SUMMARY
Accurate control of the Ran GTPase cycle depends on the regulated activity of RCC1, Ran’s nucleotide exchange factor. RanBP1 has been characterized as a co-activator of the Ran GTPase activating protein, RanGAP1. RanBP1 can also form a stable complex with Ran and RCC1, although the dynamics and function of this complex remain poorly understood. Here we show that formation of the heterotrimeric RCC1/Ran/RanBP1 complex in M-phase Xenopus egg extracts controls both RCC1’s enzymatic activity and partitioning between the chromatin-bound and soluble pools of RCC1. This mechanism is critical for spatial control of Ran-GTP gradients that guide mitotic spindle assembly. Moreover, phosphorylation of RanBP1 drives changes in the dynamics of chromatin-bound RCC1 pools at the metaphase-anaphase transition. Our findings reveal an important mitotic role for RanBP1, controlling the spatial distribution and magnitude of mitotic Ran-GTP production and thereby ensuring accurate execution of Ran-dependent mitotic events.
INTRODUCTION
The Ran GTPase that plays critical roles in multiple cellular processes including nucleocytoplasmic transport, nuclear envelope (NE) assembly, and mitotic spindle assembly (Clarke and Zhang, 2008). In interphase, GTP-bound Ran (Ran-GTP) is concentrated within the nucleus, while Ran GDP-bound (Ran-GDP) is predominant in the cytoplasm. This asymmetrical distribution drives transport between the nucleus and cytoplasm by regulating cargo binding and release of a family of Ran-GTP-binding transport receptors that are collectively called karyopherins. After mitotic NE breakdown, Ran-GTP is concentrated near mitotic chromatin, while the majority of Ran distal to chromosomes is GDP-bound. The presence of such a chromatin-centered Ran-GTP gradient has been visualized in both M-phase Xenopus laevis Egg Extracts (XEE) (Kalab et al., 2002) and mitotic somatic cells (Kalab et al., 2006). The mitotic Ran-GTP gradient guides mitotic spindle assembly by releasing spindle assembly factors (SAFs) from karyopherins in a spatially regulated manner (Clarke and Zhang, 2008).
The conversion of Ran-GDP to Ran-GTP is catalyzed by a Ran-specific guanine exchange factor (RanGEF), called RCC1 (Regulator of chromosome condensation 1), whose binding to chromatin determines the asymmetrical distribution of Ran-GTP throughout the cell cycle (Nemergut et al., 2001). The association of RCC1 to chromatin changes dramatically as XEE progress through mitosis, with large increases in the amount of chromatin-bound RCC1 shortly after the metaphase-anaphase transition (Arnaoutov and Dasso, 2003). Mammalian RCC1 also shows changes in chromatin binding during the metaphase-anaphase window (Hutchins et al., 2004). The mechanisms underlying altered association of RCC1 to chromatin during anaphase are poorly understood. Nevertheless, the fact that elevated levels of RCC1 can disrupt kinetochore structures and spindle assembly checkpoint (SAC) signaling (Arnaoutov and Dasso, 2003) suggests that the dynamics of RCC1 have important functional consequences.
RanBP1 is a Ran-GTP-binding protein (Beddow et al., 1995; Bischoff et al., 1995) whose in vivo function has been obscure. While RanBP1 is conserved between yeast and vertebrates, it is not found in some invertebrate species, such as flies and worms (Dasso, 2002). RanBP1 stimulates the enzymatic activity of Ran’s GTPase activating protein, RanGAP1, roughly ten fold within in vitro assays using purified proteins (Bischoff et al., 1995). In addition, karyopherins bind to Ran-GTP in a way that prevents its interaction with RanGAP1, but RanBP1 can release karyopherin binding and thereby allow RanGAP1-activated GTP hydrolysis on Ran (Bischoff and Gorlich, 1997; Lounsbury and Macara, 1997). RanBP1 also forms a stable heterotrimeric complex with Ran and RCC1 in vitro, strongly inhibiting RCC1’s RanGEF activity (Bischoff et al., 1995). The dynamics and potential functions of this RCC1/Ran/RanBP1 heterotrimeric complex (hereafter called the RRR complex) remain unresolved. Notably, RanBP1 is excluded from nuclei (Richards et al., 1996), preventing RRR complex formation within nucleoplasm and reducing RanBP1’s capacity to inhibit RCC1 during interphase. No such barrier prevents RRR complex formation in mitosis.
We have investigated the mitotic function and regulation of the RRR complex using XEE, a well established ex vivo model system for cell cycle studies (Arnaoutov and Dasso, 2003; Murray, 1991). We found that chromatin-based spindle assembly was defective when RCC1 was present without RanBP1, in a manner that could be corrected by restoration of RanBP1 to physiological levels. The interaction between RanBP1 and RCC1 within XEE determined both the partitioning of RCC1 between its chromatin-bound and unbound forms, as well as the level of RanGEF activity. Notably, RanBP1 was phosphorylated in a cell cycle-dependent manner, peaking in early anaphase. We mapped the mitotically phosphorylated residue of RanBP1, and tested whether this modification might control the mitotic dynamics of RCC1. Consistent with this idea, a phosphomimetic mutation of this site disrupted RRR complex assembly and promoted increased loading of RCC1 onto mitotic chromosomes, while a non-phosphorylatable RanBP1 mutant suppressed the normal fluctuations of RCC1-chromatin binding at anaphase onset. Together, our findings reveal an important mitotic role for RanBP1, controlling the spatial distribution and magnitude of the Ran-GTP gradient and thereby ensuring accurate execution of Ran-dependent mitotic events.
RESULTS
Balance between RCC1 and RanBP1 maintains normal spindle assembly
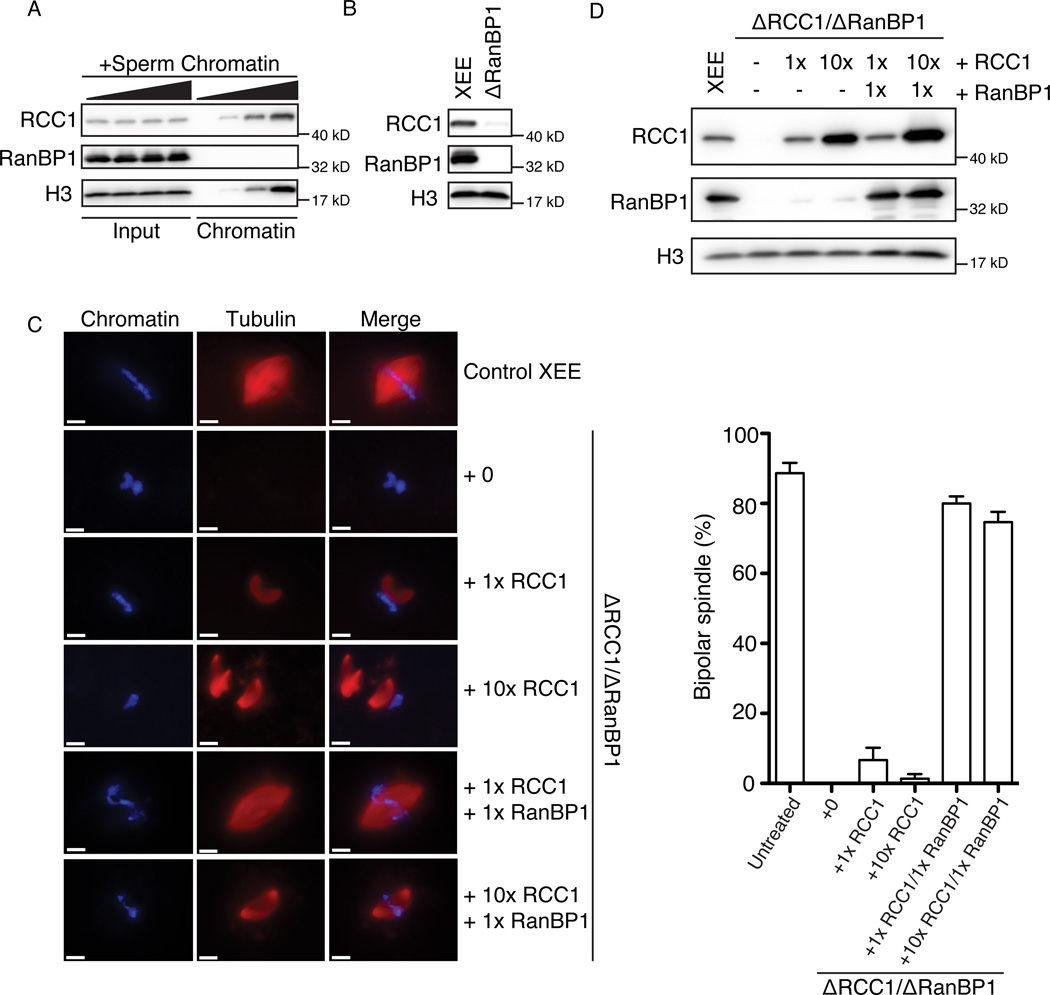
It is imaginable that somatic cells could maintain a mitotic chromosome-centered Ran-GTP gradient by simply localizing RCC1 through its binding to chromatin. On the other hand, fertilized Xenopus laevis eggs and other embryonic systems have a large stored excess of many nuclear proteins, allowing them to go through early cell cycles with only a minimal biosynthetic requirement (Dasso et al., 1992; Murray and Kirschner, 1989; Newport and Kirschner, 1982). These embryonic systems thus possess RCC1 in concentrations that grossly exceed the available chromosomal binding sites, resulting in large untethered pools of RCC1 (Dasso et al., 1992). To examine the distribution of RCC1 in metaphase-arrested XEE (CSF-XEE), we added increasing amount of demembraned sperm chromatin, which was allowed to assemble mitotic chromosomes that were later re-purified. As expected, Western blotting showed that the amount of RCC1 in the chromatin-bound fraction was proportional to the original concentration of added chromatin (Figure 1A). At low chromatin concentrations, the bulk of RCC1 was not chromatin-bound. Indeed, soluble pools of RCC1 can only be significantly depleted at very high concentrations of added sperm chromatin, comparable to chromosome-to-cytoplasm ratios attained during or after the midblastula transition (Newport and Kirschner, 1982). Chromatin binding causes a modest, two-fold stimulation of RCC1’s enzymatic activity (Nemergut et al., 2001); nevertheless, a large, unbound pool of active RCC1 should obscure a chromatin-centered Ran gradient. We therefore wondered whether the unbound pool of RCC1 might be inhibited, and postulated that the formation of the RRR complex (Bischoff et al., 1995) might be important in this context.
Figure 1. RanBP1 is required in normal spindle assembly by sequestering RCC1.
(A) Different concentrations (0, 1,000, 3,000 and 10,000 units/µl) of demembraned sperm chromatin were added and re-purified from each sample. Total CSF-XEE input (left half) and isolated chromatin (right half) were subjected to immunblottinged with antibodies against RCC1, RanBP1 and Histone H3.
(B) Mock treated CSF-XEE (left) or CSF-XEE depleted with anti-RanBP1 antibodies (right) were examined by immunoblotting with antibodies against RCC1, RanBP1 and Histone H3.
(C) Recombinant xRCC1 and/or xRanBP1 were added in to mock treated CSF-XEE or CSF-XEE depleted using anti-RanBP1 antibodies. Rhodamine-labeled α-tubulin (20 µg/ml) and demembraned chromatin (1,000 units/µl) were added. After 30 minutes at RT, aliquots of each reaction were fixed, stained with Hoechst 33342 and processed for fluorescent microscopy. Images were taken for chromatin (left, blue) and tubulin (middle, red). Scale bar, 10 µm. Spindles with chromatin in vicinity were counted as one structure. Percentage of bipolar spindles with chromatin correctly localizing at the mid-plate was plotted as mean ± SEM (N = 3 XEEs, 50 structures counted in each XEE).
(D) Samples as in (C) were subjected to immunoblotting with antibodies against RCC1, RanBP1, and Histone H3.
See also Figure S1.
The bulk of RCC1 within interphase XEE is tightly associated with RanBP1 (Pu and Dasso, 1997). We examined whether RCC1 is similarly associated with RanBP1 in CSF-XEE, and whether this might control its function in spindle assembly. We immunodepleted RanBP1 from CSF-XEE, and tested RCC1 levels before and after depletion by Western blotting. We observed that the majority of RCC1 (>90%) was co-depleted with RanBP1 (Figure 1B). Since RanBP1 stably binds RCC1 only in the presence of Ran (Bischoff et al., 1995), this finding suggests that in the absence of chromatin most RCC1 remains sequestered into the RRR complex in both interphase (Pu and Dasso, 1997) and M-phase (Figure 1B). We estimate that the endogenous RanBP1 concentration in XEE is over 40-fold (molar ratio) greater than RCC1 (Figure S1A), so that RanBP1 levels are more than sufficient to sequester RCC1 in RRR complexes. As in interphase (Pu and Dasso, 1997), less than 5% of the endogenous pool of Ran was depleted by anti-RanBP1 antibodies.
Co-depletion of RCC1 and RanBP1 allowed us to test whether the RRR complex is important for mitotic spindle assembly by adding back purified recombinant Xenopus RCC1 (xRCC1) and RanBP1 (xRanBP1) individually and together (Figure 1C,D). In control CSF-XEE, normal bipolar spindles assembled around sperm chromatin, with chromosomes aligned at a central metaphase plate (Figure 1C, row 1). CSF-XEE lacking both RanBP1 and RCC1 were incapable of assembling mitotic spindles around chromatin, as would be expected in the absence of RCC1 (Figure 1C, row 2; (Kalab and Heald, 2008)). Restoration of endogenous xRCC1 levels rescued the assembly of spindle microtubules (MTs), but these MTs consistently formed beside the chromatin, apparently without normal engagement of the MT network to the chromosomes (Figure 1C, row 3). Restoration of both xRCC1 and xRanBP1 to physiological concentrations rescued biopolar spindle assembly, with correct MT orientation and chromosomes alignment on the metaphase plate (Figure 1C, row 5). These findings confirm that RCC1 is essential for spindle assembly in CSF-XEE and further indicate an essential role of RanBP1 in spindle organization.
Undepleted CSF-XEE were relatively tolerant of excess RCC1: They assembled normal spindles when xRCC1 was added to concentrations ten-fold higher than endogenous levels (Figure S1B, row 1 to 3, S1C). When xRCC1 was added at 30-fold endogenous concentrations (Figure S1B, row 4 and 5, S1C), we observed defects in spindle organization similar to those occurring when xRCC1 was added at endogenous concentrations to depleted CSF-XEE lacking RanBP1 (Figure 1C, row 3). By contrast, depleted CSF-XEE to which we added recombinant xRCC1 at concentrations 10-fold higher than endogenous levels not only showed disengagement of spindle MTs from chromosomes but also MT asters and spindles without chromatin (Figure 1C, row 4), in a manner reminiscent of Ran-GTP-stimulated MT structures (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Zhang et al., 1999). These data suggest that CSF-XEE normally have a considerable capacity to buffer RCC1 activity in the presence of physiological levels of RanBP1, but that RCC1 can produce sufficient Ran-GTP to cause aberrant MT structures when RanBP1 is absent. Consistent with this idea, the formation of unanchored MT structures was abolished when xRanBP1 was reconstituted to physiological concentrations (Figure 1C, row 6).
A straightforward interpretation of these results is that a high concentration of RanBP1 buffers the activity of RCC1 in CSF-XEE through RRR complex assembly, so that levels of Ran-GTP sufficient to disregulate MT assembly can only be achieved at artificially high RCC1 concentrations that exceed this buffering capacity. In the absence of RanBP1, the amount of RCC1 required to cause aberrant MT assembly becomes dramatically lowered.
RCC1 is partitioned between chromatin and the RRR complex
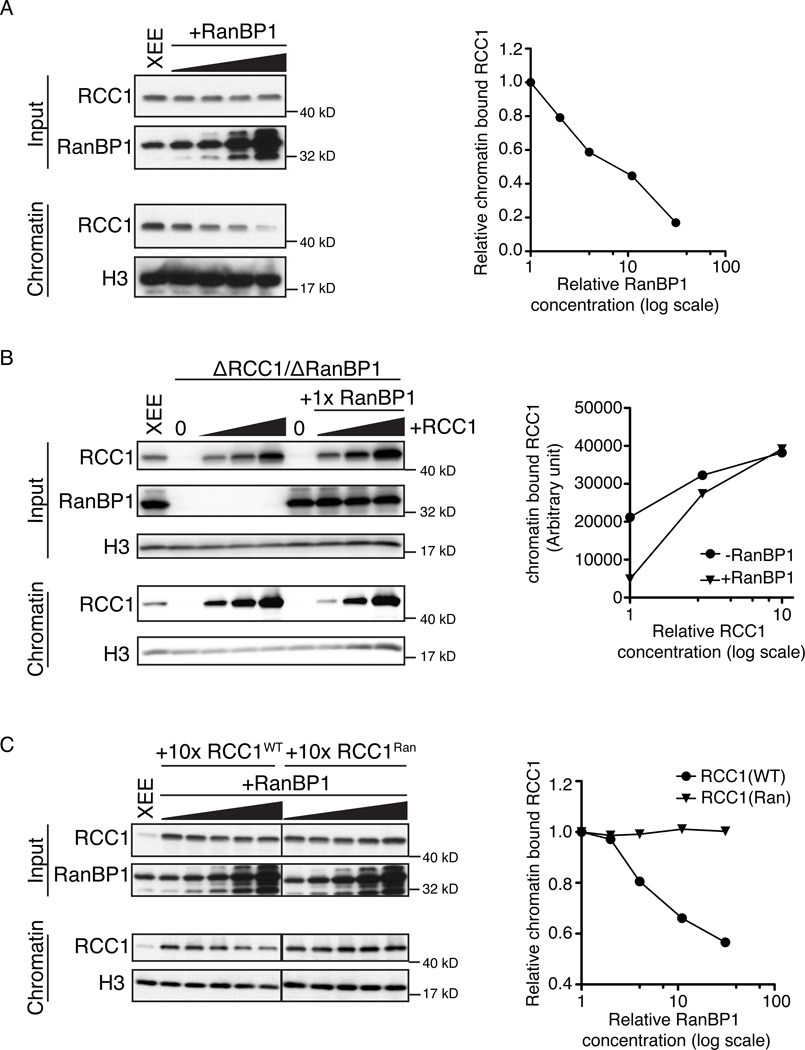
Endogenous RanBP1 did not associate with the chromatin fraction (Figure 1A), and no RanBP1 was detected on chromatin even when recombinant xRanBP1 was added to levels 10-fold higher than the endogenous RanBP1 (Figure S2A). These data suggested that the chromatin-bound and RRR-associated pools of RCC1 were mutually exclusive. To test whether RRR complex assembly and chromatin binding compete for RCC1, we added increasing amounts of xRanBP1 to otherwise untreated CSF-XEE containing a constant concentration of sperm chromatin. While the total level of RCC1 remained unchanged (Figure 2A, upper panels), progressively less RCC1 bound to chromatin as xRanBP1 concentrations increased (lower panels). To further test the capacity of RanBP1 to modulate RCC1 binding to chromatin, we compared the chromatin binding efficiency of RCC1 in the presence and absence of RanBP1, using RCC1- and RanBP1-depleted CSF-XEE. Increasing amounts of recombinant xRCC1 were added in the presence or absence of a constant level of recombinant xRanBP1 (Figure 2B, upper panel). We consistently found more RCC1 on chromatin in CSF-XEE without RanBP1 than in samples with RanBP1 (Figure 2B, lower panel), indicating that RRR complex assembly directly antagonized RCC1 chromatin binding.
Figure 2. RanBP1 competes with chromatin in binding RCC1.
(A) Increasing concentrations of recombinant xRanBP1 (0×, 1×, 3×, 10×, 30× endogenous RanBP1 level) were added to CSF-XEE. Reaction aliquots (upper panels) and isolated chromatin (lower panels) were subjected to immunoblotting with indicated antibodies. Relative chromatin bound RCC1 was plotted against RanBP1 concentration (folds of endogenous level).
(B) Recombinant xRCC1 was added at increasing concentrations (0×, 1×, 3×, 10× relative to endogenous RCC1) to CSF-XEE previously depleted using anti-RanBP1 antibodies. Each reaction was divided, and either buffer (left) or recombinant RanBP1 (right) were added. Reaction aliquots (upper panels) and isolated chromatin (lower panels) were subjected to immunoblotting with indicated antibodies. Chromatin bound RCC1 was plotted against RCC1 concentration (folds of endogenous level).
(C) Either recombinant wild type RCC1 (RCC1WT) or an RCC1 mutant that does not bind Ran (RCC1Ran) was added into CSF-XEE at 10× endogenous RCC1 levels. Recombinant xRanBP1 was added to either sample at increasing concentrations (0×, 1×, 3×, 10×, 30× relative to endogenous RanBP1). Reaction aliquots (upper panels) and isolated chromatin (lower panels) were subjected to immunoblotting with indicated antibodies. Relative chromatin bound RCC1 was plotted against RanBP1 concentration (folds of endogenous level).
In all panels, all reactions contained 10,000 units/µl demembraned sperm chromatin, and were incubated for 30 min at RT before being re-purified.
See also Figure S2.
To confirm that RanBP1 directly competes RCC1 from chromatin through RRR complex assembly, we compared the chromatin distribution of recombinant wild type xRCC1 (xRCC1WT) upon the addition of exogenous xRanBP1 to a mutant xRCC1 that does not bind Ran and that cannot form RRR complexes (xRCC1Ran; Figure S2B). xRCC1WT or xRCC1Ran were added to CSF-XEE at levels 10-fold higher than endogenous RCC1, and allowed to preload onto chromatin. Increasing amounts of recombinant xRanBP1 were then added (Figure 2C, upper panel). The amount of chromatin-bound xRCC1WT decreased with xRanBP1 addition, while the level of xRCC1Ran on chromatin remained constant (Figure 2C, lower panel), indicating that xRanBP1 caused the dissociation of RCC1 from chromatin specifically by RRR complex formation. Together, these findings indicate that formation of RRR complexes is antagonistic RCC1 chromatin binding, so that the changes in the concentration of RanBP1 modulate the partitioning of RCC1 between its chromatin bound and soluble pools in CSF-XEE.
RRR complex assembly suppresses RanGEF activity
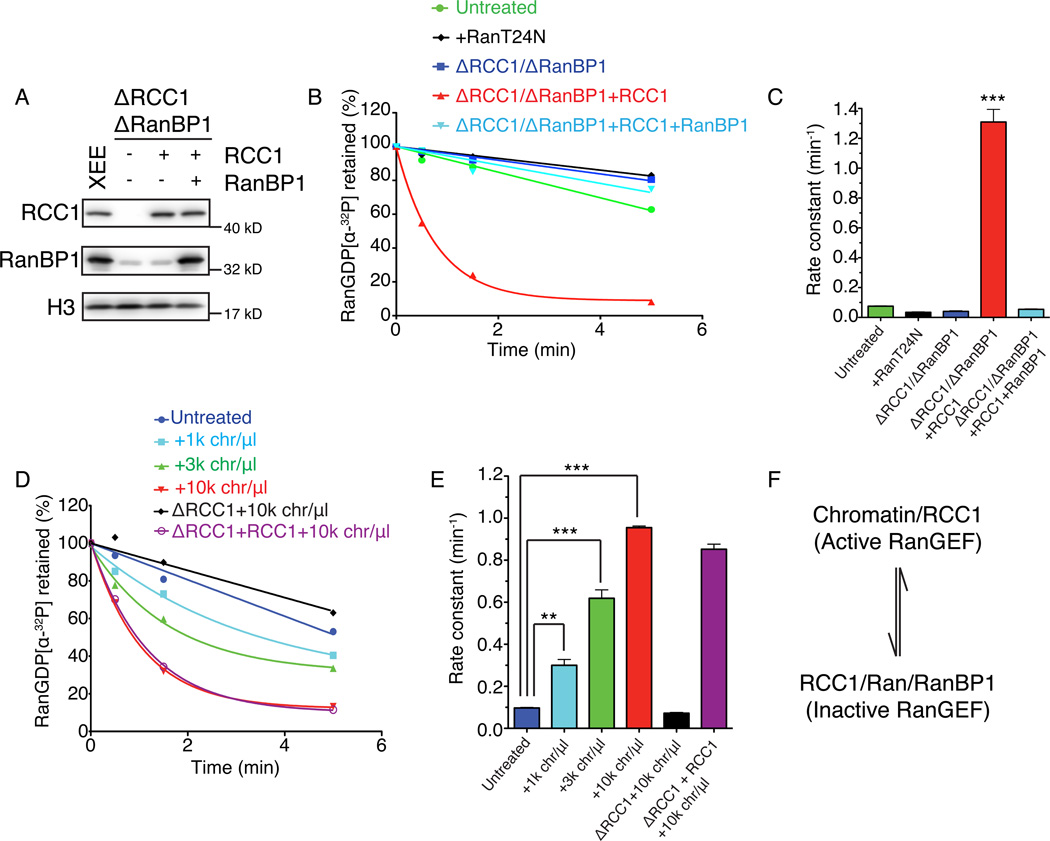
We wondered how RRR complex assembly might regulate enzymatic activity of RCC1 in egg extracts, given that RRR complex assembly inhibits RCC1’s RanGEF activity in vitro (Bischoff et al., 1995). We speculated that chromatin bound RCC1 should be active as a RanGEF, while soluble RCC1 should be inhibited within RRR complexes, and that this pattern might promote the formation of steep chromatin-centered Ran-GTP gradients in CSF-XEE (Kalab et al., 2002). To test this idea, we first added Ran-[α-32P]GDP into CSF-XEE without chromatin, and measured the kinetics of [α-32P]GDP release (Richards et al., 1995). In untreated CSF-XEE, less than 5% of [α-32P]GDP was released within the first 30 seconds (Figure 3A, 3B and 3C, green). Nucleotide release could be further inhibited through co-depletion of RCC1 and RanBP1 (Figure 3A, 3B and 3C blue) or the addition of RanT24N, a mutant that inhibits RCC1 (Figure 3B and 3C, black), indicating that the low level of nucleotide release that we observed required RCC1 activity. When we added physiological concentrations of xRCC1 to depleted CSF-XEE still lacking RanBP1, nearly 50% of [α-32P]GDP was released from Ran within the first 30 seconds (Figure 3A–C, red). This change corresponded to more than a 15-fold increase in estimated rate constant over the control CSF-XEE. The restoration of physiological levels of recombinant xRanBP1 reduced nucleotide release to a level comparable to control CSF-XEE (Figure 3A, 3B and 3C, light blue). These observations confirm that the guanine nucleotide release activity of RCC1 in CSF-XEE without chromatin was strongly inhibited by RRR complex assembly.
Figure 3. RanBP1 complex inhibited RCC1 in mitotic cytosol.
(A) RCC1 and RanBP1 were depleted from CSF-XEE using anti-RanBP1 antibodies (lane 2–4). Buffer (lane 2) or physiological concentrations of recombinant xRCC1 (lane 3) or of recombinant xRCC1 and xRanBP1 (lane 4) were added back to the depleted CSF-XEE. Total CSF-XEE was subjected to immunblotting by indicated antibodies. Lane 1 shows undepleted control CSF-XEE.
(B) 10 µM recombinant Ran charged with [α-32P]GDP was added to CSF-XEE reactions as in (A), or to undepleted CSF-XEE containing 10 µM Ran-T24N. Aliquots were taken at 0.5, 1.5, and 5 minutes after [α-32P]Ran-GDP addition, and exchange was monitored using a filter binding assay.
(C) Rate constants were determined from (B) and plotted as mean ± SEM. Significance in difference was tested by two-tailed t-test (n=3, *** p<0.0001).
(D) Demembraned sperm chromatin was added at the indicated concentrations to CSF-XEE, CSF-XEE depleted with anti-RCC1 antibodies, or RCC1-depleted CSF-XEE with recombinant xRCC1 (1× physiological concentration). Aliquots were sampled and analyzed as in (B).
(E) Rate constants were determined from (D) and plotted as mean ± SEM. Significance in difference was tested by two-tailed t-test (n=3, ** p<0.002, *** p<0.0005).
(F) RCC1 distribution equilibrium. Chromatin-bound RCC1 is an active RanGEF, while RCC1 within the RRR complex in mitotic cytosol is inactive. These two pools are in a dynamic equilibrium, determined by RanBP1 concentrations.
We predicted that the addition of chromatin to CSF-XEE should increase endogenous guanine nucleotide releasing activity by competing RCC1 from the RRR complex. To test this idea, we measured the change in [α-32P]GDP release within CSF-XEE containing chromatin. [α-32P]GDP was released at a higher rate in CSF-XEE containing chromatin than in similar reactions without chromatin, with the rates of release being directly proportional to the amount of chromatin (Figure 3D and 3E). Notably, [α-32P]GDP release could be entirely ascribed to the action of RCC1, since even the maximum concentration of chromatin was ineffective in promoting [α-32P]GDP release in CSF-XEE depleted of only RCC1 (Figure 3A, 3D and 3E, black). As expected, the restoration of to physiological RCC1 levels by addition of recombinant xRCC1 reconstituted chromatin-sensitive [α-32P]GDP release activities (Figure 3A, 3D and 3E, purple).
Taken together, our results suggest that chromatin-bound RCC1 is the primary source of RanGEF activity in CSF-XEE, with the soluble pool of RCC1 largely inhibited through the formation of RRR complexes (Figure 3F). This binary regulation of RCC1 activity should help to maintain a highly chromatin-centered Ran-GTP gradient. This model predicts that if RCC1 in the soluble fraction were not controlled by RRR complex assembly, Ran-GTP would also be produced in regions distal to chromatin, resulting in defective MT organization with respect to chromosomes. Our observations in CSF-XEE containing RCC1 but lacking RanBP1 are consistent with this prediction (Figure 1A, row 3 and 4, and 1B). It is surprising in this light that beads coated with recombinant RCC1 protein are capable of organizing spindles in CSF-XEE (Halpin et al., 2011); we would have naively predicted that RRR complex assembly should silence their capacity for Ran-GTP production. However, the tagged RCC1 protein linked to these beads does not effectively form RRR complexes (Rebecca Heald, personal communication), potentially explaining its capacity to remain enzymatically active despite the presence of excess RanBP1.
RanBP1 is phosphorylated in a cell cycle-dependent manner
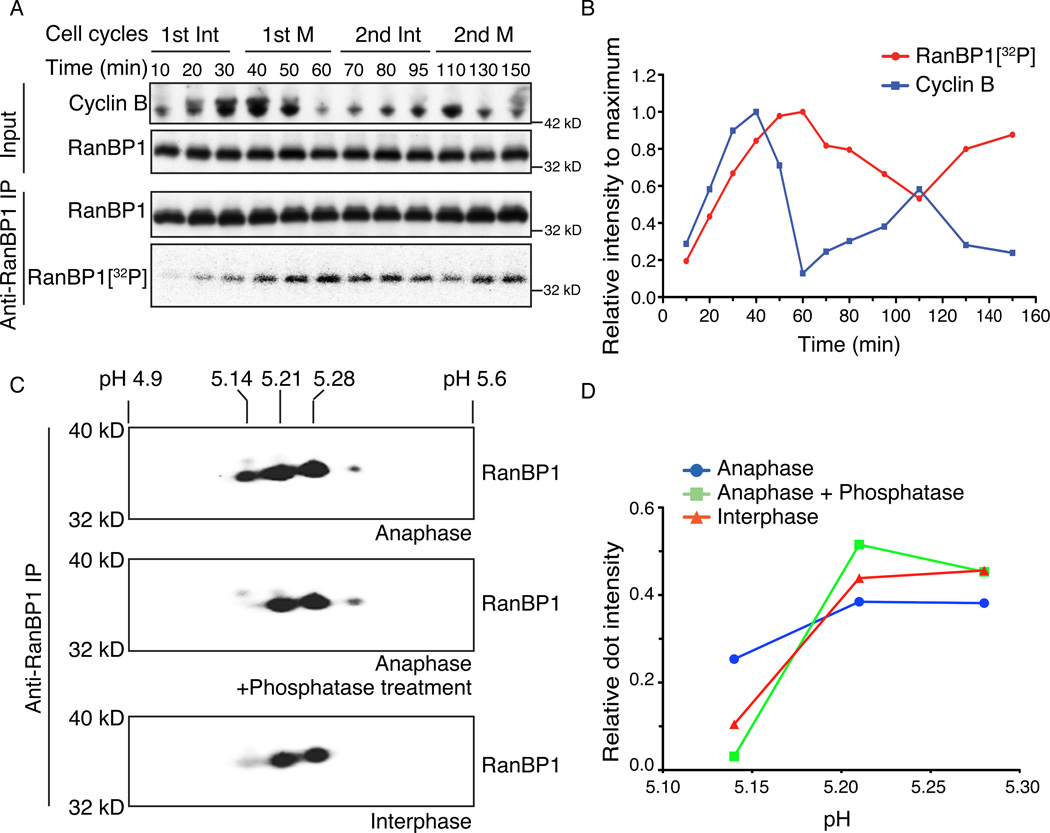
The chromatin binding dynamics of RCC1 vary during mitosis in both XEE (Arnaoutov and Dasso, 2003) and mammalian cells (Hitakomate et al., 2010; Hutchins et al., 2004). Particularly, RCC1 accumulates on chromatin at anaphase onset in cycling XEE (Arnaoutov and Dasso, 2003), and we wondered whether RRR complex-dependent partitioning of RCC1 between the chromatin-bound and -unbound pools might play a role in this phenomenon. Earlier studies have demonstrated mitotic regulation of mammalian RanBP1 through degradation and phosphorylation (Ciciarello et al., 2010; Hwang et al., 2011). We did not find any evidence that RanBP1 was subject to degradation during multiple cell cycles within cycling XEE (Figure 4A, upper panel). However, when we examined RanBP1 by immunoprecipitation at different time points after adding [γ-32P]ATP to cycling XEE to label the endogenously phosphorylated proteins, we observed that it was modified in a dynamic manner, with the highest level of RanBP1 phosphorylation (50 to 60 min. Figure 4A, lower panel) slightly after the onset of Cyclin B degradation at anaphase onset (Figure 4A and 4B).
Figure 4. RanBP1 is phosphorylated on cell cycle basis.
(A) 100 µCi/ml [γ-32P]ATP was added to cycling XEE. RanBP1 was immunoprecipitated from the reaction at different times and subjected to SDS-PAGE. XEE aliquots (upper panels) and RanBP1 immunoprecipitates (lower panel, row 1) were subjected to immunoblotting with indicated antibodies and 32P within the RanBP1 immunoprecipitate was detected by autoradiography (lower panel, row 2).
(B) The immunoblot intensity of Cyclin B for each sample from (A) was measured and normalized relative to the maximum Cyclin B intensity (blue line). 32P associated with immunoprecipitated RanBP1 for each sample was similarly measured and normalized values for each time point are shown (red line).
(C) RanBP1 was immunoprecipitated from cycling XEE at anaphase onset or interphase (bottom panel). RanBP1 precipitated from anaphase was treated with either buffer only (top panel) or alkaline phosphatase (middle panel). The RanBP1 within each sample was separated by linear pH 4–7 isoelectric focusing (IEF) followed by SDS-PAGE, and subjected to immunoblotting with antibodies against RanBP1. The numbers above show the estimated isoelectric point (pI) of individual dot.
(D) Relative RanBP1 intensity for each major spot in (C) was calculated by dividing intensity of each dot to total RanBP1 intensity. The relative RanBP1 intensity of anti-RanBP1 IP from anaphase (blue line), anti-RanBP1 IP from anaphase followed by CIP treatment (green line), and anti-RanBP1 IP from interphase (red line) were plotted against pI of each dot from (C).
See also Figure S3.
We further examined the phosphorylation dynamics of endogenous RanBP1 by two-dimension (2D) gel electrophoresis. RanBP1 was immunoprecipitated from either anaphase or interphase cycling XEE. The sample from anaphase XEE was treated with alkaline phosphatase or buffer only, and all samples were separated by isoelectric focusing (IEF) and SDS-PAGE (Figure 4C). The samples gave foci with isoelectric points (PI) near pH 5.14, pH 5.21 and pH 5.28. During anaphase, around 25% of RanBP1 was found in the most acidic fraction, pH 5.14 (Figure 4C, upper panel; Figure 4D, blue line). After phosphatase treatment, this species was depleted (Figure 4C, middle panel, and 4D, green line), suggesting that it corresponds to phosphorylated RanBP1. During interphase, only around 10% of RanBP1 focused to PI=5.14 (Figure 4C, lower panel, and Figure 4D, red line), indicating that significantly less RanBP1 is phosphorylated in interphase XEE than during anaphase. Taken together, our findings suggest that much of the endogenous pool of RanBP1 becomes phosphorylated in a cell cycle-dependent manner in cycling XEE, peaking just after the onset of anaphase.
To determine the role of RanBP1 phosphorylation, we identified the site of its modification. RanBP1 was immunopredipiated from anaphase cycling XEE and analyzed by mass spectrometry (Figure S3A). The major phosphorylated residue in this sample was Serine 60 (Ser 60) of RanBP1, which had previously been implicated as a potential site of mitotic phosphorylation in HeLa cells (Hwang et al., 2011). To further validate the Ser 60 phosphorylation on xRanBP1, a phospho-specific antibody was raised against xRanBP1 pSer60. By examining RanBP1 isolated from cycling XEE at different time points, we found that Ser 60 phosphorylation peaked at anaphase, at or slightly after the onset Cyclin B degradation (Figure S3B).
RanBP1 Ser60 phosphorylation disrupts the RRR complex
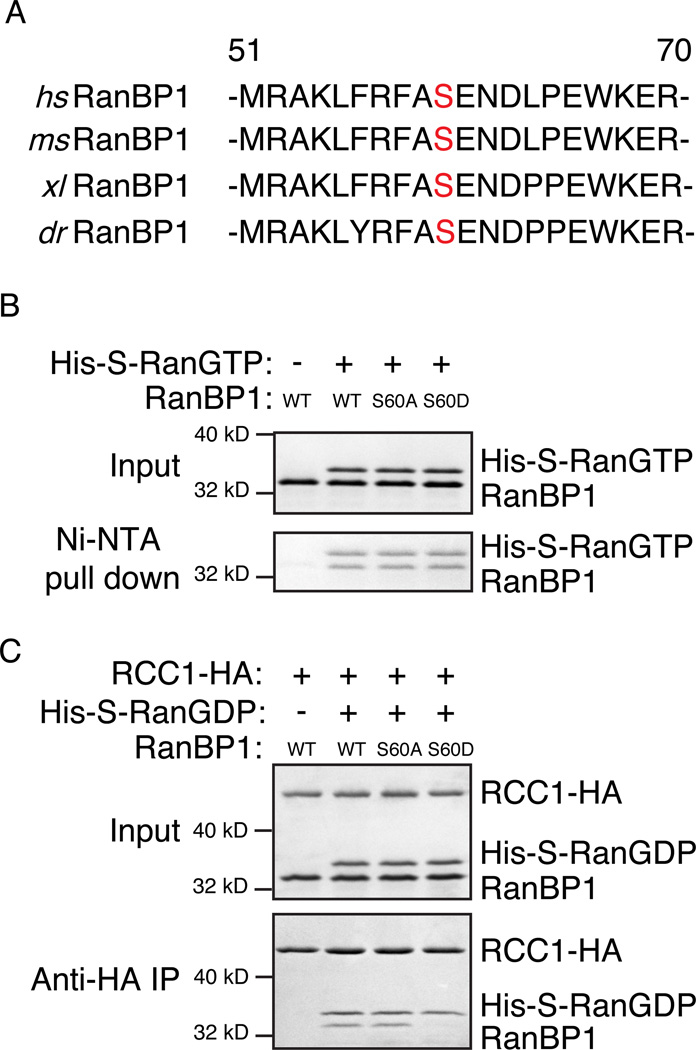
Ser 60 lies within the Ran-GTP binding domain of RanBP1, and it is conserved across a wide variety of vertebrate species (Figure 5A). This residue is not conserved in budding or fission yeast, nor is it conserved within human RanBP2 and RanBP3 (Figure S4), related proteins that share structurally similar Ran-GTP binding domains (Vetter et al., 1999). To test whether this modification can alter the formation of RRR complexes, we produced recombinant non-phosphorylatable (xRanBP1S60A) or phosphomimetic (xRanBP1S60D) mutant forms of RanBP1. We analyzed their in vitro Ran-GTP binding affinity, and observed that Ran-GTP binding was similar in xRanBP1S60A, xRanBP1S60D and wild type xRanBP1WT (Figure 5B), suggesting that Ser 60 phosphorylation does not affect the capacity of RanBP1 to bind Ran-GTP. The finding that Ran-GTP binding was similar for each of these proteins also suggested that the point mutations did not grossly disrupt xRanBP1’s structure.
Figure 5. RanBP1 Ser 60 phosphorylation releases RCC1 to bind chromatin.
(A) Protein sequence alignment of RanBP1 from H.sapiens, M.musculus, X.laevis, and D.rerio. The numbers indicate the respective positions of the first and last amino acid residue within H. sapiens RanBP1. Serine 60 is in red.
(B) Recombinant His6-S-Ran-GTP was incubated with recombinant xRanBP1WT, xRanBP1S60A, or xRanBP1S60D. Complexes formed by His6-S-Ran-GTP were precipitated using Ni-NTA resin. The original reactions (upper panel) and precipitated proteins (lower panel) were separated by SDS-PAGE and visualized by Commassie Brilliant Blue (CBB) staining. As a control, a reaction was incubated in parallel, containing recombinant xRanBP1WT but lacking His6-S-Ran-GTP (lane 1).
(C) Recombinant xRCC1-HA and His6-S-Ran-GDP were allowed to bind, followed by the addition of recombinant xRanBP1WT, xRanBP1S60A or xRanBP1S60D and further incubation. xRCC1-HA and associated proteins were precipitated using anti-HA beads. The original reactions (upper panel) and precipitated proteins (lower panel) were separated by SDS-PAGE and visualized by CBB staining. As a control, a reaction was incubated in parallel, containing xRCC1-HA and xRanBP1WT but lacking His6-S-Ran-GDP (lane 1).
See also Figure S4.
We analyzed the capacity of the mutated xRanBP1 proteins to form RRR complexes in vitro, through addition of wild type or mutated xRanBP1 to pre-formed RCC1/Ran complexes (Bischoff et al., 1995). Ran-GDP was used instead of Ran-GTP to minimize the interference from RanBP1/Ran-GTP interaction, since Ran-GDP has negligible affinity to RanBP1 without the presence of RCC1 (Bischoff et al., 1995). Interestingly, while the RRR complex was formed equally well with xRanBP1WT and xRanBP1S60A, xRanBP1S60D showed greatly reduced RRR complex levels (Figure 5C). This finding suggests that introducing negative charge at Ser 60 on RanBP1 profoundly inhibits RRR complex assembly, and that phosphorylation of RanBP1 is likely to cause dissociation of the RRR complex during anaphase in cycling XEE.
RanBP1 Ser60 phosphorylation alters RCC1 activity in XEE
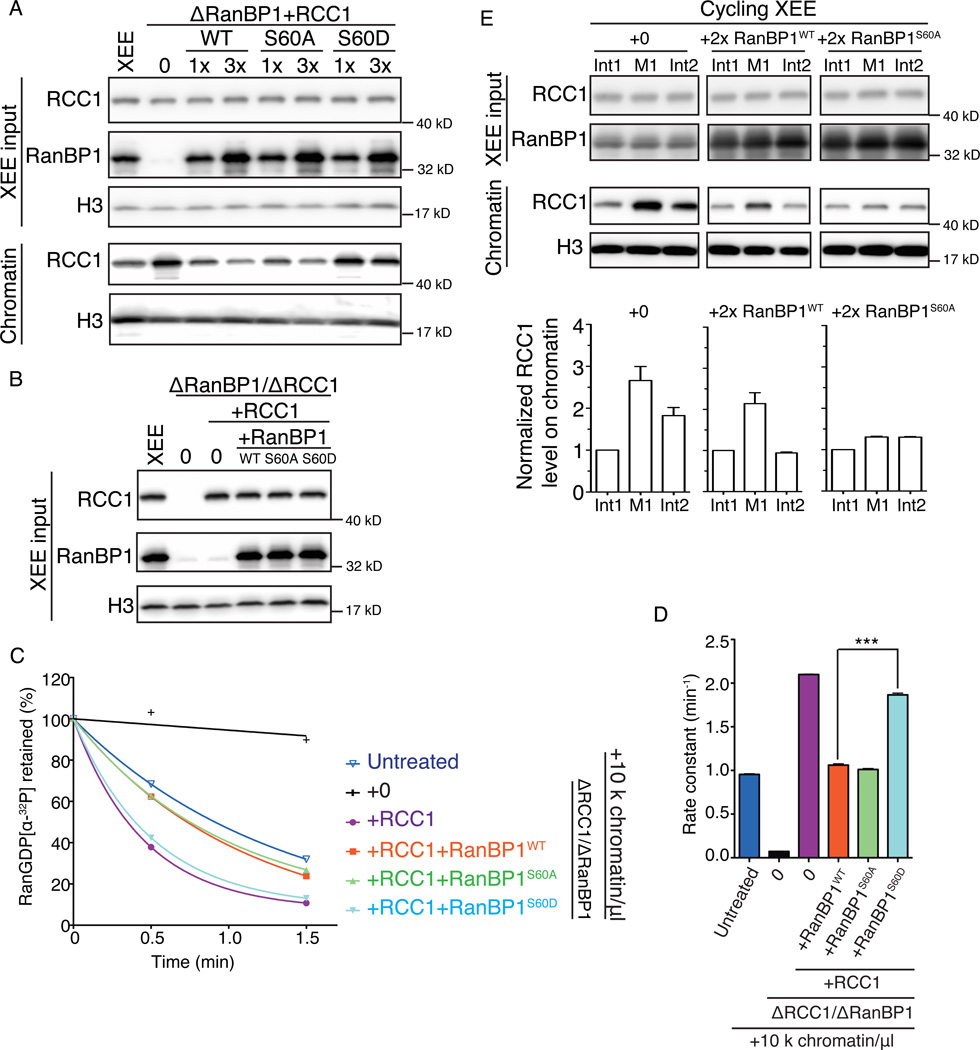
To test the effect of RanBP1 phosphorylation on the amount of chromatin-bound RCC1, we added to chromatin and physiological concentrations of xRCC1 to CSF-XEE that had been immunodepleted of endogenous RanBP1 and RCC1, along with physiological concentrations of xRanBP1WT, xRanBP1S60A or xRanBP1S60D. As before (Figure 2B), higher levels of RCC1 were detected on chromatin in the absence of RanBP1 than in untreated CSF-XEE (Figure 6A, lane 1 and 2), and this effect was reversed upon restoration of normal levels of xRanBP1WT. Recombinant xRanBP1WT at concentrations roughly 3-fold higher than endogenous levels further reduced the amount of RCC1 bound to chromatin (Figure 6A, lane 3 and 4). The reactions with xRanBP1S60A showed levels of chromatin-bound RCC1 that were very similar to those observed with xRanBP1WT (Figure 6A, lane 5 and 6). However, phosphomimetic xRanBP1S60D was far less effective in releasing RCC1 from chromatin (Figure 6A, lane 7 and 8), consistent with the idea that it could not effectively release chromatin-bound RCC1 into the soluble pool through RRR complex formation. We further analyzed the capacity xRanBP1WT, xRanBP1S60A or xRanBP1S60D to modulate RanGEF activity. Chromatin and physiological levels of recombinant xRCC1 were added to depleted CSF-XEE, together with xRanBP1WT, xRanBP1S60A or xRanBP1S60D (Figure 6B–D). RanGEF activity in each sample was estimated using a guanine nucleotide release assay (Figure 6C, D). As before (Figure 3), CSF-XEE reactions showed a chromatin- and RCC1-dependent guanine nucleotide release activity that was partially inhibited by the addition of xRanBP1WT (Figure 6C, D. Compare red and purple) and xRanBP1S60A was equally effective in inhibition of guanine nucleotide release (green). However, xRanBP1S60D was significantly less effective than xRanBP1WT (light blue), correlating well with its limited capacity to release RCC1 from chromatin (Figure 6A).
Figure 6. Endogenous RanGEF activity in CSF-XEE is increased by RanBP1 phosphorylation on Ser 60.
(A) xRCC1 was restored to physiological levels in CSF-XEE depleted using anti-RanBP1 antibodies by adding recombinant xRCC1. Recombinant xRanBP1WT, xRanBP1S60A or xRanBP1S60D were added as indicated (1× or 3× endogenous RanBP1 levels). 10,000 units/µl demembraned sperm chromatin were added to each sample and incubated for 30 min at RT before chromatin isolation. Each total reaction (upper panel) and the isolated chromatin (lower panel) was subjected to immunblotting with antibodies against RCC1, RanBP1 and Histone H3, as indicated.
(B) CSF-XEE was depleted using anti-RanBP1 antibodies, followed by addition of buffer or physiological levels of xRCC1. Where indicated, recombinant xRanBP1WT, xRanBP1S60A or xRanBP1S60D were added at concentrations equivalent to endogenous RanBP1. Each sample was subjected to immunblotting with antibodies against RCC1, RanBP1 and Histone H3.
(C) 10 µM recombinant Ran charged with [α-32P]GDP was added CSF-XEE reactions as in (B) containing demembraned sperm chromatin (10,000 units/µl). Aliquots were taken at 0.5, 1.5, and 5 minutes after [α-32P]Ran-GDP addition, and exchange was monitored using a filter binding assay, and plotted as described in Figure 3.
(D) Rate constants were determined from nucleotide release data (C) and plotted as mean ± SEM. Significance in difference was tested by two-tailed t-test (n=3, *** p<0.0001).
(E) Cycling XEE containing demembraned sperm chromatin (3,000 units/µl) was warmed to RT and allowed to initiate NE assembly. After NE closure, buffer (left), recombinant xRanBP1WT (middle) or xRanBP1S60A (right) were added at roughly twice the endogenous RanBP1 level. Chromatin was prepared from each sample at the first interphase (Int1), anaphase onset of the first mitosis (M1), and the second interphase (Int2). Total cycling XEE input at these three points was subjected to immunblotting with antibodies against RCC1 and RanBP1 (Row 1 and 2). Isolated chromatin from each sample at these three points was subjected to immunblotting by antibodies against RCC1 and Histone H3 (Row 3 and 4). Chromatin-bound RCC1 levels were normalized to the RCC1 signal intensity at Int1 for individual sample. The normalized RCC1 level on chromatin was plotted as mean ± SEM (n=3) (Lower panel).
To test whether RanBP1 phosphorylation was important for RCC1 dynamics in the context of anaphase progression, we examined the effect of RanBP1S60A on RCC1 chromatin-binding in cycling XEE (Arnaoutov and Dasso, 2003). Although treatments required to immunodeplete RanBP1 disrupted mitosis in cycling XEE, these extracts remained functionally intact when recombinant xRanBP1WT or xRanBP1S60A were added to otherwise untreated cycling XEE at levels approximately two-fold above endogenous levels. As these XEE progressed through the cell cycle, we prepared chromatin from samples taken during the first interphase, anaphase of the first M-phase and second interphase. Samples with xRanBP1WT showed lower overall levels of chromatin-bound RCC1 than buffer-treated controls (Figure 6E, left and middle panels), as might be expected (Figure 2A). Importantly, the reaction containing xRanBP1WT still showed a transient, two-fold increase in chromatin-bound RCC1 during anaphase, which was similar in magnitude to the transient increase observed in untreated XEE. By contrast, the chromatin binding dynamics of RCC1 were significantly dampened in cycling XEE with xRanBP1S60A, showing less than a 30% increase in chromatin-bound RCC1 in anaphase over interphase (Figure 6E, right panel). This small increase might be ascribed to the endogenous RanBP1 that remained in the extract and could become phosphorylated at anaphase onset. The capacity of xRanBP1S60A to suppress RCC1’s anaphase chromatin-binding dynamics strongly implies that RanBP1 Ser 60 phosphorylation is critical for this regulation.
Taken together, our results show that anaphase phosphorylation on RanBP1 Ser60 increases the partitioning of RCC1 to chromatin and thereby drives increased Ran-GTP production in late mitosis.
DISCUSSION
It has been known for nearly two decades that RCC1, Ran, and RanBP1 form a stable heterotrimeric complex (RRR complex), which inhibits RCC1’s RanGEF activity in vitro (Bischoff et al., 1995). The function of the RRR complex has remained mysterious, however, because RanBP1 is physically separated from RCC1 during interphase (Richards et al., 1996) and the formation of this complex has been presumed to have little role in regulating nuclear transport, if any. We have demonstrated that the RRR complex forms readily in M-phase CSF-XEEs (Figure 1). RCC1 binding to chromatin and RRR complex assembly were mutually exclusive, so that promoting RRR complex formation through the addition of recombinant xRanBP1 sequestered RCC1 away from chromatin (Figure 2A). Moreover, RRR complex assembly inhibited RCC1’s RanGEF activity in CSF-XEE (Figure 3), in agreement with earlier in vitro observations (Bischoff et al., 1995). Together, these findings suggest that the RRR complex plays a pivotal mitotic role in determining the partitioning of RCC1 between its active chromatin-bound and inactive soluble states, thereby setting both the location and magnitude of Ran-GTP production during mitosis.
Our observations suggest that RCC1 inhibition through RRR complex assembly helps to organize Ran-GTP production and limit it to the close vicinity of mitotic chromatin. Inappropriate Ran-GTP production in the absence of RanBP1 can lead to the assembly of defective spindles and ectopic MT assembly in XEE (Figure 1). This mitotic function of RanBP1 may be particularly critical in Xenopus embryos and extracts, given the high concentration of RCC1 relative to chromatin binding sites within the early embryo (Figure 1A, (Dasso et al., 1992)). Nevertheless, there is evidence that RanBP1 may play a similar role in somatic systems. For example, tissue culture cells depleted of RanBP1 show disorganized MT asters (Li et al., 2007), hyperstable spindle MTs and defective chromosome segregation (Tedeschi et al., 2007). Notably, RanBP1 is downregulated in aging cells (Ly et al., 2000; Pujol et al., 2002), which show relatively shallow Ran-GTP gradients (Hasegawa et al., 2013). Finally, RanBP1 expression is mis-regulated in a variety of human cancers (Rensen et al., 2008). Our results and other findings (Amato et al., 2013) suggest the possibility that this mis-regulation may contribute toward mitotic defects in these cells.
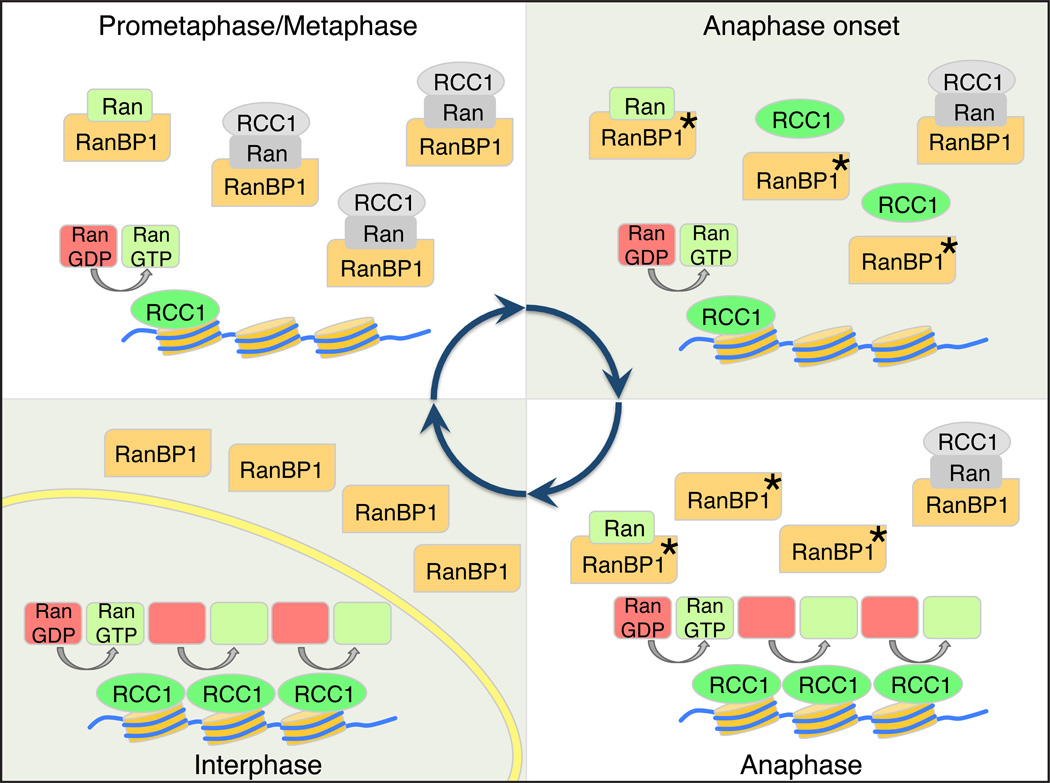
Furthermore, it has been well established that RCC1’s association to mitotic chromatin is dynamic, and that there are particularly large changes as cycling XEE (Arnaoutov and Dasso, 2003) or mammalian tissue culture cells (Hutchins et al., 2004) pass through the metaphase-to-anaphase window. While a number of post-translational modifications of RCC1 have been described (Chen et al., 2007; Hutchins et al., 2004; Li and Zheng, 2004), none of these appear to be responsible for control of RCC1 at anaphase onset. In particular, mitotic cdk1-dependent phosphorylation of RCC1 is not well correlated temporally to changes in RCC1-chromatin association, since it occurs much earlier in mitosis (Hutchins et al., 2004; Li and Zheng, 2004). It is also notable that the residues of human RCC1 that have been found as the primary targets of cdk1-dependent phosphorylation (Serine resides 2 and 11; (Hutchins et al., 2004; Li and Zheng, 2004)) are not conserved in X. laevis, so that their modifications could not modulate RCC1 association to chromatin in XEE (Arnaoutov and Dasso, 2003). We observed that RanBP1 is phosphorylated during anaphase in cycling XEE (Figure 4, S3), and that this modification disrupts RRR complex assembly (Figure 5). Our data suggest that modification of RanBP1 drives changes in RCC1 binding to chromatin, and thereby indirectly alters the rate of Ran-GTP production in anaphase (Figure 7).
Figure 7. Model for mitotic regulation of RCC1 by RanBP1.
Premetaphase: RCC1 is partitioned between an active, chromatin bound pool (green) and an inactive pool (grey), associated with RRR complexes that also contain RanBP1 and nucleotide-free Ran (grey). Anaphase Onset: Phosphorylation of RanBP1 on Ser 60 (asterisk) releases RCC1 from the RRR complex. The free RCC1 is then recruited to chromatin. Anaphase: The high level of chromatin-bound RCC1 promotes enhanced levels of Ran-GDP (red) to Ran-GTP (green) exchange on chromatin. Interphase: NE (yellow) formation physically separates RCC1 from RanBP1, preventing RRR complex assembly and inhibition of RCC1.
We speculate that this regulation may also contribute to Ran pathway organization in early interphase. Enhanced activity of RCC1 on anaphase chromatin should help provide high levels of Ran-GTP to facilitate NE reassembly (Clarke and Zhang, 2008). Moreover, physical separation of RCC1 and RanBP1 after RRR complex dissociation would allow RCC1 sequestration to chromatin within re-forming nuclei while excluding RanBP1 into the early interphase cytosol. We expect that one detrimental effect of interphase NE rupture would be the loss of RanBP1 exclusion from nucleoplasm, resulting in RRR complex formation and decreased levels of active, chromatin-bound RCC1, which in turn could decrease Ran-GTP production and contribute to the general loss of compartmental identity that has been observed under these circumstances (Hatch et al., 2013).
The conservation of the Serine 60 residue (Figure 5A) is consistent with the idea that phosphorylation of RanBP1 may play an important role in mitotic regulation of the Ran pathway in many vertebrate species. On the other hand, the absence of this residue in budding and fission yeast suggests that although RanBP1 may have mitotic roles in fungi (Ouspenski, 1998) those functions are controlled through different mechanisms. This divergence may reflect the fact that yeast do not undergo NE breakdown during mitosis and thus do not have complete mixing of their nuclear and cytoplasmic contents. More interestingly, C. elegans and D. melanogaster apparently not only lack this regulatory pathway but also appear to be devoid of RanBP1 homologues altogether, although they still possess potential homologues of RanBP2 and RanBP3 (Dasso, 2002). It is notable that the worm and fly RCC1 homologues possess extended N- or C-terminal domains, respectively, that are not found in vertebrate and fungal RCC1 homologues. In the case of Drosophila, this additional domain shares sequence homology with embryonic linker histones (Frasch, 1991). We speculate that worm and fly RCC1 may interact with chromatin in a different fashion than vertebrate RCC1 proteins, thus bypassing any requirement for RanBP1. Testing this idea in the future will be of considerable interest, and will be important for understanding how the use of the Ran pathway has evolved in different organisms.
In summary, we have documented an important mitotic role of the RanBP1 protein in controlling the localization and activity of Ran’s nucleotide exchange factor RCC1. We have shown that phosphorylation of RanBP1 during anaphase drives changes in RCC1 dynamics and allows increased Ran-GTP production. These findings resolve important and long-standing questions within the Ran field regarding the function of the heterotrimeric RanBP1/Ran/RCC1 complex and its dynamics.
EXPERIMENTAL PROCEDURES
Antibody and Immunodepletion
Rabbit polyclonal antibodies against xRCC1 were described previously (Arnaoutov and Dasso, 2003). Rabbit polyclonal antibodies against full-length xRanBP1 and phospho-specific rabbit polycolonal antibodies against a synthetic xRanBP1-based phosphorylated peptide (KMRAKLFRFApSEND) were produced by Pacific Immunology. Rabbit polyclonal antibodies against Histone H3 (#ab1791) and mouse monoclonal antibodies against Xenopus Cyclin B2 (#ab18250) were purchased from Abcam. Immunodepletions were performed by incubating XEE with antixRCC1 and anti-xRanBP1 crosslinked to protein A beads.
Xenopus laevis Egg Extract preparation and use
M-phase X. laevis egg extracts (CSF-XEE) and demembraned sperm nuclei were prepared as described (Murray, 1991). To examine spindle assembly, rhodamine-tubulin (20 µg/ml. Cytoskeleton) and sperm nuclei (1,000 unit/µl XEE) were added to CSF-XEE, and incubated at RT for 60 min. Aliquots were fixed with 11% paraformaldehyde supplemented with 10 µg/ml DAPI and processed for fluorescent microscopy. Images (Figure 1B and S1B) were taken using an ORCA-II CCD camera on a Zeiss Axioskop microscope, and the images were acquired and processed with Openlab software.
To analyze chromatin-bound proteins, demembraned sperm nuclei were added to a final concentration of 10,000 unit/µl XEE, and incubated at RT for 30 min. Reactions were diluted in dilution buffer (5 mM HEPEs, pH 7.7, 50 mM KCl, 1 mM MgCl2, 0.2% Triton-X100, and 5% (v/v) glycerol), and spun through cushion buffer (5 mM HEPEs, pH 7.7, 50 mM KCl, 1 mM MgCl2, 0.2% Triton-X100, and 30% (v/v) glycerol) at 11,000 g for 5 min. The chromatin was washed once with dilution buffer and re-centrifuged under the same conditions. The chromatin pellet was boiled in SDS sample buffer and subject to SDS-PAGE and Western blot analysis.
RanBP1 immunoprecipitation and on beads phosphatase treatment
Cycling XEE was frozen in liquid nitrogen in different time points. 50 µl XEE from anaphase XEE or interphase XEE was thawed and diluted at 1:10 (v/v) in denatured buffer (20 mM HEPEs, pH 7.7, 150 mM NaCl, 0.2% Triton-X100, 1 M urea). 25 µg anti-xRanBP1 antibodies were crosslinked on 100 µl protein A magnetic beads (Invitrogen) and incubated with each sample for 90 min at 4°C. The beads were washed three times in 1× NEBuffer 3 (NEB) and resuspended in 50 µl 1× NEBuffer 3. Either buffer or 10 units of alkaline phosphatase (NEB, M0290S) were added to the anaphase XEE beads and incubated 30 °C for 30 minutes to allow dephosphorylation of the phosphatase-treated sample. After incubation, beads were washed three times with 1× NEBuffer 3 and eluted with DeStreak Rehydration Solution (GE Healthcare). The eluted samples were separated by 2D gel electrophoresis.
Phosphorylated amino acid residue mapping
Endogenous RanBP1 from anaphase cycling XEE was immunoprecipitated using anti-xRanBP1 antibodies crosslinked to protein A Sepharose beads (GE Healthcare). After immunoprecipitation, proteins were eluted and separated by SDS-PAGE. The region of the gel containing RanBP1 was isolated and extracted. Sequence analysis was performed at the Harvard Mass Spectrometry and Proteomics Resource Laboratory (Cambridge, MA) by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry on a Thermo LTQ-Orbitrap mass spectrometer.
Endogenous Guanine Nucleotide Releasing Assay
10 µM recombinant Human Ran loaded with [α-32P]GDP was added to CSF-XEE. Aliquots were diluted in stop buffer (20 mM Tris-HCl pH 7.5, 25 mM MgCl2, 100 mM NaCl, 1mM DTT) and [α-32P]GDP binding to Ran was measured by filter retention. Triplicate samples were analyzed for each condition. Nucleotide release from the loaded Ran was plotted analyzed using Prism software. The guanine nucleotide-releasing curve showing mean ± SEM was regressed as one phase decay model. The rate constant for each reaction was also plotted as mean ± SEM and the significances in difference were analyzed by two-tailed t test.
In vitro Protein Binding Assay
In vitro protein binding was performed in protein binding buffer (20 mM HEPEs, 150 mM NaCl, 50 µg/ml Digitonin (Calbiochem), and 2 mM MgCl2). To check Ran-GTP binding affinity (Figure 5A), recombinant Xenopus RanBP1WT, RanBP1S60A, or RanBP1S60D were incubated with recombinant Human His-S-Ran-GTP and the pull down assay was done by incubating samples with Ni-NTA resin (Qiagen). To check RCC1/Ran/RanBP1 heterotrimeric complex formation (Figure 5B), recombinant WT/mutated Xenopus RanBP1 were incubated with recombinant Human His-S-Ran-GDP, and recombinant Xenopus RCC1-HA. The immunoprecipitation was done by incubating samples with anti-HA affinity gel (Sigma-Aldrich). To characterize Ran binding mutant of RCC1 (Figure S2), recombinant Xenopus RCC1WT or RCC1Ran were incubated with recombinant Human His-S-Ran-GTP. The pull down was done by incubating samples with S protein agarose (Novagen).
All bound proteins were eluted and boiled in SDS-sample buffer, separated by SDS-PAGE, and visualized by Coomassie Brilliant Blue (CBB) staining.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NICHD project no. HD008740.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes figures and Supplemental Experimental Procedures.
REFERENCES
- Amato R, Scumaci D, D'Antona L, Iuliano R, Menniti M, Di Sanzo M, Faniello MC, Colao E, Malatesta P, Zingone A, et al. Sgk1 enhances RANBP1 transcript levels and decreases taxol sensitivity in RKO colon carcinoma cells. Oncogene. 2013;32:4572–4578. doi: 10.1038/onc.2012.470. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A, Dasso M. The Ran GTPase regulates kinetochore function. Developmental cell. 2003;5:99–111. doi: 10.1016/s1534-5807(03)00194-1. [DOI] [PubMed] [Google Scholar]
- Beddow AL, Richards SA, Orem NR, Macara IG. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS letters. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. The EMBO journal. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nature cell biology. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciciarello M, Roscioli E, Di Fiore B, Di Francesco L, Sobrero F, Bernard D, Mangiacasale R, Harel A, Schinina ME, Lavia P. Nuclear reformation after mitosis requires downregulation of the Ran GTPase effector RanBP1 in mammalian cells. Chromosoma. 2010;119:651–668. doi: 10.1007/s00412-010-0286-5. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nature reviews Molecular cell biology. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Dasso M. The Ran GTPase: theme and variations. Current biology : CB. 2002;12:R502–R508. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- Dasso M, Nishitani H, Kornbluth S, Nishimoto T, Newport JW. RCC1, a regulator of mitosis, is essential for DNA replication. Molecular and cellular biology. 1992;12:3337–3345. doi: 10.1128/mcb.12.8.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M. The maternally expressed Drosophila gene encoding the chromatin-binding protein BJ1 is a homolog of the vertebrate gene Regulator of Chromatin Condensation, RCC1. The EMBO journal. 1991;10:1225–1236. doi: 10.1002/j.1460-2075.1991.tb08064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin D, Kalab P, Wang J, Weis K, Heald R. Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts. PLoS biology. 2011;9:e1001225. doi: 10.1371/journal.pbio.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Ryu SJ, Kalab P. Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. The Journal of cell biology. 2013;200:151–161. doi: 10.1083/jcb.201206142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitakomate E, Hood FE, Sanderson HS, Clarke PR. The methylated N-terminal tail of RCC1 is required for stabilisation of its interaction with chromatin by Ran in live cells. BMC Cell Biol. 2010;11:43. doi: 10.1186/1471-2121-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Moore WJ, Hood FE, Wilson JS, Andrews PD, Swedlow JR, Clarke PR. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Current biology : CB. 2004;14:1099–1104. doi: 10.1016/j.cub.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Hwang HI, Ji JH, Jang YJ. Phosphorylation of Ran-binding protein-1 by Polo-like kinase-1 is required for interaction with Ran and early mitotic progression. J Biol Chem. 2011;286:33012–33020. doi: 10.1074/jbc.M111.255620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. Journal of cell science. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Current biology : CB. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Li HY, Ng WP, Wong CH, Iglesias PA, Zheng Y. Coordination of chromosome alignment and mitotic progression by the chromosome-based Ran signal. Cell Cycle. 2007;6:1886–1895. doi: 10.4161/cc.6.15.4487. [DOI] [PubMed] [Google Scholar]
- Li HY, Zheng Y. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes & development. 2004;18:512–527. doi: 10.1101/gad.1177304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. The Journal of biological chemistry. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Ly DH, Lockhart DJ, Lerner RA, Schultz PG. Mitotic misregulation and human aging. Science. 2000;287:2486–2492. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Ouspenski II. A RanBP1 mutation which does not visibly affect nuclear import may reveal additional functions of the ran GTPase system. Experimental cell research. 1998;244:171–183. doi: 10.1006/excr.1998.4174. [DOI] [PubMed] [Google Scholar]
- Pu RT, Dasso M. The balance of RanBP1 and RCC1 is critical for nuclear assembly and nuclear transport. Molecular biology of the cell. 1997;8:1955–1970. doi: 10.1091/mbc.8.10.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol G, Soderqvist H, Radu A. Age-associated reduction of nuclear protein import in human fibroblasts. Biochemical and biophysical research communications. 2002;294:354–358. doi: 10.1016/S0006-291X(02)00492-8. [DOI] [PubMed] [Google Scholar]
- Rensen WM, Mangiacasale R, Ciciarello M, Lavia P. The GTPase Ran: regulation of cell life and potential roles in cell transformation. Frontiers in bioscience : a journal and virtual library. 2008;13:4097–4121. doi: 10.2741/2996. [DOI] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Carey KL, Macara IG. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. The Journal of cell biology. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Macara IG. The C terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. The Journal of biological chemistry. 1995;270:14405–14411. doi: 10.1074/jbc.270.24.14405. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Ciciarello M, Mangiacasale R, Roscioli E, Rensen WM, Lavia P. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. Journal of cell science. 2007;120:3748–3761. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- Zhang C, Hughes M, Clarke PR. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. Journal of cell science. 1999;112(Pt 14):2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.