Summary
The conserved protein Rap1 functions at telomeres in fungi, protozoa, and vertebrates. Like yeast Rap1, human Rap1 has been implicated in telomere length regulation and repression of non-homologous end joining (NHEJ) at telomeres. However, mouse telomeres lacking Rap1 do not succumb to NHEJ. To determine the functions of human Rap1, we generated several TALEN-mediated human cell lines lacking Rap1. Loss of Rap1 did not affect the other components of shelterin, the modification of telomeric histones, the subnuclear position of telomeres, or the 3’ telomeric overhang. Telomeres lacking Rap1 did not show a DNA damage response, NHEJ, or consistent changes in their length, arguing that Rap1 does not have an important function in protection or length regulation of human telomeres. As human Rap1, like its mouse and unicellular orthologs, affects gene expression, we propose that the conservation of Rap1 reflects its role in transcriptional regulation rather than a function at telomeres.
Keywords: telomere, shelterin, Rap1, NHEJ, DDR, transcription
Introduction
Rap1 is a component of shelterin, the protein complex that functions to protect telomeres, recruits telomerase, and regulates telomere length (reviewed in (Palm and de Lange, 2008)). Human shelterin contains two double-strand (ds) telomeric DNA binding proteins, TRF1 and TRF2, which both interact with TIN2. TIN2 in turn binds heterodimers of TPP1 and the POT1 single-stranded (ss) telomeric DNA binding factor. Mammalian Rap1 relies on TRF2 to localize to telomeres (Li et al., 2000). Similarly, the Rap1 orthologs of fission yeast and trypanosomes use a TRF1/2-like protein to accumulate at telomeres (Kanoh and Ishikawa, 2001; Yang et al., 2009). In contrast, budding yeast Rap1 binds telomeric DNA directly (Konig et al., 1996).
Rap1 was discovered as a Saccharomyces cerevisiae transcriptional regulator (repressor/activator protein) (Shore and Nasmyth, 1987). All Rap1 proteins have a N-terminal BRCT motif, a C-terminal protein interaction domain, and one or two central Myb domains (Konig et al., 1996; Liu and Lustig, 1996; Hardy et al., 1992a; Wotton and Shore, 1997; Yang et al., 2009; Kanoh and Ishikawa, 2001; Li et al., 2000). Budding yeast Rap1 binds to promoter and silencer elements and interacts with the silencing proteins Sir3p and Sir4p (Hardy et al., 1992b; Hardy et al., 1992a; Shore, 1994; Lickwar et al., 2012; Cockell et al., 1995). Although mammalian Rap1 does not interact with sirtuins, it localizes to chromosome-internal sites and controls gene expression, affecting metabolism and body weight control (Yeung et al., 2013; Martinez et al., 2013; Yang et al., 2011). Rap1 also regulates gene transcription in fission yeast and in the distantly related trypanosomes (Kanoh and Ishikawa, 2001; Yang et al., 2009).
It is unclear which telomeric functions of Rap1 are conserved between yeast and mammals. Rap1 controls telomere length in yeast, acting to inhibit inappropriate telomere elongation (Lustig et al., 1990; Conrad et al., 1990; Sussel and Shore, 1991; Kyrion et al., 1992; Kanoh and Ishikawa, 2001). This control of telomere length is largely mediated by the Rap1 interacting factors Rif1 and Rif2 (Hardy et al., 1992a; Wotton and Shore, 1997; Levy and Blackburn, 2004; Teixeira et al., 2004). In support of a role in telomere length control, shRNAs to human Rap1 induce telomere lengthening (O'Connor et al., 2004), as do overexpression of several Rap1 truncation mutants (Li and de Lange, 2003). However, Rap1 knockout mice show no change in telomere length, even after three generations (Sfeir et al., 2010). Additionally, mammalian Rif1 is not localized at telomeres (Silverman et al., 2004; Xu and Blackburn, 2004), and there is no mammalian ortholog of Rif2.
It is also unclear whether the role of Rap1 in protection of telomeres from NHEJ, which has been demonstrated in yeast (Pardo and Marcand, 2005; Miller et al., 2005), is conserved in mammals. In vitro, human Rap1 can block NHEJ when it binds to TRF2 loaded on an end-joining substrate (Bae and Baumann, 2007), and a Rap1-fusion protein can reduce telomere fusions when it is tethered to telomeres that are depleted of TRF2 (Sarthy et al., 2009). On the other hand, mouse cells lacking Rap1 show no telomere fusions and Rap1-deficient mice are alive and fertile (Sfeir et al., 2010; Martinez et al., 2010). The only telomere deprotection phenotype in Rap1-deficient mouse cells is a propensity for telomere-telomere recombination when Ku70/80 are also absent (Sfeir et al., 2010).
To determine the function(s) of human Rap1, we used Transcription Activator Like Effector Nucleases (TALENs) to knockout the TERF2IP gene. Analysis of several independent Rap1-deficient cell lines demonstrated that human Rap1 was not required for telomere protection, telomere length regulation, and other aspects of telomere function. In contrast, we document a change in the transcription of several genes upon loss of Rap1, suggesting that its transcriptional function is primarily responsible for the high degree of Rap1 conservation from unicellular organisms to mammals.
Results
Efficient TALEN-mediated Knockout of Human Rap1
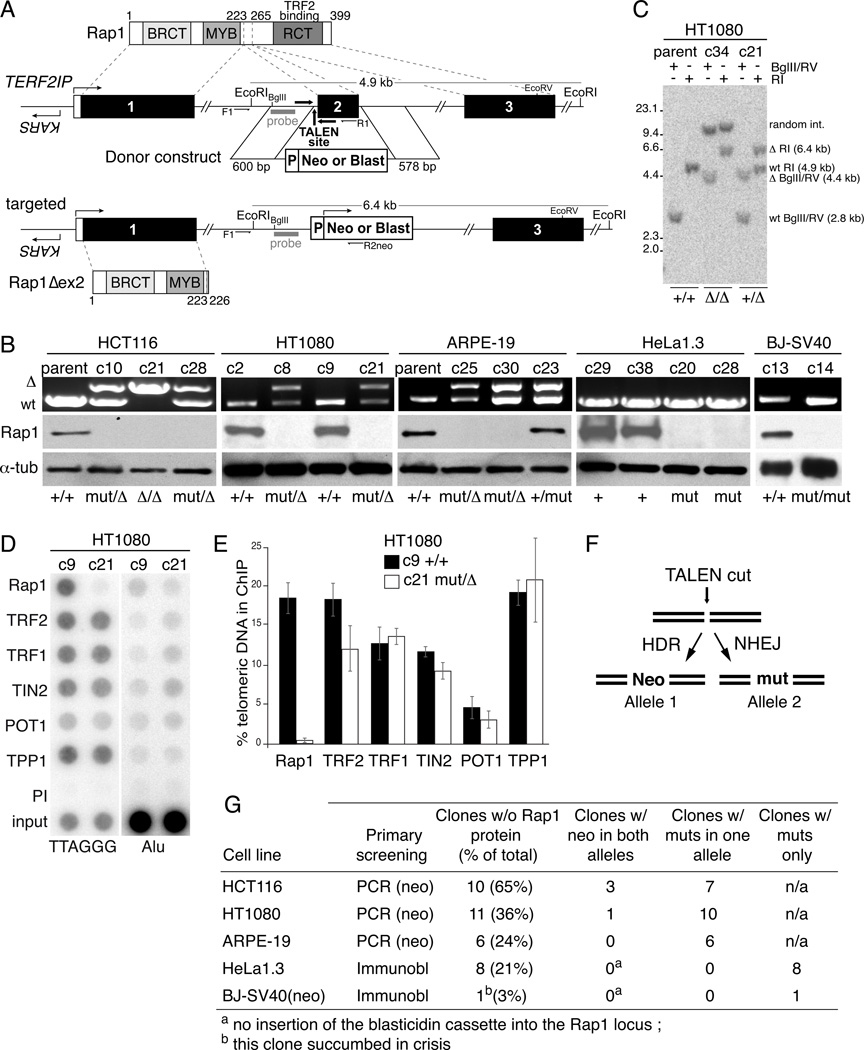
The human Rap1-encoding TERF2IP gene shares its promoter region with the essential KARS (lysyl-tRNA synthetase) gene located just upstream of exon 1 (Figure 1A). To avoid disrupting the KARS gene, we employed TALENs to delete exon 2, a targeting strategy analogous to the one used for mouse TERF2IP (Sfeir et al., 2010) (Figure 1A and S1A,B). Deletion of exon 2 should result in an mRNA encoding a 226 aa ORF that ends prematurely in a stop codon at the beginning of exon 3. As exon 3 encodes the TRF2-binding domain, the truncated Rap1 protein is not expected to localize to telomeres. Taking advantage of the small size of exon 2 (125 bp) and anticipated resection of TALEN-induced DSBs (Chen et al., 2011; Urnov et al., 2010), a neomycin donor construct was designed containing 5’ and 3’ arms homologous to the surrounding introns. Homology-directed repair (HDR) using the donor construct should result in deletion of exon 2 and insertion of the neomycin cassette.
Figure 1. TALEN-mediated inactivation of the gene for human Rap1.
(A) Schematic of human Rap1, the TERF2IP locus, the targeting construct, and the resulting knockout allele. F1, R1, and R2neo: PCR primers for genotyping. Arrows in bold: TALEN binding and cut sites.
(B) PCR genotyping of the TERF2IP gene and western blotting for Rap1 in the indicated clones. +, WT allele; Δ, targeted allele; mut, mutation resulting in loss of Rap1.
(C) Southern blot of EcoRI (RI)- or BglII/EcoRV (RV)-digested genomic DNA from targeted HT1080 clones. Probe shown in (A).
(D) Telomeric ChIP of WT and targeted HT1080 clones. Duplicate dot-blots were probed for telomeric or Alu repeats.
(E) Average percentage of telomeric DNA recovered in ChIPs with the indicated antibodies (two independent experiments). Error bars: SEMs.
(F) Schematic illustrating NHEJ and HDR after TALENs cutting.
(G) Table indicating number of Rap1 knockout clones acquired and the genetic alterations in TERF2IP.
See also Figure S1.
TERF2IP was targeted in two near-diploid cancer lines (HCT116 colorectal carcinoma and HT1080 fibrosarcoma), a subclone of the near-triploid cervical carcinoma HeLa cell line (HeLa1.3 (Takai et al., 2010)), the diploid ARPE-19 retinal pigment epithelial cell line, and primary BJ fibroblasts transformed with SV40 large T antigen (SV40LT). With the exception of SV40LT BJ, all cells expressed telomerase. For HCT116, HT1080, and ARPE-19 cells, neomycin resistant clones were obtained, analyzed by PCR, and then evaluated by Southern blotting to verify the correct neomycin insertion (Figure 1B,C). For HeLa1.3 and BJ, the blasticidin donor construct was used and clones were analyzed by immunoblotting for Rap1. This analysis identified clones that lacked the wild type TERF2IP gene and expressed no detectable Rap1 protein (Figure 1B,C and S1C). We were unable to detect the polypeptides representing the remaining ORF of the targeted TERF2IP gene (Figure S1D-F), perhaps due to nonsense-mediated decay.
Unexpectedly, immunoblotting revealed the complete loss of Rap1 in heterozygous clones with one neo insertion (Figure 1B and S1C). Telomeric ChIP of two such clones (HT1080 c21 and HeLa1.3 c28) confirmed that Rap1 was absent from telomeres (Figure 1D,E and S1G,H). Sequencing revealed small deletions close to the TALEN site in these and other Rap1-deficient clones with only one neo-containing TERF2IP gene (Figure S1B). Most mutations had ablated Rap1 by deleting the exon 2 splice acceptor site or had created a frame-shift mutation, indicating that errors generated during NHEJ had inactivated the TERF2IP gene (Figure 1F). As a result, the frequency of the TERF2IP KOs is much higher than deduced from PCR genotyping. Taking the deleterious repair events into account, the ablation of Rap1 occurred at 20–65% efficiency in HCT116, HT1080, and ARPE-19 cells (Figure 1G). The HCT116 cells showed the highest KO frequency consistent with their propensity for HDR (reviewed in (Sedivy et al., 1999)). The actual KO frequency is probably even higher as only clones with a neo cassette in the TERF2IP gene were analyzed.
The high KO frequency allowed screening of HeLa1.3 and SV40LT BJ clones by Rap1 immunoblotting. Sequencing of Rap1-negative HeLa1.3 clones revealed that all three alleles in this cell line contained inactivating mutations near the TALEN cut site (Figure S1B). Prolonged culturing of clones in blasticidin resulted in cell death, suggesting that the pEF blasticidin cassette did not confer long-term resistance. The KO frequency was lower in SV40LT BJ fibroblasts with only one blasticidin-resistant clone showing absence of Rap1 protein. This clone was not analyzed because it perished in telomere crisis, as did many of the Rap1-proficient SV40LT BJ clones.
Rap1-Deficient Cells Proliferate and Maintain Fully Protected Telomeres
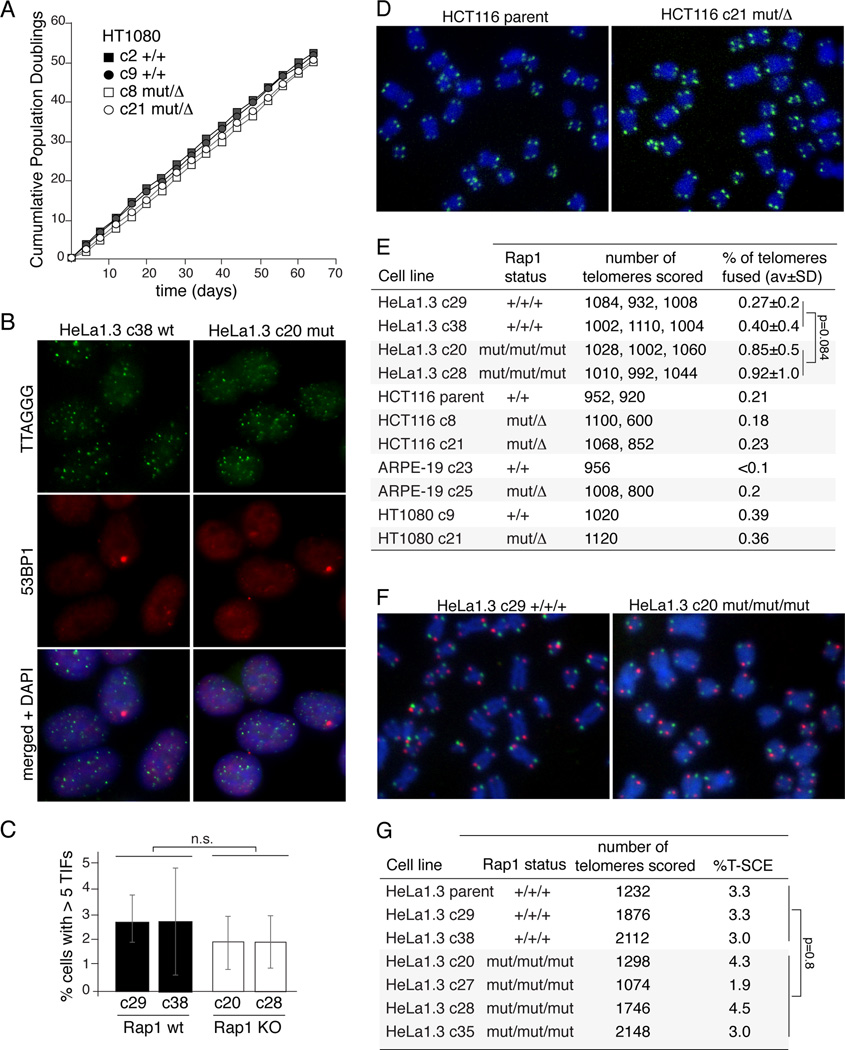
The Rap1-deficient cell lines proliferated normally (Figure 2A) and lacked a significant level of Telomere dysfunction-Induced Foci (TIFs), a read-out for telomere damage (Figure 2B,C), indicating that removal of Rap1 from telomeres does not result in a DNA damage response.
Figure 2. Telomere protection in Rap1-deficient cells.
(A) Growth curves of WT and Rap1 KO HT1080 clones.
(B) TIF assay on WT and KO HeLa1.3 clones. Green, telomeric FISH; red, IF for 53BP1; blue, DNA (DAPI).
(C) Quantification of TIFs assay (see (B)). Error bars: SDs of three independent experiments (n≥100 nuclei per clone). P values from a two-tailed paired t-test combining WT and KO datasets. n.s.: not significant.
(D) Metaphase chromosomes from the indicated WT and Rap1 KO cells. Green, telomeric FISH; blue, DNA (DAPI).
(E) Quantification of telomere fusions, detected as in (D), in the indicated clones. P values from a two-tailed paired t-test on combined WT and KO datasets.
(F) CO-FISH analysis on the indicated WT and KO HeLa clones.
(G) Table showing the percentage of telomeres showing T-SCEs as assayed in (F) in the indicated clones. P value from unpaired two-tailed t-test.
Cells lacking Rap1 also did not show a significant induction of DSB repair at telomeres (Figure 2D,E). Metaphase spreads of Rap1 KOs lacked chromosome end fusions, a read-out for telomeric NHEJ. Chromosome Orientation (CO-) FISH to monitor HDR-mediated Telomere-Sister Chromatid Exchanges (T-SCEs) established that recombination remained repressed at telomeres (Figure 2F,G). Thus, human telomeres remain protected from NHEJ and HDR in absence of Rap1.
Unaltered Telomere Length Dynamics in Absence of Rap1
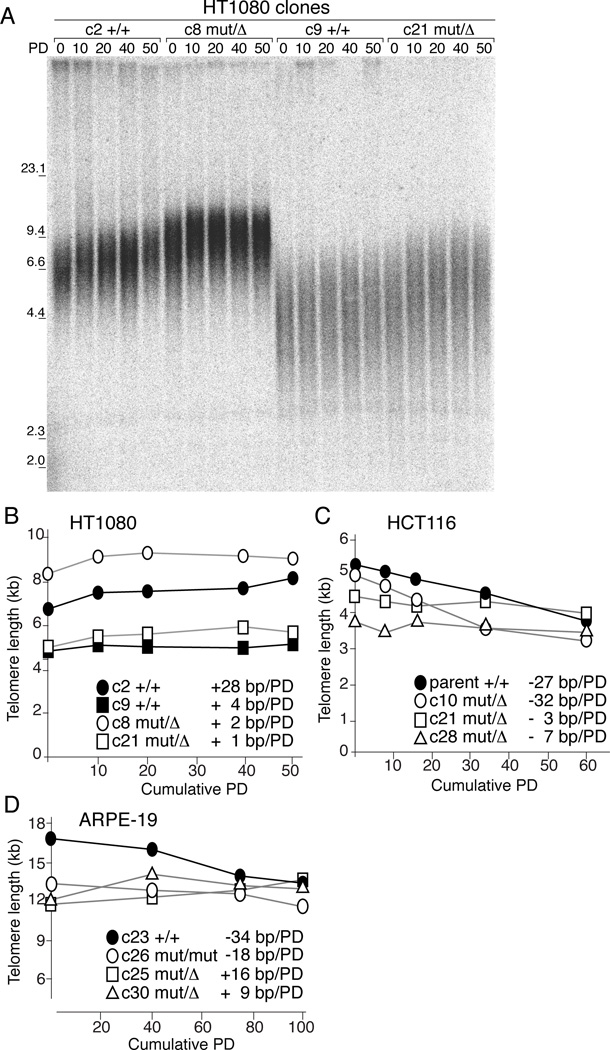
To determine whether Rap1 affected telomere length homeostasis, two HT1080 Rap1-deficient clones were cultured for 50 population doublings (PD) alongside two Rap1-proficient clones selected for their matching telomere lengths (Figure 3A). All clones exhibited a mild increase in telomere length (Figure 3B). The two Rap1-deficient clones lengthened their telomeres at a similar modest rate (1–2 bp/PD) whereas the two Rap1-proficient clones differed in the rate of telomere lengthening (28 and 4 bp/PD) (Figure 3B). Given the clonal variation and small differences in telomere length changes, the removal of Rap1 did not appear to have a strong effect on telomere length dynamics in HT1080 cells.
Figure 3. Loss of Rap1 does not affect telomere structure.
(A) Southern blot of telomeric restriction fragments from two WT and two KO HT1080 clones at the indicated PDs.
(B-D) Curves of average telomere lengths at indicated PDs in HT1080, HCT116, and ARPE-19 clones, respectively.
See also Figure S2.
Similarly, Rap1 did not affect the telomere length dynamics of HCT116 clones (Figure 3C). Two Rap1-deficient clones showed telomere shortening at variable rates (−3 to −32 bp/PD). Given that the telomere shortening in the parental cells (−27 bp/PD) is similar to that of one of the Rap1-deficient clones (c10), we conclude that also in HCT116 cells, Rap1 did not strongly affect telomere dynamics.
Finally, three Rap1-deficient ARPE-19 clones (c26, c25, and c30) showed wide variations in telomere dynamics, ranging from slight shortening (−18 bp/PD) to slight elongation (9 and 16 bp/PD) (Figure 3D). The single Rap1-proficient clone (c23) showed telomere shortening at a rate of −34 bp/PD. Thus, there is considerable variability in the telomere dynamics in ARPE-19 clones but no consistent effect of Rap1 deletion.
Given the lack of consistent shortening or lengthening phenotypes in multiple Rap1 knockouts, the simplest interpretation is that Rap1 does not play a major role in telomere length regulation. Deletion of Rap1 also did not induce an obvious change in the telomere length heterogeneity (Figure 3A and S2A), which was affected by Rap1 mutants in overexpression studies (Li and de Lange, 2003).
No Change in the Telomeric Overhang after Rap1 Loss
Removal of Rap1 from several cell lines also did not appear to affect the 3’ telomeric overhang. The amount of ss telomeric DNA was determined by in-gel hybridization of a labeled C-strand telomeric repeat probe to native telomeric restriction fragments. Quantification of normalized single-stranded telomeric DNA signals indicated that Rap1 status had no significant impact on the 3’ overhangs in HT1080, HCT116, and ARPE-19 cells (Figure S2A,B).
Unaltered Telomeric Positioning
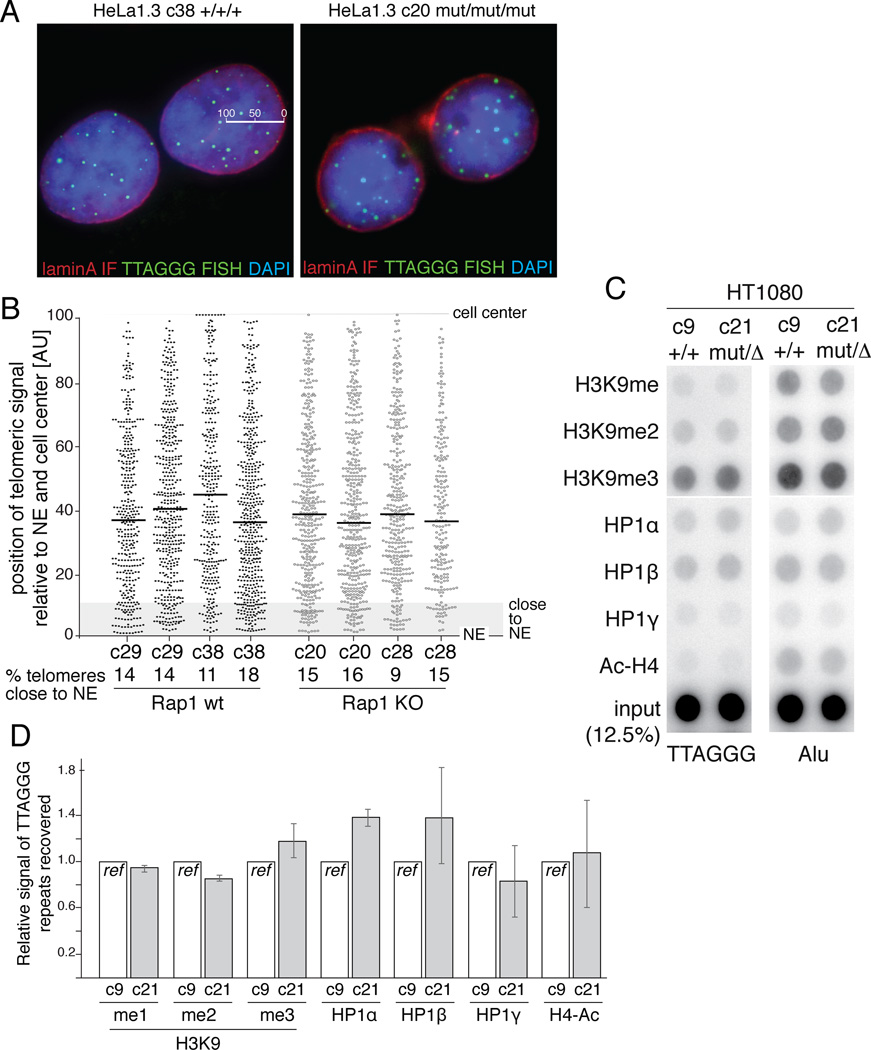
To determine whether Rap1 is involved in the peripheral positioning of telomeres in newly formed G1 cells (Crabbe et al., 2012), we used mitotic shake-off and analyzed daughter cells in early G1. Using Lamin A to mark the nuclear envelope (NE) and FISH to visualize telomeres, the subnuclear position of the telomeric signals was determined and compared between Rap1-proficient and -deficient cells (Figure 4A,B). The distribution of telomeres in the nucleus and median distance from the nuclear envelope was similar for two Rap1-proficient and -deficient clones in two independent experiments. Moreover, the percentage of telomeres present in a zone defined arbitrarily as the nuclear periphery (within 10% distance from the NE) was also similar regardless of Rap1 status, illustrating that Rap1 is not required for the more peripheral positioning of telomeres in early G1.
Figure 4. Effects of Rap1 on telomere position, chromatin modification, and gene expression.
(A) Combined IF for lamin A (red) and FISH for telomeres (green) in early G1 nuclei of WT and Rap1 KO HeLa1.3 clones. The scale in one nucleus indicates how the position of the telomeric signals was determined. Blue: DAPI DNA stain.
(B) Distance of telomeres from nuclear envelope (NE) in arbitrary units. For each nucleus imaged in a single plane, the ratio between the distance of each telomere from the center and the radius (center to NE) was plotted. Median distance for each clone is indicated by horizontal line. % telomeres at periphery reflects telomeres within 10% of the distance from the NE. Two independent experiments for each clone are shown.
(C) ChIP for modified histones at telomeres in WT and Rap1 KO cells. Duplicate blots were probed for telomeric DNA or Alu repeats.
(D) Relative telomeric ChIP signals obtained as in (C) were expressed as the ratio of signal in KO and WT clones (WT set to 1). Values represent averages of two experiments. Error bars: SEMs.
See also Table S1–3 and Figure S3.
Rap1 Affects Transcriptional Regulation, Not Telomeric Chromatin Modification
Mouse Rap1 localizes to over 8600 gene-associated loci, affecting transcription of numerous genes (Martinez et al., 2010; Yeung et al., 2013; Martinez et al., 2013), while human Rap1 is found at ~63 gene loci (Yang et al., 2011). To query the effect of Rap1 on the transcriptome, we performed microarray profiling on seven Rap1 WT and KO clones derived from three different cell lines, ARPE-19, HT1080 and HCT116. A number of differentially regulated genes were identified (Table S1–S3). The three Rap1-regulated genes in the ARPE-19 cells (LHX2, LRRC17 and CDO1) were validated by quantitative RT-PCR (qRT-PCR) and their response to Rap1 deletion was further confirmed on an additional ARPE-19 Rap1 KO clone (c26) (Figure S3A-C). The Rap1-regulated genes varied between the different cell lines, most likely due to the different origins of the cell lines. Gene ontology analysis was uninformative, because of the low number of genes identified by this limited analysis. However, one Rap1-regulated gene in the HT1080 cells was among the human Rap1-associated loci in the HT1080-derived HTC75 cell line (Yang et al., 2011). Taken together, these data are consistent with a role for Rap1 in transcriptional control in human cells.
Despite the effects of Rap1 on gene expression there was no significant effect of Rap1 on the abundance of the telomeric lncRNA called TERRA (telomeric repeat-containing RNA; reviewed in (Feuerhahn et al., 2010)) (Figure S3D,E). This finding is consistent with the unaltered TERRA levels after deletion of mouse Rap1 (Sfeir et al., 2010). Deletion of human Rap1 also had no detectable effect on general markers for the chromatin status at telomeres, as evidenced by ChIP for methylation of H3K9, acetylation of H4, and HP1β, HP1β and HP1γ (Figure 4C,D).
Discussion
Based on the genetic data presented here, we conclude that human Rap1 is not required for the protection of telomeres from NHEJ and has no obvious effect on telomere length regulation, contrary to what was anticipated from other studies (Bae and Baumann, 2007; Sarthy et al., 2009; Li and de Lange, 2003; O'Connor et al., 2004). In addition, telomeres lacking Rap1 remained protected from DNA damage signaling and HDR, and had a normal 3’ overhang. These findings are in agreement with the mouse Rap1 KO, which revealed no obvious phenotype other than that of telomeres becoming prone to undergo HDR when Ku70/80 was absent (Sfeir et al., 2010). Whether human telomeres lacking Rap1 also recombine more readily in a Ku70/80-deficient setting is difficult to assess since deletion of human Ku70/80 leads to rapid telomere loss and cell death (Li et al., 2002; Wang et al., 2009).
Despite this lack of requirement for Rap1 at telomeres, the human gene encoding Rap1 has diverged little from its chimpanzee counterpart (1 base change/100 codons; 0.25 amino acid (aa) changes/100 aa), whereas the genes for other shelterin components show much greater divergence (e.g., 3.6, 2.4, and 1.7 base changes/100 codons and 2.2, 1.1, and 0.65 aa changes/100 aa for TRF2, TRF1, and TIN2, respectively). Furthermore, assessment of a gene damage index for all protein-coding genes places TERF2IP in the top 20% of human genes with regard to mutation intolerance (Y. Itan, pers. comm.). In addition, Rap1 ranks among the top 10% of human genes in terms of ‘functional indispensability’, a characteristic that incorporates gene centrality (based on interaction data pooled from various biological systems), structural information, and evolutionary constraints (Khurana et al., 2013). In this regard, only TIN2 scores higher than Rap1, as expected based on its multiple interaction interfaces in shelterin.
These results raise the question why Rap1 is conserved. It appears unlikely that protection of telomeres from HDR is its sole raison d’etre, given the additional repression by Ku70/80. Furthermore, it seems unlikely that Rap1 has a tissue specific role at telomeres, given that mice lacking Rap1 are alive and largely normal, and the lack of telomeric phenotypes upon Rap1 deletion from four human cell lines of different tissue origin. The conservation of Rap1 is also not explained by a role in meiosis since the Rap1 KO mice are fertile and, unlike yeast lacking Rap1, form a normal meiotic bouquet (Kanoh and Ishikawa, 2001; Chikashige and Hiraoka, 2001; Chikashige et al., 2006; Sfeir et al., 2010; Scherthan et al., 2011; Shibuya et al., 2014). It is not excluded that Rap1, like TIN2, has (as yet unknown) multiple interactions within shelterin or interacts with shelterin-associated factors that explain its conservation or that its role at telomeres is redundant. However, we favor the idea that the conservation of Rap1 is due to its role in transcriptional regulation where it may have multiple distinct interaction partners that constrain its evolution. It will be of interest to identify the Rap1-interacting partners that are at gene loci since such partners may well be used by Rap1 to fulfill (as yet undefined) telomere functions.
Our data point to the difficulty in interpreting experiments in which telomeric phenotypes are observed upon overexpression of shelterin (mutant) proteins or their partial inactivation by shRNAs. We suspect that the prior finding of changes in telomere length and heterogeneity upon overexpression of Rap1 mutants (Li and de Lange, 2003) were due to nucleoplasmic titration of factors that (indirectly) influence these phenotypes. Similarly, the artificial tethering of Rap1 to telomeres may have had an effect on NHEJ that does not reflect the normal function of the protein (Sarthy et al., 2009).
These data indicate that while mammalian Rap1 has functionally diverged away from its yeast predecessors, mouse and human Rap1 are very similar. Both Rap1 knockout mice and human cells are viable, lack hallmarks of telomere dysfunction, and have no overt change in telomere length settings. While the Rap1 components of human and mouse shelterin are indistinguishable, it will be important to query the functions of other shelterin components and associated factors to gain a complete understanding of telomere maintenance and protection in human cells. Our data show that genetic approaches, such as the use of TALENs and CRISPR (Hsu et al., 2014) are versatile tools to this effect.
Experimental Procedures
Detailed experimental procedures are available in the Supplemental Information. Cell culture techniques, telomere length analysis, telomeric overhang assay, analysis of metaphase spreads, TIF analysis, ChIP, telomeric FISH, IF, and immunoblotting analysis were performed as described previously (Sfeir et al., 2010; Takai et al., 2010). TALENs knockout of TERF2IP was performed using standard protocols (Chen et al., 2011). The assay for telomeric position was published previously (Crabbe et al., 2012). Microarray analysis was performed using Whole Human Genome DNA microarrays (Illumina HumanHT-12 v4) with GeneSpring v12.6 for data analysis. Microarray data is deposited in the Gene Expression Omnibus under the accession number GSE61900.
Supplementary Material
Acknowledgements
We thank Dr. Alexandre Bolze for his guidance on the evolutionary selection pressure on Rap1. We thank Dr. Agnel Sfeir for helpful discussion. This work was supported by grants from the NIH (5R37GM49046 and 5RO1AG16642) to TdL. TdL is an American Cancer Society Research Professor. D.H. is a New Scholar in Aging of the Ellison Medical Foundation and supported by the Glenn Foundation as well as the The Shurl and Kay Curci Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
All experiments were designed by SK and TdL, executed by SK. DH designed the TALENs and provided advice. SK and TdL wrote the paper.
References
- 1.Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 2.Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 4.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 5.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 7.Crabbe L, Cesare AJ, Kasuboski JM, Fitzpatrick JA, Karlseder J. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep. 2012;2:1521–1529. doi: 10.1016/j.celrep.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 9.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992a;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 10.Hardy CF, Balderes D, Shore D. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol Cell Biol. 1992b;12:1209–1217. doi: 10.1128/mcb.12.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 13.Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput Biol. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 15.Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy DL, Blackburn EH. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol. 2004;24:10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell. 2003;14:5060–5068. doi: 10.1091/mbc.E03-06-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci USA. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, Lustig AJ. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics. 1996;143:81–93. doi: 10.1093/genetics/143.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lustig AJ, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 23.Martinez P, Gomez-Lopez G, Garcia F, Mercken E, Mitchell S, Flores JM, de Cabo R, Blasco MA. RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Rep. 2013;3:2059–2074. doi: 10.1016/j.celrep.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 27.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 28.Pardo B, Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24:3117–3127. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherthan H, Sfeir A, de Lange T. Rap1-independent telomere attachment and bouquet formation in mammalian meiosis. Chromosoma. 2011;120:151–157. doi: 10.1007/s00412-010-0295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedivy J, Vogelstein B, Liber H, Hendrickson EA, Rosmarin A. Gene Trageting in Human Cells Without Isogenic DNA. Science. 1999;283:9–9a. [Google Scholar]
- 32.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibuya H, Ishiguro K, Watanabe Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat Cell Biol. 2014;16:145–156. doi: 10.1038/ncb2896. [DOI] [PubMed] [Google Scholar]
- 34.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 35.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 36.Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sussel L, Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere Length Homeostasis Is Achieved via a Switch between Telomerase- Extendible and - Nonextendible States. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 40.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 43.Xu L, Blackburn EH. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol. 2004;167:819–830. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeung F, Ramirez CM, Mateos-Gomez PA, Pinzaru A, Ceccarini G, Kabir S, Fernandez-Hernando C, Sfeir A. Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Rep. 2013;3:1847–1856. doi: 10.1016/j.celrep.2013.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.