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki S, Franchi L, He Y, Muñoz-Planillo R, Mimuro H, Suzuki T, Sasakawa C, Núñez G. Shigella Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ. PLoS Pathog. 2014;10:e1003926. doi: 10.1371/journal.ppat.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk THC, Huizinga EG. Formation and Structure of a NAIP5-NLRC4 Inflammasome Induced by Direct Interactions with Conserved N- and C-terminal Regions of Flagellin. Journal of Biological Chemistry. 2012;287:38460–38472. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]