Abstract
Background
Genome-wide aberrations of the classic epigenetic modification 5-methylcytosine (5mC), considered the hallmark of gene silencing, has been implicated to play a pivotal role in mediating carcinogenic transformation of healthy cells. Recently, three epigenetic marks derived from enzymatic oxidization of 5mC namely 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), have been discovered in the mammalian genome. Growing evidence suggests that these novel bases possess unique regulatory functions and may play critical roles in carcinogenesis.
Methods
To provide a quantitative basis for these rare epigenetic marks, we have designed a biotin-avidin mediated Enzyme-based Immunoassay (EIA) and evaluated its performance in genomic DNA isolated from blood of patients diagnosed with metastatic forms of lung, pancreatic and bladder cancer, as well as healthy controls. The proposed EIA incorporates spatially optimized biotinylated antibody and a high degree of horseradish-peroxidase (HRP) labeled streptavidin, facilitating signal amplification and sensitive detection.
Results
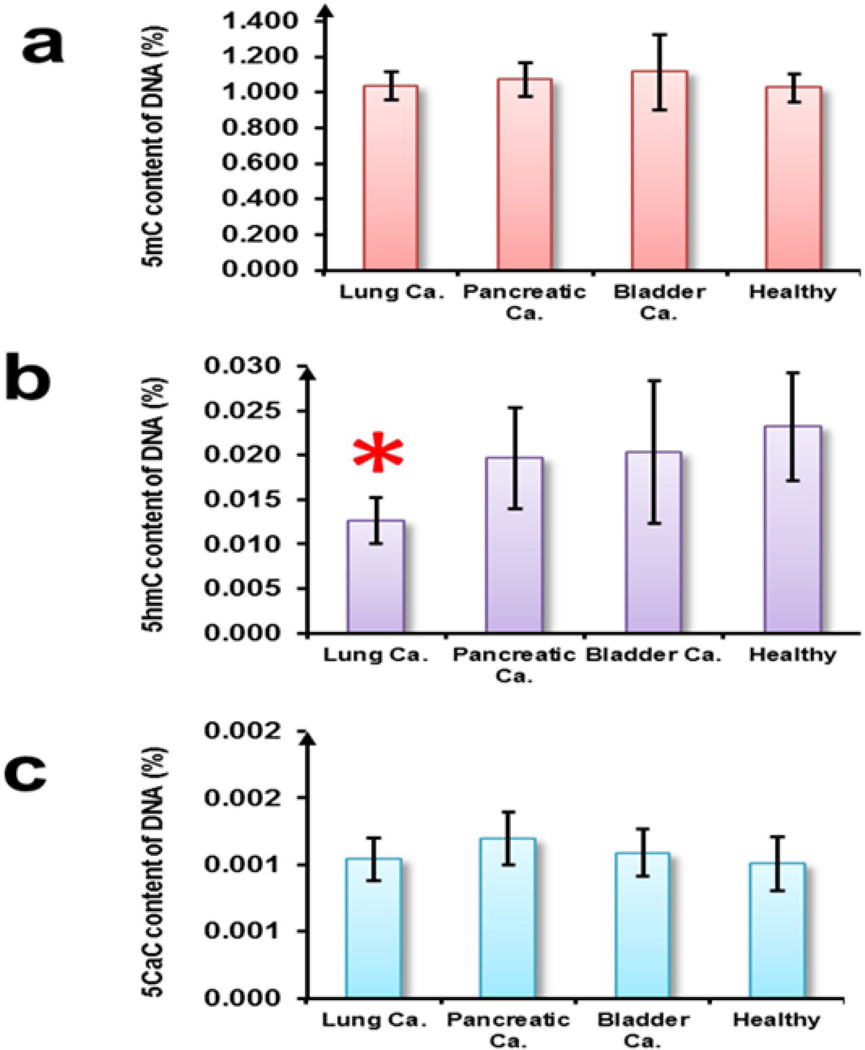
We report that the percentages of 5mC, 5hmC and 5caC present in the genomic DNA of blood in healthy controls as 1.025 + 0.081, 0.023 + 0.006 and 0.001 + 0.0002 respectively. We observed a significant (p<0.05) decrease in the mean global percentage of 5hmC in blood of patients with malignant lung cancer (0.013 + 0.003 %) in comparison to healthy controls.
Conclusion
The precise biological roles of these epigenetic modifications in cancers are still unknown but in the past two years it has become evident that the global 5hmC content is drastically reduced in a variety of cancers. To the best of our knowledge, this is the first report of decreased 5hmC content in the blood of metastatic lung cancer patients and the clinical utility of this observation needs to be further validated in larger sample datasets.
Keywords: 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), 5-carboxylcytosine (5caC), Enzyme-based Immunoassay (EIA), Cancer, Epigenetic modifications
1. Introduction
The addition of a methyl group (-CH3) at the C-5 position of the pyrimidine ring of cytosine nucleotide, mediated by a family of DNA methyltransferases, serves as an epigenetic signaling tool employed by cells to “turn off” the expression of the implicated genes [1–3]. This epigenetic modification also known as 5-methylcytosine (5mC) plays an important role in numerous cellular processes including embryonic development, genomic imprinting, cellular differentiation and preservation of genomic stability [4, 5]. Recently, three other enzymatically (TET family of dioxygenases) mediated epigenetic derivatives of 5mC namely 5-hydroxymethylcytosine (5hmC;-CH2OH group at Cytosine C-5) [6, 7], 5-formylcytosine (5fC; -CHO at Cytosine C-5) [5, 8] and 5-carboxylcytosine (5caC;-COOH at Cytosine C-5) [3, 5] have been discovered. These derivatives serve as intermediates in the DNA demethylation pathway and are believed to confer unique transcriptional potential to genes [3, 9–12].
Epigenetic alterations are very important in determining cellular fate and their aberrations are frequently observed during initiation and progression of cancers [2, 3]. Locus-specific DNA methylation markers such as DNA hypermethylation of Breast cancer type 1 susceptibility (BRCA1) gene in breast and ovarian cancer [13], Death-associated protein kinase (DAPK) gene in bladder cancer [14], O-6-methylguanine-DNA methyltransferase (MGMT) gene in colorectal and lung cancers [15, 16] have been proposed as prognostic and diagnostic biomarkers in clinical settings. Alterations of global levels of 5mC have been reported in early neoplasia and solid tumors [17–19]. For instance, there are reports suggesting a fundamental association between global methylation levels and tumor aggressiveness, regardless of the origin of cancer tissue [20–24]. Recently, extensive loss of 5hmC has been observed in multiple cancer types like lungs, prostate, breast, colon, brain and blood [3, 9–11, 25] suggesting a potential role of 5hmC in cancer development. Also in addition to 5hmC, since other critical enzymatic components (TET family of dioxygenases, Isocitrate dehydrogenase enzyme family and DNA repair enzyme thymine DNA glycosylase) of the dynamically balanced DNA demethylation have been implicated in cancers [26], the downstream components of the pathway - 5fC and 5caC - might also have different roles in cancers and need to be explored [27, 28]. Therefore, quantitative analysis on genome-wide distribution of these epigenetic marks (epimarks) has been critically considered for clinical applications. In particular, information on alterations of epimarks could prove useful in identifying cancer in its early stages for prognosis or monitor the success of administered therapies in cancer patients..
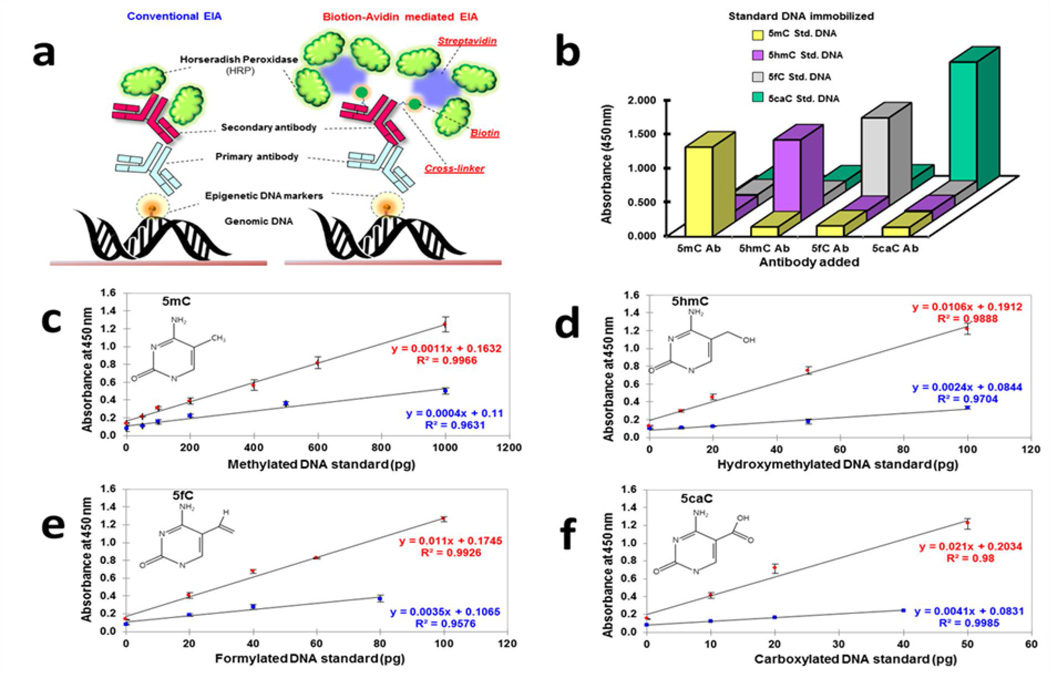
The classical method to quantify global DNA methylation is based on sophisticated instruments such as liquid chromatography and more recently accompanied with mass spectrometric techniques (LC-MS) [5, 29]. While methods based on the principles of LC-MS are quantitative and reproducible; these methods are complex, exorbitantly priced, require skilled personnel and presently unsuitable for high throughput analyses [30, 31]. Conventional immunoassays, employing HRP-conjugated-antibody, have widely been used as a quantitative method due to its analytical merits (high sensitivity, high-throughput utility, cost-effectiveness and ease of use) for the analyses of various biomarkers [29, 32]. Recent reports on the quantification of 5mC and 5hmC from different tissues and cell lines based on immunoassay principles [32] have been commercialized [33–35]. However, these immunoassays cannot be currently used for clinical diagnosis because they lack sensitivity (particularly in the assessment of 5hmC, 5fC and 5caC) and are expensive. Thus we propose a biotin-avidin mediated EIA with enhanced signal output using spatially optimized biotinylated antibody coupled with streptavidin with a high degree of HRP labeling for the detection of 5mC, 5hmC, 5fC and 5caC (refer to Figure 1a). This method was first compared with conventional immunoassay method and upon satisfactory performance, applied to detect the rare DNA epimarks in the blood of metastatic cancer patients.
Figure 1. Analytical concept of biotin-avidin mediated EIA.
(a) Schematic representation of the proposed EIA platform developed to quantitate the content of the four epimarks (5mC, 5hmC, 5fC, and 5caC) contrasted with the conventional EIA (employing HRP labeled secondary antibody). Enhanced signal detection was achieved by (i) controlling cross-linking between the Secondary antibody and Biotin and (ii) Multiple enzyme (i.e. HRP) attachment via Streptavidin conjugation. (b) Multiple Cross-Reactivity Test to determine the specificity of the proposed EIA. X axis denotes the respective primary antibody used in the EIA (anti- 5mC, 5hmC, 5fC or 5caC), Z axis denotes the positive controls (DNA standard that is either methylated, hydroxymethylated, formylated or carboxylated; Refer to the key) immobilized to the microwells and Y axis denotes the corresponding colorimetric signal (absorbance at 450 nm) generated. Other than the specific primary antibody and the type of DNA standard immobilized indicated, the rest of the EIA was performed using identical conditions. (c-f) Calibration curves for quantitation of the precise content of 5mC, 5hmC, 5fC and 5caC in the genome, respectively. Values denoted in red were obtained on performing the proposed EIA, while values denoted in blue were obtained on performing conventional EIA. Error bars indicate standard deviations between technical replicates.
2. Materials and Methods
2.1 Materials
DNA coating solution, succinimidyl-6-[biotinamido]-6-hexanamidohexanoate (NHS-LC-LC biotin), succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxy-(6-amidocaproate) (LC-SMCC), succinimidyl 6-[3(2-pyridyldithio) propionamido] hexanoate (LC-SPDP), dithiothreitol (DTT), horseradish peroxidase (HRP), Sephadex G-15, mouse anti-rabbit IgG and goat anti-mouse IgG were purchased from Pierce (Rockford, IL). Casein, Tween 20, tetramethylbenzidine (TMB), ethylenediaminetetraacetic acid disodium salt (EDTA) were obtained from Sigma (St. Louis, MO). Rabbit antibodies specific to 5hmC, 5fC and 5caC were supplied by Active Motif (Carlsbad, CA). Mouse anti-5mC was purchased from Eurogentec (Fremont, CA). Four epigenetic DNA standards to be used as positive controls: (i) 5mC and (ii) 5hmC from Zymo Research (Irvine, CA), (iii) 5fC from Epigentek (Farmingdale, NY) and (iv)5caC from Active Motif (Carlsbad, CA) were purchased. Streptavidin was purchased from Prozyme (Hayward, CA).
2.2 Clinical Sample Preparation
Biospecimens were collected from patients with lung, bladder or pancreatic cancers and healthy controls by Hoosier Oncology Group, Indianapolis, IN as part of the study entitled, “A Biological Sample Collection Protocol of Patients with and without Metastatic Solid Organ Malignancies: Hoosier Oncology Group Study BANK09-138”. Blood samples were centrifuged at 3,500 rpm for 30 minutes and stored at −70°C. All studies were approved by Institutional Review Boards (IRB) protocols at their respective institutions. Informed consent forms were obtained from all subjects. Genomic DNA was isolated from the biospecimens using QIAamp DNA Mini and Blood kit (Qiagen Inc., Valencia, CA) following manufacturer’s instructions. Information on clinical samples is presented in Supplementary Table 1. DNA was quantified by NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). Absorbance of the DNA samples was collected at 260 and 280 nm with a conventional 10 mm path length. The DNA concentration in (ng/µl) was determined by absorbance at 260 nm based on Beer’s Law. The ratio of absorbance at 260 and 280 nm was used to assess the purity of the DNA.
2.3 Preparation of Biotinylated secondary antibody
500 µg of mouse anti-rabbit or goat anti-mouse IgG secondary antibody (2 mg/ml) was labeled with 50-fold molar excess of NHS-LC-LC-biotin (dissolved in DMSO) using 10 mM phosphate buffer with 150 mM NaCl (pH 7.4) as the reaction buffer. The reaction was carried out at room temperature for 1 hour and the biotinylated antibody was purified by dialysis (dialysis buffer: 10 mM phosphate buffer contained 150 mM NaCl, pH 7.4) followed by size exclusion chromatography on Sephadex G-15 gel column where free biotin or biotin not conjugated to the secondary antibody was removed.The prepared antibodies were concentrated and stored in 10 mM phosphate buffer containing 150 mM NaCl (pH 7.4) at 4°C until further use. Bradford assay was employed to determine a concentration of labeled antibodies. The molar ratio of biotin to antibody in the biotinylated-antibody conjugates was evaluated by the HABA (4'-hydroxyazobenzene-2-carboxylic acid) Biotin quantification assay, as described previously [36].
2.4 Conjugation of streptavidin with HRP
1 mg of Streptavidin was first coupled to 50 molar excess LC-SPDP for 1 hour followed by treatment with DTT (10 mM final concentration) for reduction of dithiol groups at room temperature for 30 minutes and purified via Sephadex G-15 gel filtration column, which was pre-equilibrated in 10 mM PBS buffer containing 5 mM EDTA. By following a similar procedure, 3.7 mg HRP was activated by LC-SMCC and purified [37]. The thiolated streptavidin and maleimide-activated HRP (molar ratio 1:5) were then mixed at room temperature for 2 hours with gentle shaking to enable conjugation. This mixture was thereafter stored at 4°C until further use. The enzymatic activities of both unconjugated and streptavidin-conjugated HRP were tested via enzyme assay on microtiter plate. 0 and 0.1 fmol/mL of unconjugated as well as streptavidin-conjugated HRP were reacted with substrate solution containing hydrogen peroxide and tetramethyl benzidine.
2.5 Analytical procedure of the biotin-avidin mediated EIA
To quantitate the four target epimarks, genomic DNA (100 ng for quantification of global 5mC content and 200 ng for quantification of global 5hmC, 5fC or 5caC content) isolated from the clinical biospecimens was first coated on polystyrene surface of a microwell with DNA coating solution (100 µL) at room temperature for 2 hours with gentle agitation. After washing 3 X 5 minutes in PBS-Tween 20 and once with PBS, the residual surface was blocked with 0.5% (w/v) casein in PBS at 37°C for 1 hour. Subsequently, incubation with primary antibody (0.5 µg/mL, 100 µL) recognizing 5mC, 5hmC, 5fC or 5caC diluted in PBS containing 0.5% casein and 0.1% Tween 20 was performed for 1 hour at 37 °C. After thorough washings, biotinlyated antibodies specific to each primary antibody (0.5 µg/mL,100 µL) was added and incubated for 1 hour at 37 °C. Finally, streptavidin conjugated to HRP (0.25 µg/mL, 100 µL) was added and incubated for 1 hour at 37 °C. For signal generation, 200 µL of the colorimetric substrate mixture [1000: 10: 1 volume ratio of the following (a) 50 mM sodium acetate buffer, pH 5.1, (b) 1% (w/v) tetramethylbenzidine and (c) 3% (v/v) H2O2] was added to the microwell and maintained for 15 minutes at room temperature followed by the addition of 50 µL of 2 M sulfuric acid to stop the reaction. The optical intensity was finally measured at 450 nm with the Versamax™ absorbance microplate reader (Molecular Device; Sunnyvale, CA). To quantitatively compare with the EIA, conventional EIA was performed under the same conditions except for using secondary antibody labeled with HRP instead of using biotinylated secondary antibody and streptavidin-HRP conjugate. The Limits of Detection (LOD) have been calculated by a statistical method [38–40] based on
S.D → standard deviation of the response and
m → slope of the calibration curve (= dY/dX)
2.6 Quantitative analysis – Calibration curve
To establish calibration curve, increasing amounts of standard DNA bearing 5mC, 5hmC, 5fC or 5caC was coated on the microwell surface and immunoassay performed as described above. The signals obtained were plotted, and linear regression curve and R-square were calculated for quantification of epimarks in unknown samples as per the following equations:
| Equation (i) |
| Equation (ii) |
The normalized sample O.D450 nm was obtained from subtracting the O.D450 nm of the blank sample (no genomic/standard DNA added, but subsequent steps performed similarly) from the observed sample O.D450 nm. ‘n’ denoted a factor to normalize the respective epigenetic derivatives in the positive standard DNA to 100% (5mC, 5hmC and 5caC content in the respective positive standard DNAs being 24.9%, 24.9% and 15.7%). T-test was used to evaluate the statistical significance of the differences of levels of the DNA epimarks in the biospecimens of the metastatic cancer and healthy counterparts [41, 42].
3. Results
3.1 Analytical concept
In the proposed EIA, the obtained signal intensities were directly proportional to the amount of the epimarks. The DNA extracted from the clinical samples and subsequently used as input for EIA was of high purity (A260/280: 1.7–1.9) and yield (4–6 µg/ 200 µl of blood). As described in the Methods Section, the first procedure in the proposed platform after specific recognition of epimarks by the respective primary antibodies involved incorporation of biotinylated-secondary antibody. Here, several amine-reactive biotin cross-linkers with different lengths (ranging from 1 to 3 nm) were tested and NHS-LC-LC-biotin linker (with a length of 3 nm) was selected to be optimal. HABA (4'-hydroxyazobenzene-2-carboxylic acid) assay [36] estimated that 4.5 molecules of biotin were bound per molecule of secondary antibody. The final concentration of the biotinylated antibody was determined to 1 mg/ml by Bradford assay. The increased number of biotin on the secondary antibody facilitates multiple enzymes (i.e., HRP) tagged streptavidin to bind with the secondary antibody, which in turn remains optimally bound to the epimark recognizing primary antibody as shown in Figure 1a and thereby achieves a higher degree of labeling. For the best analytical performances, each streptavidin moiety was labelled with multiple HRP by controlling length of linkers to be extended up to 3 nm (sum of length of LC-SPDP and LC-SMCC), facilitating reduction of steric hindrance and favoring a high degree of labelling (three to four HRP per streptavidin which was verified by SDS-PAGE). The peroxidase activities of both unconjugated HRP and streptavidin-conjugated HRP were compared and did not indicate any significant difference in producing the color signal. Conventionally, since unconjugated HRP molecules do not participate in avidin-biotin reaction and are subsequently removed during the washing procedures, purification of the final conjugate is not deemed necessary.
3.2 Calibration Curve and Specificity Determination
To accurately quantitate the percentages of each epimark in genomic DNA, four types of standard curves were prepared by plotting the optical densities at 450 nm wavelength versus the amount of standard input DNA (picogram, pg unit) as shown in Figure 1 c-f. The results obtained from the proposed EIA demonstrate that the limit of detection (LOD) of each mark to be as follows: 3.802 pg for 5mC, 0.543 pg for 5hmC, 3.139 pg of 5fC standard DNA and 0.209 pg for 5caC and was suitable for detection of the rare epimarks in the scarce metastatic biospecimens. The proposed EIA examined here can be regarded as heteroscedastic because of some of the steps involved in the procedure like the NanoDrop’s UV-vis absorption spectrometry, manual handling and pipetting errors. Nevertheless, the above interpretation retains the fundamental concept of the original assumption of homoscedasticity i.e if the analyte of LOD concentration is measured, the probability of the measurements falling below the limit of decision is 5% [40]. In addition, the linearized regression curves have a high R-square value and low standard deviation, enabling reliable quantification of the corresponding marks from unknown samples (Kindly see Supplementary Tables 2–5 for inter-assay % coefficient of variation and Supplementary Tables 6–9 for intra assay % coefficient of variation). Furthermore, due to coexistence of the four epimarks in genomic DNA, it is critical to eliminate cross-reactivity which is considered as one of the major analytical factors for evaluation of the assay. Multiple cross-reactivity tests were performed to demonstrate high specificity without significant cross-reactivity in the EIA (illustrated in Figure 1b).
3.3 Clinical Application
We decided to put our proposed EIA to test on a limited dataset (n=27) of rare metastatic biospecimens and determine the content of the epigenetic derivatives in cancers. The level of 5mC was the most abundant amongst the four epimarks and constituted a mean value of 1.025 + 0.081 % of the genome in healthy controls. The 5mC content of the metastatic lung, bladder and pancreatic cancer biospecimens was not found to be significantly altered from the mean 5hmC content of healthy controls (Figure 2a). The mean 5hmC content of healthy controls was determined to be 0.023 + 0.006 % of the genome. In comparison to 5mC, inter-sample variability of 5hmC was found to be higher but consistent with earlier findings [43]. The mean 5hmC content of the lung cancer biospecimens was found to be 0.013 + 0.003 % of the genome (significantly lower at p <0.05), while no significant difference in the mean 5hmC content of pancreatic and bladder cancer biospecimens was observed in comparison to the healthy controls (Figure 2b). The precise levels of 5fC in the biospecimens could not be accurately calculated as the percentage of 5fC in the 5fC DNA standard was not disclosed by the manufacturer for reasons of proprietary and the normalization factor ‘n’ of equation (i) (described in Materials and Methods section) could not be determined. However, the normalized O.D450 nm readouts did not show any significant difference between the cancer biospecimens and their healthy counterpart. The mean 5caC content was determined to be 0.001 + 0.0002 % of the genome in healthy controls and was not found to be significantly altered in the cancer biospecimens (Figure 2c).
Figure 2. Quantification of epimarks in clinical biospecimens.
Mean global percentage of (a) 5mC, (b) 5hmC and (c) 5caC in the blood of the indicated groups of clinical samples (n=27). Error bars denote standard deviations between the clinical samples of each group. Star denotes significant difference in cancer biospecimens compared to healthy controls (p < 0.05, t-test).
4.0 Discussion
It is increasingly evident that the status of DNA methylation and its derivatives is linked to mechanisms of cancer predisposition [3, 21, 42, 44]. Thus, profiling DNA methylation derivatives in whole blood as a predictive marker for metastatic cancer is of particular interest because peripheral blood is a convenient tissue to assay and its collection is non-invasive [45]. The blood samples utilized in this study were derived from patients diagnosed at metastatic stage of cancer (Supplementary Table 1) where distant metastases was detected (M1 Stage in TNM classification scheme) and could serve as a proxy tissue for practical purposes [46]. The high affinity of avidin for biotin [47] forms the fundamental basis of the biotin-avidin mediated EIA. Multiple HRP labeled avidin can bind to biotin labeled antibody, forming the biotin-avidin complex and thereby producing enhanced intensity of the immune reaction. This biotin-avidin mediated EIA facilitates sensitive detection of the rare DNA methylation derivatives: 5mC constitutes ~1% of the genome, 5hmC (0.5 – <0.005%) [10], precise levels of 5fC and 5caC have not been well documented but in mouse ES cells it has been found to be 100-fold less frequent than 5hmC [5]. The LOD of 5mC and 5hmC reported by several commercially available immunoassay kits based solely on HRP-labeled secondary antibody is 20 and 10 pg respectively for a similar input range [10, 33, 34, 48], thus the sensitivity of the biotin-avidin mediated EIA can be considered superior in comparison (refer to Section 3.2 for the respective LODs).
To the best of our knowledge, we report for the first time a decrease in the 5hmC content (p < 0.05) in the DNA extracted from blood of patients diagnosed with metastatic lung cancer in comparison to healthy controls. Recently, it has been conclusively demonstrated by LC-MS/MS technology that the 5hmC content is decreased significantly between 2–5 fold in human squamous cell carcinomas of the lung with respect to matched tissues of healthy counterparts [42]. Using semi-quantitative immunohistochemistry (IHC) and dot-blot methodologies, a profound reduction of the 5hmC content in human lung cancer tissues and genetically engineered mouse model of lung cancer has also been reported [25]. Other than lung cancer, reduction of 5hmC content has also been demonstrated in prostate, breast, colon, brain and blood malignancies [3, 9–11, 25]. Unlike locus-specific changes, alterations of epimarks at the global level is rapidly gaining prominence as they generate additional cellular epigenetic heterogeneity, which can be used to define previously unrecognized subsets of cancer patients with distinct clinical outcomes. Further study of the loss of 5hmC in metastatic cancers may offer the promise of novel therapeutic modalities especially in lung cancers that are responsible for the maximum number of cancer deaths in US, with 2013 estimates of ~159,480 [49]. A better understanding of the role of epimarks in cancer is bound to impact clinical outcomes [20] because of their prognostic and diagnostic utility.
Thus in conclusion, due to lack of sensitivity of conventional bioanalytical techniques for the detection of the rare epigenetic modifications of the DNA (5mC, 5hmC, 5fC and 5caC), that could be integrated into high-throughput platforms; we propose a biotin-avidin mediated EIA strategy with high analytical performances. The quantitative determination of epimarks in blood, a proxy tissue of metastatic cancers, would be a practical alternative to conventional methods such as tissue biopsy, enabling diagnosis of cancers with a simple procedure that is sensitive and more economical compared to other available technologies [30, 31] (Refer to Table 1). As epigenetic modifications are potentially reversible, there is a possibility that therapies directed at modifying the epigenome might be a reality in the future [3, 50, 51] and it is probable that the investigation of global levels of methylated cytosine derivatives could be considered in the diagnostics of cancer patients especially for determining clinical prognosis. We expect this method to be applicable for diagnosis of cancer biospecimens and with minor modifications applicable for other routine assessments.
Table 1.
Comparison of the proposed EIA with LC-MS/MS and commercial EIA based kits, provides distinct advantages in terms of the optimal DNA input, Limit of Detection and cost.
| Assay | Optimal DNA input | LOD | Equipment needed |
Cost |
|---|---|---|---|---|
| LC-MS/MS | 1000–5000 ng [52] | 1.251 pg for 5mC [52], 1.411 pg for 5hmC [52], 1.55 pg for 5caC [52] |
HPLC, Mass Spectrometer |
$15 sample* |
|
Commercial EIA based Kits |
100 ng for 5mC [34], >200 ng for 5hmC [33] |
20 pg for 5mC [34], 10 pg for 5hmC [33] |
Colorimeter | $4 – 6.8/ sample [33–35] |
|
Biotin-avidin mediated EIA |
100 ng for 5mC, 200 ng for 5hmC, 200 ng for 5caC |
3.802 pg for 5mC 0.543 pg for 5hmC 0.209 pg for 5caC |
Colorimeter | $0.1/ sample |
The cost of LC-MS/MS analysis was determined based on a quote provided by the Purdue Metabolomics Core Facility (Dr. Bruce Cooper).
Supplementary Material
Highlights.
Development of Biotin-Avidin mediated Enzyme-based Immunoassay (EIA).
Quantitate epigenetic DNA methylated Cytosine derivatives (5mC, 5hmC and 5caC) in blood samples.
Ultra-sensitive assay with enhanced Limit Of Detection for the epigenetic marks.
First report of reduction in global 5hmC content in the blood of metastatic lung cancer
First quantitative report of levels of 5caC in blood.
Acknowledgements
Partial student support for BC by the National Institutes of Health, National Cancer Institute R25CA128770 Cancer Prevention Internship Program administered by the Oncological Sciences Center and the Discovery Learning Research Center at Purdue University is appreciated. This publication was made possible in part, with support from the Indiana Clinical and Translational Sciences Institute funded, in part by Grant Number (TR000006) from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Footnotes
Publisher's Disclaimer: This is a PDF .le of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 2.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 3.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature Reviews Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PA, Taylor SM. CELLULAR-DIFFERENTIATION, CYTIDINE ANALOGS AND DNA METHYLATION. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 5.Ito S, Shen L, Dai Q, et al. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He YF, Li BZ, Li Z, et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haffner MC, Chaux A, Meeker AK, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. Journal of nucleic acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nature Structural & Molecular Biology. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. Journal of the National Cancer Institute. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich MG, Weisenberger DJ, Cheng JC, et al. Detection of methylated apoptosisassociated genes in urine sediments of bladder cancer patients. Clinical Cancer Research. 2004;10:7457–7465. doi: 10.1158/1078-0432.CCR-04-0930. [DOI] [PubMed] [Google Scholar]
- 15.Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Research. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 16.Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: The time is now. Critical Reviews in Oncology Hematology. 2008;68:1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg AP, Vogelstein B. HYPOMETHYLATION DISTINGUISHES GENES OF SOME HUMAN CANCERS FROM THEIR NORMAL COUNTERPARTS. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 18.Gamasosa MA, Slagel VA, Trewyn RW, et al. THE 5-METHYLCYTOSINE CONTENT OF DNA FROM HUMAN-TUMORS. Nucleic Acids Research. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Smet C, Loriot A. DNA hypomethylation in cancer Epigenetic scars of a neoplastic journey. Epigenetics. 2010;5:8. doi: 10.4161/epi.5.3.11447. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature Medicine. 17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Mohan A, Guleria R. Biomarkers in cancer screening, research and detection: present and future: a review. Biomarkers. 2006;11:385–405. doi: 10.1080/13547500600775011. [DOI] [PubMed] [Google Scholar]
- 22.Liggett T, Melnikov A, Yi Q-l, et al. Differential Methylation of Cell-Free Circulating DNA Among Patients With Pancreatic Cancer Versus Chronic Pancreatitis. Cancer. 116:1674–1680. doi: 10.1002/cncr.24893. [DOI] [PubMed] [Google Scholar]
- 23.Tao LH, Wang W, Kramer PM, Lubet RA, Steele VE, Pereira MA. Modulation of DNA hypomethylation as a surrogate endpoint biomarker for chemoprevention of colon cancer. Molecular Carcinogenesis. 2004;39:79–84. doi: 10.1002/mc.20003. [DOI] [PubMed] [Google Scholar]
- 24.Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21:5450–5461. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Liu Y, Bai F, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song C-X, He C. Potential functional roles of DNA demethylation intermediates. Trends in Biochemical Sciences. 2013;38:480–484. doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song C-X, He C. Balance of DNA methylation and demethylation in cancer development. Genome Biology. 2012;13 doi: 10.1186/gb-2012-13-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thuc L, Kim K-P, Fan G, Faull KF. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Analytical Biochemistry. 2011;412:203–209. doi: 10.1016/j.ab.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Waterland RA. Methods of DNA methylation analysis. Current Opinion in Clinical Nutrition and Metabolic Care. 2007;10:576–581. doi: 10.1097/MCO.0b013e3282bf6f43. [DOI] [PubMed] [Google Scholar]
- 31.Gupta R, Nagarajan A, Wajapeyee N. Advances in genome-wide DNA methylation analysis. Biotechniques. 2010;49:III–XI. doi: 10.2144/000113493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leng SX, McElhaney JE, Walston JD, Xie D, Fedarko NS, Kuchel GA. ELISA and multiplex technologies for cytokine measurement in inflammation and aging research. Journals of Gerontology Series a-Biological Sciences and Medical Sciences. 2008;63:879–884. doi: 10.1093/gerona/63.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inc. EG. MethylFlash Hydroxymethylated DNA Quantification Kit (Colorimetric) Retrieved 04/16/2014 from http://www.epigentek.com/catalog/methylflash-hydroxymethylated-dna-quantification-kit-colorimetric-p-1858.html.
- 34.Inc. EG. MethylFlash Methylated DNA Quantification Kit (Colorimetric) Retrieved 04/16/2014 from http://www.epigentek.com/catalog/methylflash-methylated-dna-quantification-kit-colorimetric-p-2094.html.
- 35.Cell Biolabs I. Global DNA Methylation ELISA. Retrieved 04/16/2014 from http://www.cellbiolabs.com/global-dna-methylation-elisa.
- 36.Janolino VG, Fontecha J, Swaisgood HE. A spectrophotometric assay for biotin-binding sites of immobilized avidin. Applied Biochemistry and Biotechnology. 1996;56:1–7. [Google Scholar]
- 37.Cho I-H, Paek E-H, Kim Y-K, Kim J-H, Paek S-H. Chemiluminometric enzyme-linked immunosorbent assays (ELISA)-on-a-chip biosensor based on cross-flow chromatography. Analytica Chimica Acta. 2009;632:247–255. doi: 10.1016/j.aca.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Miller JC, Miller JN. BASIC STATISTICAL-METHODS FOR ANALYTICAL-CHEMISTRY .1. STATISTICS OF REPEATED MEASUREMENTS - A REVIEW. Analyst. 1988;113:1351–1356. [Google Scholar]
- 39.Brady TC, Yang TJ, Hyde WG, Kind AJ, Hill DW. Detection of flunixin in greyhound urine by a kinetic enzyme-linked immunosorbent assay. Journal of Analytical Toxicology. 1997;21:190–196. doi: 10.1093/jat/21.3.190. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi Y, Matsuda R, Maitani T, et al. Precision, limit of detection and range of quantitation in competitive ELISA. Analytical Chemistry. 2004;76:1295–1301. doi: 10.1021/ac0302859. [DOI] [PubMed] [Google Scholar]
- 41.Liu CC, Ou TT, Wu CC, et al. Global DNA methylation, DNMT1, and MBD2 in patients with systemic lupus erythematosus. Lupus. 2011;20:131–136. doi: 10.1177/0961203310381517. [DOI] [PubMed] [Google Scholar]
- 42.Jin S-G, Jiang Y, Qiu R, et al. 5-Hydroxymethylcytosine Is Strongly Depleted in Human Cancers but Its Levels Do Not Correlate with IDH1 Mutations. Cancer Research. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konstandin N, Bultmann S, Szwagierczak A, et al. Genomic 5-hydroxymethylcytosine levels correlate with TET2 mutations and a distinct global gene expression pattern in secondary acute myeloid leukemia. Leukemia. 2011;25:1649–1652. doi: 10.1038/leu.2011.134. [DOI] [PubMed] [Google Scholar]
- 44.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahara T, Maegawa S, Chung W, et al. Examination of Whole Blood DNA Methylation as a Potential Risk Marker for Gastric Cancer. Cancer Prevention Research. 2013;6:1093–1100. doi: 10.1158/1940-6207.CAPR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiology Biomarkers & Prevention. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 47.Barnard DL, Johnson FB, Richards DF. COMPARISON OF AN AVIDIN-BIOTIN IMMUNOASSAY WITH 3 COMMERCIALLY AVAILABLE IMMUNOFLUORESCENCE KITS FOR TYPING OF HERPES-SIMPLEX VIRUS. Journal of Clinical Pathology. 1985;38:1158–1162. doi: 10.1136/jcp.38.10.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inc. EG. DNA Methylation Quantification. Retrieved 04/16/2014 from http://www.epigentek.com/catalog/dna-methylation-quantification-c-75_21_48.html.
- 49.ACS ACS. Cancer facts and figures - 2013. Cancer Facts and Figures. 2013:60. 60. [Google Scholar]
- 50.DeWoskin VA, Million RP. The epigenetics pipeline. Nature Reviews Drug Discovery. 2013;12:661–662. doi: 10.1038/nrd4091. [DOI] [PubMed] [Google Scholar]
- 51.Singh V, Sharma P, Capalash N. DNA Methyltransferase-1 Inhibitors as Epigenetic Therapy for Cancer. Current Cancer Drug Targets. 2013;13:379–399. doi: 10.2174/15680096113139990077. [DOI] [PubMed] [Google Scholar]
- 52.Shen L, Zhang Y. Enzymatic Analysis of Tet Proteins: Key Enzymes in the Metabolism of DNA Methylation. Nuclesomes, Histones & Chromatin, Pt A. 2012;512:93–105. doi: 10.1016/B978-0-12-391940-3.00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.