Abstract
While peptides are promising as probes and therapeutics, targeting intracellular proteins will require greater understanding of highly structured, cell-internalized scaffolds. We recently reported BC1, an 11-residue bicyclic peptide that inhibits the Src homology 2 (SH2) domain of growth factor receptor-bound protein 2 (Grb2). In this work, we describe the unique structural and cell uptake properties of BC1 and similar cyclic and bicyclic scaffolds. These constrained scaffolds are taken up by mammalian cells despite their net neutral or negative charges, while unconstrained analogs are not. The mechanism of uptake is shown to be energy-dependent and endocytic, but distinct from that of Tat. The solution structure of BC1 was investigated by NMR and MD simulations, which revealed discrete water-binding sites on BC1 that reduce exposure of backbone amides to bulk water. This represents an original and potentially general strategy for promoting cell uptake.
1. Introduction
Constrained peptides are an important class of biologically-inspired molecules with utility for inhibiting protein-protein interactions. A growing body of work has shown that precisely designed covalent constraints can improve peptide affinity, selectivity, metabolic stability, and in some cases, cell penetration.1 Cell internalization via endocytosis is frequently correlated to amphipathic character and positive charge, both of which promote association with the plasma membrane and induce various endocytic pathways.2,3 By contrast, there are strikingly few reports of conformationally constrained peptides with neutral or net negative charge that are taken up by mammalian cells.4-6 While the mechanisms of internalization for polycationic sequences, including the HIV-derived Tat peptide, have been richly explored,3,7-9 comparatively little is known about the precise mechanisms by which non-cationic, constrained peptides enter the cell.5,10,11 Herein, we report a series of neutral and negatively charged constrained peptides that are internalized by mammalian cells. We also report details of uptake mechanism and identify unique structural features that may promote cellular uptake.
The head-to-tail cyclic peptide HT1 and bicyclic peptide BC1 (Figure 1) were designed as inhibitors of the SH2 domain of Grb2.12 They were derived from disulfide- and thioether-bridged macrocycles originally described by Roller and co-workers.13,14 BC1, which has an IC50 of 350 nM for Grb2 inhibition in biochemical assays, did not show anti-proliferative activity up to 30 μM (Supplementary Figure S1). This was not surprising, since small molecules that target Grb2-SH2 produced by Burke and colleagues inhibited the protein with potencies in the 1-20 nM range, but only affected cells in the 0.8-25 μM range.15,16 These inhibitors of the SH2 domain have shown low-nanomolar affinity in vitro and yet were unable to disrupt the Grb2-pErbB2 interaction in live cells.15 This lack of cellular activity can be attributed to the high degree of negative charge characteristic of these phosphotyrosine isosteres and mimetics. Because our peptides did not demonstrate Grb2-dependent effects on cultured cells, we were interested in whether the primary barrier was uptake of BC1 by mammalian cells or subsequent endosomal release. If BC1 was taken up by mammalian cells, even to a small extent, it would be surprising since most cell-internalized peptides are highly cationic. Among peptides with less overall charge, such as amphipathic helices, a single negative charge is typically enough to completely prevent internalization.5,17-19 Thus, we designed a series of experiments to explicitly test the uptake of BC1 and analogs in cultured cells, in order to begin to elucidate how structured peptides could be designed to maximize cell uptake.
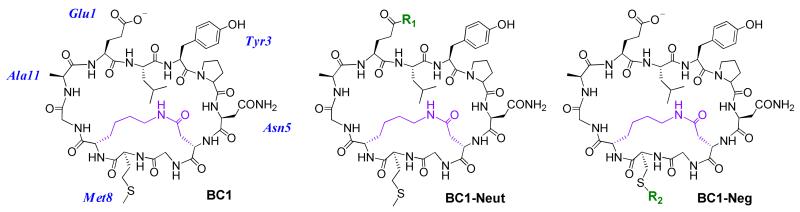
Figure 1.
Chemical structures of the BC1 scaffold and BODIPY-FL-labeled analogs. R1 is the adduct of hydrazine-functionalized BODIPY-FL, and R2 is the adduct of maleimide-functionalized BODIPY-FL. Fluorescein-labeled peptides were also prepared and tested to rule out dye-dependent effects on cell uptake. HT1-Neut and HT1-Neg are head-to-tail cyclic analogs of BC1-Neut and BC1-Neg, and Lin-Neut and Lin-Neg are linear analogs of BC1-Neut and BC1-Neg. See Supplementary Information for complete structural and synthesis details.
2. Results and Discussion
2.1. Cell uptake of dye-labeled cyclic and bicyclic peptides
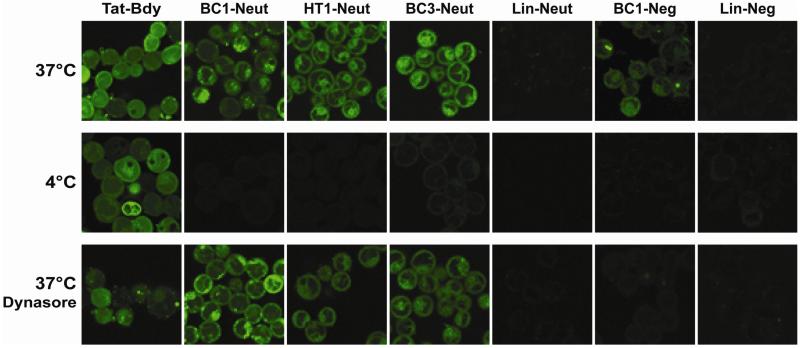
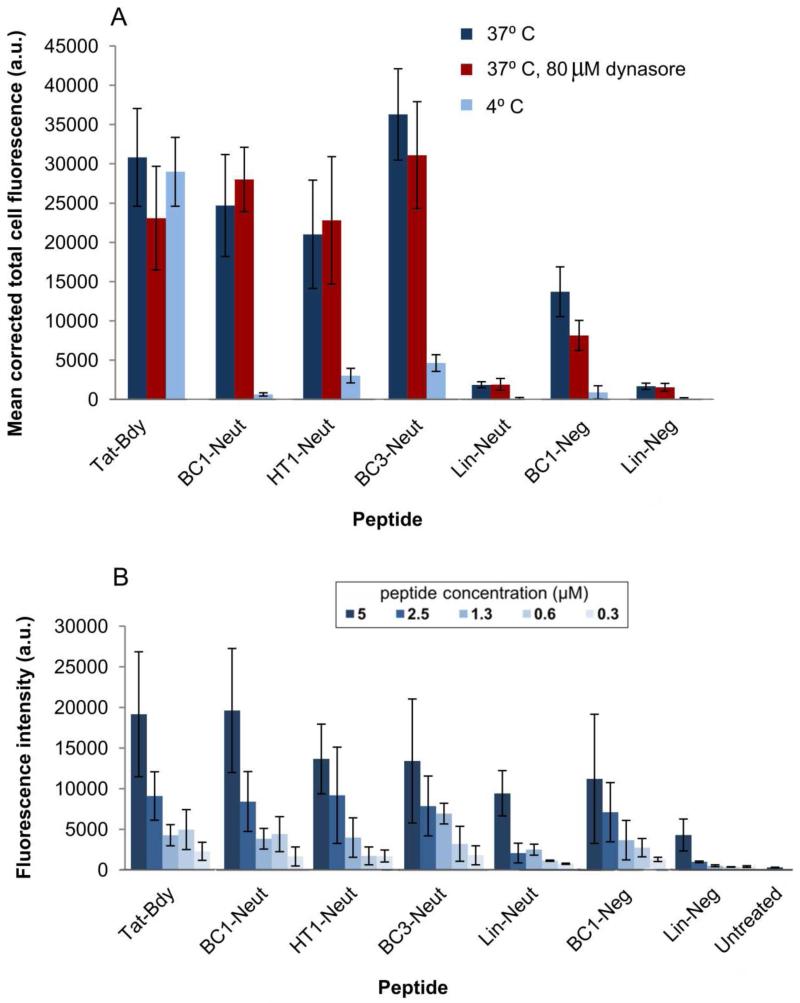
Our first variants were designed to explore the role of the negative charge. Specifically, we prepared variants of HT1 and BC1 with BODIPY-FL conjugated to Glu1 to eliminate the negative charge (HT1-Neut and BC1-Neut), or with the dye conjugated to a Cys residue substituted for Met8 to maintain a net charge of-1 (BC1-Neg; see Figure 1 and Supplementary Figure S2). To explore the role of conformational constraint, we also prepared dye-labeled linear analogs, as well as a dye-labeled bicyclic scaffold with a different lactam cross-link (BC3-Neut, Supplementary Figure S2). BODIPY-FL-labeled Tat peptide (Tat-Bdy) was used as a positive control. We used confocal fluorescence microscopy to investigate the cellular uptake of these dye-labeled peptides using MDA-MB-453 cells, a metastatic human epithelial cancer line. Confocal microscopy revealed that both BC1-Neg and BC1-Neut were internalized by MDA-MB-453 cells (Figure 2). Uptake was quantified using two independent measurements: ImageJ analysis of confocal microscopy images, and direct fluorometry of cell lysates following incubation and washing (Figure 3). Both techniques showed similar trends. The neutral BC1-Neut was internalized as effectively as Tat-Bdy, and, surprisingly, the anionic BC1-Neg was internalized almost as effectively. Interestingly, BC3-Neut was internalized to a similar extent, implying that multiple bicyclic structures can be efficiently taken up by cells. The less rigid, monocyclic peptide HT1-Neut was taken up by cells to a similar extent as BC1-Neut, but linear peptides labeled at Glu1 (Lin-Neut) or at the N-terminus (Lin-Neg) were not effectively. In the case of these linear peptides, the discrepancy between the fluorescence exhibited by cell lysates and the quantification of confocal microscopy images suggests that some fluorescence was associated with cell lysates at high concentrations (5 μM peptide). In fact, visualization by confocal microscopy shows dim fluorescence localized to the exterior of the cell (experiments performed with 10 μM peptide).. However, this association diminishes dramatically at lower peptide concentrations. As shown in Figure 3b and Supplementary Figure S8, cell lysate fluorescence decreases very rapidly for these linear peptides at lower concentrations compared to lysates from cells incubated with bicyclic and cyclic scaffolds. Thus, for these scaffolds we observed a general correlation between structural rigidity and cell uptake for peptide concentrations below 10 μM. Similar results were observed for fluorescein-labeled peptides as well as for peptides with the dye attached at a third location (Supplementary Figure S4). We concluded that the observed uptake is an intrinsic property of these constrained scaffolds.
Figure 2.
Analysis of peptide internalization by confocal fluorescence microscopy. Mammalian breast epithelial cancer cells (MDA-MB-453) were grown to 95% confluence in a 24-well glass-bottom plate and incubated with 10 μM fluorescently-labeled peptide in PBS for 1 hour at 37°C or 4°C.8 Low-temperature conditions were used to demonstrate the extent to which the uptake process was energy-dependent. Alternatively, cells were pre-treated with 80 μM dynasore (a dynamin inhibitor that prevents clathrin-mediated endocytosis)20 for 30 min at 37° C prior to incubation with dye-labeled peptides at the same temperature. Cells were washed thoroughly in PBS prior to imaging by confocal microscopy. While peptides were incubated in PBS, we also verified their stability in buffered human serum as described previously (Supplementary Figure S3).12
Figure 3.
Quantification of cell fluorescence. (A) Corrected total cell fluorescence (CTCF) of individual cells for each experimental condition tested. Images collected by confocal fluorescence microscopy were background-subtracted using the ImageJ software suite to compute CTCF values for individual cells. The mean CTCF of 5 representative cells within each image is plotted. Error bars denote standard deviation. (B) Quantitation of cellular internalization by direct fluorometry.31 MDA-MB-453 cells were seeded in a 96 well plate at 105 cells/ml and grown for several hours prior to incubation with the indicated concentrations of peptide for 1 hour at 37° C in PBS. Cells were then washed and lysed in 1% Triton x-100 prior to fluorometric analysis at 485 nm excitation, 535 nm emission. Supplementary Figure S8 shows the same data but with a narrower y-axis.
Some previous work has correlated degree of structure to cell penetration for other classes of constrained peptides.2,5,11 However, the precise mechanisms by which constrained peptides are taken up by the cell are still poorly understood. We sought to begin to characterize the mechanism of uptake for our cyclic and bicyclic scaffolds. The patterns of fluorescence associated with BC1-Neut and BC1-Neg contain a few bright punctate signals as well as more diffuse fluorescence that permeates the cytoplasm, with less fluorescence in the nucleus. HT1-Neut and BC3-Neut have more of their fluorescence distributed throughout the cytoplasm, with fewer visible puncta. These results are consistent with endocytic entry for these peptides, with some extent of endosomal escape. At 4° C, overall fluorescence intensity was dramatically reduced for all internalized cyclic and bicyclic scaffolds, but less reduced for Tat-Bdy, as expected due to the multiple modes by which Tat is internalized.3 The role of clathrin-mediated endocytosis in facilitating cellular uptake was studied using the small-molecule dynamin inhibitor dynasore.20 Pre-treatment with dynasore resulted in increased formation of punctate fluorescence for Tat-Bdy, but resulted in increased cell-surface localization for cyclic and bicyclic scaffolds (Figure 2, quantified in Supplementary Figures S5-S7). Taken together, these data implicate an energy-dependent, endocytic pathway for uptake of these constrained peptides that differs substantially from the uptake mechanism of Tat. Future work will apply emerging label-free methods, pathway-specific assays, and assays for quantitating endosomal escape to further refine our understanding of these peptides’ surprising cellular internalization.8,19,21
2.2. NMR structure and molecular dynamics of bicyclic peptide BC1 in aqueous solution
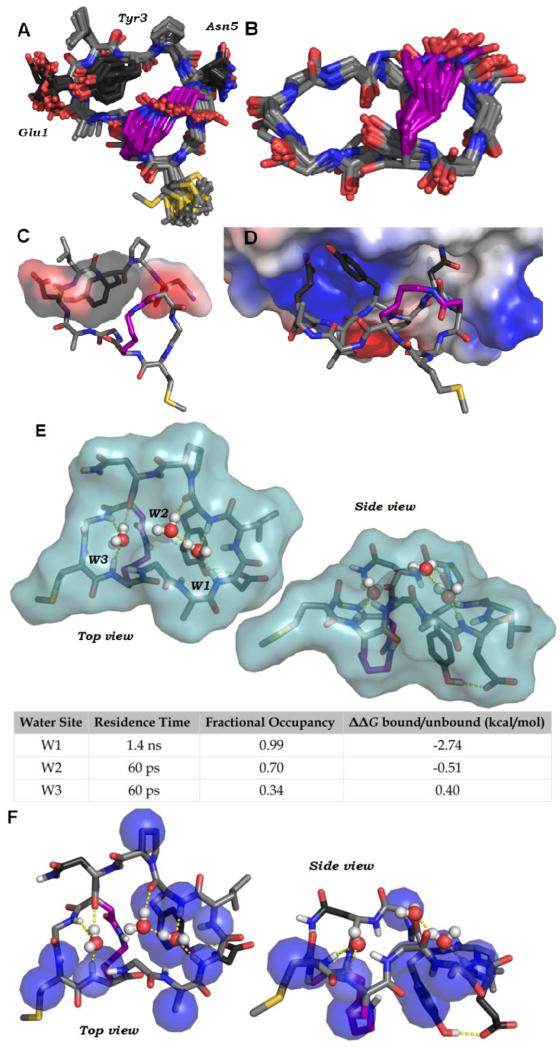
While the uptake of BC1 and related scaffolds is clearly dependent on constrained structure, prior work has demonstrated that rigidification does not automatically confer increased uptake.2,17,22 We hypothesized that specific structural features of BC1 promote its association with the plasma membrane, leading to uptake via endocytosis. To further explore this hypothesis, we determined the NMR structure of BC1 in aqueous solution. TOCSY, NOESY, ROESY and 1H-13C HSQC NMR spectra were obtained for 0.76 mM BC1 in 9:1 H2O:d6-DMSO. The recorded spectra were well-resolved and allowed complete resonance assignments (Supplementary Table S1). The Hα and amide proton chemical shifts of most residues differed significantly from typical values for random-coil structures, indicating the peptide adopts a well-defined structure (Supplementary Figure S9). 90 NOE-derived distance restraints were compiled (Supplementary Table S2), and these were used in multi-stage simulated annealing calculations to produce a structural ensemble of thirty lowest-energy structures (Figure 4A-B).
Figure 4.
Solution structure of BC1. (A) NMR-derived structural model of BC1. 90 NOE-derived distance constraints were used in two-stages simulated annealing and energy minimization using CNSolve.32 The 30 lowest-energy structures are shown. Carbons are shown in light gray, side chain carbons of the binding epitope are shown in dark grey, and side chain carbons of the cross-link are shown in purple. (B) The atoms of the backbone and cross-link, with all other side chain atoms omitted. Backbone RMSD is 0.33 Å. (C) A representative structure of BC1 showing the putative binding epitope (transparent surface). A pTyr-mimicking epitope is formed by Gly1 and Tyr3. Pro4 organizes a turn that positions Asn5 on the same face as Glu1 and Tyr3, consistent with established binding modes for peptide ligands of Grb2-SH2.30 (D) A representative solution structure of BC1 was docked into the phosphotyrosine-binding site of the Grb2-SH2 domain (PDB ID: 1JYR)33 using Molecular Operating Environment (Chemical Computing Group) with the OPLS-AA forcefield. The protein is depicted as an electrostatic potential surface. Following energy minimization, Glu1 and Tyr3 form similar interactions with the Grb2-SH2 domain as pTyr-containing peptides.30 Asn5 is able to access a known specificity pocket. (E) A 100 ns molecular dynamics simulation of BC1 in explicit water revealed three distinct water-binding pockets formed by backbone carbonyls and amide protons. Surface represents the Van der Waals surface of BC1. The table shows approximate residence time, fractional occupancy (percentage of the simulation steps in which a water occupied the pocket) and calculated δδG of binding for each water. (F) Representative NMR structure of BC1 showing heavy atoms (blue spheres) that have protons with cross-relaxation peaks with water protons. For these protons, the water cross-peak in the ROESY was opposite in sign to the same cross-peak in the NOESY, which is characteristic of direct magnetization transfer to water (Supplementary Figure S11).28,29
Figure 4B shows the solution structure of the BC1 backbone (RMSD 0.33 Å). The backbone of Ala11 through Tyr3 forms an unusual turn structure that points amide protons inward and carbonyl oxygens out towards solvent. This effectively shields the hydrogen bond donors from aqueous solvent. To explore how BC1 might interact with water, we performed all-atom molecular dynamics simulations on a tethered model of the BC1 solution structure. During a 100 ns MD simulation in TIP4P explicit water using the OPLS/AA forcefield,23,24 we observed three distinct water molecules that bind three separate pockets formed by backbone amides and carbonyls (Figure 4E, Supplementary Video Files). The most tightly bound water molecule in the simulation binds in a pocket defined by the inward-facing amide protons of Ala11, Glu1, Leu2, and Tyr3, providing a unique explanation for the conformation of this loop. Over the course of the 100 ns simulation, this pocket exhibited 99% occupancy, with an average water molecule residence time of 1.4 ns and a calculated binding δδG of−2.74 kcal/mol. Discrete, bound waters are typical for protein surfaces,25-27 but this behavior is unusual for small, constrained peptides. To find experimental evidence of water-binding sites, we further analyzed the NOESY and ROESY data. Specifically, we looked for cross-peaks between BC1 protons and the water line at 4.78 ppm, which occur when discrete water molecules persist for 100-300 ps or longer.28,29 Direct magnetization transfer to water was observed from several protons within BC1, including protons within Ala11, Glu1, Leu2, Tyr3, Pro4, Met8, and Lys9 (Supplementary Figure S11). These protons map closely to predicted water-binding sites suggested by MD simulations (Figure 4F), providing experimental evidence that the unusual backbone of BC1 binds discrete waters. This appears to reduce the overall number of solvent-exposed H-bond donors, because several amide protons are shielded from bulk solvent upon the binding of a single water molecule. Future work will explore whether this property is directly or indirectly responsible for cell uptake, and whether this property can be generalized for the design of constrained peptides and small molecule macrocycles.
2.3. Observation and proposed binding mode of a conformation-dependent phosphotyrosine mimic
Finally, prior work established that BC1 competes for the phosphotyrosine (pTyr) binding site on Grb2-SH2, and that BC1 is recognized by anti-phosphotyrosine antibodies.12 Still, the precise nature of the phosphotyrosine-like epitope was not established. The solution structure of BC1 allowed direct observation of the pTyr mimic. Many of the strongest NOEs were between side-chain protons of Glu1 and Tyr3, providing direct evidence that these side chains are in close contact. In the refined ensemble of structures, the side chains of Glu1 and Tyr3 form a discontinuous epitope that resembles pTyr (Figure 4C). To further validate that these side chains form a pTyr-mimicking epitope, we tested BC1 variants with the conservative changes Glu1 to Asp, or Tyr3 to Phe (Supplementary Figure S12). These variants had no binding activity in a fluorescence polarization competitive binding assay (Supplementary Figure S13). Thus, we conclude that Glu1 and Tyr3 form a discontinuous, pTyr-mimicking epitope. To provide a more complete model for Grb2 binding, a representative BC1 structure was docked into the pTyr-binding site of Grb2-SH2 (Figure 4D). This model demonstrated that BC1 is pre-organized in a manner consistent with direct binding to the pTyr-binding pocket of Grb2-SH2, and is capable of making many of the same contacts observed in crystal structures with pTyr-containing peptides.30
3. Conclusion
In summary, our structural and computational data provide unique molecular-level explanations for the pTyr mimicry and cell uptake of the bicyclic scaffold BC1. Because scaffolds HT1 and BC3 show similar uptake profiles, diverse classes of neutrally charged, constrained scaffolds may be more readily designed than previously assumed. Future work will seek to diversify these scaffolds, clarify their uptake mechanisms, maximize endosomal escape, and apply them in selections and de novo design. Ultimately, these studies will reveal general principles for promoting high degrees of structure and cell penetration for non-helical, non-cationic peptides.
Supplementary Material
Acknowledgments
This work was supported in part by a Smith Family Award for Excellence in Biomedical Research and by NIH DP2-0D007303 to JAK, Tufts start-up fund and the Knez Family Faculty Investment Fund to YSL, and by NIH F31-CA180566 to JSQ. The authors are grateful to S. Pochapsky (Brandeis University Department of Chemistry) for her kind assistance in optimizing NMR experiments. The authors also thank T. Corlin (Tufts University Department of Biomedical Engineering) and Prof. R. Scheck for helpful conversations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Synthesis and structures of BC1 analogs, complete experimental descriptions, NMR Spectroscopic information, MD simulation data, and videos of MD trajectories.
References
- (1).Bock JE, Gavenonis J, Kritzer JA. ACS Chem. Biol. 2013;8:488–499. doi: 10.1021/cb300515u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Walensky LD, Bird GH. J. Med. Chem. 2014 doi: 10.1021/jm4011675. doi 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- (4).Frank AO, Vangamudi B, Feldkamp MD, Souza-Fagundes EM, Luzwick JW, Cortez D, Olejniczak ET, Waterson AG, Rossanese OW, Chazin WJ, Fesik SW. J. Med. Chem. 2014;57:2455–2461. doi: 10.1021/jm401730y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wallbrecher R, Depré L, Verdurmen WPR, Bovée-Geurts PH, van Duinkerken RH, Zekveld MJ, Timmerman P, Brock R. Bioconjug. Chem. 2014;25:955. doi: 10.1021/bc500107f. [DOI] [PubMed] [Google Scholar]
- (6).Madden MM, Muppidi A, Li ZY, Li XL, Chen JD, Lin Q. Synthesis of cell-permeable stapled peptide dual inhibitors of the p53-Mdm2/Mdmx interactions via photoinduced cycloaddition. Bioorg. Med. Chem. Lett. 2011;21:1472–1475. doi: 10.1016/j.bmcl.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Vives E, Brodin P, Lebleu B. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- (8).Appelbaum JS, LaRochelle JR, Smith BA, Balkin DM, Holub JM, Schepartz A. Chem. Biol. 2012;19:819–830. doi: 10.1016/j.chembiol.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lorents A, Kodavali PK, Oskolkov N, Langel U, Hallbrink M, Pooga M. J. Biol. Chem. 2012;287:16880–16889. doi: 10.1074/jbc.M111.318063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. J. Am. Chem. Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- (11).Cascales L, Henriques ST, Kerr MC, Huang Y-H, Sweet MJ, Daly NL, Craik DJ. J. Biol. Chem. 2011;286:36932–36943. doi: 10.1074/jbc.M111.264424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Quartararo JS, Wu P, Kritzer JA. ChemBioChem. 2012;13:1490–1496. doi: 10.1002/cbic.201200175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Oligino L, Lung FDT, Sastry L, Bigelow J, Cao T, Curran M, Burke TR, Wang SM, Krag D, Roller PP, King CR. J. Biol. Chem. 1997;272:29046–29052. doi: 10.1074/jbc.272.46.29046. [DOI] [PubMed] [Google Scholar]
- (14).Lung FDT, King CR, Roller PP. Development of non-phosphorylated cyclic thioether peptide binding to the Grb2-SH2 domain. Lett. Pept. Sci. 1999;6:45–49. [Google Scholar]
- (15).Gao Y, Voigt J, Wu J,X, Yang D, Burke TR. Bioorg. Med. Chem. Lett. 2001;11:1889–1892. doi: 10.1016/s0960-894x(01)00316-x. [DOI] [PubMed] [Google Scholar]
- (16).Lee K, Zhang M, Liu H, Yang D, Burke TR. J. Med. Chem. 2003;46:2621–2630. doi: 10.1021/jm030049q. [DOI] [PubMed] [Google Scholar]
- (17).Okamoto T, Zobel K, Fedorova A, Quan C, Yang H, Fairbrother WJ, Huang DCS, Smith BJ, Deshayes K, Czabotar PE. ACS Chem. Biol. 2013;8:297–302. doi: 10.1021/cb3005403. [DOI] [PubMed] [Google Scholar]
- (18).Bird GH, Bernal F, Pitter K, Walensky LD. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yu P, Liu B, Kodadek T. Nat. Biotechnol. 2005;23:746–751. doi: 10.1038/nbt1099. [DOI] [PubMed] [Google Scholar]
- (20).Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dev. Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- (21).Ochocki JD, Mullen DG, Wattenberg EV, Distefano MD. Bioorg. Med. Chem. Lett. 2011;21:4998–5001. doi: 10.1016/j.bmcl.2011.04.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kwon Y-U, Kodadek T. Chem. Biol. 2007;14:671–677. doi: 10.1016/j.chembiol.2007.05.006. [DOI] [PubMed] [Google Scholar]
- (23).Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- (24).Jorgensen WL, Maxwell DS, Tirado-Rives J. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- (25).Dastidar S, Mukhopadhyay C. Phys. Rev. E. 2003;68:021921. doi: 10.1103/PhysRevE.68.021921. [DOI] [PubMed] [Google Scholar]
- (26).Smolin N, Winter R. J. Phys. Chem. B. 2004;108:15928–15937. [Google Scholar]
- (27).Sheu S-Y, Yang D-Y. J. Phys. Chem. B. 2010;114:16558–16566. doi: 10.1021/jp105164t. [DOI] [PubMed] [Google Scholar]
- (28).Otting G, Wuethrich K. J. Am. Chem. Soc. 1989;111:1871–1875. [Google Scholar]
- (29).Otting G, Liepinsh E, Wuthrich K. Science. 1991;254:974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- (30).Rahuel J, Gay B, Erdmann D, Strauss A, GarciaEcheverria C, Furet P, Caravatti G, Fretz H, Schoepfer H, Grutter M. Nat. Struct. Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- (31).Holm T, Johansson H, Lundberg P, Pooga M, Lindgren M, Langel U. Nat Protoc. 2006;1:1001–1005. doi: 10.1038/nprot.2006.174. [DOI] [PubMed] [Google Scholar]
- (32).Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- (33).Nioche P, Liu W-Q, Broutin I, Charbonnier F, Latreille M-T, Vidal M, Roques B, Garbay C, Ducruix A. J. Mol. Biol. 2002;315:1167–1177. doi: 10.1006/jmbi.2001.5299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.