Abstract
In recent years, some substantial advances in understanding human (and non-human) brain organization have emerged from a relatively unusual approach: the observation of spontaneous activity, and correlated patterns in spontaneous activity, in the “resting” brain. Most commonly, spontaneous neural activity is measured indirectly via fMRI signal in subjects who are lying quietly in the scanner, the so-called “resting state”. This Primer introduces the fMRI-based study of spontaneous brain activity, some of the methodological issues active in the field, and some ways in which resting state fMRI has been used to delineate aspects of area-level and supra-areal brain organization.
Keywords: fMRI, resting state, functional connectivity, intrinsic, brain
Introduction
A rapidly expanding approach to understanding neural organization is to observe spontaneous neural activity, and particularly the spatial and temporal patterns of this activity, in living animals. Spontaneous activity can be assayed using many techniques, ranging from single unit recordings to electroencephalography (EEG) to calcium imaging, each with particular spatial and temporal resolution and tradeoffs. In humans, most studies of spontaneous activity utilize functional magnetic resonance imaging (fMRI), a non-invasive technique that typically permits a whole-brain image every few seconds with a spatial resolution of a few millimeters. fMRI measures neural activity via the following mechanism: neural activity increases local blood flow, which changes the ratio of oxy- to deoxyhemoglobin, which alters the magnetic properties of tissue in ways that are detectable with MRI (the Blood Oxygen Level Dependent (BOLD) signal). Thus, the principal technique used to measure human spontaneous brain activity does so indirectly, via metabolic and hemodynamic processes elicited by neural activity (see (Logothetis, 2008) for review).
The study of spontaneous fMRI signal represents a paradigm shift in human neuroimaging. In the first years of fMRI research, nearly all studies involved some behavioral manipulation, and any signal fluctuations unrelated to the manipulation were removed (as far as possible) by complex analyses. Understandably, many investigators suspected that the signal in task-free periods was driven largely by noise (and unconstrained, underspecified neural activity), and that there was little to be gained by studying such data. A publication by Biswal and colleagues in 1995 changed this perspective by demonstrating that fluctuations in the fMRI signal, in the absence of a task, were highly and specifically correlated among functionally related brain regions. In particular, the same brain regions that were active during finger tapping also displayed coherent low-frequency spontaneous fMRI signal when the subject was asked to lie quietly in the scanner for several minutes (Figure 1A) (Biswal et al., 1995). Over the next 8 years, a handful of publications extended this line of inquiry, demonstrating that the correlated fluctuations were most prominent at low frequencies (< 0.08 Hz), that they were unlikely to be explained as artifactual byproducts of motion, cardiac, or respiratory factors, and that spontaneous low-frequency fMRI signal was highly and specifically correlated at rest among auditory processing regions, visual processing regions, and other brain regions (Cordes et al., 2000; Hampson et al., 2002; Lowe et al., 1998; Stein et al., 2000; Xiong et al., 1999).
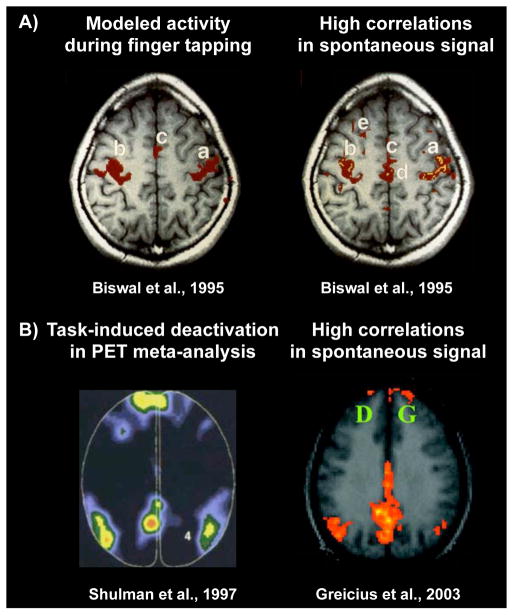
Figure 1. Spatial correspondence between task-evoked activity patterns and patterns in spontaneous fMRI signal correlations.
A) Modeled BOLD activity during finger tapping, and a seed correlation map with location (a) as the seed. B) Locations in a PET meta-analysis where deactivations are seen across tasks, and a seed correlation map with the posterior cingulate as the seed. Images modified from the indicated publications.
These initial studies were met with caution or indifference by many neuroscientists. Widespread interest was generated, however, by a report in 2003 that regions of the default mode network displayed correlated spontaneous fMRI signal (Figure 1B) (Greicius et al., 2003). The default mode network consists of several brain regions that were first reported in meta-analyses of PET and then fMRI data for their puzzling but highly reproducible tendency to deactivate during task performance (Mazoyer et al., 2001; Raichle et al., 2001; Shulman et al., 1997). Unlike motor, visual, or auditory cortex, the default mode network involved large swathes of “association cortex” and was thought to subserve “higher-order”, “cognitive” operations such as introspection or autobiographical memory (see (Buckner et al., 2008) for review). Shortly after the report by Greicius and colleagues, more publications emerged detailing high and specific correlations within other “cognitive” systems (that were also first defined by task-evoked activity in PET and fMRI studies), such as attention systems (Fox et al., 2006a; Laufs et al., 2003) and executive systems (Dosenbach et al., 2007; Seeley et al., 2007).
Resting state analyses also began to reveal unanticipated aspects of brain organization. For example, studies of interactions among systems revealed that low-frequency fMRI signal in the default mode network was negatively correlated with low-frequency fMRI signal in attention and executive systems (Fox et al., 2005; Fransson, 2005). It was already known that default regions deactivated during most tasks, and that attention and executive regions activated during most tasks, but it was not known that in the absence of tasks these regions also displayed anticorrelated signals. Thus, resting state fMRI data suggested that certain portions of the brain might be organized into anticorrelated networks, perhaps operating in ongoing competition. The functional characterizations of these networks – default mode regions supporting “internally-oriented” processes, attention/executive regions supporting “externally-oriented” processes – fueled such speculation. In this manner, by reproducing and then extending features of brain organization that were salient to cognitive neuroscientists, the study of spontaneous fMRI signal began to attract a much broader following.
In the decade since these landmark studies, the pace of research in the field of “functional connectivity MRI” (referring to correlated fMRI signal, also called “intrinsic” or “resting state” connectivity when the signal is acquired in the resting state) has steadily accelerated. The field now encompasses studies of correlated fMRI signal at rest, in sleep, under anesthesia, during tasks, and in animal models including macaques, marmosets, mice, rats, and pigeons. Substantial clinical and lifespan literatures exist.
The purpose of this Primer is to introduce the technique of functional connectivity MRI to a nonspecialist audience. In many neuroscientific fields, experimental design is of primary importance and data analysis techniques may be relatively well-established and uncontroversial. In functional connectivity MRI, the situation is nearly reversed: there is relatively little experimental design to discuss (at least within the scope of this Primer), but the data can be analyzed in a huge variety of ways to investigate different questions. This Primer will first outline some basics of resting state datasets and analysis and then build to some of the more advanced techniques currently practiced in the field. The presentation of these methods will illustrate how functional connectivity MRI has augmented our understanding of brain organization. Because this article is a Primer, it is aimed at introducing techniques and concepts of analysis, not at comprehensively surveying the field. The focus will be on the healthy, adult, human brain; clinical, lifespan, and non-human findings are mentioned only insofar as they advance the issues at hand. A deeper discussion is favored over a broader discussion at several points of the article. Certain emerging topics, such as dynamics in the resting state, are not discussed. For further information the reader is referred to: (Raichle, 2010) for a historical and metabolic perspective; (Deco et al., 2011; Hutchison et al., 2013) for dynamical perspectives; (Bullmore and Sporns, 2012; Sporns, 2014) for network perspectives; (Murphy et al., 2013) for a methods perspective; (Lee et al., 2013) for a clinical perspective; and (Buckner et al., 2013; Craddock et al., 2013) for general perspectives.
Obtaining correlates of spontaneous neural activity with fMRI
The basics of a resting state dataset
A resting state dataset is typically acquired by asking a subject to lie quietly in an fMRI scanner for 5 or more minutes. Many groups ask the subject to fixate a crosshair, but other groups do not pose this requirement. Some groups ask the subject to lie with the eyes closed. fMRI signal correlations from these conditions are largely comparable but differences can be detected (Patriat et al., 2013; Van Dijk et al., 2010). The minimal requirements of a resting state study make it easy to scan a wide variety of populations, but the cost is subject boredom and a possibility that the subject may fall asleep. Indeed, a recent analysis of publicly available datasets indicates that substantial portions of eyes-closed subjects fall asleep, that fewer eyes-open subjects fall asleep, and that fixating subjects rarely fall asleep (Tagliazucchi and Laufs, 2014). Since sleep entails changed patterns of neural activity, well-characterized by EEG, that are likely also detectable in resting state fMRI data (see (Picchioni et al., 2013) for review), it is important to avoid inadvertently including sleep-related state changes in data. In eyes-open and fixation scans it may be adequate simply to watch subjects to ensure that the eyes remain open. If eyes-closed scans are used, periodic verbal interactions (which complicate the “resting” state) or other external measures of sleep state such as heart rate variability or EEG may be useful. The issue of inadvertent sleep has only recently gained prominence, and the field has not yet developed consensus on how to deal with this issue.
An fMRI dataset consists of a series of three-dimensional images (also called volumes or frames), where each image is composed of tens to hundreds of thousands of voxels (volumetric pixels). A certain amount of time is required to acquire signal, meaning that temporal resolution is in tension with both spatial resolution (voxel size) and image size (i.e., whether the image covers the entire brain or only part of the brain). In principle, voxel sizes can be sub-millimeter, and temporal resolution can be sub-second. Smaller voxels better separate tissue types and even layers of cortex, but have smaller signal-to-noise ratios. Increased temporal resolution aids statistical power, but the information gained by increased sampling rates is fundamentally limited by the fact that fMRI measures a slow hemodynamic response. In recent years, a typical dataset might consist of whole-brain images with voxel sizes of ~2–4 mm and intervals between images of ~2 seconds. Recently developed acceleration techniques permit more rapid image acquisition (whole-brain images every few hundred ms), which can then be traded for increased spatial resolution or image size (see (Feinberg and Yacoub, 2012) for review).
Voxel size (and any explicit blurring in addition to the blurring inherent in data realignment and resampling) governs the ability to detect particular aspects of brain organization. For example, larger voxels (e.g., 4 mm) can simultaneously sample tissue on opposing banks of a sulcus, or tissue in the occipital lobe and the cerebellum (such phenomena are called partial volume effects). Activity that is shared across conditions in a larger voxel, when re-examined at a higher resolution, may turn out to reflect blurring across separate (and separable) neural populations (Beauchamp et al., 2004). Fine-scale organization in V1 such as ocular dominance columns (Menon et al., 1997) or orientation hypercolumns are resolvable by fMRI (Yacoub et al., 2008), but will not be detected with the “typical” datasets of recent years.
Two types of limitations of these fMRI images should be mentioned. The first type of limitation is that, for practical purposes, most fMRI datasets have incomplete images of the brain. Due to the proximity of sinuses (air pockets) in the cranium, fMRI signal dropout occurs in the inferior temporal, orbitofrontal, and lateral mid-temporal cortex. Most resting state datasets have poor signal quality in those regions. For example, in the ICA images of Figure 2, the underlying brain template is the average fMRI signal across subjects, and the dropout of orbitofrontal and lateral temporal signal intensity is visible in certain slices. Specific scanning procedures can reduce signal dropout in such regions. In addition to signal dropout, and for the same underlying reasons, geometric image distortion can occur in these same regions. Image distortion can be largely corrected using techniques such as field maps (which measure distortion).
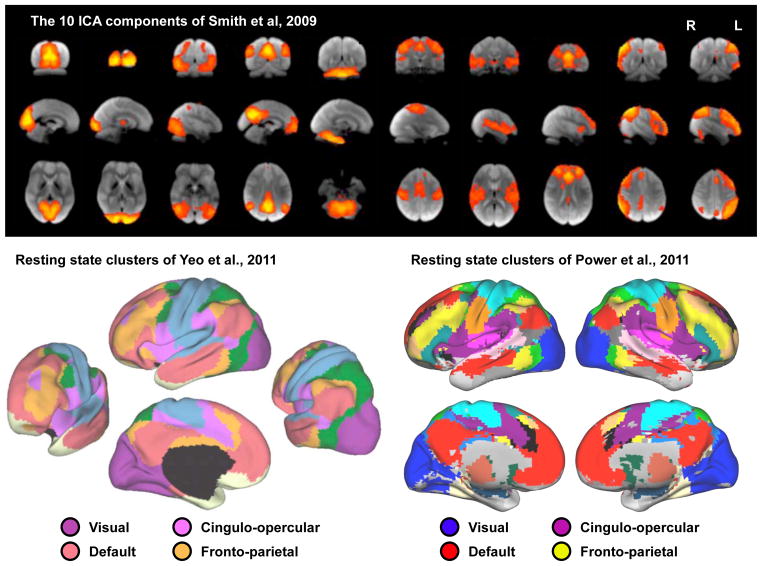
Figure 2. Large-scale correlation patterns in resting state fMRI data.
Data are shown from 3 reports on the spatial patterns of correlated BOLD signal: a 10-component ICA analysis, a surface-based analysis of surface vertex clustering, and a volume-based analysis of voxelwise clustering. Images modified from the indicated publications.
The second type of limitation is that fMRI signal (technically, T2*-weighted signal, which is MRI signal optimized to detect BOLD changes) reflects many factors, only some of which relate to neural activity. First, the signal reflects neurally-driven changes in oxy- to deoxyhemoglobin ratios, which is the BOLD signal of interest. But the depth and rate of respiration also influence oxy- to deoxyhemoglobin ratios, modulating BOLD signal in non-neural ways (Birn et al., 2006; Wise et al., 2004). Head motion, present in virtually all scans, can produce large signal disruptions that can take many seconds to resolve (Friston et al., 1996; Power et al., 2014; Satterthwaite et al., 2013). Cardiac and respiratory cycles can also modulate signal via several mechanisms. A fuller accounting of the various sources of artifact is beyond the scope of this review (see (Murphy et al., 2013)), but the magnitude of the problem is captured by the following statistic: in Human Connectome Project data, which are high-quality and undergo an advanced denoising procedure, of the roughly 200 signals identified per subject, only ~23 on average are considered to reflect neural activity, constituting ~4% of the variance (Marcus et al., 2013).
Isolating neural signal from artifactual signal
Accurate estimation of neurally-driven covariance in resting state fMRI signals thus depends on removal of artifactual influences on the signals. Several major denoising approaches are mentioned here. Some investigators obtain relevant physiologic recordings during resting state scans, such as cardiac rate, respiratory-belt traces, or end-tidal pCO2 recordings, and use regression models to reduce physiology-related variance in the dataset (Birn et al., 2008; Chang and Glover, 2009b; Glover et al., 2000). Nearly all investigators regress motion estimates from datasets, and some withhold motion-contaminated images from analysis (Lemieux et al., 2007; Power et al., 2014). Some investigators regress signals found in white matter or the ventricles, which are presumably not-of-interest, from the gray matter voxels (Behzadi et al., 2007; Jo et al., 2010; Weissenbacher et al., 2009). Still other investigators regress the average signal from the whole brain from the dataset (global signal regression) (Fox et al., 2009). Another technique is to apply independent component analysis (ICA) to the entire dataset to derive a subset of signals, classify them as signals-of-interest and signals-of-noninterest, and then remove the uninteresting signals (Beckmann et al., 2005; Griffanti et al., 2014; van de Ven et al., 2004) (a version of this process was used to derive the ~200 signals in Human Connectome Project data mentioned above). Some techniques, such as multi-echo scanning, leverage the physical principles underlying the fMRI signal to help distinguish BOLD signal (neural- and pCO2-related signal) from non-BOLD signal (Bright and Murphy, 2013; Kundu et al., 2013). Other methods exist or are emerging in the rapidly evolving resting state denoising literature.
The level of technical detail just given about denoising may be unusual for a general introduction to functional connectivity MRI. However, to understand the field, it is important to know that a wide variety of denoising techniques are used, and that these techniques are not equally efficacious (that is, the choice of denoising techniques is not trivial and can have substantial effects on findings). Two examples, both active issues in the field, should illustrate why denoising methodology deserves close attention.
The first example relates to the anticorrelations between the default mode and attention networks mentioned previously, first highlighted in two functional connectivity MRI studies that used global signal regression (Fox et al., 2005; Fransson, 2005). Beginning in 2009, several groups reported that they could not detect anticorrelations between these networks under their preferred processing strategy and/or without global signal regression (Anderson et al., 2011; Hampson et al., 2010; Jo et al., 2010; Murphy et al., 2009; Weissenbacher et al., 2009). On conceptual grounds, two groups raised the possibility that the anticorrelations were a byproduct of global signal regression rather than a true neurobiological phenomenon (Murphy et al., 2009; Weissenbacher et al., 2009). Subsequently, the anticorrelations have been found in several functional connectivity MRI studies that address the criticisms made against the original papers (Carbonell et al., 2011, 2014; Chai et al., 2012; Chang and Glover, 2009a; Fox et al., 2009; He and Liu, 2012; Keller et al., 2013; Marx et al., 2013; Power et al., 2014). And, importantly, the anticorrelations have also been detected with electrophysiologic techniques (Keller et al., 2013). One possibility for the lack of anticorrelations in some fMRI studies is that the data contained artifactual signals that spuriously increased covariance to the point where no negative correlations remained. Consistent with this possibility, unremoved physiologic artifact is sufficient to mask the presence of anticorrelations (Chang and Glover, 2009a), and anticorrelations are stronger in lower-motion subjects compared to higher-motion subjects (in whom the anticorrelations are almost absent) (Power et al., 2014). Thus, the extent of artifact in a dataset, and the extent of artifact removal, influences the ability to detect anticorrelations in resting state fMRI.
The second example concerns a set of motion-related findings that impact all resting state studies, but which will be illustrated with the developmental literature. In the mid-2000s, several groups, including ourselves, began to report that short-distance functional connectivity MRI correlations were strong in children and tended to weaken over development, while long-distance correlations were weak in children and tended to strengthen over development (Fair et al., 2009; Fair et al., 2007; Kelly et al., 2009; Supekar et al., 2009). In 2011, we and two other groups reported a previously unrecognized aspect of motion artifact, which is that spurious motion-related variance tends to be more similar at nearby voxels than at distant voxels (Power et al., 2012; Satterthwaite et al., 2012; Van Dijk et al., 2012). These studies demonstrated that even relatively small amounts of motion cause these effects, and that several common denoising techniques did not adequately remove motion-related variance. These studies indicated that, all other things being equal, a higher-motion group will have higher short-distance correlations than a lower-motion group (and, depending on the denoising strategy employed, the higher-motion group can also have lower long-distance correlations). It is widely known that children tend to move more than young adults, raising the possibility that the previously-described developmental effects were at least partially due to motion. Indeed, several studies with improved removal of motion-related variance now find much smaller developmental effects (Power et al., 2012; Satterthwaite et al., 2012; Satterthwaite et al., 2013). Increased scrutiny of motion artifact in resting state fMRI is also causing reappraisal of previously-reported aging and clinical effects (e.g., (Tyszka et al., 2014; Van Dijk et al., 2012)). The literature on techniques to identify and remove motion artifact is currently one of the most rapidly evolving fronts of functional connectivity MRI (e.g., (Bright and Murphy, 2013; Kundu et al., 2013; Power et al., 2014; Yan et al., 2013)), and multiple methods now exist that have evidence for removing much or nearly all motion artifact from resting state datasets. The problems caused by small movements are unfortunately not limited to functional connectivity MRI: previously unrecognized effects of small movements, capable of causing spurious group differences, have recently been reported in the structural connectivity MRI literature (e.g., (Koldewyn et al., 2014; Yendiki et al., 2013)).
As these examples show, denoising is a critical issue in resting state MRI studies. Inadequate removal of artifact can fundamentally change the conclusions one draws from a dataset. In the remainder of this Primer, we will mainly focus on studies of healthy young adults, who generally move little. Hereafter, when we speak of denoised fMRI signal, it should be understood that we are ideally speaking of neurally-driven BOLD signal, though perfect isolation of such signal from other signals does not occur in practice.
Measuring brain relationships via spontaneous BOLD signal
Common analysis techniques
After a dataset has undergone preliminary processing and denoising, a wide variety of analyses can occur, including analyses of signal similarity, frequency content, dynamics, and causal influences among signals. This Primer is mainly concerned with “functional connectivity”, or the observed statistical dependencies between signals, such as those measured by a correlation. “Effective connectivity” concerns models of causal influences consistent with the observed functional connectivity, and is discussed later in the Primer.
The simplest measure of signal similarity is the Pearson correlation coefficient (hereafter, unless specified, “correlation” refers to Pearson correlation), which measures the linear dependence between two signals. For example, one voxel’s signal can be correlated with another voxel’s signal to measure a pairwise relationship. Similarly, the signal averaged across some group of voxels (a region of interest (ROI)) can be correlated with the signal in another group of voxels. Commonly, a “seed” voxel signal or “seed” ROI signal is correlated with the signal at every other voxel to map out pairwise relationships throughout the entire brain. These correlations are often represented as a heat map (a “seed correlation map”) superimposed on a high-resolution brain image; weaker correlations are often not shown. The images in Figure 1 follow this convention, using a seed in motor cortex in Figure 1A, and a seed in the posterior cingulate in Figure 1B. When many seeds (voxels or ROIs) are studied simultaneously, a common technique is to measure all pairwise correlations and to represent these correlations in a seed by seed matrix. This matrix defines a network – the set of pairwise relationships between a set of nodes – and can be used for several types of analysis, including methods derived from network science, such as graph theoretic analyses (discussed later).
Another common technique for measuring signal similarity involves independent component analysis (ICA), already mentioned as a tool for denoising. ICA is a matrix decomposition technique, which, given a set of voxel signals, will attempt to determine a subset of maximally spatially independent signals that can be linearly combined to form the original signals. An intuitive analogy is to imagine a cocktail party, where, at any given location in the room, the sound heard (the actual signal) comprises several voices (the neural sources) and perhaps music or street noise (artifactual sources). ICA aims to separate these various sources into components starting from the actual signal. In functional connectivity studies, once the components are determined, the spatial locations of the signal can be represented as maps, similar in principle to the seed maps, showing the correlations of voxel signals with the signal of a particular component.
The seed correlation and ICA methods represent distinct but related approaches to analyzing functional connectivity data. Hundreds of thousands of neurons are represented in a voxel, and these neurons participate in multiple processes at a variety of spatial scales and frequencies, some of which have low-frequency representations in the BOLD signal. A seed map represents all of these processes at once, whereas ICA attempts to separate these processes into components. It is important to note that the accuracy of a seed correlation depends upon the success of prior denoising and correct placement of the seed, while the accuracy of an ICA component depends upon successfully separating artifactual variance from neurally-driven BOLD variance in the components and also upon choosing a proper number of components to model relative to the number of signals that actually exist (the number of components generated by ICA must be prespecified, but can be optimized using cost functions). Additionally, note that we have described only one (common) way in which ICA is applied to resting state data; other possibilities are reviewed in (Beckmann, 2012).
On the meaning of resting state BOLD signal similarity
The reader has seen some of the findings that stimulated interest in functional connectivity MRI and the basics of resting state datasets and analysis (e.g., Figure 1). Before turning to some of the more recent applications of resting state fMRI, it is appropriate to consider a set of bedrock questions for the field: What brain property is being in captured these studies? Up until this point, we have purposefully used the vague term “relationship” to describe similarities in low-frequency BOLD signal. What is the nature of these “relationships”? What causes them? What do they mean? What is actually being measured by signal similarity? A full answer cannot be given, but several relevant points follow.
At the most basic level, high BOLD signal similarity creates a strong suspicion that the signals represent related processes across regions. Two decades of resting state studies have now built a substantial empirical basis for the claim that brain regions that are coactive during a task tend to have correlated low-frequency fMRI signal (e.g., (Smith et al., 2009)). An influential model of cognition, proposed by Donald Hebb, is that mental representations (elements of a sensation, an emotion, a thought, etc.) occur via activity in “ensembles” of neurons, and, critically, that these neurons are organized such that they can reinforce their tendency to coactivate by recurrent, self-excitatory strengthening of connectivity (thus refining and distinguishing the ensemble) (Hebb, 1949). The flexibility and variety of mental representations is thought to arise by combining ensembles in different ways. Over time, such a general mechanism yields a hierarchy of ensembles, representing function, sculpted by histories of coactivation, and capable of efficient but flexible mental representations. A possible hypothesis is that statistical histories of coactivation between neurons shape functional connectivity MRI relationships via Hebbian mechanisms (Fair et al., 2007). Consistent with this hypothesis, practice-induced changes in resting state functional connectivity that correlate with behavioral performance have been observed between task-involved regions after motor learning (Vahdat et al., 2011), perceptual learning (Powers et al., 2012), reasoning training (Mackey et al., 2013), and in other contexts (see (Guerra-Carrillo et al., 2014) for review).
In the first years of functional connectivity MRI, it was natural to suspect that the stream-of-consciousness thoughts that we all have when resting quietly (about eating, errands, plans, worries, etc.) substantially shaped the signal fluctuations. This concern is lessened by several findings across states of consciousness and animal models. First, in humans, the low-frequency correlation structure is similar at rest and in the early stages of sleep (Horovitz et al., 2009; Larson-Prior et al., 2011). Second, organized low-frequency fluctuations in fMRI signal are also found in awake and anesthetized macaques (Hutchison et al., 2011; Moeller et al., 2009; Vincent et al., 2007), marmosets (Liu et al., 2013), rats (Hutchison et al., 2010; Liang et al., 2011), mice (Jonckers et al., 2013; Jonckers et al., 2011; Sforazzini et al., 2014), and awake pigeons (De Groof et al., 2013). In animal models, different anesthetics have different effects on functional connectivity, as does the depth of anesthesia, but in general, light anesthesia produces relatively little alteration in patterns of functional connectivity. Findings in humans under anesthesia are mixed across studies and resting state networks, with some studies reporting little change in correlations under light anesthesia, and others reporting more substantially changed functional connectivity (see (Heine et al., 2012) for review; note that most rodent and primate studies utilize head posts or bite bars to immobilize the head, whereas in human studies the head is almost never immobilized). Deep anesthesia unambiguously alters patterns of functional connectivity. Third, there are suggestive similarities between the spatial organization of the correlations in macaques and humans, such as putative homologues of the default mode network and the attention networks (Hutchison et al., 2012; Vincent et al., 2007). Several groups now believe they have identified a default-mode-like network in both rats (Lu et al., 2012) and mice (Sforazzini et al., 2014). Collectively, these observations indicate that low-frequency resting state fluctuations are at best weakly related to consciousness or conscious thought, since they are present and similar in light sleep and light anesthesia. These observations also raise the possibility that low-frequency fluctuations may share an evolutionarily conserved mechanistic basis and/or function across clades spanning humans, rodents, and birds (though it is possible the fluctuations arose independently or have different bases or functions across species).
Measures of functional connectivity MRI are related to but distinct from measures of structural connectivity. High correlations are often found between brain regions known to be anatomically connected, but high correlations also occur between regions that are not monosynaptically connected. For example, in macaques there are high correlations between the eccentric representations in V1 in both hemispheres, which have no known direct physical projections (Vincent et al., 2007). An additional example is found in the cerebellum, which displays correlated activity with cortical regions that are separated from the cerebellum by several synapses (Krienen and Buckner, 2009). Lesions at the mediating synapses, for example in the pons, perturb corticocerebellar correlations in predictable ways (Lu et al., 2011). The observed correlation between any two regions likely reflects weighted representation of many or all possible pathways between those regions, many of them mediated by other brain regions (Adachi et al., 2012). Indeed, some of the most successful computational models that predict observed functional connectivity from known structural connectivity do so by considering paths other than the shortest anatomical pathway between two brain regions (whether measured by distance or synapses) (Goni et al., 2014).
Ultimately, low-frequency fluctuations in BOLD signal are of interest for what they reveal about neural activity. The mechanisms linking neural activity to hemodynamic responses are not fully known, but some salient points are mentioned here. Simultaneous intracortical electrophysiological recording and fMRI in macaques reveals that both spiking activity and synaptic activity (measured by local field potentials (LFPs)) can predict BOLD responses, with a lag of several seconds between neural activity and the BOLD response (Logothetis et al., 2001). LFPs, loosely speaking, represent the aggregate membrane potentials of cells near an electrode (subject to cell orientation, location, type, size, and other factors). Importantly, at times where LFPs and spiking become dissociated, LFPs predict BOLD signal in the absence of spiking. These results therefore indicate that BOLD signal at a region reflects inputs and local processing more than outputs of the region (see (Logothetis, 2008) for review). Local processing (e.g., neurotransmitter recycling, etc.) is energetically costly and is intimately linked with metabolic processes that modulate blood flow (Raichle and Mintun, 2006). With regard to ongoing spontaneous fMRI signal, several studies have found positive correlations between the BOLD signal and fluctuations in band limited power in the upper gamma range of LFPs (~50–90 Hz) (Leopold et al., 2003; Niessing et al., 2005; Scholvinck et al., 2010; Shmuel and Leopold, 2008). Thus, when BOLD signal increases synchronously at multiple locations, one contribution to such correlations may be a shared increase of LFP amplitude at particular frequencies, which may reflect increased synchrony of membrane potential modulation at those frequencies (and vice versa for decreases). Other links between electrophysiology and BOLD signal exist and are reviewed in (Scholvinck et al., 2013).
We have surveyed evidence that spontaneous low-frequency fMRI correlations are modified, to some extent, by experience, that they are present in some sleep and unconscious states, that they are found in many animals, and that they reflect certain aspects of ongoing neural activity. Additionally it should be mentioned that the fluctuations underlying these correlations are comparable in magnitude with task-evoked activations (Damoiseaux et al., 2006; Fox et al., 2006b). Further, much of the organization of the correlations persists, largely unchanged, in a variety of task states (Cole et al., 2014) (though changes do occur, see (Rehme et al., 2013; Sepulcre et al., 2010)). Interestingly, ongoing signal fluctuations appear – on a moment-to-moment basis – to influence whether and how stimuli are perceived, as well as reaction times and motor responses (see (Sadaghiani et al., 2010) for review). Although the ultimate function of these ongoing fluctuations remains unknown, they are energetically costly, and it is tempting to speculate that they serve some basic and important role in the brain (Buzsaki et al., 2013; Raichle, 2010).
Using resting state relationships to explore brain organization
Refining system-level descriptions
Once it was realized that targeted studies could be used to map out targeted systems (e.g., the motor, visual, auditory, and default mode systems (Biswal et al., 1995; Cordes et al., 2000; Greicius et al., 2003; Lowe et al., 1998)), a next logical step was to map out relationships across the brain in a data-driven, untargeted fashion. The development of ICA methods enabled whole-brain partitioning of variance into components. Initial studies determined low numbers of components but as data quality and analysis methods improved, the number of components has increased (e.g., the number of “neural” compared to “determined” (neural + artifactual) components is 10 of 25 in (Damoiseaux et al., 2006), 8 of ~30 in (Sorg et al., 2007), 10 of 20 and 45 of 70 in (Smith et al., 2009), and ~23 of ~200 in (Marcus et al., 2013)). An illustration of a whole-brain partitioning of variance using ICA is shown in Figure 2. Another method of data-driven analysis involves calculating pairwise correlations between all voxels and using clustering algorithms to identify groups of highly correlated voxels. Note that this approach is a seed-based approach, in contrast to the ICA-based approach above. Following such approaches, the brain has been partitioned into somewhere around 15–20 large-scale clusters (e.g., (Power et al., 2011; Yeo et al., 2011), shown in Figure 2). There is evident similarity in the clustering structure found across these two studies, and many of these clusters correspond to ICA components identified in other studies.
In many cases, functional connectivity reflects relationships between brain regions that have long been known to be related. For example, it is not surprising that visual processing regions are grouped, or that auditory processing regions are grouped. Other sets of functional relationships, reflected in functional connectivity correlations, would have been unknown twenty years ago. For example, a shared functional characteristic of the default mode regions (task-induced deactivation) was only discovered in 1997. Other recognitions of functional commonalities among brain regions are even more recent, such as the recognition in 2006 of shared executive characteristics between anterior insula and anterior cingulate cortex (that distinguished these regions from other executive regions in lateral frontal and parietal cortex) (Dosenbach et al., 2006). Both default mode relationships and a distinction between cingulo-opercular and fronto-parietal regions are reflected in the resting state (Dosenbach et al., 2007; Greicius et al., 2003; Seeley et al., 2007). In the examples just mentioned, unifying functional characteristics were known for each of the systems prior to the discovery that the systems exhibited correlated low-frequency BOLD signal, lending intelligibility to the observed patterns of correlations.
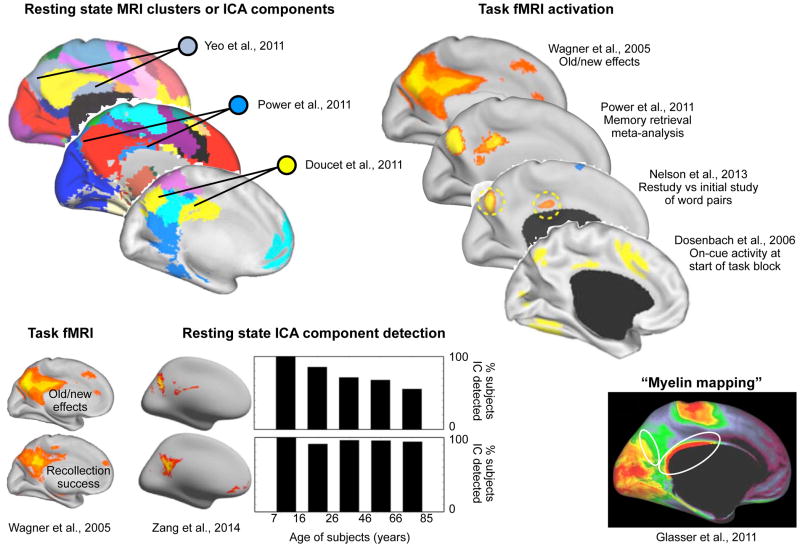
In some cases, however, functional connectivity has grouped regions that were not so widely recognized as a functional system. A medial parietal cluster encompassing the precuneus is an example of this phenomenon (see Figure 3; this cluster also has lateral parietal representation, not shown). In 2011, several resting state studies, both seed-based and ICA, grouped a set of medial parietal regions (Doucet et al., 2011; Power et al., 2011; Yeo et al., 2011). In light of this grouping, it became easier to recognize the significance of patterns in prior task studies and other concurrent and subsequent literature. For example, spatially similar regions had exhibited differential responses to old versus new stimuli in previous studies (Konishi et al., 2000; Shannon and Buckner, 2004; Wagner et al., 2005), and memory-related effects have been seen in these regions in more recent studies (Nelson et al., 2013; Power et al., 2011). Additionally, spatially similar regions have exhibited strong activation at the beginning of task blocks, a signal consistent with processes instantiating task parameters (Dosenbach et al., 2006). Spatially similar regions also appear to have higher myelin content than surrounding tissue (Glasser and Van Essen, 2011). And interestingly, this group of regions, unlike nearby tissue in the default mode system, seems to lose its distinctness in terms of resting state correlations with age (Yang et al., 2014). Thus, resting state correlations grouped a set of regions and made it easier to recognize that they shared a variety of specific characteristics, bolstering the case that these regions form a functional system.
Figure 3. A medial parietal resting state network exhibits specific functional, structural, and lifespan properties.
A grouping of regions mainly in medial parietal cortex that has been identified in multiple resting state analyses exhibits memory-related and oldness-related activations, activity at the beginning of a task block, specific age-related decreases in resting state correlations, and increased putative myelin content relative to surrounding tissue. Images modified from the indicated publications. Note that the illustration from (Yeo et al., 2011) is the 17-cluster partitioning, not the 7-cluster partitioning shown in Figure 2.
In all of these examples, the pattern of resting state correlations respects many distinctions in function and structure. The large-scale (system-level) patterns in resting state activity therefore serve as a useful organizing framework for interpreting results and patterns in other modalities. After becoming familiar with the brain-wide patterns of resting state fMRI correlations, it is now difficult for us to read the literature without noticing spatial patterns congruent with the resting state patterns.
Refining area-level descriptions
The structure (and therefore function) of neural tissue is organized at multiple spatial scales. At the level of millimeters to centimeters, the resolution of fMRI, “area-level” organization is evident in the cortex, at least in many locations. Areas are sections of the cortex, much like patches in a quilt, that contain specific sets of neurons, arranged with specific layering, with specific sets of incoming and outgoing projections from and to other locations in the brain and body. Areas therefore are expected to exhibit specific functional capabilities. Some areas, especially those most proximal (in terms of synapses) to sensory and motor organs, have a readily mapped topographic organization: neuron response properties form retinotopic maps in several early visual areas, tonotopic maps in early auditory areas, or maps of the body in primary motor and somatosensory areas.
With great effort, roughly 100 areas per hemisphere have been defined after a century of intense study in the macaque (see Figure 4) (Van Essen et al., 2012a). The best-defined areas are visual areas in occipital, parietal, and temporal cortex, auditory areas in temporal cortex, and somatosensory and motor areas near the central sulcus. But even in this best-studied model, the area-level organization in much of the macaque brain is only somewhat understood, especially in frontal cortex or other locations where well-behaved maps are difficult to define.
Figure 4. Area-level mapping of cortex in macaques and humans.
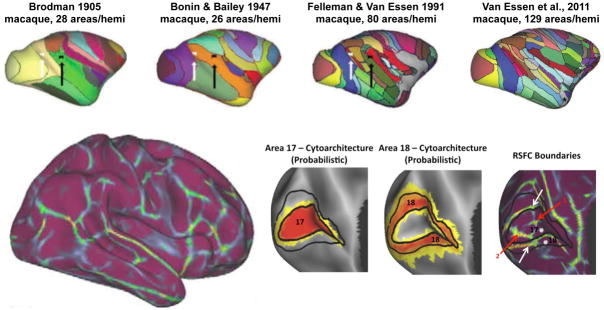
At top, several macaque cortical parcellation schemes show the refinement of area-level maps over the last century, modified from (Van Essen et al., 2012a). At bottom, a parcellation of the human cortex based on resting state functional connectivity. The insets show postero-medial views of the occipital lobe of the left hemisphere, with cytoarchitectonic locations for Brodmann areas 17 and 18, and corresponding resting state boundaries. The white arrows denote borders that align well with the predicted area borders, and the red arrows denote boundaries that may be byproducts of unremoved artifact at the occipital pole. Modified from (Wig et al., 2014b).
In humans, considerably less is known about area-level organization than in macaques, though certain sensory and motor areas have been defined by multiple criteria. It is expected that humans will exhibit more areas than macaques, perhaps on the order of 200 areas per hemisphere (Van Essen et al., 2012b). Many of the techniques used to study macaque area-level organization are inapplicable to humans (e.g., tracer injection), and human studies are limited mainly to post-mortem studies, non-invasive imaging such as MRI, and a relatively small number of neurosurgical cases. Because resting state correlations reflect, to some extent, anatomical connectivity, and are strong between functionally-related tissue, it is possible that these correlations could be used in ways analogous to tracers to identify a “connectivity fingerprint” of tissue, and that this “fingerprint” could be used to help delineate areas in the brain.
A major effort of several groups has been to use resting state functional connectivity to map out area-level distinctions in the human cortex. For brevity, we will only describe the approach that we have taken, emphasizing that many other approaches have been used (e.g., (Blumensath et al., 2013; Craddock et al., 2012; Shen et al., 2010)). The general idea is that signals within an area should be relatively homogeneous, and that signals in different areas should be somewhat different. Topographic influences are a complicating factor, because they should increase signal similarity, across areas, in topographically-corresponding locations.
To identify borders between areas, we have used gradient-based techniques that measure the rate of change of signal similarity between nearby locations. The underlying presumption is that signals should change little and slowly within an area, but rapidly at a border between areas (again, topographic influences challenge this procedure, since adjacent maps sometimes have mirrored orientations with corresponding topography immediately on either side of the border). This boundary-mapping approach, based on local changes in connectivity, was first developed in structural connectivity data (Johansen-Berg et al., 2004) and was later adapted to and refined in resting state fMRI data in a series of studies (Barnes et al., 2012; Cohen et al., 2008; Nelson et al., 2010a; Nelson et al., 2010b; Wig et al., 2014a; Wig et al., 2014b).
The boundary-mapping technique defines roughly 200 regions in each hemisphere and has yielded several notable results. First, the boundaries defined in the resting state recapitulate some boundaries of functional distinctions during tasks (Nelson et al., 2010a; Wig et al., 2014a; Wig et al., 2014b). Second, the borders respect the large-scale systems described in the previous section, but subdivide large, contiguous swathes of those systems into multiple putative areas, as expected (Wig et al., 2014a; Wig et al., 2014b). Third, the boundaries seem to respect some cytoarchitectonic distinctions between well-known cortical areas in humans, such as those between V1 and V2 (Figure 4) (Wig et al., 2014b). Fourth, there are instances where borders are probably being driven by artifact (e.g., borders running on the crowns of gyri, borders outlining regions of signal dropout, etc.) (Wig et al., 2014b). Corroboration of borders with distinctions in other modalities, such as structural connectivity, cytoarchitectonics, or function will help clarify the neurobiological meaning of these borders (Amunts et al., 2013). Such multimodal analyses are a key aim of the Human Connectome Project.
Resting state correlations are modulated, within known areas, by topographic representations. One example of this is that correlations across visual areas are linked by their eccentricity – foveal representations in V1 exhibit increased correlations with foveal representations in other areas such as V2, MT, etc., and likewise for peripheral representations (Yeo et al., 2011). Another example is in somatosensory (and motor) cortex, where Brodmann areas 1–3 occupy the posterior bank of the central sulcus in long, narrow, vertically-oriented strips that run from lateral cortex to the midline. Within these areas exist well-known representational maps of the body, organized such that the legs are on the midline, the hands are superior, and the face is inferior and lateral. The functional connectivity signal along the central sulcus is modulated strongly by its position in the body representation in two notable ways. First, the signal in facial representations is sufficiently distinct from the signal in the rest of the body that these signals form separate clusters (see the orange versus cyan modules of (Power et al., 2011) in Figure 2), potentially distinguishing the dorsal column/medial lemniscal system from the trigeminal lemniscal system. Second, the homotopic correlations (between mirrored locations in each hemisphere) between face representations are higher than those between the leg representations, which are higher than those between the hand representations (Yeo et al., 2011). These apparent within-area distinctions are reminiscent of “domain” distinctions recently described by Kaas and colleagues for separate reach and grasp regions within primary motor cortex (Kaas, 2012).
These 3 examples of topographic distinctions can be interpreted in terms of the topographic specificity of projections among different visual areas, in terms of input from different thalamic nuclei (the face and body representations receive projections from different thalamic nuclei), and in terms of differing densities of callosal crossing fibers between representations of various body parts. But these topographic distinctions are also amenable to speculative interpretations based more on statistical histories of coactivation than on anatomical projections (remember that correlations can be modulated, at least for several days, by statistical experience (Guerra-Carrillo et al., 2014)). For example, when considering homotopic correlations, the two hands are relatively independent compared to the halves of the tongue or face. Or, part of the reason that signals at facial representations differ from signals in representations of the rest of the body is that facial signals correlate strongly with signals at auditory regions. This finding that may reflect physical proximity and fMRI signal blurring to some extent, but it also may reflect the fact that facial and auditory representations must be frequently coordinated in the service of language.
Further studies will be needed to determine the dominant influences on within-area and between-area resting state signal differences. But the discussion thus far should demonstrate that resting state signals can yield information likely relevant to the area-level and within-area organization of the brain.
Building network representations of the brain
Network studies of the brain represent a departure from the analyses mentioned thus far. When viewing a complex system as a network, the focus is less on the properties of a single element of the system, and more on the role and contextualization of that element within the larger system. Additionally, overall properties of the system that emerge from the arrangement of interactions among elements are of interest. The brain, itself a network of neurons organized at many levels, is a natural candidate for this type of analysis.
Many aspects of network analysis are intuitive to anyone who rides a subway. The network is defined by a set of nodes (the elements of the system, e.g., stations) and a set of edges (the pairwise relationships between nodes, e.g., the tracks between the stations). Nodes with many or strong connections (many tracks, a train every 6 minutes) are generally more important than nodes with few or weak connections (few tracks, a train every 20 minutes). Nodes that connect distant parts of a network (stations with express lines) or that link distinct parts of the network (stations with multiple lines) or that define bottlenecks (stations connecting a tunnel under a bay) are especially important for network traffic. All of these properties, intuitive to someone studying a subway map, can be determined algorithmically. The mathematical study of networks is called graph theory, and graph is another name for a network, when represented as a set of nodes and a set of edges.
The prospect of creating comprehensive network representations of the brain is very attractive, as evidenced by large federal initiatives such as the Human Connectome Project or the Brain Activity Map Project. At present, large-scale networks with whole-brain coverage, at least in humans, are only feasible via MRI-based techniques, such as diffusion tensor imaging (DTI) (which measures diffusion of water, presumably along axons) or resting state functional connectivity. Arguably, at the moment, resting state methods yield the most complete and fine-grained network representations available, due to well-known difficulties in DTI fiber reconstruction techniques when mapping long fibers, and the tendency for fiber reconstruction techniques to funnel fibers to the tips of gyral blades and not the banks of gyri (Johansen-Berg and Behrens, 2013). There are, however, many difficulties with forming network representations using resting state MRI data. Difficulties exist with node definition, edge definition, and the interpretation of network measures. Each of these topics represent issues that can fundamentally alter the results or interpretation of a study.
Node definition is a major challenge for human neuroimaging network studies. Unlike the subway stations in the example above, the appropriate nodes of the human brain are not obvious. As mentioned, area-level organization is amenable to fMRI resolution, but the area-level organization of the human brain is largely unknown. Investigators must therefore choose nodes in the absence of ground truth. Common node choices include voxels, random parcellations, anatomically-driven brain atlases (e.g., parcellations based on gyral anatomy), or functionally-defined ROIs (e.g., task-defined ROIs). These node choices, like the denoising choices mentioned earlier, can fundamentally alter the results of a study. For example, one common node definition scheme is the Automated Anatomical Labeling (AAL) atlas, a set of 90 parcels defined in large part by gyral anatomy. The parcels of the AAL atlas are large and most parcels likely traverse multiple true underlying areas. Figure 5 shows, as an example, some of the signals within the left pre-central and left middle temporal parcels of the atlas. Most nodes in the AAL atlas probably blend several signals (and, by extension, divide signals), rendering these representations both inaccurate and relatively non-specific. The blending of signals is evidenced by the fact that AAL-based networks usually contain 3–5 clusters (e.g., (Gratton et al., 2012; He et al., 2009; Meunier et al., 2009; Power et al., 2011)), often not reflecting the specific patterns known from ICA or seed correlation maps. In contrast, clustering in voxelwise and functionally-defined networks returns more clusters, most of which visibly respect correlation patterns known from ICA or seed map analyses (Figure 5). This issue of misdefining nodes has elicited unusually strong language from several groups (e.g., (Power et al., 2011; Smith et al., 2011; Wig et al., 2011)), perhaps best encapsulated by Stephen Smith and colleagues: “The most striking result [of our study] was that the use of functionally inaccurate ROIs (when defining the network nodes and extracting their associated timeseries) is extremely damaging to network estimation; hence, results derived from inappropriate ROI definition (such as via structural atlases) should be regarded with great caution.”
Figure 5. Considerations in resting state networks: node definition.
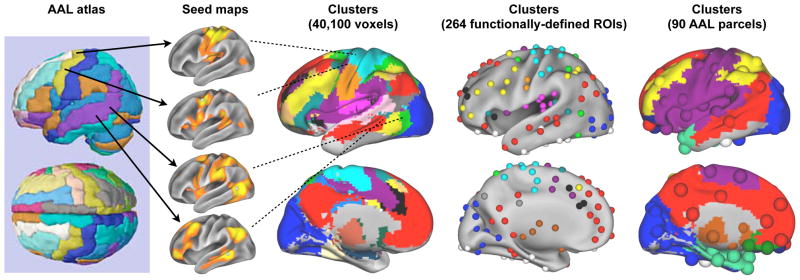
The 90-parcel AAL atlas is shown, as are seed maps from sites within two of the parcels. The clustering structure of resting state networks with nodes of voxels, functionally-defined ROIs, and the AAL parcels are shown. Image at left is modified from www.prefrontal.org and the images at right are modified from (Power et al., 2011).
Even if perfectly defined nodes were available, investigators would still face difficult decisions about edge definition in resting state fMRI. The basic issue is this: Pearson correlations are widely-used and widely-understood, and are measurable in all datasets and for large numbers of nodes. But Pearson correlations cause difficulties of interpretation since they represent both direct and indirect associations of nodes. However, other measures that might better isolate direct associations between nodes (“effective connectivity” methods such as dynamic causal modeling, Granger causality, etc.) often scale poorly with network size. Many of these techniques do not estimate connectivity well between a dozen nodes, much less hundreds of nodes. And those techniques that appear able to handle larger networks (e.g., partial correlations) cannot practically be used in many datasets because the timeseries are not long enough to support the technique (e.g., to isolate the unique variance between two nodes, the signals at other nodes are regressed, and thus the demand for degrees of freedom scales with network size). Some groups have advocated using techniques that use a penalty factor (akin to a threshold) to estimate sparse inverse covariance matrices (i.e., partial correlations) from data with limited degrees of freedom. The graphical lasso method is one such approach. However, such methods require signals with normal distributions (i.e., the values of all timepoints behave as if sampled from a random variable). Due to the slow hemodynamic response, fMRI signal is autocorrelated for several seconds, leading to a non-normal distribution of signal. To satisfy requirements for signal normality, data can be “prewhitened” by using autoregressive approaches to eliminate the dependency between successive timepoints However, it is not easy to implement such approaches without also distributing the effects of (temporally limited) artifact throughout the timeseries. Additionally, since low-frequency modulation of BOLD is the signal of interest, care must be taken that removing the effects of prior timepoints on a current timepoint does not also remove the low-frequency modulations of interest. It should be noted that the effects of autocorrelation impact the statistics of all edge measures since autocorrelation decreases the actual (effective) degrees of freedom relative to the apparent degrees of freedom. For example, if 600 seconds of data are obtained, but hemodynamic autocorrelation spans 6 seconds, there are roughly 100 degrees of freedom in the data, regardless of how rapidly the data are sampled. Clearly, complicated and difficult issues arise when defining network edges. Because most work on large-scale brain networks has used Pearson correlations, we will focus on issues related to edges defined by this measure.
Interpreting network representations of the brain
The point of a network analysis is to discover properties of the system, presumably ones that would have been difficult or impossible to find using other approaches. A large number of properties can be calculated in graph theory (see (Rubinov and Sporns, 2010) for review). Of these, we consider 3 for the purposes of discussion: node degree (the number of edges on a node), the participation coefficient of a node (the extent to which a node’s edges are distributed among many clusters versus limited to the node’s own cluster), and the path length between two nodes (how many nodes lie between two nodes). In the context of the subway system, the meaning of these measures is unambiguous: a station with high degree has many sets of tracks or trains, a station with high participation coefficient links many segregated parts of the subway system (clusters, probably different subway lines), and a pair of stations with a high path length are separated by many stations (and likely much track).
Interpreting these properties in a Pearson correlation network is not straightforward. Recall that the Pearson correlation between the resting state fMRI signals of two brain regions very likely reflects both directly shared and many mediated processes involving the regions. On this view, two of the measures, degree and path length, become hard to interpret. Path length is supposed to describe the distance between two nodes, measured by the number (or weights) of edges that must be traversed to travel between nodes. If the correlation coefficient is substantially affected by mediated relationships, the idea of measuring serial steps between nodes becomes problematic. Indeed, the correlation itself can be viewed as a path length of sorts, though the existence of negative correlations complicates this view, since one would not view negative relationships as “less” than nothing (and actually, one suspects that negatively correlated regions are more related than regions with correlations near zero). In short, though it is not unreasonable to think that information might pass serially along chains of highly correlated nodes, path length measures are not nearly as easy to interpret in resting state correlation networks as they are in many other types of networks.
Node degree is used in many networks to identify influential nodes: an airport with many flights is likely more important than one with few flights, and a person with many sexual partners is more likely to acquire and spread disease than a person with few partners. Node degree, however, can also be complicated to interpret in resting state correlation networks. The interpretation is complex because, unlike in transportation or communication networks, the degree of a node correlates with cluster size in correlation networks (because most nodes in a cluster all correlate with each other) (Power et al., 2013). In other words, in a correlation network, a node with high degree may not be an especially influential node, but rather may just be part of a large cluster. At the voxel level, voxel degree will be influenced by area size, topographic influences, and system size. In accord with this view, the voxels with the highest degree in resting state fMRI data are found in the posterior cingulate, the angular gyrus, and ventromedial prefrontal cortex, all members of the largest resting state cluster in the brain, representing the default mode system (Buckner et al., 2009; Power et al., 2013).
In contrast with the previous two measures, the meaning of participation coefficient seems interpretable in resting state data. A node with a low participation coefficient correlates strongly mainly with other nodes in its cluster, whereas a node with high participation coefficient correlates strongly with nodes in multiple clusters. Nodes with high participation coefficients would therefore seem to be participating in several types of processes that are segregated into and represented within different systems. In contrast, nodes with low participation coefficients would seem to be participating in a more restricted set of processes that are found mainly in a single system. Note that participation coefficients depend upon cluster definition, and that various network definitions can yield quite different clustering (Figure 5).
From the discussion above, it should be clear that complicated issues arise both in the formation and interpretation of resting state networks. A great variety of measures can be and have been calculated in resting state networks, with many reported features of the networks (e.g., “hubs” measured by degree centrality or other centrality measures) and many reported differences across clinical diagnoses and the lifespan (see (Bullmore and Sporns, 2012; Sporns, 2014) for review). Most of these measures have no proven biological interpretation. A critical challenge for the field is to move from simply reporting mathematical features of resting state networks to translating these features into the realities of behavior and neurobiology. For example, given a “hub”, what are the expected properties of the processing performed at that region? What are the behavioral correlates of these processes? What specific predictions do these measures make that can be falsified or verified? More links between behavior, processing operations, and network properties are needed to refine our understanding of how to interpret network properties reported in resting state networks.
Conclusions
Resting state functional connectivity analysis has grown from an unexpected observation in fMRI “noise” into a major area of human neuroimaging. Acquiring resting state data is relatively uncomplicated, but analysis of the data is not. Major questions remain: What is the origin, and what are the functions, of the spontaneous activity reflected in low-frequency BOLD signal? How does the low-frequency activity relate to instantaneous activity, perceptions, thoughts, and behavior? What are the genetic influences on the spatial and temporal organization of resting state signals? How do the signals evolve over the lifespan? Partial answers to some of these questions exist, but much work remains.
Highlights.
Spontaneous fMRI signal correlations reflect aspects of brain organization
These signals can be used to characterize aspects of area-level brain organization
These signals can be used to delineate some aspects of supra-areal organization
These signals enable construction of network representations of much of the brain
Acknowledgments
We thank Tim Laumann and Steve Nelson for comments and help with Figures. This work was supported by NIH F30 MH940322 (JDP) and by the Intellectual and Developmental Disabilities Research Center at Washington University (NIH/NICHD P30 HD062171).
Footnotes
The authors have no conflict of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jonathan D Power, Email: jonathan.power@nih.gov.
Bradley L Schlaggar, Email: schlaggarb@neuro.wustl.edu.
Steven E Petersen, Email: sep@npg.wustl.edu.
Bibliography
- Adachi Y, Osada T, Sporns O, Watanabe T, Matsui T, Miyamoto K, Miyashita Y. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb Cortex. 2012;22:1586–1592. doi: 10.1093/cercor/bhr234. [DOI] [PubMed] [Google Scholar]
- Amunts K, Lepage C, Borgeat L, Mohlberg H, Dickscheid T, Rousseau ME, Bludau S, Bazin PL, Lewis LB, Oros-Peusquens AM, et al. BigBrain: an ultrahigh-resolution 3D human brain model. Science. 2013;340:1472–1475. doi: 10.1126/science.1235381. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Druzgal TJ, Lopez-Larson M, Jeong EK, Desai K, Yurgelun-Todd D. Network anticorrelations, global regression, and phase-shifted soft tissue correction. Hum Brain Mapp. 2011;32:919–934. doi: 10.1002/hbm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes KA, Nelson SM, Cohen AL, Power JD, Coalson RS, Miezin FM, Vogel AC, Dubis JW, Church JA, Petersen SE, Schlaggar BL. Parcellation in left lateral parietal cortex is similar in adults and children. Cereb Cortex. 2012;22:1148–1158. doi: 10.1093/cercor/bhr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp MS, Argall BD, Bodurka J, Duyn JH, Martin A. Unraveling multisensory integration: patchy organization within human STS multisensory cortex. Nat Neurosci. 2004;7:1190–1192. doi: 10.1038/nn1333. [DOI] [PubMed] [Google Scholar]
- Beckmann CF. Modelling with independent components. NeuroImage. 2012;62:891–901. doi: 10.1016/j.neuroimage.2012.02.020. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos T Roy Soc B. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. NeuroImage. 2008;40:644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Blumensath T, Jbabdi S, Glasser MF, Van Essen DC, Ugurbil K, Behrens TE, Smith SM. Spatially constrained hierarchical parcellation of the brain with resting-state fMRI. NeuroImage. 2013;76:313–324. doi: 10.1016/j.neuroimage.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright MG, Murphy K. Removing motion and physiological artifacts from intrinsic BOLD fluctuations using short echo data. NeuroImage. 2013;64:526–537. doi: 10.1016/j.neuroimage.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Yeo BT. Opportunities and limitations of intrinsic functional connectivity MRI. Nat Neurosci. 2013;16:832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Logothetis N, Singer W. Scaling brain size, keeping timing: evolutionary preservation of brain rhythms. Neuron. 2013;80:751–764. doi: 10.1016/j.neuron.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell F, Bellec P, Shmuel A. Global and system-specific resting-state fMRI fluctuations are uncorrelated: principal component analysis reveals anti-correlated networks. Brain Connect. 2011;1:496–510. doi: 10.1089/brain.2011.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell F, Bellec P, Shmuel A. Quantification of the impact of a confounding variable on functional connectivity confirms anti-correlated networks in the resting-state. NeuroImage. 2014;86:343–353. doi: 10.1016/j.neuroimage.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Chai XJ, Castañón AN, Ongür D, Whitfield-Gabrieli S. Anticorrelations in resting state networks without global signal regression. NeuroImage. 2012;59:1420–1428. doi: 10.1016/j.neuroimage.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Effects of model-based physiological noise correction on default mode network anti-correlations and correlations. NeuroImage. 2009a;47:1448–1459. doi: 10.1016/j.neuroimage.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. NeuroImage. 2009b;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Wendt GJ, Turski PA, Moritz CH, Quigley MA, Meyerand ME. Mapping functionally related regions of brain with functional connectivity MR imaging. AJNR Am J Neuroradiol. 2000;21:1636–1644. [PMC free article] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, 3rd, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33:1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock RC, Jbabdi S, Yan CG, Vogelstein JT, Castellanos FX, Di Martino A, Kelly C, Heberlein K, Colcombe S, Milham MP. Imaging human connectomes at the macroscale. Nat methods. 2013;10:524–539. doi: 10.1038/nmeth.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groof G, Jonckers E, Gunturkun O, Denolf P, Van Auderkerke J, Van der Linden A. Functional MRI and functional connectivity of the visual system of awake pigeons. Behav Brain Res. 2013;239:43–50. doi: 10.1016/j.bbr.2012.10.044. [DOI] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci. 2011;12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Dosenbach NU, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RA, Fox MD, Snyder AZ, Vincent JL, Raichle ME, et al. Distinct brain networks for adaptive and stable task control in humans. Proc Natl Acad Sci U S A. 2007;104:11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NU, Visscher KM, Palmer ED, Miezin FM, Wenger KK, Kang HC, Burgund ED, Grimes AL, Schlaggar BL, Petersen SE. A core system for the implementation of task sets. Neuron. 2006;50:799–812. doi: 10.1016/j.neuron.2006.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet G, Naveau M, Petit L, Delcroix N, Zago L, Crivello F, Jobard G, Tzourio-Mazoyer N, Mazoyer B, Mellet E, Joliot M. Brain activity at rest: a multiscale hierarchical functional organization. J Neurophysiol. 2011;105:2753–2763. doi: 10.1152/jn.00895.2010. [DOI] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Power JD, Dosenbach NUF, Church JA, Miezin FM, Schlaggar BL, Petersen SE. Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol. 2009;5:e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Dosenbach NUF, Church JA, Cohen AL, Brahmbhatt S, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. Development of distinct control networks through segregation and integration. Proc Natl Acad Sci U S A. 2007;104:13507–13512. doi: 10.1073/pnas.0705843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg DA, Yacoub E. The rapid development of high speed, resolution and precision in fMRI. NeuroImage. 2012;62:720–725. doi: 10.1016/j.neuroimage.2012.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci U S A. 2006a;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Zacks JM, Raichle ME. Coherent spontaneous activity accounts for trial-to-trial variability in human evoked brain responses. Nat Neurosci. 2006b;9:23–25. doi: 10.1038/nn1616. [DOI] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J Neurophysiol. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P. Spontaneous low-frequency BOLD signal fluctuations: an fMRI investigation of the resting-state default mode of brain function hypothesis. Hum Brain Mapp. 2005;26:15–29. doi: 10.1002/hbm.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R. Movement-related effects in fMRI time-series. Magn Reson Med. 1996;35:346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Goni J, van den Heuvel MP, Avena-Koenigsberger A, Velez de Mendizabal N, Betzel RF, Griffa A, Hagmann P, Corominas-Murtra B, Thiran JP, Sporns O. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci U S A. 2014;111:833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C, Nomura EM, Perez F, D’Esposito M. Focal brain lesions to critical locations cause widespread disruption of the modular organization of the brain. J Cogn Neurosci. 2012;24:1275–1285. doi: 10.1162/jocn_a_00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L, Salimi-Khorshidi G, Beckmann CF, Auerbach EJ, Douaud G, Sexton CE, Zsoldos E, Ebmeier KP, Filippini N, Mackay CE, et al. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. NeuroImage. 2014;95:232–247. doi: 10.1016/j.neuroimage.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Carrillo B, Mackey AP, Bunge SA. Resting-State fMRI: A Window into Human Brain Plasticity. Neuroscientist. 2014 doi: 10.1177/1073858414524442. [DOI] [PubMed] [Google Scholar]
- Hampson M, Driesen N, Roth JK, Gore JC, Constable RT. Functional connectivity between task-positive and task-negative brain areas and its relation to working memory performance. Magn Reson Med. 2010;28:1051–1057. doi: 10.1016/j.mri.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Peterson BS, Skudlarski P, Gatenby JC, Gore JC. Detection of functional connectivity using temporal correlations in MR images. Hum Brain Mapp. 2002;15:247–262. doi: 10.1002/hbm.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Liu TT. A geometric view of global signal confounds in resting-state functional MRI. NeuroImage. 2012;59:2339–2348. doi: 10.1016/j.neuroimage.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, Zhu C, Gong Q, Zang Y, Evans AC. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PloS One. 2009;4:e5226. doi: 10.1371/journal.pone.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb DO. The organization of behavior: a neuropsychological theory. Wiley; 1949. [Google Scholar]
- Heine L, Soddu A, Gomez F, Vanhaudenhuyse A, Tshibanda L, Thonnard M, Charland-Verville V, Kirsch M, Laureys S, Demertzi A. Resting state networks and consciousness: alterations of multiple resting state network connectivity in physiological, pharmacological, and pathological consciousness States. Front Psychol. 2012;3:295. doi: 10.3389/fpsyg.2012.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Braun AR, Carr WS, Picchioni D, Balkin TJ, Fukunaga M, Duyn JH. Decoupling of the brain’s default mode network during deep sleep. Proc Natl Acad Sci U S A. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Gallivan JP, Culham JC, Gati JS, Menon RS, Everling S. Functional connectivity of the frontal eye fields in humans and macaque monkeys investigated with resting-state fMRI. J Neurophysiol. 2012;107:2463–2474. doi: 10.1152/jn.00891.2011. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Leung LS, Mirsattari SM, Gati JS, Menon RS, Everling S. Resting-state networks in the macaque at 7 T. NeuroImage. 2011;56:1546–1555. doi: 10.1016/j.neuroimage.2011.02.063. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Mirsattari SM, Jones CK, Gati JS, Leung LS. Functional networks in the anesthetized rat brain revealed by independent component analysis of resting-state FMRI. J Neurophysiol. 2010;103:3398–3406. doi: 10.1152/jn.00141.2010. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J, et al. Dynamic functional connectivity: promise, issues, and interpretations. NeuroImage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. NeuroImage. 2010;52:571–582. doi: 10.1016/j.neuroimage.2010.04.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE, Robson MD, Drobnjak I, Rushworth MF, Brady JM, Smith SM, Higham DJ, Matthews PM. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc Natl Acad Sci U S A. 2004;101:13335–13340. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TEJ. Diffusion MRI: From Quantitative Measurement to In vivo Neuroanatomy. Academic Press; 2013. [Google Scholar]
- Jonckers E, Delgado YPR, Shah D, Guglielmetti C, Verhoye M, Van der Linden A. Different anesthesia regimes modulate the functional connectivity outcome in mice. Magn Reson Med. 2013 doi: 10.1002/mrm.24990. [DOI] [PubMed] [Google Scholar]
- Jonckers E, Van Audekerke J, De Visscher G, Van der Linden A, Verhoye M. Functional connectivity fMRI of the rodent brain: comparison of functional connectivity networks in rat and mouse. PloS One. 2011;6:e18876. doi: 10.1371/journal.pone.0018876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH. Evolution of columns, modules, and domains in the neocortex of primates. Proc Natl Acad Sci U S A. 2012;109(Suppl 1):10655–10660. doi: 10.1073/pnas.1201892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CJ, Bickel S, Honey CJ, Groppe DM, Entz L, Craddock RC, Lado FA, Kelly C, Milham M, Mehta AD. Neurophysiological investigation of spontaneous correlated and anticorrelated fluctuations of the BOLD signal. J Neurosci. 2013;33:6333–6342. doi: 10.1523/JNEUROSCI.4837-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AM, Di Martino A, Uddin LQ, Shehzad Z, Gee DG, Reiss PT, Margulies DS, Castellanos FX, Milham MP. Development of anterior cingulate functional connectivity from late childhood to early adulthood. Cereb Cortex. 2009;19:640–657. doi: 10.1093/cercor/bhn117. [DOI] [PubMed] [Google Scholar]
- Koldewyn K, Yendiki A, Weigelt S, Gweon H, Julian J, Richardson H, Malloy C, Saxe R, Fischl B, Kanwisher N. Differences in the right inferior longitudinal fasciculus but no general disruption of white matter tracts in children with autism spectrum disorder. Proc Natl Acad Sci U S A. 2014;111:1981–1986. doi: 10.1073/pnas.1324037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Wheeler ME, Donaldson DI, Buckner RL. Neural correlates of episodic retrieval success. NeuroImage. 2000;12:276–286. doi: 10.1006/nimg.2000.0614. [DOI] [PubMed] [Google Scholar]
- Krienen FM, Buckner RL. Segregated fronto-cerebellar circuits revealed by intrinsic functional connectivity. Cereb Cortex. 2009;19:2485–2497. doi: 10.1093/cercor/bhp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Brenowitz ND, Voon V, Worbe Y, Vértes PE, Inati SJ, Saad ZS, Bandettini PA, Bullmore ET. Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proc Natl Acad Sci U S A. 2013;110:16187–16192. doi: 10.1073/pnas.1301725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior LJ, Power JD, Vincent JL, Nolan TS, Coalson RS, Zempel J, Snyder AZ, Schlaggar BL, Raichle ME, Petersen SE. Modulation of the brain’s functional network architecture in the transition from wake to sleep. Prog Brain Res. 2011;193:277–294. doi: 10.1016/B978-0-444-53839-0.00018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs H, Krakow K, Sterzer P, Eger E, Beyerle A, Salek-Haddadi A, Kleinschmidt A. Electroencephalographic signatures of attentional and cognitive default modes in spontaneous brain activity fluctuations at rest. Proc Natl Acad Sci U S A. 2003;100:11053–11058. doi: 10.1073/pnas.1831638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Smyser CD, Shimony JS. Resting-state fMRI: a review of methods and clinical applications. AJNR Am J Neuroradiol. 2013;34:1866–1872. doi: 10.3174/ajnr.A3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux L, Salek-Haddadi A, Lund TE, Laufs H, Carmichael D. Modelling large motion events in fMRI studies of patients with epilepsy. Magn Reson Imaging. 2007;25:894–901. doi: 10.1016/j.mri.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Leopold DA, Murayama Y, Logothetis NK. Very slow activity fluctuations in monkey visual cortex: implications for functional brain imaging. Cereb Cortex. 2003;13:422–433. doi: 10.1093/cercor/13.4.422. [DOI] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N. Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J Neurosci. 2011;31:3776–3783. doi: 10.1523/JNEUROSCI.4557-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JV, Hirano Y, Nascimento GC, Stefanovic B, Leopold DA, Silva AC. fMRI in the awake marmoset: somatosensory-evoked responses, functional connectivity, and comparison with propofol anesthesia. NeuroImage. 2013;78:186–195. doi: 10.1016/j.neuroimage.2013.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412:150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Lowe MJ, Mock BJ, Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. NeuroImage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Lu H, Zou Q, Gu H, Raichle ME, Stein EA, Yang Y. Rat brains also have a default mode network. Proc Natl Acad Sci U S A. 2012;109:3979–3984. doi: 10.1073/pnas.1200506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Liu H, Zhang M, Wang D, Cao Y, Ma Q, Rong D, Wang X, Buckner RL, Li K. Focal pontine lesions provide evidence that intrinsic functional connectivity reflects polysynaptic anatomical pathways. J Neurosci. 2011;31:15065–15071. doi: 10.1523/JNEUROSCI.2364-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey AP, Miller Singley AT, Bunge SA. Intensive reasoning training alters patterns of brain connectivity at rest. J Neurosci. 2013;33:4796–4803. doi: 10.1523/JNEUROSCI.4141-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Harms MP, Snyder AZ, Jenkinson M, Wilson JA, Glasser MF, Barch DM, Archie KA, Burgess GC, Ramaratnam M, et al. Human Connectome Project informatics: quality control, database services, and data visualization. NeuroImage. 2013;80:202–219. doi: 10.1016/j.neuroimage.2013.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx M, Pauly KB, Chang C. A novel approach for global noise reduction in resting-state fMRI: APPLECOR. NeuroImage. 2013;64:19–31. doi: 10.1016/j.neuroimage.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazoyer B, Zago L, Mellet E, Bricogne S, Etard O, Houde O, Crivello F, Joliot M, Petit L, Tzourio-Mazoyer N. Cortical networks for working memory and executive functions sustain the conscious resting state in man. Brain Res Bull. 2001;54:287–298. doi: 10.1016/s0361-9230(00)00437-8. [DOI] [PubMed] [Google Scholar]
- Menon RS, Ogawa S, Strupp JP, Ugurbil K. Ocular dominance in human V1 demonstrated by functional magnetic resonance imaging. J Neurophysiol. 1997;77:2780–2787. doi: 10.1152/jn.1997.77.5.2780. [DOI] [PubMed] [Google Scholar]
- Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. NeuroImage. 2009;44:715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- Moeller S, Nallasamy N, Tsao DY, Freiwald WA. Functional connectivity of the macaque brain across stimulus and arousal states. J Neurosci. 2009;29:5897–5909. doi: 10.1523/JNEUROSCI.0220-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Bandettini PA. Resting-state fMRI confounds and cleanup. NeuroImage. 2013;80:349–359. doi: 10.1016/j.neuroimage.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? NeuroImage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Arnold KM, Gilmore AW, McDermott KB. Neural signatures of test-potentiated learning in parietal cortex. J Neurosci. 2013;33:11754–11762. doi: 10.1523/JNEUROSCI.0960-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, Velanova K, Donaldson DI, Phillips JS, Schlaggar BL, Petersen SE. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010a;67:156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Dosenbach NU, Cohen AL, Wheeler ME, Schlaggar BL, Petersen SE. Role of the anterior insula in task-level control and focal attention. Brain Struct Funct. 2010b;214:669–680. doi: 10.1007/s00429-010-0260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing J, Ebisch B, Schmidt KE, Niessing M, Singer W, Galuske RA. Hemodynamic signals correlate tightly with synchronized gamma oscillations. Science. 2005;309:948–951. doi: 10.1126/science.1110948. [DOI] [PubMed] [Google Scholar]
- Patriat R, Molloy EK, Meier TB, Kirk GR, Nair VA, Meyerand ME, Prabhakaran V, Birn RM. The effect of resting condition on resting-state fMRI reliability and consistency: a comparison between resting with eyes open, closed, and fixated. NeuroImage. 2013;78:463–473. doi: 10.1016/j.neuroimage.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picchioni D, Duyn JH, Horovitz SG. Sleep and the functional connectome. NeuroImage. 2013;80:387–396. doi: 10.1016/j.neuroimage.2013.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Lessov-Schlaggar CN, Petersen SE. Evidence for hubs in human functional brain networks. Neuron. 2013;79:798–813. doi: 10.1016/j.neuron.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AR, 3rd, Hevey MA, Wallace MT. Neural correlates of multisensory perceptual learning. J Neurosci. 2012;32:6263–6274. doi: 10.1523/JNEUROSCI.6138-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. Two views of brain function. Trends Cogn Sci. 2010;14:180–190. doi: 10.1016/j.tics.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Mintun MA. Brain work and brain imaging. Annu Rev Neurosci. 2006;29:449–476. doi: 10.1146/annurev.neuro.29.051605.112819. [DOI] [PubMed] [Google Scholar]
- Rehme AK, Eickhoff SB, Grefkes C. State-dependent differences between functional and effective connectivity of the human cortical motor system. NeuroImage. 2013;67:237–246. doi: 10.1016/j.neuroimage.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sadaghiani S, Hesselmann G, Friston KJ, Kleinschmidt A. The relation of ongoing brain activity, evoked neural responses, and cognition. Front Syst Neurosci. 2010;4:20. doi: 10.3389/fnsys.2010.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. NeuroImage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Ruparel K, Erus G, Elliott MA, Eickhoff SB, Gennatas ED, Jackson C, Prabhakaran K, Smith A, et al. Heterogeneous impact of motion on fundamental patterns of developmental changes in functional connectivity during youth. NeuroImage. 2013;83:45–57. doi: 10.1016/j.neuroimage.2013.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholvinck ML, Leopold DA, Brookes MJ, Khader PH. The contribution of electrophysiology to functional connectivity mapping. NeuroImage. 2013;80:297–306. doi: 10.1016/j.neuroimage.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci U S A. 2010;107:10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Liu H, Talukdar T, Martincorena I, Yeo BT, Buckner RL. The organization of local and distant functional connectivity in the human brain. PLoS Comput Biol. 2010;6:e1000808. doi: 10.1371/journal.pcbi.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sforazzini F, Schwarz AJ, Galbusera A, Bifone A, Gozzi A. Distributed BOLD and CBV-weighted resting-state networks in the mouse brain. NeuroImage. 2014;87:403–415. doi: 10.1016/j.neuroimage.2013.09.050. [DOI] [PubMed] [Google Scholar]
- Shannon BJ, Buckner RL. Functional-anatomic correlates of memory retrieval that suggest nontraditional processing roles for multiple distinct regions within posterior parietal cortex. J Neurosci. 2004;24:10084–10092. doi: 10.1523/JNEUROSCI.2625-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Papademetris X, Constable RT. Graph-theory based parcellation of functional subunits in the brain from resting-state fMRI data. NeuroImage. 2010;50:1027–1035. doi: 10.1016/j.neuroimage.2009.12.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel A, Leopold DA. Neuronal correlates of spontaneous fluctuations in fMRI signals in monkey visual cortex: Implications for functional connectivity at rest. Hum Brain Mapp. 2008;29:751–761. doi: 10.1002/hbm.20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GL, Fiez JA, Corbetta M, Buckner RL, Miezin FM, Raichle ME, Petersen SE. Common Blood Flow Changes across Visual Tasks: II. Decreases in Cerebral Cortex. J Cogn Neurosci. 1997;9:648–663. doi: 10.1162/jocn.1997.9.5.648. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. NeuroImage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Sorg C, Riedl V, Mühlau M, Calhoun VD, Eichele T, Läer L, Drzezga A, Förstl H, Kurz A, Zimmer C, Wohlschläger AM. Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17:652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- Stein T, Moritz C, Quigley M, Cordes D, Haughton V, Meyerand E. Functional connectivity in the thalamus and hippocampus studied with functional MR imaging. AJNR Am J Neuroradiol. 2000;21:1397–1401. [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Musen M, Menon V. Development of large-scale functional brain networks in children. PLoS Biol. 2009;7:e1000157. doi: 10.1371/journal.pbio.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H. Decoding Wakefulness Levels from Typical fMRI Resting-State Data Reveals Reliable Drifts between Wakefulness and Sleep. Neuron. 2014;82:695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Tyszka JM, Kennedy DP, Paul LK, Adolphs R. Largely typical patterns of resting-state functional connectivity in high-functioning adults with autism. Cereb Cortex. 2014;24:1894–1905. doi: 10.1093/cercor/bht040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahdat S, Darainy M, Milner TE, Ostry DJ. Functionally specific changes in resting-state sensorimotor networks after motor learning. J Neurosci. 2011;31:16907–16915. doi: 10.1523/JNEUROSCI.2737-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven VG, Formisano E, Prvulovic D, Roeder CH, Linden DEJ. Functional connectivity as revealed by spatial independent component analysis of fMRI measurements during rest. Hum Brain Mapp. 2004;22:165–178. doi: 10.1002/hbm.20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. NeuroImage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J. Cortical parcellations of the macaque monkey analyzed on surface-based atlases. Cereb Cortex. 2012a;22:2227–2240. doi: 10.1093/cercor/bhr290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J, Coalson T. Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb Cortex. 2012b;22:2241–2262. doi: 10.1093/cercor/bhr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Wagner AD, Shannon BJ, Kahn I, Buckner RL. Parietal lobe contributions to episodic memory retrieval. Trends Cogn Sci. 2005;9:445–453. doi: 10.1016/j.tics.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C. Correlations and anticorrelations in resting-state functional connectivity MRI: a quantitative comparison of preprocessing strategies. NeuroImage. 2009;47:1408–1416. doi: 10.1016/j.neuroimage.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Cohen AL, Power JD, Nelson SM, Glasser MF, Miezin FM, Snyder AZ, Schlaggar BL, Petersen SE. Parcellating an Individual Subject’s Cortical and Subcortical Brain Structures Using Snowball Sampling of Resting-State Correlations. Cereb Cortex. 2014a;24:2036–2054. doi: 10.1093/cercor/bht056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Petersen SE. An approach for parcellating human cortical areas using resting-state correlations. NeuroImage. 2014b;93(Pt 2):276–291. doi: 10.1016/j.neuroimage.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Schlaggar BL, Petersen SE. Concepts and principles in the analysis of brain networks. Ann N Y Acad Sci. 2011;1224:126–146. doi: 10.1111/j.1749-6632.2010.05947.x. [DOI] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. NeuroImage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Xiong J, Parsons LM, Gao JH, Fox PT. Interregional connectivity to primary motor cortex revealed using MRI resting state images. Hum Brain Mapp. 1999;8:151–156. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<151::AID-HBM13>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub E, Harel N, Ugurbil K. High-field fMRI unveils orientation columns in humans. Proc Natl Acad Sci U S A. 2008;105:10607–10612. doi: 10.1073/pnas.0804110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CG, Cheung B, Kelly C, Colcombe S, Craddock RC, Di Martino A, Li Q, Zuo XN, Castellanos FX, Milham MP. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. NeuroImage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Chang C, Xu T, Jiang L, Handwerker DA, Castellanos FX, Milham MP, Bandettini PA, Zuo XN. Connectivity trajectory across lifespan differentiates the precuneus from the default network. NeuroImage. 2014;89:45–56. doi: 10.1016/j.neuroimage.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendiki A, Koldewyn K, Kakunoori S, Kanwisher N, Fischl B. Spurious group differences due to head motion in a diffusion MRI study. NeuroImage. 2013;88C:79–90. doi: 10.1016/j.neuroimage.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]