SUMMARY
It is well understood that antigen-presenting cells (APC) within tumors typically do not maintain cytotoxic T cell (CTL) function, despite engaging them. Across multiple mouse tumor models and human tumor biopsies, we have delineated the intratumoral dendritic-cell (DC) populations as distinct from macrophage populations. Within these, CD103+ DCs are extremely sparse and yet remarkably capable CTL stimulators. These are uniquely dependent upon IRF8, Zbtb46 and Batf3 transcription factors and generated by GM-CSF and Flt3L cytokines. Regressing tumors have higher proportions of these cells, T-cell dependent immune clearance relies upon them, and abundance of their transcripts in human tumors correlates with clinical outcome. This cell type presents opportunities for prognostic and therapeutic approaches across multiple cancer types.
Keywords: Tumor, CTL, Dendritic Cell, Macrophage, IRF4, IRF8, GM-CSF, CSF1, Adoptive T cell Immunotherapy
INTRODUCTION
In immunoevasive tumors, a complex microenvironment develops alongside the lesion and despite the recruitment of CD8 T cells, there is no effective control of the developing mass. This microenvironment is prominently composed of the mononuclear phagocytic lineage (MPS) in addition to tumor-associated fibroblasts (TAF) and a variety of additional immune infiltrates including neutrophils and tumor specific T cells (Hanahan and Weinberg, 2011; Kraman et al., 2010). A primary conundrum at present is to understand why the latter cells, which include CD8+ T cells with the potential to kill the tumor, ultimately fail to do so.
The MPS-lineage typically should have the potential to present antigens to T cells by virtue of their phagocytic capacity. In tumors the infiltrating MPS lineage has been described as comprising tumor-associated macrophages (TAM: see Lewis and Pollard, 2006), tumor DC as well as monocytes. While monocytes do not clearly function as APC, they may act at minimum as precursors to TAMs and DCs in normal and transformed tissues (Cheong et al., 2010; Cortez-Retamozo et al., 2012; Geissmann et al., 2010). A number of studies have implicated specific MPS-lineage derived cells, particularly immature monocytes, in dampening the responsiveness of CTLs in tumors (Kusmartsev et al., 2005). Through intravital imaging, we and others have found that antigen-specific CD8 T cells are initially captured in prolonged interactions with myeloid cells, along the tumor border (Boissonnas et al., 2013; Engelhardt et al., 2012). In those experiments, the myeloid cells that phagocytosed tumor antigens and crosspresented them, when purified in aggregate, failed to stimulate T cells in vitro. Thus, by all criteria to date, the immune microenvironment is a combination of poorly stimulatory and/or actively inhibitory APC partners for CTLs. While depletion of regulatory T cells and checkpoint blockades are suggested to broadly license tumor APCs (Curran et al., 2010), there has been no evidence of strongly stimulatory APCs within the native tumor.
As immunotherapies targeting costimulatory blockade (Leach et al., 1996) move to the forefront of cancer therapeutics, it becomes increasingly important to understand the spatial and temporal context of costimulation and antigen-presentation. Antigen presentation at the lymph node (LN) for priming of tumor reactive T cell expansion is clearly critical, and as such has been successfully targeted therapeutically using GM-CSF (Dranoff, 2002) to increase presentation in the LN. As such, much of the focus has remained on the LN despite our clear understanding that antigen-presentation also occurs within the tumor itself and likely influences the functions of tumor CTLs.
It was therefore our goal to dissect the distinct composition of the myeloid tumor microenvironment across a broad range of tumors, with the purpose of understanding the lineage relationships amongst these populations and how each influenced tumor T cell responses and outcome.
RESULTS
Surface Markers Delineate Rare Tumoral DC Subsets from Abundant Macrophages
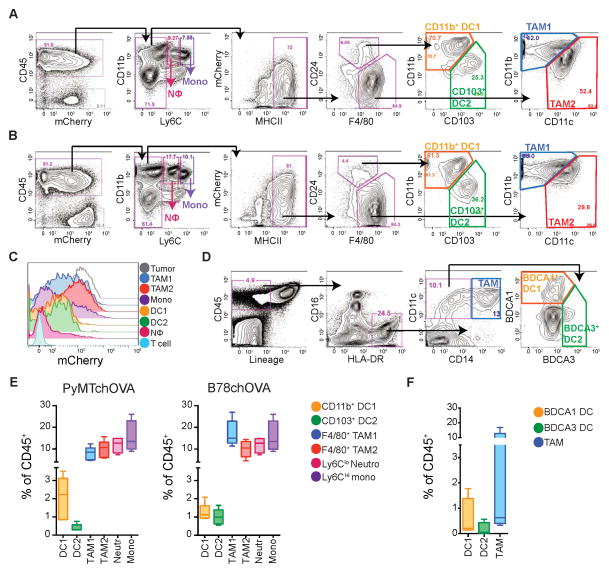
To dissect the tumor infiltrating myeloid populations, we devised an 11-color flow cytometry panel and progressive gating strategy using a spontaneous breast tumor model, PyMTChOVA (Engelhardt et al., 2012), engineered along with the initiating oncogene to independently co-express fluorescent mCherry protein and ovalbumin. We profiled the tumoral CD45+ compartment, many of which had phagocytosed tumor antigen and thus exhibit mCherry fluorescence (Figure 1A). Subgating all hematopoietic cells by the myeloid-specific marker CD11b and the monocyte-marker Ly6C allowed removal of neutrophils and monocytes (see Figure S1A). Within the MHCII+ cells, DCs were distinguished from Macrophages based on CD24hi and F4/80lo expression, neither of which, alone, is sufficient to make this distinction. Subsequently, DCs were found to parse into two populations based on differential expression of CD11b and CD103, as has been observed in healthy peripheral tissues (Hashimoto et al., 2011). We found these populations in two mouse models of melanoma (B78ChOVA (a variant of B16 expressing mCherry and OVA), Figure 1B and BRAF V600E, Figure S1B), across mouse strains (e.g. FVB PyMT; Figure S1B), and in ectopic tumors (Lewis Lung Carcinoma; Figure S1B). We refer to these DC populations as “CD11b+ DC1” and “CD103+ DC2” henceforth for ease of discrimination and discussion.
Figure 1. Rare DC and abundant Macrophages in mouse and human tumors.
(A) Flow cytometry and gating of tumor APC populations from digested and CD45 enriched PyMTchOVA tumors. A–C: Representative of greater than 5 independent experiments.
(B) Cytometry of tumor APC populations in ectopic B78ChOVA tumors.
(C) Histogram of tumor-derived mCherry fluorescence, by tumor-infiltrating immune cells in B78chOVA.
(D) Representative cytometry of digested human melanoma metastatic biopsy identifying corollary DC and TAM populations defined by CD45+ Lin− (CD3e, CD56, CD19) HLA-DR+ and split by CD14, BDCA1 and BDCA3. Double negative cells likely reflect B cells escaping lineage gate, immature monocytes or pDC.
(E) Relative proportions of tumor infiltrating myeloid cells as a % of total CD45+ cells for PyMTchOVA and B78chOVA models. Pooled data from individual tumors, presented as mean ± SEM from (n=5) mice.
(F) Frequency of DC and TAM populations infiltrating human metastatic melanoma presented as a % of total CD45+ cells. Pooled data from multiple patients, presented as mean ± SEM from (n=4) biopsies.
See also Figure S1.
Parsing of the F4/80hi CD24lo compartment also revealed two types of macrophages, identified by differential expression of CD11c and CD11b. CD11clo CD11bhi (heretofore “TAM1”) and CD11chi CD11blo cells (“TAM2”) appear to broadly correspond to similarly delineated MHCIIhi and MHCIIlo populations (Movahedi et al., 2010) (see Figure 5C below). While CD11c, otherwise a ‘protoypical’ DC marker, was highest on DCs it was highly expressed in TAM2 and to a lesser extent in TAM1 (Figure S1C). These populations existed across all models examined although the prevalence of each and their ability to be unambiguously distinguished varied slightly (Figure 1A–B, S1B). For the rest of this report, we therefore applied our lineage and function studies to one example of spontaneous (PyMTChOVA) and ectopic tumor model (B78ChOVA), except where indicated.
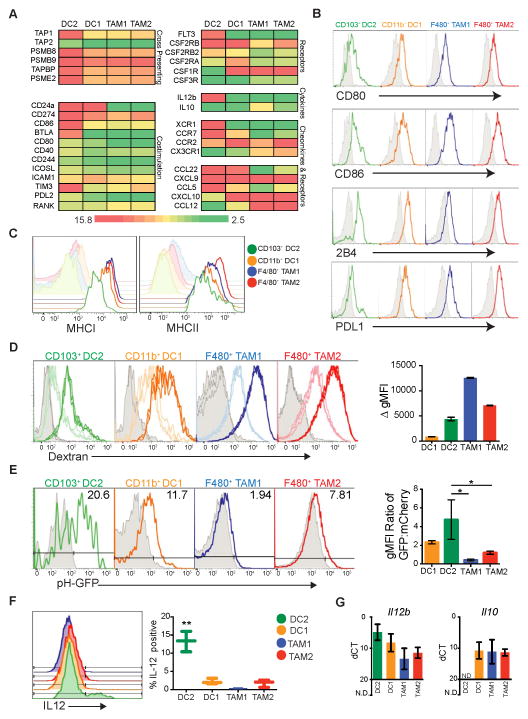
Figure 5. Unique antigen processing and presentation capabilities of CD103+ DC2.
All data (A–G) is from the ectopic B78chOVA tumor model.
(A) Heat map of Log2 transformed expression from RNAseq across populations for selected genes involved in cross presenting, cytokine and chemokine production, and costimulation. Color scale defined as, green=bottom 20th percentile, red=top 80th percentile, with 20th–80th percentile graduated and centered at yellow (50th percentile). Data from biological triplicates of sorted cells.
(B) Cytometry of surface protein levels of ligands for T cell regulatory molecules (colored) as compared to respective isotypes (grey).
(C) Cytometry of MCHI and MHCII (colored) expression compared to respective isotype (shaded).
(D) Cytometry of ex vivo dextran uptake across populations. Grey=no dextran, light histogram=dextran binding at 4° C, and dark histogram=dextran uptake at 37° C, displayed in triplicate. Delta geometric Mean Fluorescence Intensity (gMFI) for each population plotted as mean ± SEM. Data are representative of 2 independent experiments (n=6).
(E) Cytometry analysis of relative pH of endocytic compartments across populations. B78 tumor cells were transfected with the ratiometric pH construct, N1-mCherry-pHlourin. Representative histograms show florescence of pHluorin in mCherry+ cells, where less pH-GFP represents a more acidic environment. Grey histograms are respective populations from a non-pHluorin expressing control tumor (B78 parental). Data summarized as the ratio of gMFI between GFP and mCherry fluorescence. Data presented as mean ratio ± SEM, pooled from 3 independent experiments.
(F) Intracellular cytokine stain of IL12 in populations. % of IL12+ cells quantified across each population, data pooled from 2 independent experiments, (n=3), plotted as mean ± SEM. Statistical significance indicated by *p<0.05.
(G) Il12b and Il10 transcript levels, measured by qPCR. Data presented as mean ΔCt ± SEM calculated from biological triplicates (n=3) of individual tumors, (N.D. not detected).
See also Figure S4.
mCherry loading and retention, derived from the tumor, was assessed for each of these populations. This revealed that the uptakehi cells, localized to the tumor margin in our previous report and then identified only by CD11c (Engelhardt et al., 2012), were best captured in the TAM1 and TAM2 gates (Figure 1C and Figure S1D). Comparatively, CD11b+ DC1s and CD103+ DC2s took up or retained less mCherry while some monocytes but few neutrophils showed evidence of modest antigen loading. CD11b+ and CD103+ DC subsets have been found in many peripheral mouse tissues and their counterparts have been identified in peripheral human tissues, defined by expression of BDCA1 and BDCA3, respectively (Dzionek et al., 2000; Haniffa et al., 2012). We found that an equivalent TAM/DC distinction was also possible in human metastatic melanoma samples using these markers (Figure 1D). CD16− HLADR+ CD11c+CD14+ cells representing all TAMs were distinct from CD16− HLADR+ CD11c+CD14− DC populations, which were in turn parsed by differential expression of BDCA1 (“DC1”) and BDCA3 (“DC2”). Common across mouse models (Figure 1E) and human melanoma biopsies (Figure 1F) is the presence and rarity of the CD11b+/BDCA1 DC1 and CD103+/BDCA3 DC2 populations, with DC2 being particularly sparse.
Protein and Transcriptional Delineation of Tumor DCs and Macrophages
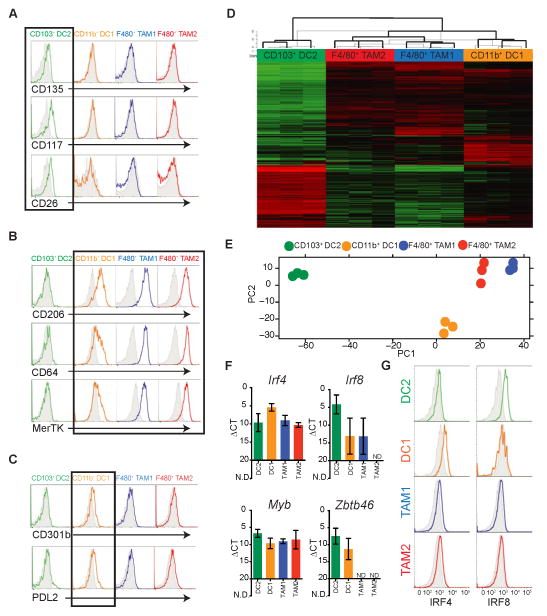
To validate our gating strategies we applied panels of antibodies defined by the ImmGen consortium (Gautier et al., 2012; Miller et al., 2012). Consistent with our assignment of “DC”, CD103+ DC2 expressed CD135 (Flt3), CD117 (cKit) and CD26 whereas both TAM populations did not in the B78chOVA and PyMTchOVA models. (Figure 2A and Figure S2A). Surprisingly, CD11b+ DC1 did not express detectable levels of DC markers and actually segregated more with TAM1 and TAM2 by virtue of expression of several “macrophage” markers including CD206, CD64 and MerTK (Figure 2B and Figure S2B). CD11b+ DC1, however, slightly expressed CD301b and PDL2, both of which have been used to define IRF4 dependent “DCTh2” populations found in the skin (Figure 2C and Figure S2C) (Gao et al., 2013; Kumamoto et al., 2013).
Figure 2. Surface and transcriptional profiling highlights distinct lineages of tumor DCs and Macrophages.
All data (A–G) is from the ectopic B78chOVA tumor model. Cell lineages are defined as per Figure 1.
(A) Expression of a panel of DC specific markers compared to respective isotype (grey shaded). A black box outlines the CD103+ DC2 population.
(B) Differential expression of Macrophage specific markers (colored) with corresponding isotypes (grey shaded). A black box outlines the CD11b+ DC1, TAM1, and TAM2 populations.
(C) Specific expression of DC-Th2 makers (colored), by CD11b+ DC1 populations compared to respective isotype (grey shaded). A black box outlines CD11b+ DC1.
(D) Global transcriptional profiles revealed by RNAseq of FACS-purified populations from biological triplicates. Data displayed as a heat map of Log2 fold change relative to the global average of the top 1000 genes by maximum variance between DC1, DC2, TAM1, and TAM2.
(E) PCA of DC1, DC2, TAM1, and TAM2 populations based on RNAseq global transcriptional profiles.
(F) qRT-PCR analysis of expression of Irf4, Irf8, Myb, and Zbtb46 (zDC) from sorted APC populations. Data presented as mean ΔCt ± SEM calculated from biological triplicates (n=3), (N.D. not detected).
(G) Intracellular staining for IRF4 and IRF8 in tumor APC populations as compared to the respective isotype (grey).
See also Figure S2.
To further delineate these APCs, we analyzed the gene expression profiles of sorted cells from B78chOVA tumors using RNAseq. As shown in Figure 2D, blocks of genes clearly segregate the four populations, with TAM1, TAM2 and CD11b+ DC1 being the most similar by PCA analysis (Figure 2E) and CD103+ DC2 the most distinct. Amongst the genes most differentially expressed, DC lineage-defining transcription factors Irf8 (Tamura et al., 2005) and Zbtb46 (zDC) (Meredith et al., 2012) were specific for CD103+ DC2 alone, or both DCs respectively, whereas Irf4 was modestly enriched in CD11b+ DC1 and all of which were validated by RT-qPCR (Figure 2F). This was also confirmed at the protein level by intracellular flow cytometry for IRF4/8 (Figure 2G and Figure S2D). All populations expressed Myb, which indicates hematopoietic stem cell origin as opposed to deriving from tissue precursors, seeded from the yolk-sac (Schulz et al., 2012).
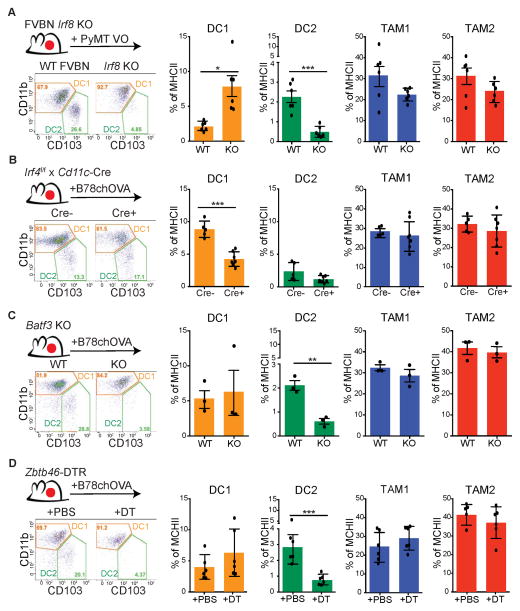
As these intratumoral populations may derive through distinct tumor-specific mechanisms and not rely on these transcription factors as they do in some normal tissues, we investigated IRF8, IRF4, Batf3 and zDC dependency using knockout or transcription factor-driven Diphtheria toxin receptor (DTR) mice. We took advantage of various ectopic tumors, due to the vagaries and length of breeding these alleles to a spontaneous model. Using an ectopic PyMT breast tumor model, we found that loss of Irf8 specifically ablated the CD103+ DC2s but did not affect TAM1 or TAM2 and mildly enriched the percentage of CD11b+ DC1, perhaps as a result of compensation (Figure 3A). Conversely, conditional deletion of Irf4, driven by CD11c-Cre (Williams et al., 2013) resulted in the specific reduction in CD11b+ DC1 with little change in the others in the B78chOVA model (Figure 3B). In agreement with RNAseq data, Batf3 deficient animals also lacked tumoral CD103+ DC2 populations in a B78chOVA model, without effect on CD11b+ DC1, TAM1 or TAM2 proportions (Figure 3C). Finally, when a zDC-driven DTR allele was used, we somewhat unexpectedly found a specific and significant reduction in CD103+ DC2 with little or no changes in the CD11b+ DC1 or TAM1/TAM2 populations in B78chOVA tumors (Figure 3D). This may represent vagaries of the DTR allele or subtle but significant variations in zDC expression. Taken together, we conclude that CD103+ DC2 represent a distinct lineage of APC as compared to CD11b+ DC1 and the highly abundant TAM1/TAM2 in the tumor.
Figure 3. Differential IRF4, IRF8 and Batf3 requirements for tumor infiltrating APC populations.
All data is representative flow cytometric analysis of CD11b+DC1 and CD103+DC2 populations (gated on CD45+, Ly6C−, MHCII+, and CD24+) Data shown as mean ± SEM. Statistical significance indicated by *p<0.05, **p<0.01, ***p<0.001; ns=not statistically significant.
(A) Ectopic PyMT-VO tumors from an Irf8−/−(KO) compared to control (WT). Relative cell proportions as a % of total MHCII+ cells. Data pooled from individual mice (n=6) from 2 independent experiments.
(B) Ectopic B78chOVA tumors in Irf4f/f x CD11c-CRE+ host compared to Cre-negative littermates. Relative cell proportions as a % of total MHCII+ cells. Data pooled from individual mice (n=7) from 2 independent experiments.
(C) Ectopic B78chOVA tumors in Batf3 KO, compared to WT. Relative cell proportions graphed as a % of total MHCII+. Data pooled from individual mice (n=6).
(D) Ectopic B78chOVA tumors in Zbtb46-DTR mice, receiving acute 24 hour depletion with DT or PBS. Relative cell proportions graphed as a % of total MHCII+ cells. Data pooled from individual mice (n=6) from 2 independent experiments.
CD103+DC2 are Programmed by Distinct Cytokines
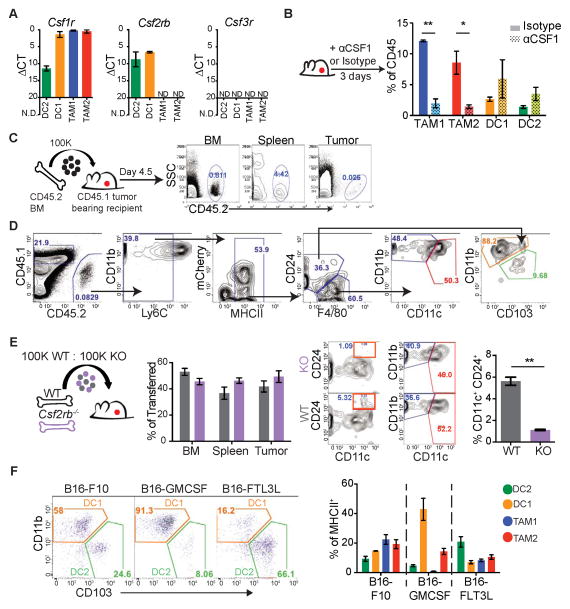
APCs derive from bone marrow (BM) precursors and their differentiation into DC/macrophage subsets depends on specific cytokines. To determine the cytokines driving differentiation into these populations we queried Colony Stimulating Factor (CSF) receptor expression across models by qPCR. Whereas Csf1r (M-CSFR) was found exclusively in TAM1, TAM2 and CD11b+ DC1, Csf2rb (GM-CSFR) was uniquely expressed in the DC1 and DC2 subsets, and Csf3r (G-CSFR) was absent in all (Figure 4A). Using either neutralizing antibody treatment or cytokine receptor deficient mice with ectopic tumors, we functionally tested CSF cytokine reliance of the APCs at the tumor.
Figure 4. Differential reliance on M-CSF and GM-CSF cytokines by tumor-infiltrating APC populations.
(A) qPCR of Csf1r, Csf2rb, and Csf3r expression from sorted APCs. Data presented as mean ΔCt ± SEM calculated from biological triplicates (n=3) of individual B78chOVA tumors (N.D. not detected).
(B) Cytometry of tumor APCs after 3 days of αCSF1 (αCSF1, dotted) compared to isotype (filled) treated tumor animals. Quantified as % of total tumor CD45+ cells, pooled from individual mice (n=6) from 2 independent experiments shown as mean ± SEM. Statistical significance indicated by *p<0.05, **p<0.01, ***p<0.001; ns=not statistically significant
(C) Schematic of BM progenitor adoptive transfer and contributions to BM, spleen, and tumor.
(D) Representative cytometry of tumor arriving congenic cells. Gated on CD45.2 and following the gating strategy of Figure 1A.
(E) Competitive BM adoptive transfer of WT vs. Csf2rb KO GMP progenitors into B78chOVA tumor recipients. Repopulation efficiency plotted as % of total transferred cells. Representative gating of tumor arriving GMP cells, WT (grey), KO (purple). Quantification of tumor arriving DCs, defined by CD24+ CD11c+. Data pooled from 2 independent experiments, plotted as mean ± SEM from individual tumors (n=6).
(F) Cytometry of CD11b+ DC1 and CD103+ DC2 populations (gated on CD45+, Ly6C− MHCII+, CD24+) between ectopic B16-F10, B16-GMCSF and B16-FLT3L cytokine expressing tumors. Populations presented as % of total MHCII+ cells for each tumor. Data are pooled from 3 independent experiments, plotted as mean ± SEM from individual tumors (n=6).
See also Figure S3.
While TAM1 and TAM2 cells critically relied upon CSF1 for their maintenance, as has been shown previously (Wyckoff et al., 2004), CD11b+ DC1 and CD103+ DC2 populations were uniquely independent of CSF1 (Figure 4B). For use of cytokine receptor deficient mice, we developed a congenic adoptive transfer model, whereby Granulocyte Macrophage Progenitors (GMP) were transferred into ectopic tumor-bearing hosts and repopulation was tracked in the BM, spleen and tumor (Figure 4C). At the tumor GMP-derived cells populated all myeloid compartments, confirming GMP origin of CD11b+ DC1, CD103+ DC2, TAM1, and TAM2 (Figure 4D). By use of the GMP adoptive system with a competitive transfer, we found a selective inability of Csf2rb−/− cells to reconstitute DCs at the tumor, here defined as the sum of DC1/DC2 using CD24+ CD11c+ gating. We found no effect on TAM1 and TAM2 repopulation, suggesting a unique requirement of CSF2 (GM-CSF) for tumoral DC development (Figure 4E) while no requirement for CSF-3 was found for any of the four APCs (Figure S3).
As DCs are prototypically driven by GM-CSF or FLT3-ligand (FLT3L), we assessed cytokine sufficiency to drive DC populations at the tumor using B16 melanoma tumor models engineered to express GMCSF or FLT3L. While GMCSF expression by the tumor drastically skewed the proportion of CD11b+ DC1, FLT3L expressing tumors drove unique expansion of the rare CD103+ DC2 at the tumor (Figure 4F).
Unique Antigen Processing and Presentation Capabilities of CD103+ DC2
Having established the lineage requirements of the different APCs we then assessed their ability to initiate, engage and sustain T cell responses. To parse the cells with regard to antigen processing, presentation, and costimulation we analyzed transcript and protein levels of genes involved in these pathways using RNASeq data from Figure 2. Differences were considerable, across broad swaths of potential APC function (Figure 5A). Notably, while surface levels of molecules involved in regulating T cell responses including CD80, CD86 and 2B4 were comparable between populations, CD103+ DC2s showed distinct transcriptional signatures consistent with heightened cross presentation, enhanced costimulation, and increased expression of chemokines that would be expected to enhance T cell interactions (Figure 5A, B and Figure S4A). There were no major differences in MHCI and MHCII expression between the APCs with the exception of slightly reduced MHCI on CD103+ DC2 (Figure 5C). However, significant differences in phagocytic capacity were observed in CD103+ DC2s compared to TAM1/TAM2, measured exogenously by ex vivo dextran uptake from ectopic tumors (Figure 5D).
As DC maturation and phagocytic capacity are often inversely correlated, we hypothesized that the decreased phagocytic capacity of CD103+ DC2 might correspond to a more mature DC with increased cross-presentation of antigen (Guermonprez et al., 2002). Efficient cross presentation of antigen in DCs relies upon NOX2 to regulate phagasomal pH thereby preventing destruction of T cell peptides, which can be determined using a ratiometric assay comparing intracellular fluorescence intensity of a pH-sensitive and pH-insensitive fluorophore following phagocytosis (Savina et al., 2006). We therefore generated a B78 tumor line expressing a fusion of a pH-sensitive GFP (pHluorin, quenched below pH 6.5) and a pH insensitive fluorophore (mCherry). By analyzing pHluorin intensity alone within the mCherry+ compartment of each population, we found that only the ‘DC’ populations maintained pHluorin in an alkaline (fluorescent) environment; comparing the ratio of pHluorin and mCherry signals showed that CD103+ DC2 maintained the most basic endocytic compartment while TAM1 and TAM2 populations displayed highly acidic and therefore degradative phagocytic pathways (Figure 5E). In addition to the increased alkaline phagosomal lumen of CD103+ DC2, these cells demonstrated differential expression of the pro-inflammatory cytokine IL-12 and absence of anti-inflammatory IL-10 (Figures 5F, G and Figure S4B). Together, all of these features suggest CD103+ DC2 are highly poised for efficient antigen cross-presentation to CD8+ T cells.
CD103+ DC2 are Superior Stimulators of Naïve and Activated CD8+ T cells
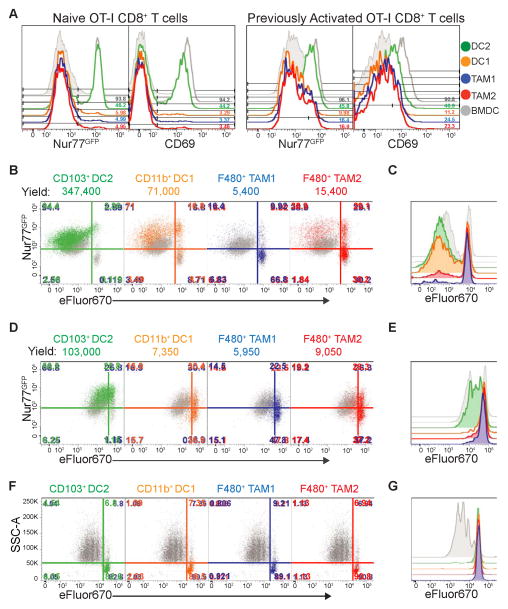
Previously, we found that the aggregate antigen-ingesting myeloid compartment could stimulate naïve but not previously activated CD8+ T cells when taken directly from tumors (Engelhardt et al., 2012). However, based on the unique cross-presentation phenotype of CD103+ DC2, we sought to test the T cell stimulatory capacity of each population, freshly isolated from tumors. After 12 hours of coculture with ovalbumin-specific OT-I CD8+ T cells, the CD103+ DC2 population was the only population capable of robustly inducing TCR signaling, measured by GFP expression driven by a Nur77 reporter (Nur77GFP) and CD69 levels in both naïve and previously activated OT-I CD8+ T cells. Importantly, this was consistent in both ectopic and spontaneous mouse models (Figure 6A and Figure S5A). Extended coculture of dye-labeled OT-I CD8+ T cells revealed that CD11b+ DC1 and CD103+ DC2 populations were the most robust stimulators of naive CD8+ T cell proliferation, and demonstrated that nearly the entire stimulatory capacity previously identified in phagocytosing tumor myeloid cells lies within these DC (Figure 6B–C, Figure S5B, and Figure S5C). Interestingly, CD103+ DC2 were uniquely capable of inducing strong proliferation of established CTLs, which were not stimulated by the other populations, indicating CD103+ DC2 were superior cross presenting stimulators of CTLs in the tumor (Figure 6D–E and Figure S5D, respectively).
Figure 6. CD103+ DCs are Superior T cell stimulators for naïve and activated CD8+ T cells.
All data (A–G) is from the ectopic B78chOVA tumor model. T cells+BMDC (shaded grey), T cells+BMDC+ SL8 (unshaded grey), T cells+tumor APCs (respective colored histograms). Plated at 20,000 T cells: 4,000 APC ratio. Representative flow plots from 4 independent experiments, unless noted.
(A) Flow cytometry of early activation markers, Nur77GFP and CD69 (12 hr) on naïve or previously activated OT-I CD8+ T cells cultured on sorted APC populations directly from tumors
(B) Representative cytometry of Naïve OT-I CD8+ T cell proliferation, measured by dye dilution of eFluor670 plotted against Nur77GFP (as measure of TCR triggering), at 72 hours following co-culture with tumor APC populations. Total cell yield counts listed above graphs.
(C) Histogram overlay of Naive T cell proliferation between tumor APCs.
(D) Representative cytometry of T cell proliferation, measured by dye dilution of eFluor670 plotted against Nur77GFP, at 72 hours for previously activated OT-I CD8+ T cell blasts cultured on tumor APC populations. Total cell yield counts listed above graphs.
(E) Histogram overlay of previously activated OT-I CD8+ T cell proliferation across tumor APCs.
(F) Representative cytometry of T cell proliferation, measured by dye dilution of eFluor670, at 72 hours for Naive OT-II CD4+ T cells cultured on tumor APC populations. Representative flow plots from 2 independent experiments.
(G) Histogram overlay of Naïve OT-II CD4+ T cell proliferation across tumor APCs.
See also Figure S5.
Ultimately, at their normally low frequencies in total tumor isolate, CD103+DC2 remain unable to drive proliferation of CTLs (Figure S5E (Engelhardt et al., 2012)). Additionally, none of the APC subsets induced CD4+ T cell proliferation directly from the tumor. (Figure 6F–G and Figure S5F). However exogenous peptide did restore DC1 and DC2 capacity to stimulate proliferation, suggesting these DCs may not be inherently incapable of CD4 T cell stimulation (Figure S5G). Critically, this identifies the unique capacity of CD103+ DC2 within the tumor to uptake, process, and cross-present tumor antigen to robustly stimulate CTLs. This challenges the simple concept that tumors contain only weak or suppressive myeloid populations.
CD103+ DC2 Localization and T cell Interactions Revealed by Intravital Imaging
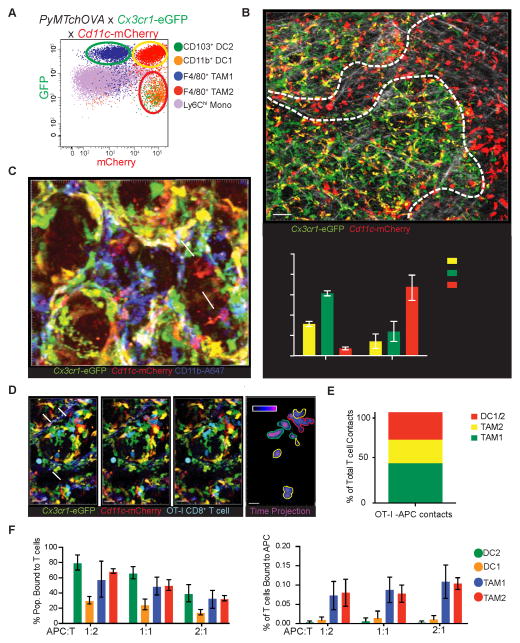
Given the unique ability of the rare CD103+ DC2s to stimulate T cells, we sought to understand the spatial organization of these cells within tumor and their interaction dynamics with T cells both in vivo and in vitro. To differentiate these populations in living spontaneous tumors in vivo, the PyMTchOVA allele was crossed on to Cx3cr1-eGFP and Cd11c-mCherry alleles, generating three uniquely fluorescent populations in the myeloid compartment (Figure 7A). Both DC1 and DC2 subsets were marked red (mCherry only), while TAM1 and TAM2 populations were green (eGFP only) and yellow (mCherry and eGFP), respectively. Using this model, with 2-photon intravital imaging, we observed that TAM1 and TAM2 populations are preferentially marginating tightly on tumoral lesions. This zone is one where we had previously found T cells to be preferentially captured (Engelhardt et al., 2012). In contrast, DC subsets, typically, were found in separate collagen-rich zones distal to the tumor lesions, making up nearly 70% of all distally localized APC (Figure 7B).
Figure 7. Intravital and slice imaging reveals CD11b+ DC1 and CD103+ DC2 are sparse near tumor margins yet can interact with T cells when present there.
(A) Representative cytometry of tumor APCs in PyMTchOVA x Cx3cr1-eGFP x Cd11c-mCherry. Populations as previously defined, are plotted as mCherry vs. GFP. Green, Yellow and Red circles indicate the fluorescent profile each population displays in this model. Red (mCherry only cells), Yellow (mCherry and GFP double positive cells), and Green (GFP only cells). By flow cytometry DC1/DC2 populations fall in the Cherry-only population, while TAM1 and TAM2 comprise the yellow and green populations respectively.
(B) Intravital 2-photon representative still image of an early carcinoma lesion from a PyMTchOVA x Cx3cr1-eGFP x Cd11c-mCherry reporter. Regions indicated with dashed line, marked either distal or marginating to lesions, were determined with a combination of mCherry fluorescence and collagen structure. Collagen fibers marked (white) by 2nd harmonic generation. Scale bar 50 μm. Inset: Quantification of Proximal/distal location of the APCs within the tumor. Data pooled from independent imaging runs, presented as mean ± SEM.
(C) Representative confocal still image from live tumor slices in PyMTchOVA x Cx3cr1-eGFP x Cd11cmCherry tumors, stained with CD11b-A647 antibody. mCherry only cell (arrowhead DC2, red) and mCherry+ CD11b+ cell (arrow DC1, purple) in the tumor. Scale bar 15 μm.
(D) Representative image sequence of CFP expressing OT-1 CD8+ T cells (blue) dynamically interacting with APC cells in the PyMTchOVA x Cx3cr1-eGFP x Cd11c-mCherry model by live slice confocal imaging 4 days after T cell transfer at 0, 30 and 60 min. Arrows indicate T cell interactions with Red (DC1/DC2), Green (TAM1) or Yellow (TAM2) cells. Scale bar 30 μm. Last panel displays time projection of CFP expressing T cells through 60 min imaging timeframe, with outline color dictated by APC of contact.
(E) APC-T cell contacts in vivo as a % of total T cell couples observed. Accumulated data of 4 different positions imaged for 30 minutes in 2 independent intravital 2 photon imaging runs. Contacts were scored manually by counting physical contact made between T cells and red, yellow and green APCs. Color of bar represents the APC of contact (Red: CD103+, CD11b+ DC1, Green: TAM1, yellow: TAM2).
(F) Ex vivo T cell coupling assay with digested tumor positively selected for CD45+ cells with previously activated OT-I CD8+ T cell. Data calculated as % of T cells couples within each of the populations (left), and as a total % of T cell couples (right). Data pooled from 2 independent experiments, plotted as mean ± SEM.
Since this approach did not fully differentiate between CD11b+ DC1 and CD103+ DC2 cells amongst those on the margins of tumor foci, we sought to determine whether the few red DCs might preferentially represent exclusively one or the other subset. To delineate the subsets in situ, we utilized live tumor slice imaging, with anti-CD11b antibody staining. Using this, we could distinguish CD11b+ DC1 from CD103+ DC2 subsets in situ in the presence of the red/green fluorescent reporters and found that, both CD11b+ and CD11b− DCs were present at these locations (Figure 7C and Movie S1). We conclude that while TAMs generally represent the dominant cell type at the tumor margin, pro-CTL stimulatory APCs nevertheless can be found there, albeit in very low numbers.
Our previous data demonstrated that incoming CTLs engaged in arrest behavior at the tumor margin and we sought to determine whether these might be taking place with DCs or TAMs or both. In vivo T cell dynamics were analyzed in the red/green reporter system by adoptive transfer of CFP expressing OT-I CD8+ T cells into spontaneous breast tumor bearing mice, for either intravital or live slice imaging. We observed stable T cell interactions largely confined to the tumor margins, as previously described (Boissonnas et al., 2013; Engelhardt et al., 2012) (Figure 7D–E and Movie S2). Although TAM1 interactions dominated all interactions scored, DCs and TAM2s were also well represented in T cell arrests. This demonstrates that DC1/DC2 in the tumor-proximal regions are not incapable nor physically excluded from engaging T cells within tumors but did raise a fundamental question of whether either is intrinsically more capable of engaging T cells.
To answer this, we divorced APC selection from the physical constraints of the tissue and digested tumor to make single-cell suspension and introduced in vitro activated OT-I CTLs and allowed them to form antigen-specific couples. We then quantified the percentage of each APC population that was occupied with a T cell by flow cytometry. This revealed that OT-I T cells couple preferentially with CD103+ DC2 and TAM1/TAM2 subsets (Figure 7F left panel). However, due to the high frequency of TAM1/TAM2, most T cell-APC couples are formed with TAM1/TAM2 cells (Figure 7F right panel). We conclude that DC2 contribute to T cell interactions in tumors and, when present near the margin are capable of competing for T cell occupancy.
Rare Tumor CD103+ DC2 are Required for Efficient Adoptive T cell Therapy
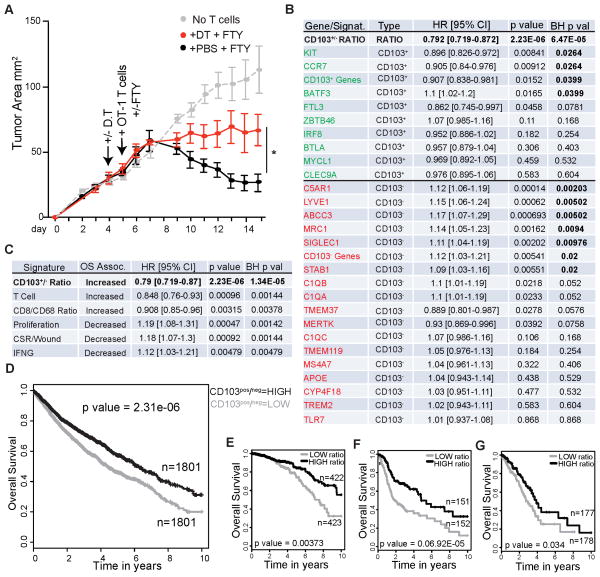
We were surprised to find that the proportions of CD11b+ DC1 and CD103+ DC2 were nearly inverted in a spontaneously regressing EG7 tumor model, hereto after referred to as EG7.2, as compared to a fully aggressive and outgrowing line EG7.1. While the aggressively growing tumors maintained the relative proportions of DCs we observed in all other aggressive tumors (Figure S6A), the spontaneously regressing model contained unusually high numbers of the CD103+ DC2 (Figure S6B). We also observed increased tumor growth in the Irf8 KO tumor model, which lack CD103+ DC2, but not in the Irf4 conditional KO model (Figure S6C–D). These together suggest that DC2 tumoral abundance may play an important role in tumor control, however the differences in outgrowth may be caused by many variances in these tumors beyond their populations of myeloid cells and their ability to stimulate CTLs. To formally test whether the CD103+ DC2 are necessary for efficient CTL mediated tumor regression, we turned to the outgrowing EG7.1 tumor model and performed adoptive T cell therapy of activated tumor specific T cells (Helmich and Dutton, 2001). We performed these experiments in zDC-DTR mice, which permitted us to specifically ablate CD103+ DC2 in the tumor (Figure 3D). In order to isolate the effect of the CD103+ DC2 to the site of the tumor, and eliminate any effect of LN priming, we designed the experiment to include two strategies, (1) use of activated OT-I CD8+ CTL blasts, which do not require priming in the LN and typically do not traffic there, and (2) treatment of animals with the SIP1R antagonist FTY-720, which prevents LN exit of rare transferred CTL T cells that traffic to the LN. The effect of FTY-720 alone had minimal effects on transferred CTLs to mediate tumor regression (Figure S6E). However, we found that ablation of CD103+ DC2s in the context of FTY-720 had a significant effect on the ability of CTLs to mediated efficient tumor regression, massively slowing T cell mediation tumor regression (Figure 8A).
Figure 8. Rare CD103+ DC2 population at the tumor is required for efficient adoptive CTL therapy.
(A) Tumor growth curve plotted as tumor area (mm2) over time for EG7.1 in zDC-DTR hosts. Arrows indicate time of i.p. D.T./PBS administration, and i.v. transfer of 5×106 previously activated OT-I CD8+ T cells. DT/PBS was subsequently administered every 3rd day and FTY-720/Saline was subsequently administered every other day throughout time course. Representative data presented as mean tumor area ± SEM (n=4) from 2 independent experiments. Statistical significance indicated by *p<0.05.
(B) Comparison of prognostic value of CD103+/CD103− Ratio Gene Signal as compared to the individual genes (either CD103+ specific, green, or TAM1/TAM2/CD11b DC1 specific genes, red) using TCGA datasets in a multivariate COX proportional hazards survival analysis adjusting for cancer type as a covariate. Data expressed as Hazard Ratio (HR) with 95% confidence intervals, where a value <1 means increased Overall Survival (OS); >1 means decreased OS for genes with BH p values<0.05 (bolded values).
(C) Comparison of the prognostic value of the CD103+/CD103− Ratio Gene Signal with several published prognostic gene signatures using TCGA datasets in a multivariate COX proportional hazards survival analysis adjusting for cancer type as a covariate. Data expressed as Hazard Ratio (HR) with 95% confidence intervals, where a value <1 means increased Overall Survival (OS); >1 means decreased OS for genes with BH p values<0.05.
(D) K-M plot across all 12 cancer types in Human TCGA data sets, adjusting for cancer type based on HIGH CD103+/CD103− gene Ratio and LOW CD103+/CD103− Ratio expressers (median split/cancer).
(E) K-M plot for overall survival of Breast Cancer patients in TCGA data set. Data Parsed on HIGH CD103+/CD103− gene Ratio and LOW CD103+/CD103− Ratio expressers.
(F) K-M plot for overall survival of Head and Neck Squamous Cell Carcinoma patients in TCGA data set. Data Parsed on HIGH CD103+/CD103− gene Ratio and LOW CD103+/CD103− Ratio expressers.
(G) K-M plot for overall survival of Lung Adenocarcinoma patients in TCGA data set. Data Parsed on HIGH CD103+/CD103− gene Ratio and LOW CD103+/CD103− Ratio expressers.
See also Figure S6.
Signatures of Intratumoral CD103+ DC2 Abundance Predict Outcome Across Human Cancer
To determine if a critical role for CD103+ DC2 abundance translated to human tumors, we took advantage of TCGA data (Cancer Genome Atlas Research et al., 2013; Hoadley et al., 2014) that quantifies relative gene expression from numerous human cancer types with matched outcome data. We used our RNAseq data to select for high level transcripts that characterized CD103+ DC2 and also selected a subset of genes that characterized TAM1/TAM2/CD11b+DC1 cells but were deficient in CD103+ DC2. We identified human homologs of those mouse genes and assayed expression of these genes in TCGA data from all cancer types to assess prognostic associations. In a Proportional Hazards survival analysis, adjusting the model for cancer type as a co-variate, we observed that the individual genes from these populations had only modest prognostic benefits (expressed as Hazard Ratio (HR)). In order to represent the relative proportion of the two cell types, we defined a ratio of the CD103+ and CD103− gene expression data and used this as a continuous variable within the Cox analysis. High expression of this ratio was significantly associated with increased overall survival (BH p=0.00019), (Figure 8B).
This analysis shows that the cell type we identified, when ratioed with its functional opposite, generates a very strong prognostic value for outcome across human cancers. Comparing this ‘signature’ to other previously described ‘immune scores’ shows that the ratio of CD103+/CD103− genes provides the strongest pro-immune survival signal compared to other current analyses of TCGA data including those based on total T cell abundance (Palmer et al., 2006) and that made by bulk ratio of CD8 T cells to macrophages (CD8/CD68 DeNardo et al., 2011) (Figure 8C). Our score also compares favorably, though opposite in prognosis, for those immune scores associated with poor outcome. It is also notable that CSF1 expression in tumors in these patients also anti-correlates with the CD103/BDCA3 gene ratio measure, although it likewise anti-correlates with total tumor Flt3L levels (Figure S6F–G).
Finally, we sought to analyze the TCGA data within individual cancer types. Adjusting for cancer type, a Kaplan-Meier (K-M) plot for all 12 cancers in this dataset shows the overall benefit in tumors with a high CD103+/CD103− gene-expression profile (Figure 8D; unadjusted plot in Figure S6H). The extent of this association is particularly profound in Breast Cancer, Head-Neck Squamous Cell Carcinoma, and Lung Adenocarcinoma (Figure 8E–G). Overall, this represents an unexpectedly strong immune signature, the more so as it was derived entirely from empirical immunoprofiling in mouse tumor models.
DISCUSSION
A critically important feature of this work is that within the diverse array of myeloid cells at the tumor, a rare population of pro-immune DCs exist even in immunoevasive tumors. This contrasts with previous characterizations of the myeloid lineage in tumors that have highlighted their immunosuppressive functions. This study puts a face and a name upon a specific subset of intratumoral DCs whose functions one would wish to enhance as part of immunotherapy and serves to begin to demystify the complexity of this critical compartment.
This work provides an understanding of the tumoral myeloid environment as having lineage parallels to other non-transformed tissues. Significant confusion in the current literature of myeloid subpopulations results from inappropriate grouping of cells (e.g. CD11b+) or from lack of a common method for distinguishing the various subpopulations (e.g. CD11c expression). Recent additions to the repertoire of antibody markers and total expression-array analyses of DCs versus macrophages versus monocytes provided significant clarity to this situation (Gautier et al., 2012; Miller et al., 2012). In particular, while conventional DCs are seen to express one or more of markers such as CD24, TAMs are better described through surface expression of F4/80, CD64 and MerTK. To that end, CD11b+ DC1s in tumors appear, by RNA expression and surface expression, to be more closely allied to macrophages. This proximity of these cells has been observed in other peripheral sites(Gautier et al., 2012). In general, a remarkable similarity in IRF4, IRF8 and Batf3 dependent origins and surface phenotypes suggests that the overall origins and distinctions of tumor-infiltrating myeloid cells is quite similar to counterparts in normal tissues.
Our studies confirm earlier work showing that mice deficient of Batf3 failed to spontaneously clear highly immunogenic tumors (Hildner et al., 2008). However, our studies provide the additional insight that the key populations defined by Batf3 and IRF8, but not IRF4 expression, are not only present and functional within tumors but in fact are required for productive responses to adoptively transferred CTLs generated in vitro, after T cells are already primed and in the absence of profound LN involvement. This places at least some of the key Batf3- and now IRF8-dependent cells as playing key roles in repriming within the tumor. Thus our understanding shifts emphasis from the LN to the tumor for T cell control. Our work also puts these cells in context in the tumor and shows, while they may be sporadically present on the tumor margins where T cells will encounter their APC, they are very sparse there. Clinically, this suggests that enhancement of the intratumoral load of these cells will be an important co-factor defining the success of adoptive T cell therapies and, broadly, that providing restimulation within the tumor represents an important requirement for T cell function at that site. That the requirement for CD103 cells is intratumoral is further supported by our TCGA analysis in which gene-expression data used to assess prognostic value derives exclusively from mRNA taken from the tumor but not the LN.
While our data did not show an absolute dependence on CD103+ DC2 to facilitate adoptive CTL control of tumor outgrowth, the effect was profound. It is not clear whether residual T cell-dependent control in the DT-treated cohort might represent myeloid-independent activity of these cells, a compensatory role by other myeloid cells (e.g. CD11b+ DC1, which stimulate CTL extremely weakly in vitro) or merely our inability to fully eliminate the CD103+ DC2 population. To that last possibility, it will be increasingly important to more effectively manipulate the relative population densities of myeloid cells, sparing the CD103+ DC2 or even providing means to enhance them. Our demonstration of enhanced CD103+ DC2 generation in Flt3L expressing tumors provides a compelling rationale as to why such therapy may work and indeed may be synergistic with T cell therapies such as anti-CTLA4 (Curran and Allison, 2009). Conversely, the efficacy of α-CSF1 therapies (Ries et al., 2014; Strachan et al., 2013) may be in part due to the sparing of CD103+ DC2 under such blockade, as demonstrated in our work.
Despite significantly increased clarity of the identities of intratumoral myeloid populations and their similarities to those in normal tissues, much remains to be elucidated concerning the additional functional diversity of the intratumoral APCs. In particular, the role of CD11b+ DC1 in the tumor remains obscure. Their transcriptional profiling and surface markers may place them closer, in identity, to TAMs as compared to CD103+ DC2, which is found in other macrophage populations from healthy peripheral sites(Gautier et al., 2012). We also note that these highly resemble recently described “DC-Th2”, defined by their reliance on the transcription factor IRF4, expression of CD301b and PDL2 and ability to effectively prime Th2 responses (Gao et al., 2013; Kumamoto et al., 2013; Williams et al., 2013). At present, we failed to find a DC population that could robustly stimulate CD4 T cells when taken directly from the tumor, however DC1 and DC2 could be rescued upon add back of peptide, suggesting either that the MHCII processing pathway is not highly active in these cells or that our digest conditions affect MHCII antigen loading. Regardless, this may reinforce the hypothesis that the absence of effective T cell help and its attendant γc cytokine production are a critical missing link in the tumor microenvironment.
Our study of these cell types now brings to the forefront many spatio-temporal issues about how the various myeloid populations interface with tumors, with one-another, and with T cells. While it is clear from our imaging that the marginating cells are dominated by TAM1 and TAM2, the nature of ‘APC selection’ by T cells is not fully resolved; do CTLs with particular activation choose particular subsets of myeloid cells and, conversely, does the in situ interaction of T cells with CD103+ DC2 give them abilities to kill, so long as they do not encounter a TAM in the interim? Many of these types of questions will require elaboration of spectral labeling methods. In addition, these types of approaches will require significant adoption and/or development of biosensor-like reporters to determine where and when complete TCR signaling is taking place.
Finally, a very important finding of this study relates to the applicability of the myeloid delineation to multiple human tumors. Using bioinformatics based on these populations, we observed that CD103+ DC2 enriched transcripts, taken from mouse models and expressed as a ratio with an equivalent selection from the TAM/DC1 populations provides a strong prognostic signal in TCGA data, across multiple tumor types. The fact that this ‘signature’ correlates with patient survival better than other published signatures provides an additional and compelling reason to suggest that this population is critical for robust tumor control in mice and humans. Clearly, additional profiling of these populations in context of immunotherapies will be required to test this further and should be undertaken alongside all further immunotherapy trials. It will be particularly interesting to determine whether patients having CD103/BDCA3 ‘High’ tumors will represent better responders to checkpoint blockade. In sum, it is clear that these rare cells should now be a target to augment their numbers as well as a biomarker that may define those whose immune response is well positioned to eliminate cancers.
EXPERIMENTAL PROCEDURES
Mouse Tumors
PyMT-ChOVA transgenic C57BL/6 founder mice were as described (Engelhardt et al., 2012) and offspring were screened for the PyMT-ChOVA transgene by PCR and monitored for tumors and used at 20–30 weeks of age. B78ChOVA is a variant of B78 (Graf et al., 1984), generated and used as described in supplemental methods. All additional strain information can be found in supplemental methods. All mice were maintained under SPF conditions and treated in accordance with the regulatory standards of the NIH and American Association of Laboratory Animal Care standards, and consistent with the UCSF Institution of Animal Care and Use Committee (IACUC approval: AN106779-01A)
Flow Cytometry
All antibodies were purchased from BD Pharmingen, eBioscience, Invitrogen, Biolegend, the UCSF hybridoma core, or were produced in the Krummel Lab. For surface staining cells were incubated with anti-Fc receptor antibody (clone 2.4G2) and stained with antibodies in PBS + 2 % FCS for 30 min on ice. Viability was assed by staining with fixable Live/Dead Zombie (Biolegend) or DAPI. For intracellular staining, mice were injected with 10 ug/gram of body weight with Brefeldin A (Cayman) 6 hr prior to harvest, cells were stained with antibodies against surface markers, then fixed with 2 % PFA for 10 min at 25 °C and permeabilized with 0.2 % Saponin then stained with target antibody. All flow cytometry was performed on a BD Fortessa flow cytometer. Analysis of flow cytometry data was done using Flowjo (Treestar). Cell sorting was performed using a BD FACS Aria II.
Human samples
Tissue was vigorously minced with surgical scissors and transferred to a 25 ml Erlenmeyer with magnetic stir bar with 3 mg/ml Collagenase A (Roche) and 50 U/ml DNase I (Roche) per 0.3 g of tissue for 1 hr at 37° C and 5 % CO2 with constant agitation. Samples are then filtered though a 70 um filter, spun down and resuspended for staining. For all human samples, informed consent was obtained from all subjects and work was performed in accordance with IRB approval (IRB number 13-12246, 12/06/2013-12/05/2014).
TCGA Bioinformatics Analysis
Clinical expression analysis uses genome-wide mRNA levels (Illumina mRNA-seq) from 3602 patient tumor samples representing 12 cancer types (845 breast, 265 ovarian, 303 head & neck squamous, 122 bladder, 168 glioblastoma, 190 colon, 173 AML, 72 rectal, 355 lung adenocarcinoma, 259 lung squamous, 480 kidney, and 370 uterine cancers), normalized and combined into a single dataset by the TCGA PanCancer working group as published (Cancer Genome Atlas Research et al., 2013; Hoadley et al., 2014) (data is in the TCGA Data Portal https://tcga-data.nci.nih.gov/tcga/ and available as syn1715755 on https://www.synapse.org/). The CD103+/CD103− ratio signature is calculated as the log of the mean expression of CD103+ DC genes divided by the mean expression of the CD103− DC genes, followed by zscore standardization (mean=0, sd=1; gene list in Figure 8C). We also evaluate published T cell (Palmer 17 et al., 2006), proliferation (Wolf et al., 2014), CSR/wound (Chang et al., 2005), and gamma interferon (Viigimaa et al., 2010) signatures as published, along with a CD8/CD68 expression ratio (DeNardo et al., 2011). Overall survival data was obtained from the TCGA portal (downloaded 6/2013) (Cancer Genome Atlas Research et al., 2013) and survival analysis performed using Cox Proportional Hazards modeling in a multivariate model adjusting for cancer type. Log rank p values are used to assess significance after adjusting for multiple comparisons using the Benjamini-Hochberg method (Bejamini and Hochberg, 1995). Kaplan-Meier survival plots are generated using the Survival package in R. In the all-data KM plot (Figure 8E), we ‘adjusted’ for cancer type by classifying each sample as ‘high’ or ‘low’ using that cancer types’ median value of the CD103+/CD103− ratio signature.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software. Unless specifically noted all data are representative of >3 separate experiments. Error bars represent SEM calculated using Prism, and are derived from triplicate experimental conditions. Specific statistical tests used were paired and unpaired T tests and all p values less than 0.05 were considered statistically significant.
Supplementary Material
SIGNIFICANCE.
Current cancer immunotherapies are based on enhancing the ability of host or introduced T cells to reject tumors. However, efficient CTL function requires frequent repriming and abundant tumor macrophages, which capture CTL at the tumor margin, either fail to achieve this and/or actively inhibit T cell responses. Here, we show that the abundant macrophages in tumors have a functional opposite, in the form of antigen-presenting CD103+ DC. These cells efficiently cross-present tumor antigens and are differentially distributed within the tumor microenvironment compared to tolerizing APC. We describe how intratumoral CD103+ DC are uniquely targetable, how their abundance is required for T cell therapy in mice, and how their transcript prevalence predicts outcome in human cancers.
HIGHLIGHTS.
Mouse and human tumors contain rare DCs that are stimulatory for T cells
Stimulatory DCs are programmed through unique cytokines and transcription factors
CD103+ DCs are sparse proximal to tumor margins but plentiful in distal regions
Tumor DCs are necessary for T cell mediated tumor rejection and predict survival
Acknowledgments
We thank L. Lanier, J. Roose and L. Fong for advice and C. Guillard, S. Kogan for the generous sharing of materials. This work was supported by NIH grants U01CA105379, U54 CA163123. M. Broz was supported by the Genentech Predoctoral Research Fellowship, the Margaret A. Cunningham Immune Mechanisms in Cancer Research Fellowship, and the ARCS Scholarship. We thank K. Corbin for Imaging assistance, M. Headley for critical reading of the manuscript and C. Lin for administrative assistance.
Footnotes
AUTHOR CONTRIBUTIONS
M. Broz designed and performed experiments unless specified. M. Binnewies participated in depletions and cell purifications for RNAseq; B.B. performed imaging experiments and analyses; A.N. performed some flow cytometry; J.P., D.E. and A.B. performed RNAseq and analysis; M.R and A.D. supplied clinical samples; D.B. supplied critical reagents; S.A. supplied critical input on analysis of cross-presentation, A.S. supplied mice for IRF4 analysis; D.W. performed analysis of TCGA data. M.F.K. conceived and participated in design and interpretation of experiments. M.B and M.F.K. wrote, revised and edited the manuscript and M. Binnewies and B.B. edited the manuscript.
Accession Numbers
RNAseq data titled “Delineating Tumor-infiltrating Antigen Presenting Cells” was uploaded to the NCBI GEO repository under accession number: GSE61462.
The authors do not have competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bejamini Y, Hochberg Y. Controlling the False Discovery Rate – a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Boissonnas A, Licata F, Poupel L, Jacquelin S, Fetler L, Krumeich S, Thery C, Amigorena S, Combadiere C. CD8+ tumor-infiltrating T cells are trapped in the tumor-dendritic cell network. Neoplasia. 2013;15:85–94. doi: 10.1593/neo.121572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van’t Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, Jeffrey KL, Anthony RM, Kluger C, Nchinda G, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez-Retamozo V, Etzrodt M, Newton A, Rauch PJ, Chudnovskiy A, Berger C, Ryan RJ, Iwamoto Y, Marinelli B, Gorbatov R, et al. Origins of tumor-associated macrophages and neutrophils. Proc Natl Acad Sci U S A. 2012;109:2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran MA, Allison JP. Tumor vaccines expressing flt3 ligand synergize with ctla-4 blockade to reject preimplanted tumors. Cancer Res. 2009;69:7747–7755. doi: 10.1158/0008-5472.CAN-08-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer discovery. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranoff G. GM-CSF-based cancer vaccines. Immunol Rev. 2002;188:147–154. doi: 10.1034/j.1600-065x.2002.18813.x. [DOI] [PubMed] [Google Scholar]
- Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, Egeblad M, Krummel MF. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 2012;21:402–417. doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Nish SA, Jiang R, Hou L, Licona-Limon P, Weinstein JS, Zhao H, Medzhitov R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf LH, Jr, Kaplan P, Silagi S. Efficient DNA-mediated transfer of selectable genes and unselected sequences into differentiated and undifferentiated mouse melanoma clones. Somatic cell and molecular genetics. 1984;10:139–151. doi: 10.1007/BF01534903. [DOI] [PubMed] [Google Scholar]
- Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S. Antigen presentation and T cell stimulation by dendritic cells. Annu Rev Immunol. 2002;20:621–667. doi: 10.1146/annurev.immunol.20.100301.064828. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P, Wasan PS, Wang XN, Malinarich F, Malleret B, et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Miller J, Merad M. Dendritic cell and macrophage heterogeneity in vivo. Immunity. 2011;35:323–335. doi: 10.1016/j.immuni.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmich BK, Dutton RW. The role of adoptively transferred CD8 T cells and host cells in the control of the growth of the EG7 thymoma: factors that determine the relative effectiveness and homing properties of Tc1 and Tc2 effectors. J Immunol. 2001;166:6500–6508. doi: 10.4049/jimmunol.166.11.6500. [DOI] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MD, Niu B, McLellan MD, Uzunangelov V, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, Gopinathan A, Tuveson DA, Fearon DT. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330:827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- Kumamoto Y, Linehan M, Weinstein JS, Laidlaw BJ, Craft JE, Iwasaki A. CD301b(+) dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 2013;39:733–743. doi: 10.1016/j.immuni.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmartsev S, Nagaraj S, Gabrilovich DI. Tumor-associated CD8+ T cell tolerance induced by bone marrow-derived immature myeloid cells. J Immunol. 2005;175:4583–4592. doi: 10.4049/jimmunol.175.7.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, Nussenzweig MC. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, Van den Bossche J, Mack M, Pipeleers D, In’t Veld P, De Baetselier P, Van Ginderachter JA. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, et al. Targeting Tumor-Associated Macrophages with Anti-CSF-1R Antibody Reveals a Strategy for Cancer Therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Dumenil AM, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Strachan DC, Ruffell B, Oei Y, Bissell MJ, Coussens LM, Pryer N, Daniel D. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8 T cells. Oncoimmunology. 2013;2:e26968. doi: 10.4161/onci.26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O’Shea JJ, Singh H, Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viigimaa M, Vaverkova H, Farnier M, Averna M, Missault L, Hanson ME, Dong Q, Shah A, Brudi P. Ezetimibe/simvastatin 10/20 mg versus rosuvastatin 10 mg in high-risk hypercholesterolemic patients stratified by prior statin treatment potency. Lipids in health and disease. 2010;9:127. doi: 10.1186/1476-511X-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JW, Tjota MY, Clay BS, Vander Lugt B, Bandukwala HS, Hrusch CL, Decker DC, Blaine KM, Fixsen BR, Singh H, et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nature communications. 2013;4:2990. doi: 10.1038/ncomms3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DM, Lenburg ME, Yau C, Boudreau A, van ‘t Veer LJ. Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLoS One. 2014;9:e88309. doi: 10.1371/journal.pone.0088309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.