Summary
Mitotic arrest deficient 1 (Mad1) plays a well-characterized role in the major cell cycle checkpoint that regulates chromosome segregation during mitosis, the mitotic checkpoint (also known as the spindle assembly checkpoint). During mitosis, Mad1 recruits Mad2 to unattached kinetochores [1, 2], where Mad2 is converted into an inhibitor of the anaphase promoting complex/cyclosome bound to its specificity factor Cdc20 [1, 3–6]. During interphase, Mad1 remains tightly bound to Mad2 [2, 3, 7, 8] and both proteins localize to the nucleus and nuclear pores [9, 10], where they interact with Tpr (Translocated Promoter Region). Recently, it has been shown that interaction with Tpr stabilizes both proteins [11], and that Mad1 binding to Tpr permits Mad2 to associate with Cdc20 [12]. However, interphase functions of Mad1 that do not directly affect the mitotic checkpoint have remained largely undefined. Here we identify a previously unrecognized interphase distribution of Mad1 at the Golgi apparatus. Mad1 colocalizes with multiple Golgi markers and cosediments with Golgi membranes. Although Mad1 has previously been thought to constitutively bind Mad2, Golgi-associated Mad1 is Mad2-independent. Depletion of Mad1 impairs secretion of α5 integrin and results in defects in cellular attachment, adhesion, and FAK activation. Additionally, reduction of Mad1 impedes cell motility, while its overexpression accelerates directed cell migration. These results reveal an unexpected role for a mitotic checkpoint protein in secretion, adhesion and motility. More generally, they demonstrate that, in addition to generating aneuploidy, manipulation of mitotic checkpoint genes can have unexpected interphase effects that influence tumor phenotypes.
Results and discussion
An unexpected, perinuclear localization of Mad1 (Fig. S1A–B) was identified in interphase HeLa cells after immunofluorescence using an affinity purified rabbit anti-Mad1 antibody, which produces a single band on immunoblots [Fig. S1C; [13]]. A similar perinuclear localization was observed in primary Murine Embryonic Fibroblasts (MEFs) and the breast cancer cell line MDA-MB-231 (Fig. S1A, B). To biochemically confirm the existence of a cytoplasmic pool of Mad1, a fractionation experiment was performed to separate nuclear from cytoplasmic extract. Three nuclear markers, histone H3, lamin A and lamin C, as well as a cytoplasmic marker (tubulin), were used to confirm that appropriate fractionation was achieved. HeLa cells, MEFs, MDA-MB-231 cells and an additional breast cancer cell line, Cal51, all contained a cytoplasmic pool of Mad1 (Fig. S1D–E).
Multiple experiments were performed to test the specificity of anti-Mad1 antibodies. First, Mad1 was transiently depleted in HeLa cells using siRNA. Fractionation followed by immunoblotting using the rabbit anti-Mad1 antibody revealed that total, nuclear, and cytoplasmic pools of Mad1 were depleted (Fig. S1F). Second, an additional antibody [14], was used to confirm the identity of Mad1. This mouse monoclonal antibody also recognizes a single band of roughly 85 kDa by immunoblotting (Fig. S1C) that is reduced following siRNA mediated depletion of Mad1 (Fig. S1F). Third, stable HeLa cell lines in which Mad1 expression was knocked down constitutively (to be referred to hereafter as Mad1-KD) were generated by retroviral infection of three distinct shRNA sequences followed by antibiotic selection. Mad1-KD cell lines grew at rates comparable to control cells, and did not have obvious delays in any stage of the cell cycle (Fig. S1G–I). Mad1 levels were diminished, but not absent, in all three cell lines (Fig. S1J). In Mad1-KD cell lines, the cytoplasmic pool of Mad1 became undetectable by immunofluorescence (Fig. S1K). Fourth, fractionation experiments in parental and Mad1-KD cell lines #1–3 showed a reduction in the cytoplasmic pool of Mad1 (Fig. S1L), as detected by both Mad1 antibodies. Fifth, both Mad1 antibodies showed a cytoplasmic fraction of YFP-tagged Mad1 (Fig. S1M). We conclude that interphase cells contain a cytoplasmic pool of the mitotic checkpoint protein Mad1.
Cytoplasmic Mad1 is localized to the Golgi
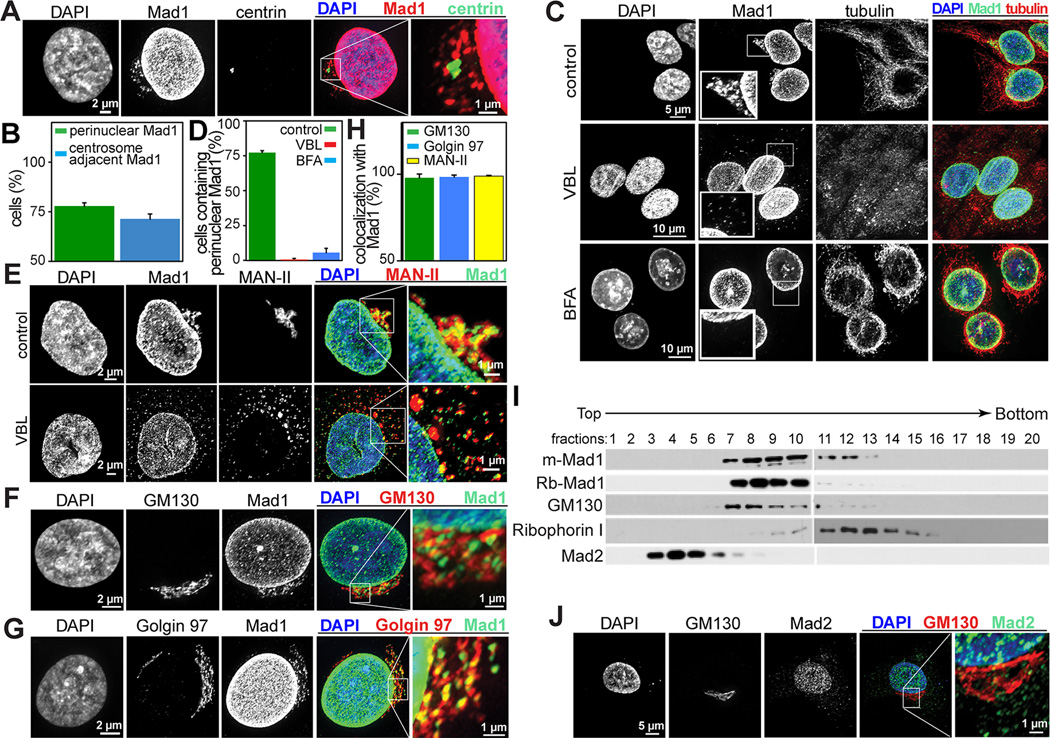
Perinuclear Mad1 was coincident with the centrosome (Fig. 1A–B), as is the Golgi apparatus [15]. Golgi organization is dependent on polymerized microtubules. Microtubule depolymerization with 12 (Fig. 1C–D) or 4 (Fig. S2A) hours of vinblastine treatment cause the Golgi to disassemble and perinuclear Mad1 to disperse.
Figure 1. The cytoplasmic pool of Mad1 is localized to the Golgi.
(A) Cytoplasmic Mad1 in HeLa cells colocalizes with the centrosome, identified with centrin. The boxed region is enlarged in the far right panel.
(B) Quantification +/− SD of Mad1 colocalization with centrosomes. n=200 cells from each of 3 independent experiments.
(C) Perinuclear cytoplasmic Mad1 staining in HeLa cells is dispersed in response to 12 hours of 100 ng/mL vinblastine (VBL) or 30 minutes of 10 µg/mL BFA, as are Golgi proteins. The boxed regions are enlarged in the insets.
(D) Quantification +/− SD of cells with perinuclear cytoplasmic Mad1 after treatment with vinblastine (VBL) or BFA. n=200 cells from each of 3 independent experiments.
(E–G) Cytoplasmic Mad1 colocalizes with the Golgi proteins MAN-II (E), GM130 (F) and Golgin 97 (G). Like MAN-II, Mad1 disperses in response to treatment with the microtubule destabilizing drug vinblastine (E). The boxed regions are enlarged in the far right panels.
(H) Quantification of colocalization of Mad1 with the Golgi proteins MAN-II, GM130 and Golgin 97 in HeLa cells. Quantification indicates the percentage of cells in which Mad1 colocalized with the indicated Golgi protein at 40× magnification +/− SD. n=200 cells from each of 3 independent experiments.
(I) Cytoplasmic Mad1 cofractionates with the Golgi in an Optiprep gradient. HeLa cell extracts were fractionated as described in the Experimental Procedures. Mad1, as detected using both mouse monoclonal and rabbit polyclonal antibodies, cosedimented with the Golgi protein GM130 and is largely separated from the ER (Ribophorin I) and the Mad1 binding partner Mad2.
(J) The Mad1 binding partner Mad2 does not colocalize with the Golgi (marked by GM130) in HeLa cells. The boxed region is enlarged in the far right panel.
The fungal metabolite Brefeldin A (BFA) causes rapid disassembly of the Golgi without gross perturbation of microtubules [16–18]. BFA treatment also resulted in dispersal of the perinuclear Mad1 signal (Fig. 1C–D), strongly suggesting it was localized to the Golgi. To confirm this, Mad1 was tested for colocalization with several Golgi antigens. Cytoplasmic Mad1 exhibited a similar distribution as three Golgi markers, alpha-mannosidase II (MAN-II), GM130 and Golgin 97 (Fig. 1E–H). Colocalization of cytoplasmic Mad1 with Golgi markers was essentially complete when all three Golgi antigens were visualized in the same cell (Fig. S2B).
Mad1 did not colocalize with early or late endosomes, labeled with Rab5A and Rab7A, respectively (Fig. S2C–D). Nor did Mad1 colocalize with RINT-1, which cycles between the Golgi and the ER (Fig. S2E). However, partial colocalization was observed between Mad1 and Rab6A (Fig. S2F), which accumulates in both the trans-Golgi network and endosomes.
To further establish the Golgi localization of Mad1, cytoplasmic extract was fractionated over an OptiPrep density gradient. Mad1 was enriched in fractions containing the Golgi marker GM130, but largely absent from fractions containing the endoplasmic reticulum marker Ribophorin I (Fig. 1I). These data support the conclusion that the perinuclear cytoplasmic pool of Mad1 localizes to the Golgi apparatus.
A Mad2-free pool of Mad1
Mad1 and Mad2 are thought to interact throughout the cell cycle. Mad2 is expressed in excess of Mad1 in yeast, Xenopus extracts, mouse and human cells [19–22]. Gel filtration analysis has revealed that Mad1 elutes in a single peak that also contains Mad2. However, Mad2 also fractionates into a second peak of the expected size for monomeric Mad2 [7, 19, 23]. Depletion of Mad2 removes all detectable Mad1 in numerous systems, while substantial Mad2 remains after immunoprecipitation with Mad1 [2, 13, 19]. These data suggest that the entire pool of Mad1 is bound to Mad2 throughout the cell cycle. Interestingly, however, we found that Mad2 did not cofractionate with Mad1 after sedimentation on an Optiprep gradient (Fig. 1I). Nor did Mad2 colocalize with Golgi markers by immunofluorescence (Fig. 1J). Thus, unlike kinetochore- and nuclear pore-bound pools of Mad1, Golgi localized Mad1 appears free of Mad2.
Mad1 functions in the secretion of α5 integrin
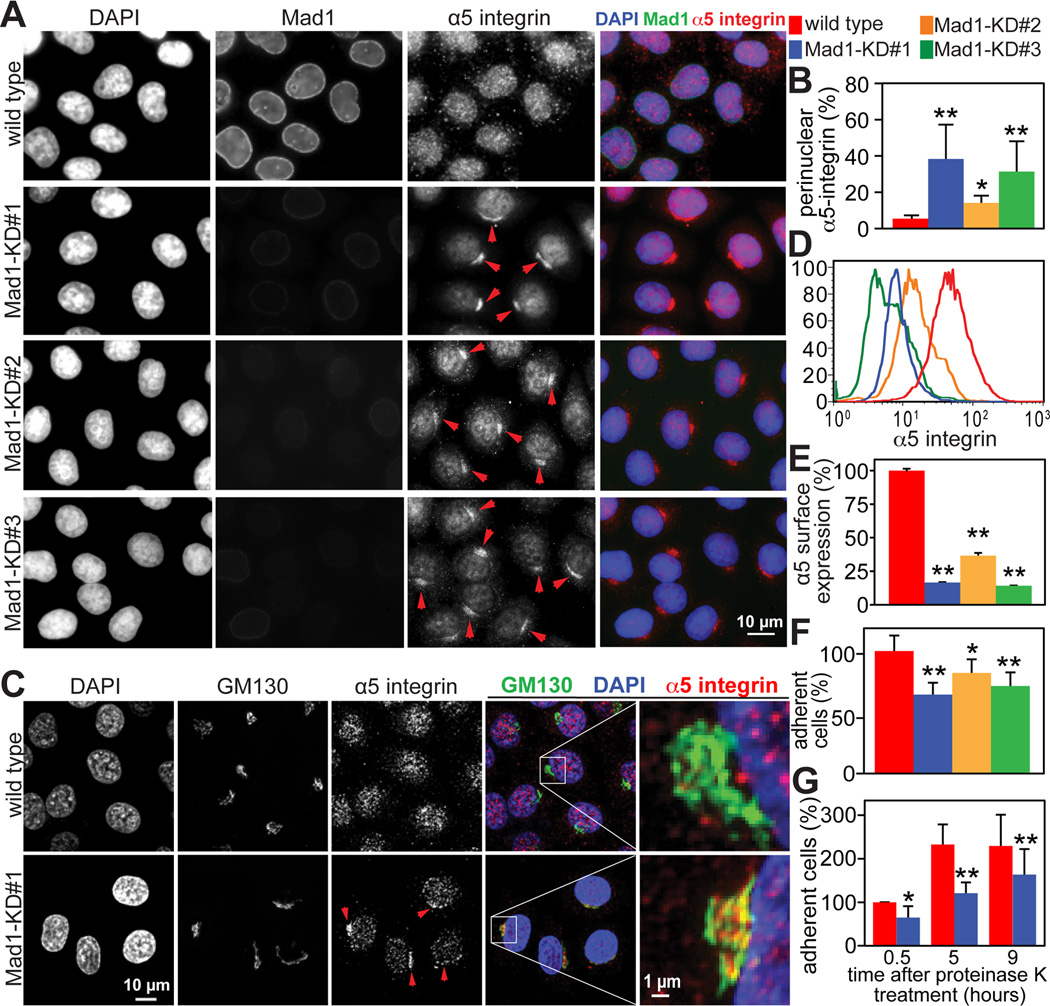
To determine whether Mad1 functions in protein trafficking, secretion of a variety of proteins was analyzed. Plasma membrane accumulation of the epidermal growth factor receptor (EGFR) was not affected in Mad1 knockdown cells (Fig. S2G–H). Nor was accumulation of the vesicular stomatitis virus envelope glycoprotein VSVG at the plasma membrane delayed due to reduction of Mad1 (Fig. S2I–J and Supplemental Movie 1). These data suggest that there is no global defect in protein secretion after Mad1 depletion. However, an intracellular epitope of α5 integrin coalesced near the nucleus in Mad1 knockdown cell lines #1–3 after fixation and permeabilization (Fig. 2A arrowheads). To confirm that this antibody was indeed recognizing α5 integrin, single z planes from the bottom of the cell were imaged. These demonstrated the expected cell surface staining, which was coincident with the focal adhesion marker paxillin (Fig. S3A). Co-staining with GM130 revealed that the perinuclear pool of α5 integrin was localized to the Golgi (Fig. 2C, arrowheads).
Figure 2. Golgi localized Mad1 participates in α5 integrin secretion and cell adhesion.
(A) Perinuclear accumulation of α5 integrin in Mad1-KD HeLa cell lines #1–3, indicated by arrowheads.
(B) Quantification +/− SD of perinuclear accumulation of α5 integrin, as depicted in A. n=200 cells from each of 3 independent experiments.
(C) Perinuclear α5 integrin in Mad1-KD#1 HeLa cells accumulates in the Golgi, shown by colocalization with GM130. Arrowheads indicate Golgi accumulation of α5 integrin. The boxed regions are enlarged in the far right panels.
(D) Flow cytometry analysis demonstrating that surface expression of an extracellular epitope of α5 integrin is reduced on Mad1-KD cell lines #1–3 as compared to wild type HeLa cells.
(E) Quantification +/− SD of flow cytometry measurements in D. n=10,000 events from each of 2 independent experiments.
(F) Reduced expression of Mad1 results in defective cellular attachment. Wild type and Mad1-KD HeLa cell lines #1–3 were plated on 5 µg/mL fibronectin and incubated at 37°C for 30 minutes before removing unattached cells. The average number of attached cells per 10× field +/− SD is shown. n=3.
(G) Mad1-mediated α5 integrin secretion promotes cell adhesion. Cell surface proteins were cleaved with 500 µg/mL proteinase K for 30 minutes on ice. After adding PMSF, cells were washed and permitted to recover at 37°C for the indicated amounts of time before plating 200,000 cells and allowing them to adhere to fibronectin coated dishes for 30 minutes. The average number of attached cells per 10x field +/− SD (normalized to the number of wild type cells at 0.5 hours) from 3 independent experiments is shown.
*=p<0.05. **=p<0.001.
To further examine whether α5 integrin secretion was inhibited in cells depleted of Mad1, non-permeabilized cells were labeled with an antibody recognizing an extracellular epitope of α5 integrin. Flow cytometry revealed that substantially less α5 integrin localized to the cell surface in Mad1-KD cell lines #1–3 as compared to wild type (Fig. 2D–E).
Secretion of other integrin subunits was then examined. Subtle perturbations in the trafficking of α1, α2 and β1 integrins were also observed in Mad1-KD cells (Fig. S3B–F), but were less pronounced than effects on α5. mRNA levels of all four integrin subunits tested were unaffected in Mad1-KD cells (Fig. S3G), as were protein levels of α5 (Fig. S3H).
α and β integrin subunits associate in the ER before transiting the Golgi en route to the cell surface [24]. α5 subunits exclusively pair with β1, but β1 subunits dimerize with numerous alpha chains, including α1, α2, α3, α4, and α6 [25]. Mad1 participation in the secretion of only a subset of alpha subunits may explain why α5 integrin dramatically accumulated in the Golgi after Mad1 depletion, but effects on its binding partner β1 integrin were less conspicuous.
Two cleavage events are required to produce the mature α5 integrin on the cell surface. First, the 44 amino acid signal peptide is cleaved from the N-terminus. The second cleavage event produces mature products of 98 and 18 kD, and occurs in either the trans Golgi network or during exocytosis [27]. To determine whether maturation of α5 integrin was affected in Mad1 depleted cells, α5 integrin-3xFLAG was introduced into control and Mad1-KD#1 cells. Conversion of the precursor into the 18 kD mature form was notably impaired in Mad1 knockdown cells (Fig. S3J), further supporting the conclusion that Mad1 participates in α5 integrin trafficking.
Reduction of Mad1 impairs cellular attachment and spreading on fibronectin
α5β1 heterodimers anchor cells to the ECM by binding fibronectin [28]. The accumulation of α5 integrin in the Golgi in Mad1-KD cells suggested that depletion of Mad1 may cause deficits in cell adhesion. To test this hypothesis, wild type and Mad1-KD cell lines #1–3 were plated and permitted to attach to fibronectin coated dishes. Significantly fewer Mad1-KD cells were adherent after 30 minutes as compared to control cells (Fig. 2F). To determine whether decreased secretion was responsible for the attachment defect, cell surface proteins were removed with proteinase K before plating on fibronectin. Mad1-KD#1 cells again exhibited a defect in attachment (Fig. 2G), confirming that impaired biosynthetic transport of newly translated α5 integrin leads to delays in cellular adhesion. In contrast, attachment to collagen, which is mediated by a variety of integrins including α1β1 and α2β1, was not affected by reduction of Mad1 (Fig. S3K).
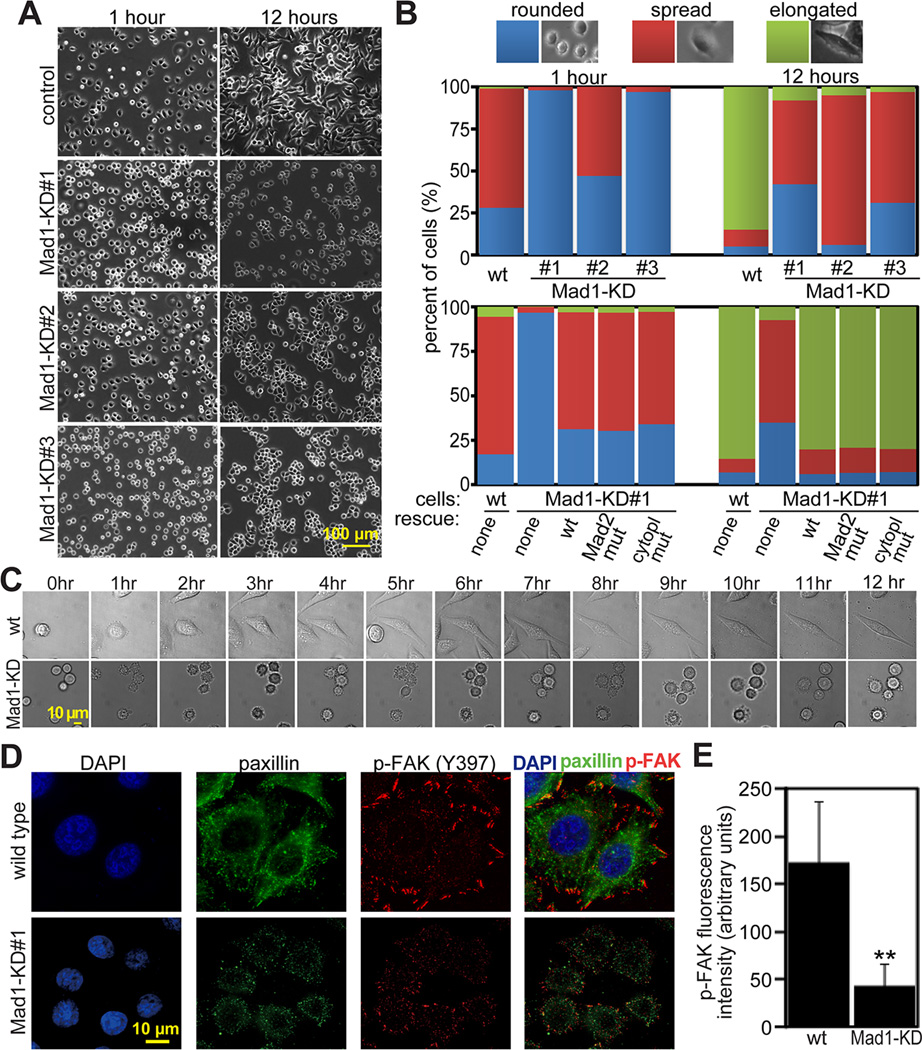
To determine effects of Mad1 depletion on cell spreading, wild type and Mad1-KD HeLa cell lines #1–3 were plated on fibronectin coated coverslips. Within 1 hour after plating, most wild type cells had attached to the substrate and begun spreading, while many more Mad1-KD cells remained rounded (Fig. 3A–B). After 12 hours, 85% of wild type HeLa cells had elongated, as compared to 8%, 5% and 3% of Mad1-KD cell lines #1–3, respectively (Fig. 3A–B). To further characterize this defect, wild type and Mad1-KD#1 cells were plated on fibronectin coated coverslips and observed using phase contrast timelapse microscopy (Fig. 3C and Supplemental Movie 2). Wild type cells blebbed and began extending and retracting filopodia-like protrusions almost immediately. Mad1-KD#1 cells blebbed, but did not extend protrusions. Wild type cells were noticeably flattened and elongated at 3 hours, while Mad1-KD#1 cells remained rounded even at 12 hours.
Figure 3. Mad1 depletion impairs cellular spreading and FAK-Tyr 397 phosphorylation.
(A) Mad1-KD cell lines #1–3 are substantially less well spread than wild type HeLa cells 1 and 12 hours after plating on fibronectin.
(B) Quantification of cellular phenotype 1 and 12 hours after plating on fibronectin coated dishes. n>300 cells from each of 3 independent experiments. Top, wild type and Mad1-KD HeLa cell lines 1–3. Bottom, wild type and Mad1-KD#1 cells transfected with the indicated shRNA-resistant rescue construct. “Mad2 mut” indicates Mad1 containing the mutations K541A/L543A, which is unable to bind Mad2 [26]. “Cytopl mut” indicates a cytoplasmic Mad1 mutant (aa 180–718) lacking the nuclear import signal [12]. All rescue contructs were GFP-tagged.
(C) Still images from timelapse analysis of wild type and Mad1-KD#1 HeLa cells showing that wild type cells begin spreading by 3 hours while Mad1-KD cells remain rounded 12 hours after plating on fibronectin. See also Supplemental Movie 2.
(D) Reduction of Mad1 results in reduced phosphorylation of FAK on Tyr 397 4 hours after plating on fibronectin coated coverslips.
(E) Quantification of fluorescence intensity +/− SD of phospho-FAK (Tyr 397) in wild type and Mad1-KD#1 cells, as pictured in D. n=40 cells from each of 3 independent experiments. **=p<0.001.
ECM binding by integrins results in recruitment and activation of signaling molecules such as focal adhesion kinase (FAK), which becomes phosphorylated at tyrosine 397 when active [29]. Phospho-FAK levels were significantly diminished in Mad1-KD#1 cells (Fig. 3D–E), consistent with a deficit in integrin mediated signaling.
Mad1 functions in secretion and attachment are independent of Mad2
To test whether earlier mitotic defects could cause deficits in cell attachment and spreading, cells expressing reduced levels of the mitotic checkpoint components BubR1 and Mad2 were examined. In both cases, α5 integrin did not accumulate in the Golgi (Fig. S4A–D). Stable reduction of Mad2 also caused no delay in cell spreading (Fig. S4E). Together, these data support the conclusions that 1) Mad1 facilitates secretion of α5 integrin during interphase and 2) this function of Mad1 is independent of Mad2.
To further test a possible role of Mad2 in integrin secretion, Mad1 knockdown cells were reconstituted with shRNA-resistant Mad1 constructs. Expression of wild type Mad1 prevented accumulation of α5 integrin in the Golgi (Fig. S4F–G), and permitted Mad1 knockdown cells to attach and spread on fibronectin coated dishes with normal kinetics (Fig. 3B). Importantly, the Mad1 mutant that cannot bind Mad2 also rescued α5 integrin secretion and cellular spreading, as did the cytoplasmic Mad1 mutant (Fig. 3B and S4F–G). These data demonstrate that the defects in α5 integrin secretion in Mad1 knockdown cells are a consequence of an interphase, cytoplasmic function of Mad1 that is independent of Mad2.
Mad1 promotes directional cell motility
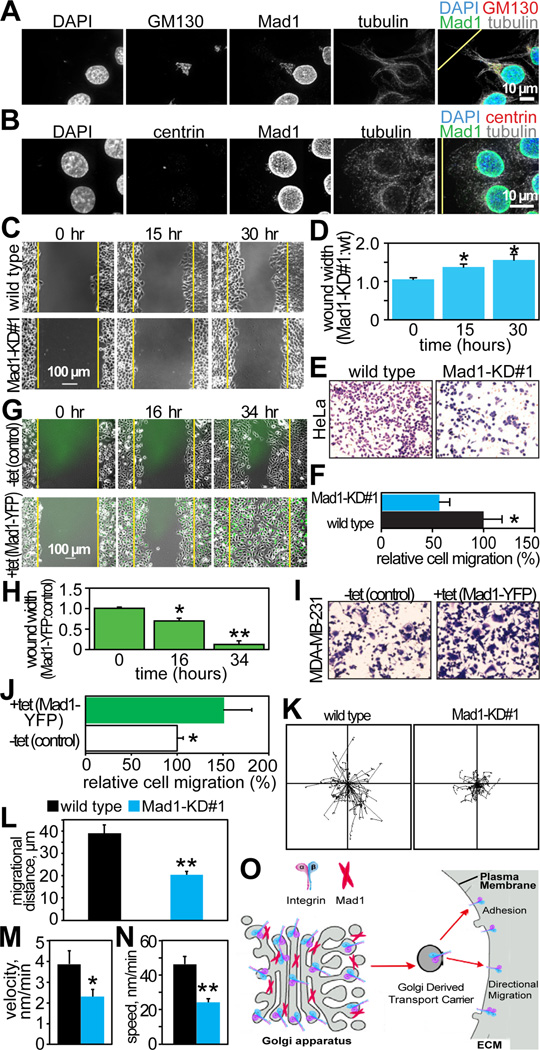
During directional cell migration, the centrosome orients toward the leading edge, as does the Golgi [30, 31]. After wounding, Mad1 immunoreactivity was observed on the side of the cell adjacent to the wound edge in a pattern strikingly similar to that of the Golgi and the centrosome (Fig. 4A–B).
Figure 4. Mad1 is required for directional cell migration.
(A–B) Mad1 orients with the Golgi (GM130; A) and centrosomes (centrin; B), towards the direction of cell migration 6 hours after wounding confluent HeLa cells. The yellow lines in the overlaid images indicate the location of the wound edge.
(C–F) Reduction of Mad1 inhibits directed cell migration. (C) Still images from timelapse analysis showing delayed wound healing in Mad1-KD#1 HeLa cells as compared to wild type. Yellow lines indicate wound edges at 0 hours. See also Supplemental Movie 3.
(D) Quantification of the rate of wound healing in wild type and Mad1-KD#1 HeLa cells. The ratio of the wound size in Mad1-KD#1 versus wild type cells at the indicated times is shown. Data indicate the mean ± SD from 3 independent experiments.
(E) Transwell assay showing reduced migration of Mad1-KD#1 HeLa cells as compared to wild type.
(F) Quantification of the number of cells that migrated in the transwell assay shown in E. Error bars represent SD of three independent experiments.
(G–J) Overexpression of Mad1 accelerates directed cell migration. (G) Still images from timelapse analysis showing control (−tet) or Mad1-YFP expressing (+ tet) MDA-MB-231 cells 0, 16 and 34 hours after wounding. Yellow lines indicate wound edges at 0 hours. Related to Supplemental Movie 4.
(H) Quantification of the rate of wound closure in G. The ratio of the size of the wound in Mad1 overexpressing cells to the size of the wound in control cells is shown. Data indicate the mean ± SD from 3 independent experiments.
(I) Increased migration of Mad1-YFP expressing cells in a transwell assay.
(J) Quantification of cell migration in the transwell assay depicted in I, normalized to control. Error bars represent SD of 3 independent experiments.
(K–N) Wild type and Mad1-KD#1 HeLa cells were plated in one side of a microfluidics channel in media containing 0.1% serum. 12 hours later, 20% FBS was added to the opposite side of the chamber. Cells were imaged every 5 minutes for 14 hours and single cell motility was tracked as in [35]. n=60 cells per condition. (K) Tracks of individual cells. (L) Migrational distance +/− SEM. (M) Velocity (displacement towards final destination/time) +/− SEM of individual cells. (N) Speed (total distance/time) +/− SEM of individual cells.
(O) Model depicting Mad1 localizing to the Golgi where it facilitates secretion of α5 integrin, which is required for cellular attachment, adhesion, and directed cell motility.
*=p<0.05. **=p<0.001.
To directly test the role of Mad1 in cell migration, control and Mad1-KD#1 cells were observed by timelapse microscopy after wound formation. Wound closure was substantially delayed in Mad1-KD cells relative to wild type HeLa cells (Fig. 4C–D and Supplemental Movie 3). Consistent with these results, transwell experiments revealed that Mad1-KD cells were significantly impaired in their migratory ability as compared to wild type cells (Fig. 4E–F).
We were unable to generate HeLa cells uniformly overexpressing Mad1. Therefore, MDA-MB-231 cells stably expressing Mad1-YFP in response to tetracycline were used to perform the converse experiments. Cells with elevated levels of Mad1 repaired wounds more rapidly than control cells (Fig. 4G–H and Supplemental Movie 4). Similarly, in transwell assays, Mad1-YFP expressing cells exhibited enhanced migratory ability as compared to controls (Fig. 4I–J). These results demonstrate that the level of Mad1 is directly related to the rate of cell motility.
We then examined the motility of single cells. Mad1-KD#1 cells exhibited shortened migrational distance (Fig. 4K–L), lower directional velocity (Fig. 4M), and reduced overall speed (Fig. 4N) as compared to controls. Thus, individual cells exhibit impaired migration after reduction of Mad1.
Prior to this study, interphase roles of Mad1 remained largely unknown. Here, we identify a novel interphase function of Mad1 that does not involve Mad2. Endogenous Mad1 localizes to the Golgi in multiple mammalian cell types. The pool of Mad1 in the Golgi facilitates secretion of α5 integrin, which promotes cellular adhesion, spreading, and motility (Fig. 4O). These functions are compromised in cells with reduced levels of Mad1. Heightened expression of Mad1 is common in breast tumors where it serves as a marker of poor prognosis [13]. It will now be of interest to determine whether increased Mad1 in cancer cells potentiates migration and metastasis.
Supplementary Material
Acknowledgements
We thank Dr. Andrea Musacchio and Dr. Patricia Keely for antibodies, Dr. Amber Schuh for assistance with Golgi purification, and Eric Britigan and Dr. Scott Nelson for assistance with microscopy. This work was supported in part by the National Institutes of Health through R01CA140458 (BAW), R01GM088151 (AA), T32 CA009135 (LMZ) and R01EB010039 (DJB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint. Embo J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. Embo J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 5.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 6.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fava LL, Kaulich M, Nigg EA, Santamaria A. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J. 2011;30:3322–3336. doi: 10.1038/emboj.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- 10.Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweizer N, Ferrás C, Kern DM, Logarinho E, Cheeseman IM, Maiato H. Spindle assembly checkpoint robustness requires Tprmediated regulation of Mad1/Mad2 proteostasis. J Cell Biol. 2013;203:883–893. doi: 10.1083/jcb.201309076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. Nuclear pores protect genome integrity by assembling a premitotic and mad1-dependent anaphase inhibitor. Cell. 2014;156:1017–1031. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, Roopra A, Weaver BA. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci U S A. 2012;109:E2205–E2214. doi: 10.1073/pnas.1201911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 15.Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Experimental cell research. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- 16.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- 19.Chung E, Chen RH. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol Biol Cell. 2002;13:1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 21.Heinrich S, Geissen EM, Kamenz J, Trautmann S, Widmer C, Drewe P, Knop M, Radde N, Hasenauer J, Hauf S. Determinants of robustness in spindle assembly checkpoint signalling. Nat Cell Biol. 2013;15:1328–1339. doi: 10.1038/ncb2864. [DOI] [PubMed] [Google Scholar]
- 22.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 23.Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. Embo J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heino J, Ignotz RA, Hemler ME, Crouse C, Massagué J. Regulation of cell adhesion receptors by transforming growth factor-beta. Concomitant regulation of integrins that share a common beta 1 subunit. J Biol Chem. 1989;264:380–388. [PubMed] [Google Scholar]
- 25.Hemler ME, Huang C, Schwarz L. The VLA protein family. Characterization of five distinct cell surface heterodimers each with a common 130,000 molecular weight beta subunit. J Biol Chem. 1987;262:3300–3309. [PubMed] [Google Scholar]
- 26.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol. 2011;13:475–482. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lissitzky JC, Luis J, Munzer JS, Benjannet S, Parat F, Chrétien M, Marvaldi J, Seidah NG. Endoproteolytic processing of integrin pro-alpha subunits involves the redundant function of furin and proprotein convertase (PC) 5A, but not paired basic amino acid converting enzyme (PACE) 4, PC5B or PC7. Biochem J. 2000;346(Pt 1):133–138. [PMC free article] [PubMed] [Google Scholar]
- 28.Humphries M. Integrin structure. Biochemical Society Transactions. 1999;28:311–339. [PubMed] [Google Scholar]
- 29.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Magdalena J, Millard TH, Machesky LM. Microtubule involvement in NIH 3T3 Golgi and MTOC polarity establishment. Journal of cell science. 2003;116:743–756. doi: 10.1242/jcs.00288. [DOI] [PubMed] [Google Scholar]
- 31.Cau J, Hall A. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. Journal of cell science. 2005;118:2579–2587. doi: 10.1242/jcs.02385. [DOI] [PubMed] [Google Scholar]
- 32.Cairo Lucas V, Ptak C, Wozniak Richard W. Mitosis-Specific Regulation of Nuclear Transport by the Spindle Assembly Checkpoint Protein Mad1p. Molecular Cell. 2013;49:109–120. doi: 10.1016/j.molcel.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Zasadil LM, Britigan EM, Weaver BA. 2n or not 2n: Aneuploidy, polyploidy and chromosomal instability in primary and tumor cells. Semin Cell Dev Biol. 2013;24:370–379. doi: 10.1016/j.semcdb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young EW, Pak C, Kahl BS, Yang DT, Callander NS, Miyamoto S, Beebe DJ. Microscale functional cytomics for studying hematologic cancers. Blood. 2012;119:e76–e85. doi: 10.1182/blood-2011-10-384347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.