Summary
Receptor tyrosine kinase signaling is critical for mammalian craniofacial development, but the key downstream transcriptional effectors remain unknown. We demonstrate that SRF is induced by both PDGF and FGF signaling in mouse embryonic palatal mesenchyme cells, and Srf neural crest conditional mutants exhibit facial clefting accompanied by proliferation and migration defects. Srf and Pdgfra mutants interact genetically in craniofacial development, but Srf and Fgfr1 mutants do not. This signal specificity is recapitulated at the level of cofactor activation: while both PDGF and FGF target gene promoters show enriched genome-wide overlap with SRF ChIP-seq peaks, PDGF selectively activates a network of MRTF-dependent cytoskeletal genes. Collectively, our results identify a novel role for SRF in proliferation and migration during craniofacial development and delineate a mechanism of receptor tyrosine kinase specificity mediated through differential cofactor usage, leading to a unique PDGF-responsive SRF-driven transcriptional program in the midface.
Keywords: SRF, PDGF, FGF, MRTF, neural crest, craniofacial development
Introduction
Receptor tyrosine kinases (RTKs) engage shared signaling effectors, such as extracellular signal related kinase (ERK) and phosphatidylinositol 3-kinase (PI3K), but the in vivo phenotypes associated with different RTK mutants can be quite distinct (Lemmon and Schlessinger 2010). A central question revolves around how signal specificity arises from a seemingly general set of transduction pathways. At a transcriptional level, RTK signaling classically modulates the expression of immediate early genes (IEGs) (Cochran et al. 1984; Lau and Nathans 1987). While different RTK pathways, such as platelet derived growth factor (PDGF) and fibroblast growth factor (FGF) signaling, induce similar sets of IEGs in cultured cells (Fambrough et al. 1999), genetic experiments in mice suggest a degree of IEG specificity downstream of PDGF signaling (Schmahl et al. 2007). Thus, a major goal remains to characterize the key transcriptional mediators regulated by RTK signaling and determine their specificity downstream of different receptors.
Development of the mammalian face comprises derivatives from all three germ layers, including a unique contribution from the neural crest. Many components of RTK signaling are linked to craniofacial syndromes and phenotypes in both mice and humans (Newbern et al. 2008; Bentires-Alj et al. 2006). Mice harboring neural crest cell (NCC) conditional loss of PDGF receptor α (PDGFRα) using the Wnt1-Cre transgene exhibit cleft face and palate (Tallquist and Soriano 2003). Combined loss of both PDGFRα-specific ligands, PDGFA and PDGFC, results in facial clefting (Ding et al. 2004). In humans, mutations in and around PDGFC (Choi et al. 2009; Calcia et al 2013) and PDGFRα (Rattanasopha et al. 2012) have been associated with cleft lip and palate (CL/P), reflecting a conserved role for PDGF signaling in mammalian midface development. Interestingly, NCC conditional loss of FGF receptor 1 (FGFR1) also results in craniofacial defects (Trokovic et al. 2003; Wang et al. 2013), indicating a requirement for both PDGF and FGF signaling in NCCs for craniofacial morphogenesis.
Serum response factor (SRF) is a transcription factor critical for coupling actin dynamics and signaling pathways to gene expression (Posern and Treisman 2006; Olson and Nordheim 2010). SRF was identified as a regulator of the serum response in fibroblasts (Treisman 1987), and more recent work has focused on understanding the mechanisms of SRF specificity at the transcriptional level (Gineitis and Treisman 2001), particularly in regard to interactions with its two major cofactor families: ternary complex factors (TCFs) and myocardin related transcription factors (MRTFs) (Esnault et al. 2014). SRF can be activated in response to many extracellular stimuli, including PDGF and FGF (Treisman 1996; Wang et al. 2004). However, the specificity of SRF activation at a receptor level is unclear, and a direct comparison of SRF function downstream of multiple RTKs has not been carried out.
SRF is essential across many developmental and physiological contexts, including mesoderm formation (Arsenian et al. 1998), cardiac development (Parlakian et al. 2004), angiogenesis (Franco et al. 2008), oligodendrocyte differentiation (Stritt et al. 2009), neuronal migration (Alberti et al. 2005), and circadian regulation (Gerber et al. 2013). SRF was first implicated in neural crest development through an in situ hybridization screen (Adams et al. 2008), and neural crest conditional Srf mouse mutants show defects in dorsal root ganglion (DRG) formation (Wickramasinghe et al. 2008), cardiac outflow tract development, and mandible formation (Newbern et al. 2008). No facial clefting phenotypes have been previously reported, and the role of SRF in midface development remains unknown.
In the present study, we report that SRF is required for craniofacial development and responds differentially to PDGF and FGF signaling through selective interactions with MRTF and TCF cofactors. Wnt1-Cre; Srffl/fl mutants exhibit overt facial clefting as well as proliferation and migration deficits in the cranial neural crest and its derivatives. We find that Srf and Pdgfra double mutants (Wnt1-Cre; Srf+/fl; Pdgfra+/fl) display varying degrees of craniofacial defects, but Srf and Fgfr1 (Wnt1-Cre; Srf+/fl; Fgfr1+/fl) do not interact genetically, indicating that SRF function downstream of these two RTKs is not identical. We demonstrate that this specificity is encoded at the level of MRTF-SRF activation and recapitulated in the genome wide binding profile of SRF and MRTF at the promoters of PDGF target genes, particularly those involved in cytoskeletal organization. Taken together, our studies illustrate a novel role for SRF in controlling proliferation and migration during craniofacial development and uncover an example of RTK specificity mediated by a common transcription factor through differential cofactor usage and unique output gene expression signatures.
Results
PDGF activates SRF in MEPMs, and PDGFRα and SRF are coexpressed during craniofacial development
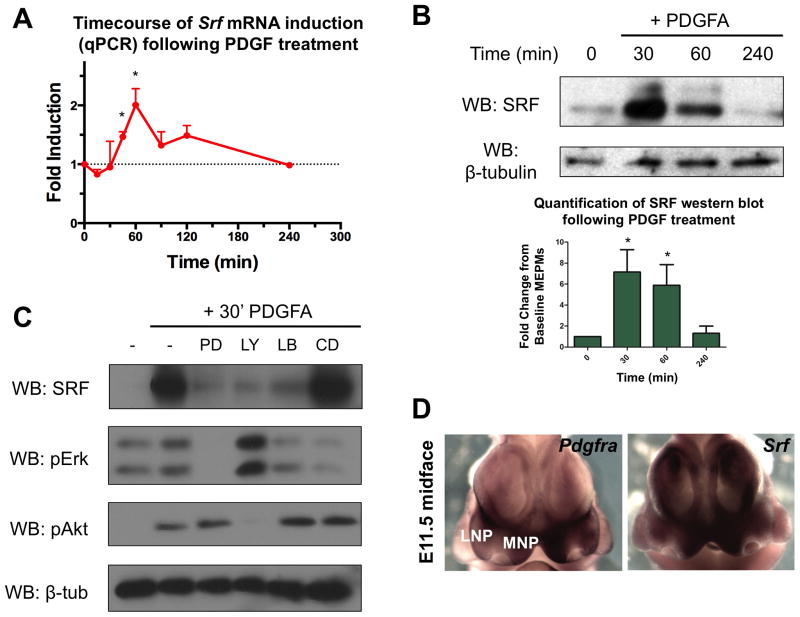
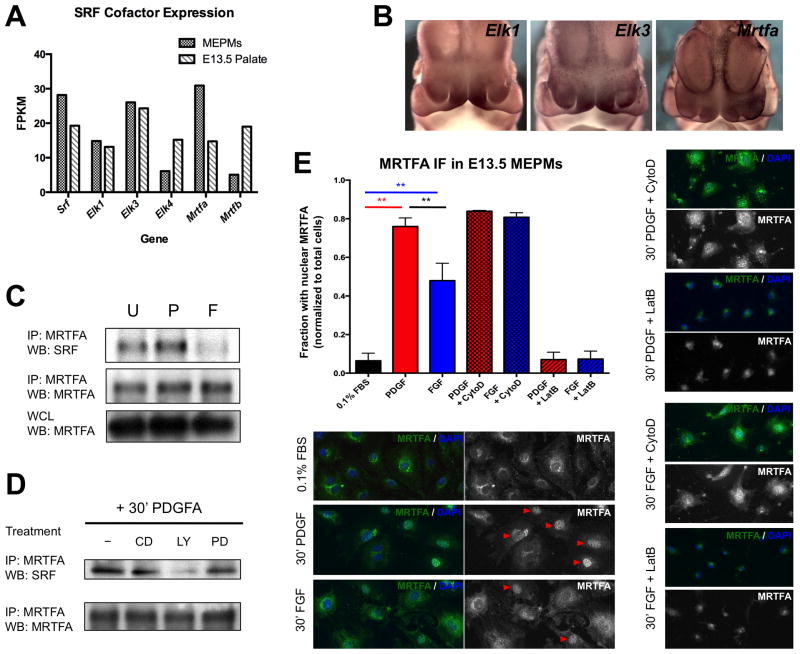
To identify transcriptional targets of PDGF signaling in the midface, RNA-seq was carried out in E13.5 mouse embryonic palatal mesenchyme (MEPM) cells treated with PDGFA (which specifically activates PDGFRα), identifying Srf as a PDGF target gene (Table S2, GSE61755). MEPMs express many palatal mesenchyme markers, including Pdgfra, and have been used to study PDGF (Fantauzzo and Soriano 2014) and Ephrin signaling (Bush and Soriano 2010). A quantitative PCR (qPCR) timecourse revealed the peak of Srf mRNA induction to occur at 60 minutes following PDGF treatment (Fig. 1A), and Western blot confirmed this increase at the protein level (Fig. 1B). The increase in SRF protein prior to Srf mRNA is likely due in part to post-transcriptional regulation of IEG induction (Avraham and Yarden 2011). We observed the appearance of a shifted band following PDGF treatment; indeed, SRF is phosphorylated at multiple residues in response to growth factor treatment, and previous work has shown these modifications can affect SRF activity in vitro (Rivera et al. 1993; Iyer et al. 2006). Thus, we treated PDGF stimulated MEPM lysates with calf intestinal phosphatase (CIP) which resulted in loss of the upper band (Fig. S1A), indicating PDGF treatment promotes SRF phosphorylation. To determine the signaling dependence of SRF induction, we performed Western blots following PDGF treatment in the presence of PD325901 (MEK inhibitor), LY294002 (PI3K inhibitor), latrunculin B (MRTF inhibitor), and cytochalasin D (MRTF activator). We found that PDGF mediated SRF induction requires both PI3K and ERK signaling as well as MRTF activity (Fig. 1C).
Figure 1.
SRF is a target of PDGF signaling in craniofacial development. (A–B) In E13.5 MEPMs, PDGF stimulation increases (A) Srf mRNA (2-fold peak induction) and (B) protein (7-fold peak induction) (n=3). Data plotted as mean ± SEM. *p<0.05. (C) SRF induction following PDGF stimulation requires ERK, PI3K, and MRTF activity, as evidenced by inhibition of these pathways. Cells treated with 30 ng/mL PDGFAA for desired duration. PD = PD325901, LY = LY294002, LB = Latrunculin B, CD = cytochalasin D. (D) At E11.5, Pdgfra and Srf mRNA are coexpressed in the developing medial nasal process (MNP) and less robustly in the lateral nasal process (LNP). See also Fig. S1 and Table S2.
Next, we analyzed the expression pattern of Srf and Pdgfra during craniofacial development. Whole mount in situ hybridization (WISH) revealed both genes are expressed in the E11.5 medial nasal process (MNP) (Fig. 1D), and we confirmed protein co-expression in the developing MNP and maxillary process (MxP) with anti-SRF immunofluorescence on Pdgfra+/GFP reporter embryos (Hamilton et al. 2003) (Fig. S1B). At E13.5, both Srf and Pdgfra were present broadly in the craniofacial region, with expression noted in the anterior palate at both the mRNA and protein levels (Fig. S1C–D). These experiments show that PDGF induces SRF in an ERK, PI3K, and MRTF dependent manner, and SRF is co-expressed with PDGFRα in the midface.
Srf conditional mutants exhibit overt facial clefting and interact genetically with Pdgfra mutants but not Fgfr1 mutants
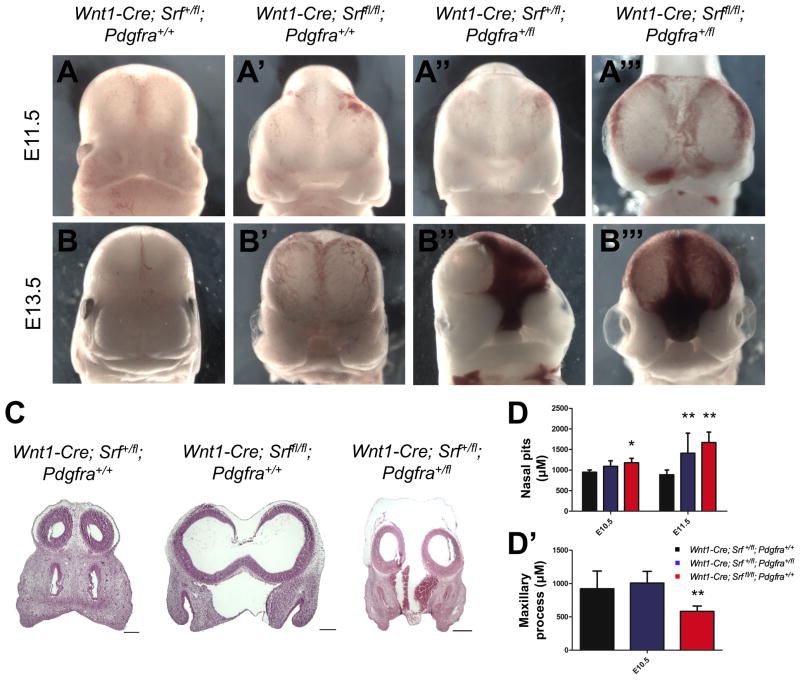
While cardiac, neuronal, and mandibular defects have been observed in NCC conditional Srf mutants (Newbern et al. 2008; Wickramasinghe et al. 2008), detailed analysis of the craniofacial phenotypes in these mice has not been carried out. We therefore conditionally disrupted Srf in NCCs using the Wnt1-Cre driver (Danielian et al. 1998). We found fully penetrant facial clefting in Wnt1-Cre; Srffl/fl mutants compared to heterozygous Wnt1-Cre; Srf+/fl controls, which appear grossly normal (Fig. 2A–2B′). Further, Wnt1-Cre; Srf+/fl; Pdgfra+/fl double heterozygotes exhibit partially penetrant facial clefting (Fig. 2A″, 2B″), and infrequently recovered Wnt1-Cre; Srffl/fl; Pdgfra+/fl mutant embryos display even more severe phenotypes characterized by gross midline hemorrhage and blistering in the cephalic region (Fig. 2A‴, 2B‴) (Table S1). The defects observed in Pdgfra/Srf double mutants are reminiscent of Pdgfra−/− knockouts, which also exhibit facial clefting, hemorrhaging, and blisters (Soriano 1997). We next performed hematoxylin and eosin (H&E) staining, confirming that the facial clefting extends through the midline in both Wnt1-Cre; Srffl/fl and Wnt1-Cre; Srf+/fl; Pdgfra+/fl mutants at E11.5 (Fig. 2C). We carried out morphometric analysis to quantify the relative severity of clefting across genotypes. We found significantly increased distances between the nasal pits (Fig. 2D) as well as reduced maxillary process length (Fig. 2D′) in Srf mutants beginning as early as E10.5. No differences in mandibular morphogenesis were detected at these stages (data not shown). Thus, SRF function is required in NCCs for craniofacial development, and Srf/Pdgfra compound heterozygotes exhibit phenotypes despite the grossly normal appearance of Srf or Pdgfra heterozygotes, suggesting these two genes may function within a common network
Figure 2.
Wnt1-Cre; Srffl/fl mutants exhibit facial clefting and interact genetically with Pdgfra. (A–B) Neural crest cell conditional deletion of Srf results in fully penetrant facial clefting at both (A, A′) E11.5 and (B, B′) E13.5. (A″, B″) Embryos heterozygous for both Srf and Pdgfra display a partially penetrant clefting phenotype while (A‴, B‴) Srf homozygous mutants missing one copy of Pdgfra display exacerbated phenotypes, including severe midline hemorrhage and blebbing. (C) Frontal sections in E11.5 embryos show clefting and forebrain expansion in both the Wnt1-Cre; Srffl/fl and Wnt1-Cre; Srf+/fl; Pdgfra+/fl mutants as well as midline hemorrhage in the double heterozygous condition. (D) Morphometry reveals differences in the distance between nasal pits and (D′) maxillary process length in Wnt1-Cre; Srffl/fl mutants (n=5 at E10.5, n=11 at E11.5) and Wnt1-Cre; Srf+/fl; Pdgfra+/fl embryos (n=4 at E10.5, n=9 at E11.5) compared to controls (n=10 at E10.5, n=10 at E11.5). *p<0.05. **p<0.01. Scale bars: 200 μm. All data plotted as mean ± SEM. See also Fig. S2 and Table S1.
Given the activation of SRF in response to many extracellular signals (including FGF), we hypothesized that SRF may also function downstream of FGF signaling during craniofacial development. Indeed, FGF stimulation induces Srf mRNA in E13.5 MEPMs; however, in contrast to PDGF signaling, FGF mediated SRF induction required ERK signaling but not PI3K or MRTF activity (Fig. S2A–B). Further, no interaction between Fgfr1 and Srf conditional mutants (Wnt1-Cre; Srf+/fl; Fgfr1+/fl) was observed (Fig. S2C), despite the fact that FGFR1 is the primary FGF receptor in the neural crest and craniofacial mesenchyme (Trokovic et al. 2003, Park et al. 2008). These results suggest that activation of SRF by PDGF and FGF signaling is fundamentally different, and more broadly, that these two receptors perform at least a subset of non-overlapping functions.
We recently showed that the original Wnt1-Cre results in Wnt1 overexpression and enlargement of the midbrain and therefore generated a Wnt1-Cre2 transgenic line as an alternative without these caveats (Lewis et al. 2013). Facial clefting phenotypes obtained with this new Cre driver were similar to those observed with the original Wnt1-Cre (Fig. S3A).
SRF mutants display cell proliferation and migration deficits during craniofacial development
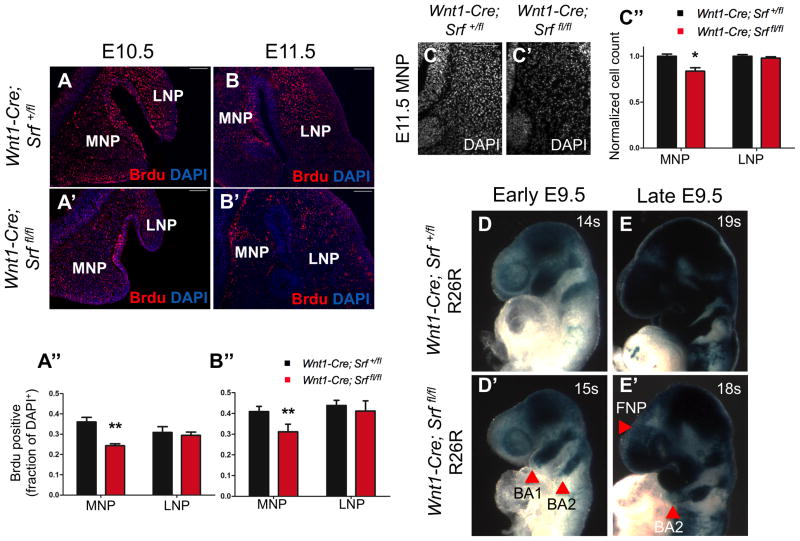
SRF is known to control a diverse range of cellular outcomes, including cell proliferation, migration, survival, and differentiation. Thus, we examined each of these processes in Wnt1-Cre; Srffl/fl mutants to determine the basis for the observed clefting phenotypes. We found reduced proliferation in the MNP of Srf mutants (Fig. 3A–B″) at both E10.5 and E11.5. Similarly, we found fewer cells specifically in the MNP of E11.5 Wnt1-Cre; Srffl/fl mutant embryos (Fig. 3C) although no such reduction in total cell number was observed at E10.5 (data not shown). This spatiotemporally specific proliferation defect in Wnt1-Cre; Srffl/fl mutants is consistent with previous work showing that Wnt1-Cre; Pdgfrafl/fl mutants also exhibit reduced MNP proliferation (He and Soriano 2013). We did not find any difference in apoptosis between control and Srf mutant embryos in the MNP or LNP, and we did not detect any change in the expression of MNP marker genes, such as Alx3 (data not shown).
Figure 3.
Wnt1-Cre; Srffl/fl mutants display proliferation and lineage tracing defects in vivo. (A, B) Srf mutants exhibit decreased proliferation specifically in the MNP at (A-A″) E10.5 (n=5) and (B-B″) E11.5 (n=7). **p<0.001. (C) The MNP of Srf conditional mutants is hypocellular, with significantly fewer cells at E11.5 (n=7). Cell counts were normalized to number of cells in littermate control. *p<0.005. (D, E) Lineage tracing using the ROSA26 reporter (R26R) reveals reduced contribution of neural crest cells to (D, D′) the first and second branchial arches (BA1, BA2) and (E, E′) frontonasal prominence (FNP) at E9.5 (somite number indicated). Blue (lacZ positive) cells are generated by Wnt1Cre mediated recombination and thus label the neural crest and its derivatives. Scale bars: 100 μm. All data plotted as mean ± SEM.
Classic studies have shown that many craniofacial structures are predominantly derived from the neural crest (Couly et al. 1993; Chai et al. 2000). To visualize defects in migration and population of the craniofacial mesenchyme by NCCs, we crossed Srf conditional mutants to R26R-lacZ mice (Soriano 1999). At E9.5, Wnt1-Cre; Srffl/fl; R26R+/− mutant embryos showed impaired neural crest contribution to the frontonasal prominence (FNP), first branchial arch (BA1), and second branchial arch (BA2) (Fig. 3D–E). We often observed BA1 and BA2 defects in early E9.5 embryos (<18 somites, 7/9 mutant embryos with phenotype) and FNP and BA2 defects in late E9.5 embryos (≥18 somites, 6/9 mutant embryos with phenotype). The combination of lineage tracing and proliferation defects reflects the requirement of SRF activity in the neural crest to both fully populate the craniofacial mesenchyme and respond to proliferative signals.
SRF is required for cellular responses to RTK signaling in facial prominence cells
Since Wnt1-Cre; Srffl/fl embryos are recovered below Mendelian ratios at E13.5, we turned to an earlier stage of craniofacial development to further investigate SRF function. Given the in vivo defects observed in the E11.5 midface, we established facial prominence cells (FPCs) from E11.5 embryos as a primary cell culture model to study the effects of SRF loss; control FPCs show robust expression of Srf, Pdgfra, and Fgfr1, and Wnt1-Cre2; Srffl/fl mutant FPCs express almost no Srf mRNA (Fig. S3B) or protein (Fig. S3C).
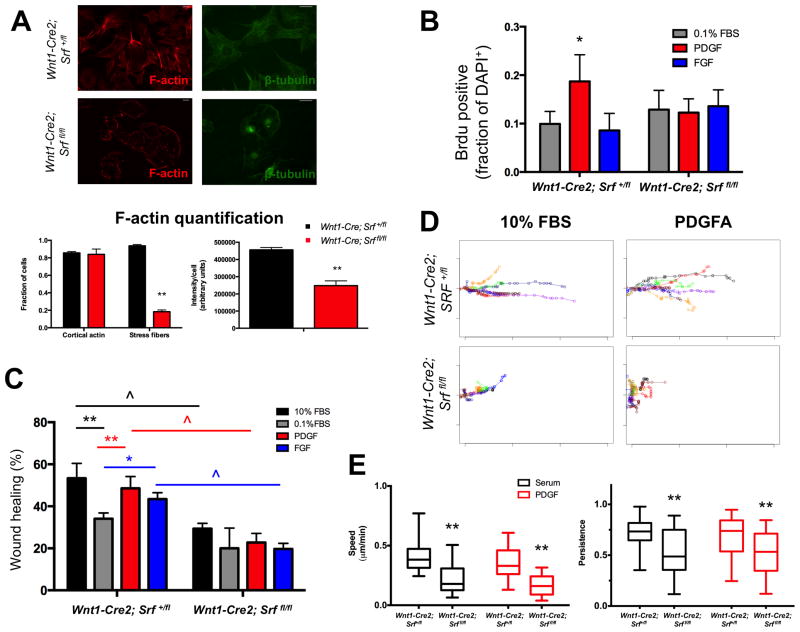
SRF is critical for maintaining proper cytoskeletal morphology (Schratt et al. 2002). Thus, we stained Srf mutant FPCs for F-actin and β-tubulin and observed gross defects in actin stress fiber formation and microtubule organization (Fig. 4A). Next, we tested the proliferative response of FPCs to PDGF and FGF stimulation. Surprisingly, only PDGF induced proliferation in control FPCs while Srf mutant FPCs fail to proliferate following PDGF treatment, reflecting the requirement of SRF function for PDGF-dependent cell proliferation (Fig. 4B). The selective response of FPCs to PDGF may partially explain the phenotypic interactions observed between Srf and Pdgfra conditional mutants but not Srf and Fgfr1 conditional mutants. Finally, we performed scratch assays to compare the response of control and Srf mutant FPCs. In Wnt1-Cre2; Srf+/fl FPCs, 10% FBS, PDGF, and FGF all induced significant wound closure, but this response was abrogated under all conditions in Wnt1-Cre2, Srffl/fl mutant FPCs, reflecting an intrinsic defect in Srf mutant cells (Fig. 4C, Fig. S3D). Knockdown of SRF is crucial for both cell motility and directional persistence (Medjkane et al. 2009), and we therefore performed time-lapse microscopy and single cell tracking to better understand this deficit. These experiments revealed that although a subset of Wnt1-Cre2; Srffl/fl mutant FPCs do move efficiently (Fig. 4D, Fig. S3E), Srf mutant FPCs overall are significantly slower and exhibit decreased directional persistence compared to control cells (Fig. 4E). In sum, Wnt1-Cre2; Srffl/fl FPCs display proliferation and motility defects in response to growth factor stimulation, linking the observed in vivo proliferation and lineage tracing defects in Srf mutants to RTK signaling and demonstrating the functional relationship between these pathways in the midface.
Figure 4.
Srf mutant E11.5 facial prominence cells (FPCs) do not proliferate in response to PDGF and exhibit defective wound healing. (A) Srf mutant FPCs lack actin stress fibers, show decreased total F-actin staining, and display gross microtubule disorganization. (n>100 cells/condition) (B) Control FPCs exhibit a modest proliferative response to PDGF stimulation, but Srf mutant FPCs fail to proliferate in response to PDGF (n=4). (C) Although control FPCs show significant wound healing when treated with 10% FBS, PDGF, or FGF compared to 0.1% FBS, Srf mutant FPCs fail to show significant closure when compared to 0.1% FBS starved cells (n=3). Further, Srf mutant FPCs show significant decreases in wound healing across all growth factor conditions when compared to control FPCs. (D) Representative trajectories from 10 cells tracked during wound healing in response to either 10% FBS or PDGF. Srf mutant FPCs show both decreased directionality and total distance traveled, although some mutant cells move relatively efficiently (blue trajectory in 10% FBS condition). The heterogeneous migration properties were not due to incomplete loss of SRF (Fig. S3B–C). (E) Srf mutant cells show decreased speed and persistence in response to both 10% FBS and PDGF when compared to control cells (quartile plot with whiskers spanning 5%–95%). Scale bars: 25 μm. Data in A–C plotted as mean ± SEM. **p<0.001. ^p<0.001. *p<0.05. Cells treated with either 30 ng/mL PDGFAA or 50 ng/mL FGF1 + 1 μg/mL heparin. See also Fig. S3.
PDGF mediates MRTFA-SRF complex formation and uniquely activates a set of MRTF-SRF associated cytoskeletal genes
In addition to direct transcriptional induction, many mechanisms have been described to regulate SRF function, including alternative splicing (Belaguli et al. 1999), direct nuclear translocation (Camoretti-Mercado et al. 2000), and differential cofactor usage (Posern and Treisman 2006). Therefore, we investigated how these parameters are modulated by RTK activation and whether signal specificity was encoded through these mechanisms. We did not detect alternative splicing of SRF (Fig. S4A–C), and PDGF treatment did not significantly alter the cellular localization of SRF in MEPMs (Fig. S4D). Similarly, we did not observe changes in SRF splicing or localization following FGF treatment (data not shown), suggesting these mechanisms are not utilized by either RTK in this context.
The two major cofactor families utilized by SRF are the TCFs (Elk1, Elk3/Net, and Elk4/Sap1) and MRTFs (MRTFA/Mkl1 and MRTFB/Mkl2). MRTF-dependent SRF activity occurs downstream of changes in actin concentration predominantly mediated by Rho-family small GTPases (Miralles et al. 2003; Vartiainen 2007). In contrast, TCF-dependent SRF activation lies downstream of ERK signaling (Posern and Treisman 2006). The distinction between the two mechanisms of SRF activation has a functional consequence, as unique SRF regulated gene sets are controlled through each of these pathways (Gineitis and Treisman 2001). Although RTK signaling is traditionally associated with robust activation of ERK, PDGF and FGF have also been shown to modulate small GTPase function in many contexts, including the midface (He and Soriano 2013). We began by screening the expression of SRF cofactor genes in MEPM RNA-seq and published E13.5 palate RNA-seq data (www.facebase.org, accession FB00000278.2); only Elk1, Elk3, and Mrtfa are expressed above a FPKM threshold of 10 in both datasets (Fig. 5A). We next performed WISH in E11.5 embryos to determine the expression pattern of these cofactors and compared them to Srf and Pdgfra. Both Elk1 and Elk3 show strong expression in the MNP, but Mrtfa is also expressed throughout the craniofacial region, albeit more diffusely (Fig. 5B). Thus, based on expression pattern alone, any of these cofactors may synergize with SRF in the midface.
Figure 5.
Both MRTF and TCF cofactors play roles downstream of RTK signaling in craniofacial development. (A) Of the five major TCF and MRTF cofactor family members, only Elk1, Elk3, and Mrtfa are expressed above a threshold of 10 fragments per kilobase of transcript per million mapped reads (FPKM) in both E13.5 MEPMs and E13.5 palate. (B) Whole mount in situ hybridization reveals Elk1 and Elk3 mRNA are enriched in the E11.5 medial nasal process (MNP). Mrtfa mRNA expression in the midface is more diffuse but shares expression domains with Pdgfra and Srf. (C) PDGF modestly increases SRF-MRTFA association while FGF reduces SRF-MRTFA complex formation. MRTFA levels are not modulated by PDGF or FGF treatment, thus serving as an additional loading control. (D) MRTFA-SRF association following 30 minutes PDGF stimulation requires PI3K activity. All biochemistry performed in E13.5 MEPMs. CD = cytochalasin D, PD = PD325901, LY = LY294002. U = untreated cells, P = 30 minutes 30 ng/mL PDGFAA, F = 30 minutes 50 ng/mL FGF1 + 1 μg/mL heparin. (E) MRTFA immunofluorescence shows greater nuclear accumulation of MRTFA in response to PDGF compared to FGF, although a significant number of FGF treated cells contain nuclear MRTFA. Red arrowheads mark cells counted as containing nuclear MRTFA. Cytochalasin D and Latrunculin B were used as a positive and negative control, respectively. ** p < 0.05. Data plotted as mean ± SEM. See also Fig. S4.
In order to directly test SRF-cofactor complex formation, we treated E13.5 MEPMs with either PDGF or FGF, performed immunoprecipitation (IP) for either Elk1 or MRTFA, and then Western blotted for SRF. Although both PDGF and FGF promoted formation of an SRF-Elk1 complex, only PDGF treatment resulted in SRF-MRTFA association; conversely, FGF stimulation reduced the amount of SRF-MRTFA complex (Fig. 5C, Fig. S4E). This PDGF mediated SRF-MRTFA association required PI3K (Fig. 5D) activity while SRF-Elk1 association required both ERK and PI3K signaling (Fig. S4E), consistent with previous work implicating PI3K as the key effector of PDGFRα signaling during craniofacial development (Klinghoffer et al. 2002; Fantauzzo and Soriano 2014). Since MRTF activation results in shuttling of the protein from the cytoplasm to the nucleus (Miralles et al. 2003), we next performed MRTFA immunofluorescence in MEPMs (Fig. 5E). While MRTFA is predominantly cytoplasmic in starved cells, PDGF stimulation increases nuclear MRTFA. FGF also induces MRTFA translocation, but to a lesser extent than PDGF. Indeed, PDGF induced MRTFA shuttling occurs at comparable ratios to cytochalasin D treated MEPMs, consistent with a full MRTFA response to PDGF. The observed heterogeneity of the MRTFA response in MEPMs may be in part a result of our primary cell culture system, and quantitatively similar changes in nuclear MRTFA have been observed in other studies (Ho et al. 2013).
These results raised the possibility that PDGF signaling preferentially drives MRTFA dependent SRF activity while both PDGF and FGF activate Elk1 mediated SRF function. In order to gain insight toward the genome wide role of SRF downstream of these pathways, we integrated our MEPM RNA-seq data with SRF chromatin immunoprecipitation (ChIP) sequencing data from mouse C2C12 (ENCODE 2012; GSM915168) and 3T3 (Esnault et al. 2014) cells. Although SRF binding events are unlikely to be fully conserved across different contexts, a previous study estimated ~60% of SRF binding events in the proximal promoter are shared between cell types (Sullivan et al. 2011), supporting our correlative approach. We also recently generated an analogous RNA-seq dataset for FGF treated MEPMs, allowing comparison of the transcriptional outputs from both PDGF and FGF signaling (Table S2). First, we plotted the distribution of SRF ChIP-seq peaks from the transcriptional start site (TSS) of all genes upregulated at one hour by either PDGF (125 significant genes) or FGF (135 significant genes) (Fig. 6A–B). We found enrichment of SRF ChIP-seq peaks upstream from the TSS of RTK regulated genes, suggesting SRF mediated transcription plays a key role in the genome-wide response to both PDGF and FGF. A full list of these peaks and genes is provided (Table S3). No such enrichment was observed in randomly selected, expression matched control genes (Fig. 6A–B, black lines) unresponsive to growth factor treatment or when plotting the peak distribution from ChIP-seq data for Jun (induced upon PDGF treatment in MEPMs), Pax5 (not expressed in MEPMs), or p300 (a transcriptional co-activator sampled in the E13.5 palate) (Fig. S5A–C).
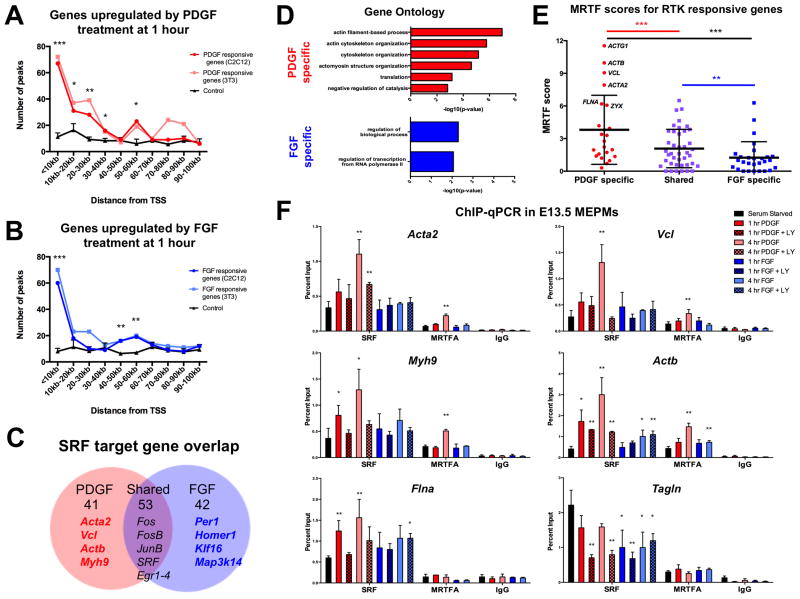
Figure 6.
Both PDGF and FGF responsive genes correlate with SRF binding genome-wide, but only PDGF target gene promoters are enriched for MRTF. (A, B) Both PDGF (red) and FGF (blue) responsive genes show enrichment for SRF binding events from C2C12 (ENCODE 2012) and 3T3 (Esnault et al. 2014) SRF ChIP-seq data. Randomly sampled expression matched control genes (black, n=3 random sets) show no such enrichment. A total of 67 PDGF responsive genes have an SRF ChIP-seq peak within 10 kb of the TSS while 52 FGF responsive genes have an SRF ChIP-seq peak within 10 kb of the TSS. *p<0.1, **p<0.05, ***p<0.01. (C) Closer inspection of SRF target genes downstream of each RTK reveals 56% overlap, with many classic IEGs (Fos, Jun, Egr) activated jointly by both PDGF and FGF. (D) PDGF-SRF targets show enrichment for actin cytoskeletal elements while FGF-SRF targets show minimal functional organization. (E) PDGF-SRF targets show significantly increased MRTF scores compared to FGF-SRF targets, consistent with PDGF-specific activation of an MRTF associated transcriptional program. (F) SRF and MRTFA ChIP-qPCR in E13.5 MEPMs reveals increased binding of these factors at the promoters of cytoskeletal genes in response to PDGF (red) in contrast to FGF (blue). Inhibition of PI3K signaling (patterned bars) reduces PDGF stimulated SRF binding, although a significant response is still observed at some promoters. *p<0.1, **p<0.05, compared to serum starved. (n=3). Cells treated with 30 ng/mL PDGFAA or 50 ng/mL FGF1 + 1 μg/mL heparin. See also Fig. S5 and Table S3.
A total of 94 PDGF and 95 FGF target genes contain an SRF ChIP-seq peak within 70 kb of the TSS in either C2C12 or 3T3 cells; over half of these genes are induced by both pathways (56% shared) (Fig. 6C). Many classic IEGs (such as Fos, Jun, and Egr) fall into the group of 53 genes with SRF binding events that are jointly induced by both PDGF and FGF signaling. Interestingly, a high percentage of genes (56–75%) were conserved in both ChIP-seq datasets (Table S3). Gene ontology analysis revealed that PDGF-responsive SRF targets show overrepresentation of genes associated with the actin cytoskeleton while FGF-mediated SRF targets show no such relationship (Fig. 6D). To visualize these target genes at the level of cofactor specificity, we next integrated our datasets with recently published MRTF and TCF ChIP-seq datasets in 3T3 cells that assigned a score for each target gene reflecting the relative binding of these cofactors (Esnault et al. 2014) (Table S4). We found that many shared RTK targets possess high TCF scores, but a subset of PDGF specific genes show high MRTF scores (Fig. S5D–E). Indeed, MRTF scores for PDGF-SRF target genes are significantly increased compared to MRTF scores or FGF-SRF target genes (Fig. 6E), but no such difference is observed for TCF scores (Fig. S5F). This correlation between PDGF target genes and SRF-MRTF binding genome-wide may reflect a PDGF-MRTF-SRF circuit not regulated by FGF signaling.
SRF regulates the expression of a cytoskeletal network critical for craniofacial development
Our genomic analyses suggest PDGF mediated SRF activation results in activation of a key cytoskeletal transcriptional program; however, given the ChIP-seq and RNA-seq data were generated from different cell types, our correlative approach alone does not delineate whether these binding events and gene expression changes are functional. Thus, we probed this network in more detail using a candidate-based approach. We selected eight genes for further study based on published mouse craniofacial phenotypes and reported disease associations in humans (Fig. S6A). Many of these genes show high MRTF scores and, in the case of Acta1 and Myh9, have been previously identified as MRTF target genes (Sun et al. 2006; Medjkane et al. 2009). We performed endogenous SRF ChIP in E13.5 MEPMs at previously identified SRF binding sites in these eight target promoters, finding six (Vcl, Acta2, Myh9, Actb, Tgln, Flna) to show SRF binding (Fig. S6B). Five of these six promoters (Acta2, Vcl, Myh9, Actb, Flna) exhibit increased SRF binding following PDGF stimulation; comparing the two RTKs, all six targets show significantly greater SRF binding in response to PDGF, while only one promoter (Actb) exhibits increased binding following FGF stimulation (Fig. 6F). In contrast, SRF binding at the promoter of the shared target Fos is induced by both PDGF and FGF signaling, and low occupancy was observed in the promoter of Arid5b, an IEG not bound by SRF in either ChIP-seq dataset (Fig. S6C).
Given the importance of PI3K signaling downstream of PDGF in the midface, we assayed the effect of PI3K inhibition on SRF binding. We found decreased PDGF mediated SRF binding at all six targets following pretreatment with LY294002; however, two target promoters (Acta2 and Actb) still showed significant responses, indicating PI3K signaling is not always required for SRF binding (Fig. 6F). In addition, PI3K inhibition significantly decreased SRF binding at the Tagln promoter across all conditions, suggesting PI3K promotes SRF maintenance at this locus. Next, we carried out endogenous MRTFA ChIP and demonstrate significantly increased MRTFA binding at four loci (Acta2, Vcl, Myh9, Actb) in response to PDGF treatment (Fig. 6F). In contrast, FGF induced significant MRTFA binding at only the Actb promoter, consistent with the increased SRF binding at this region. No MRTFA binding was observed at either the Flna or Fos (Fig. S6C) promoter. Collectively, these results indicate PDGF and FGF differentially modulate SRF and MRTFA binding at target gene promoters, in part through PI3K signaling.
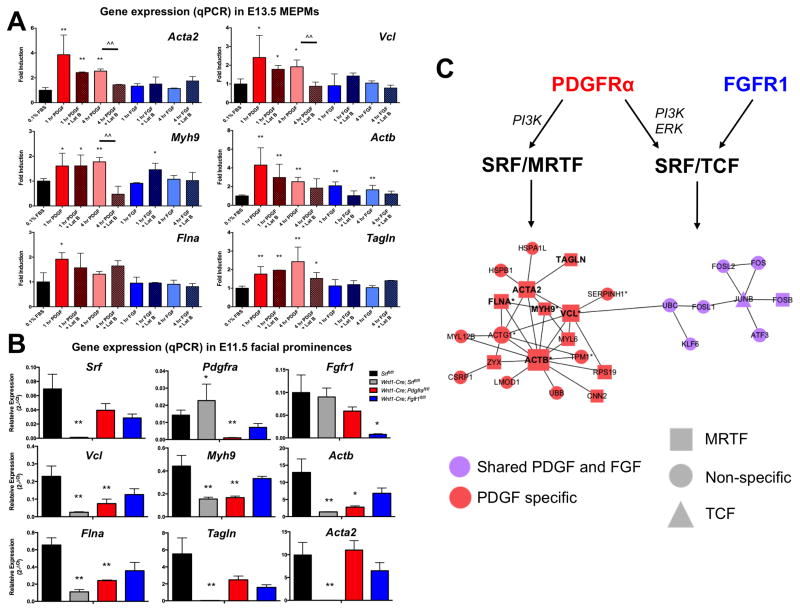
To determine the MRTF dependence of these genes, we stimulated MEPMs with PDGF or FGF in the presence of Latrunculin B (Fig. 7A). All six genes are selectively induced by PDGF. Further, Latrunculin B inhibits the PDGF mediated expression of five genes (Acta2, Vcl, Myh9, Actb, Tagln), confirming these targets are indeed MRTF dependent. While MRTFA nuclear accumulation is observed at 30 minutes following PDGF treatment, both the repressive effect of Latrunculin B and observed MRTFA binding at target gene promoters is more pronounced at four hours, suggesting MRTF mediated changes in gene expression may be a delayed response. The induction of Fos by both PDGF and FGF is not affected by Latrunculin B (Fig. S6D). We then measured the expression of these six genes in E11.5 facial prominences (MNP, LNP, and MxP) dissected from Srf, Pdgfra, and Fgfr1 conditional mutants. As expected, the expression of Srf, Pdgfra, and Fgfr1 were decreased in the corresponding mutants; further, we observed downregulation of all six targets in Srf mutant facial prominences (Fig. 7B). Finally, we found significantly decreased expression of four PDGF-SRF targets (Vcl, Myh9, Actb, and Flna) in Pdgfra mutants, but not Fgfr1 mutants (Fig. 7B). Interestingly, a modest increase in Pdgfra expression was observed in Srf mutants, possibly indicating a compensatory feedback mechanism. These results reflect the perturbation of a PDGF responsive, MRTF dependent cytoskeletal circuit specifically in Pdgfra and Srf mutants.
Figure 7.
A PDGF-MRTF-SRF axis controls expression of key cytoskeletal regulators in craniofacial development.. (A) PDGF (red bars) robustly activates expression of cytoskeletal target genes in MEPMs while FGF (blue bars) does not. Latrunculin B treatment (patterned bars) inhibits PDGF mediated gene expression, indicating induction of these shared PDGF-SRF targets is MRTF dependent. (B) The expression of all six cytoskeletal SRF target genes is reduced in E11.5 Wnt1-Cre; Srffl/fl mutant facial prominences (grey bars). While none of these genes show reduced expression in Wnt1-Cre; Fgfr1fl/fl mutant facial prominences (blue bars), four of six genes (Vcl, Myh9, Actb, and Flna) show downregulation in Wnt1-Cre; Pdgfrafl/fl mutant facial prominences (red bars). (C) Protein-protein interaction (PPI) network constructed from PDGF regulated SRF target genes (red) and FGF regulated SRF target genes (purple) recapitulates unique SRF functions downstream of PDGF signaling. Furthermore, the PDGF specific SRF network contains an enrichment of MRTF target genes (squares) compared to the shared network, which has equal TCF (triangle), MRTF (square), and non-specific (circles) genes. All six genes with altered expression in SRF mutants (bold) fall under the PDGF specific network, and many PDGF-SRF target genes have known roles in craniofacial development (* asterisk; Source: www.informatics.jax.org, www.omim.org). Data plotted as mean ± SEM. *p<0.1, **p<0.05 compared to serum starved or wildtype control, ^^p<0.05 compared to Latrunculin B treatment (n=3). See also Fig. S6 and Table S4.
Our data suggest the following model: PDGF mediates MRTFA-SRF association and binding at select target gene promoters to drive MRTF dependent expression of key actomyosin cytoskeleton elements (such as Vcl, Actb, Acta2, and Myh9). In contrast, both PDGF and FGF signaling increase Elk1-SRF complex formation to modulate the classic IEG signature (including Fos, Fosb, and Junb) observed downstream of these pathways. By building a protein-protein interaction (PPI) network of the targets downstream of PDGF, FGF, and SRF (Chen et al. 2012), we can better visualize this specificity at both the RTK and cofactor levels (Fig. 7C). Consistent with our framework, the PDGF specific PPI network shows strong correlation with MRTF target genes. Our studies imply that loss of this PDGF-MRTF-SRF axis explains in part both the craniofacial phenotypes of Pdgfra and Srf conditional mutants as well as the observed phenotypic interaction between these two genes.
Discussion
SRF is a classic regulator of the transcriptional response to growth factor signaling. In the present study, we find that neural crest conditional loss of SRF results in facial clefting accompanied by proliferation and migration defects. By analyzing SRF activation downstream of both PDGF and FGF signaling, we uncover a PDGF-MRTF-SRF circuit critical for cytoskeletal gene expression in the midface. We conclude that SRF is required for craniofacial development, and RTK signaling encodes the specificity of SRF mediated gene expression at the level of cofactor recruitment in this developmental context.
Many phenotypes have been reported in SRF neural crest conditional mutants (Newbern et al. 2008, Wickramasinghe et al. 2008), and we now describe gross facial clefting in these mutants, which had not been previously appreciated. We further demonstrate SRF drives proliferation of the NCC derived MNP mesenchyme. MNP cell proliferation is also decreased in Wnt1-Cre; Pdgfrafl/fl mice (He and Soriano 2013), suggesting a common mechanism underlying both PDGF and SRF activity. Consistent with this notion, SRF mutant FPCs fail to proliferate in response to PDGF stimulation. Thus, PDGF signaling acts through SRF to drive a functional proliferation program in the midface. In the epidermis, loss of Srf leads to cell proliferation defects due to abnormalities in the actomyosin network (Luxenburg et al. 2011); many targets of the PDGF-MRTF-SRF axis elucidated in our study (such as Actb, Myh9, Flna, and Actg1) were also implicated in the proliferation defects observed in these epidermis-specific Srf mutants, reflecting the importance of these genes. We further show that Wnt1-Cre; Srffl/fl embryos exhibit lineage tracing defects characterized by decreased infiltration of NCCs into the FNP and first two branchial arches, raising the possibility that the facial clefting may be in part the result of an insufficient number of neural crest derived progenitors populating the craniofacial mesenchyme. To understand the basis for these deficits, we analyzed SRF deficient primary cells isolated from the cranial NCC derived facial prominences and demonstrated decreased speed and directional persistence in wound healing assays. Upon delamination from the dorsal neural tube, neural crest cells migrate in stereotypic streams throughout the embryo to populate a diverse range of derivatives, responding to guidance cues as they migrate collectively to their final destinations (Theveneau and Mayor 2012). Many PDGF regulated SRF target genes in our study are involved in force generation (Myh9, Myl6, Myl12b), focal adhesion formation (Vcl, Flna, Zyx) and cytoskeletal organization (Actb, Actg1, Acta2), key processes for cell migration. In addition, MRTFs have been shown to mediate cell motility and directionality in other contexts (Medjkane et al. 2009), suggesting the observed defects may be MRTF-dependent.
Upon activation, RTKs become phosphorylated and engage downstream effectors, which in turn activate many shared intracellular pathways. One outcome of this signaling cascade is the transcription of IEGs. Many RTKs induce overlapping sets of IEGs, leaving open the question of how transcriptional specificity is generated. We demonstrate that PDGF, but not FGF, selectively promotes recruitment of SRF and MRTFA to target gene promoters, leading to induction of a unique gene expression program. This PDGF-MRTF-SRF axis is enriched for cytoskeletal regulators, many of which exhibit MRTF dependent induction and decreased expression in Srf and Pdgfra mutants. On the other hand, both PDGF and FGF increase Elk1-SRF complex formation, in line with our observation that over half of the identified SRF target genes are jointly activated by both PDGF and FGF. The targets downstream of this shared RTK-Elk1-SRF axis, such as Fos, Jun, and Egr, comprise a classic set of IEGs activated by multiple pathways (Fambrough et al. 1999), suggesting other stimuli can compensate for loss of Elk1-SRF mediated transcription. Consistent with this notion, Srf null embryos express residual amounts of Fos and Egr1 but show complete loss of Acta1 expression (Arsenian et al. 1998). We propose that the PDGF-MRTF-SRF axis has unique roles in the developing midface, an assertion supported by our epistasis results and gene expression studies.
Two critical points merit further discussion. First, what are the key signaling parameters encoding differential cofactor activation and SRF-mediated gene expression downstream of PDGF and FGF signaling? A potential answer lies in the importance of PI3K activity in the formation of an MRTFA-SRF complex and in driving maximal SRF binding at target gene promoters in response to PDGF stimulation. Previous studies have shown PI3K to be the primary effector downstream of PDGF signaling during midface development, closely mirroring the craniofacial phenotypes observed in PDGFRα mutants (Klinghoffer et al. 2002; Fantauzzo and Soriano 2014). Thus, one explanation is that PDGF activated PI3K signaling selectively promotes MRTFA-SRF association, perhaps through Rho-family small GTPases, which modulate actin dynamics and MRTF activity in other contexts (Pipes et al. 2006). Both Rac1 and Cdc42 are required in the neural crest for craniofacial development (Thomas et al. 2010; Fuchs et al. 2009) and have been implicated downstream of PDGF signaling in MEPMs (He and Soriano 2013; Fantauzzo and Soriano 2014). Although FGF signaling can modulate both Rac1 and Cdc42 in other systems (Fera et al. 2004; Clark et al. 2011), it is unclear whether this relationship is conserved in the midface. Alternatively, while both PDGF and FGF signal through common kinase cascades, the magnitude and duration of this induction can be quite different. Indeed, PDGF and FGF mediated pERK activation patterns are markedly different in MEPMs, with PDGF stimulation resulting in a transient pERK pulse but FGF treatment driving sustained pERK activation (unpublished data). Consistent with this observation, a recent study showed that sporadic pERK pulses drive SRF mediated transcription more efficiently than sustained pERK activity (Aoki et al. 2013), reflecting yet another layer of control. In sum, a combination of both qualitative and quantitative differences in signaling parameters likely accounts for the observed SRF specificity.
Second, is this specificity of SRF activation ‘hard-wired’ into the PDGF and FGF signaling networks or is it context dependent? The answer is almost certainly the latter, as the magnitude and kinetics of activation downstream of even the same RTK can vary depending on many parameters, including expression level (Traverse et al. 1994). The expression pattern of many signaling components is restricted over the course of development, necessitating diverse, context-specific control systems. The neural crest itself is a multipotent population with many sublineages, all expressing different combinations and amounts of receptors and signaling effectors. In the case of PDGFRα and SRF, we describe a PDGF-SRF circuit in the midface. However, many of the observed hemorrhaging and blistering phenotypes in these mutants may be due in part to requirements for PDGF signaling and SRF activity in neural crest derived vascular components, such as smooth muscle cells and pericytes (Etchevers et al. 2001). The PDGF-SRF signaling axis may be wired differently in these cells, particularly at the levels of receptor activation, effector requirements, and cofactor recruitment. In DRG sensory neurons, SRF activity downstream of nerve growth factor (NGF) is dependent on ERK mediated MRTFA activity, but not on TCF-SRF complex formation (Wickramasinghe et al. 2008). Thus, SRF function can be controlled by different RTKs in divergent neural crest lineages through a common set of cofactors and signaling effectors. The degree of sophistication and stimulus-dependent utilization of SRF regulatory mechanisms is remarkable, and determining the exact rules governing SRF activity following receptor activation across these diverse developmental contexts will be rewarding. Our studies provide one such example of the intricate control systems in place to encode transcriptional specificity downstream of two different RTKs at the level of SRF cofactor recruitment.
Experimental Procedures
Mouse strains
All animal experiments were approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai. Srftm1Rmn mice (Ramanan and Ginty 2005) referred to as Srffl/fl in the text, were a gift from Dr. Xin Sun, University of Wisconsin-Madison, and were backcrossed a minimum of three generations to 129S4 mice prior to all experiments included in this study. Tg(Wnt1-Cre)2Sor mice (Lewis et al. 2013), referred to as Wnt1-Cre2 in the text, were backcrossed to 129S4 mice for four generations prior to all experiments in this study. Pdgfratm11(EGFP)Sor (Hamilton et al. 2003), referred to as PdgfraGFP/+ in the text, and Gt(ROSA)26Sortm1Sor mice (Soriano 1999), referred to as R26R in the text, were maintained on a C57BL/6 background. PDGFRαtm8Sor mice (Tallquist and Soriano 2003), referred to as Pdgfrafl/fl, in the text, FGFR1tm5.1Sor mice (Hoch and Soriano 2006), referred to as Fgfr1fl/fl in the text, and Tg(Wnt1-Cre)11Rth mice (Danielian et al. 1998), referred to as Wnt1-Cre in the text, were all maintained on a 129S4 genetic background.
Tissue culture and proliferation/scratch assays
Primary mouse embryonic palatal mesenchyme (MEPM) cells were isolated from E13.5 secondary palatal shelves (day of plug: E0.5) as previously described (Fantauzzo and Soriano 2014). Primary mouse facial prominence cells (FPCs) were generated from E11.5 mouse facial prominences (Fig. S3B), but we were unable to passage these FPCs in culture. Therefore, MEPMs were used for further experiments investigating RTK mediated control of SRF function, such as Western blots and ChIP. For proliferation assays, cells were starved overnight and then incubated with 10 μm BrdU for 4 hours (Bush and Soriano 2010) with either 30 ng/mL PDGFAA or 50 ng/mL FGF1 + 1 μg/mL heparin. For wound healing assays, cells were grown to confluence, starved overnight, and then scratched. Details are available in Supplemental Procedures.
Histology, In situ hybridization, and Immunofluorescence
Embryos were dissected and embedded in either paraffin or optimal cutting temperature (OCT) compound for sectioning. In situ hybridization and immunofluorescence were performed according to standard protocols. See Supplemental Procedures for further details.
qPCR
For analysis of SRF induction, E13.5 MEPMs were starved overnight and then treated with PDGF or FGF for the desired time duration. For analysis of SRF target gene expression in mutant embryos, E11.5 facial prominence lysates were harvested and RNA was extracted directly from tissue. All experiments were conducted on litters from three independent biological replicates. Statistical analysis was performed using a two-tailed, paired Student’s t-test for MEPM timecourses, in which cells from the same embryo were considered paired. For comparison of expression between facial prominence lysates from different genotypes, a two-tailed, unpaired Student’s t test was used. Further details are available in Supplemental Procedures.
Time lapse imaging
FPCs were prepared and scratch assays performed as described above. Live cell imaging was carried out on an Olympus IX-70 widefield epi-fluorescence system with a stage-top incubation chamber to maintain cell viability. Images were taken with a 10x lens every 5 minutes across 250 minutes, for a total of 50 images per field of view. Four fields of view per condition per embryo were imaged, and two independently dissected control and mutant embryos were analyzed. Ten cells were randomly selected in each field of view for tracking and calculation of migration parameters, and thus, a total of 40 cells per condition per embryo were analyzed. Image analysis was performed in ImageJ (v 1.47; NIH, Bethesda, MD, USA) using the Manual Tracking plugin. Calculation of trajectories, speed and persistence were implemented through custom code in R (www.R-project.org/).
IP and Western blot
E13.5 MEPMs were serum starved for 24 hours in 0.1% FBS and stimulated with PDGF or FGF for desired duration. When applicable, cells were pretreated for 1 hr with 10 μM of desired inhibitor. IPs and Western blots were performed according to standard protocols using HRP-conjugated secondary antibodies and quantified in ImageJ (v 1.47; NIH, Bethesda, MD, USA). A minimum of two biological replicates were performed for each set of IP experiments. Further details are provided in Supplemental Procedures.
ChIP
E13.5 MEPMs were isolated and stimulated with PDGF or FGF as described above. ChIP was performed as previously described (Fantauzzo and Soriano 2014) to test occupancy in input, IgG, and antibody (anti-SRF or anti-MRTFA) precipitated samples. qPCR was carried out as described above, and statistics were performed using a two-tailed, paired Student’s t-test, in which cells from the same embryo were considered paired. Data presented are from three independent biological replicates. Further details are provided in Supplemental Procedures.
ChIP-seq and RNA Seq data integration
C2C12 SRF ChIP-seq peak data was downloaded directly from the ENCODE consortium (www.encodeproject.org; GEO GSM915168) in the ‘narrowPeaks’ format, which lists significant peaks identified by the ENCODE consortium. 3T3 SRF ChIP-seq data was similarly obtained (Esnault et al. 2014). Chromosomal coordinates for each of these peaks were then compared to the transcriptional start site (TSS) for each gene regulated by PDGF or FGF signaling at 1 hour, and a frequency distribution was generated by counting the number of peaks within successive 10kb bins of the TSS. Strategy was implemented through custom code in R. Similar distributions for control gene sets (randomly selected, expression matched genes that are not regulated by RTK activation) were generated to assess baseline ChIP-seq peak enrichment. A similar approach was implemented for the following control ChIP-seq datasets:
Jun (GEO: GSM9212901)
Pax5 (GEO: GSM923584)
E13.5 palate p300 (www.facebase.org; Accession FB00000263)
Supplementary Material
Supplemental Figure 1, related to Figure 1. SRF phosphorylation and PDGFRα and SRF coexpression in the developing mouse face. (A) PDGFAA treatment results in SRF phosphorylation, as CIP treatment of SRF immunoprecipitated lysates followed by anti-SRF Western blot results in loss of the upper band in the SRF doublet. (B) SRF protein is broadly expressed throughout the E11.5 neural crest derived craniofacial mesenchyme, including the MNP, LNP, and MxP. At high magnification, strong SRF (red) co-expression was noted with PDGFRα (green) in the (B′) MNP as well as the (B″) MxP (which eventually gives rise to the secondary palate), as shown in sections from PDGFRαGFP/+ embryos. Weaker co-expression is also visible in the (B‴) LNP. (C) Pdgfra and (C′) Srf mRNA are both expressed in the anterior E13.5 palate along with (D, D′) SRF protein (green). Note PDGFRα expression is strictly mesenchymal but SRF expression extends to both the mesenchyme and epithelium.
MNP = medial nasal process, LNP = lateral nasal process. MxP = maxillary process
IF scale bar (B, D): 100 μm
ISH scale bar (C): 200 μm
Supplemental Figure 2, related to Figure 1 and 2. FGF signaling activates Srf, but Fgfr1 conditional mutants do not interact genetically with Srf conditional mutants. (A) Timecourse of Srf mRNA expression following FGF1 treatment reveals a peak of Srf expression at 60 minutes (n=3, plotted as mean ± SEM, * p < 0.05). (B) FGF mediated SRF induction requires Erk signaling but not PI3K or MRTF activity. PD = PD325901, LY = LY294002, LB = Latrunculin B, CD = cytochalasin D. (C) Wnt1-Cre; Srf+/fl; Fgfr1+/fl double heterozygotes are overtly normal. Cells treated with 50 ng/mL FGF1 + 1 μg/mL heparin for desired duration.
Supplemental Figure 3, related to Figure 4 Wnt1-Cre2; Srffl/fl mice exhibit overt facial clefting similar to Wnt1-Cre; Srffl/fl mice. (A) Wnt1-Cre2; Srffl/fl mutant embryos show overt facial clefting at E11.5. Both Wnt1-Cre and Wnt1-Cre2 mediated deletion of Srf results in fully penetrant facial clefting phenotypes and embryonic lethality by E13.5. No Wnt1-Cre Srffl/fl or Wnt1-Cre2; Srffl/fl mutants are recovered postnatally. (B) FPCs were generated from E11.5 embryos by dissecting the indicating regions (red), comprising the medial nasal process (MNP), lateral nasal process (LNP), and maxillary process (MxP). Wnt1-Cre2; Srffl/fl FPCs express very low Srf mRNA levels but show robust expression of both Pdgfra and Fgfr1 compared to control embryos. (n=3, plotted as mean ± SEM) (C) Immunofluorescence staining shows reduced levels of SRF protein in Srf mutant FPCs. (D) Static images taken at 2 hours and 24 hours post-wounding in both control Wnt1-Cre2; Srf+/fl and mutant Wnt1-Cre2; Srffl/fl FPCs show increased closure with 10% FBS, 0.1% FBS, and PDGF in control compared to mutant cells. (E) Trajectories for 10 representative cells in both control Wnt1-Cre2; Srf+/fl and mutant Wnt1-Cre2; Srffl/fl FPCs shows decreased motility in mutant FPCs. Interestingly, a minority of SRF deficient cells (red arrowhead) exhibit normal trajectories.
Supplemental Figure 4, related to Figure 5. SRF is not alternatively spliced and does not undergo nuclear translocation following PDGF stimulation. (A) Schematic representation of the SRF locus and protein domains depicts the two major isoforms identified in our RNA-seq data: full length SRF (SRFfull) and a dominant negative form of SRF (SRFΔex5) which lacks the transactivation domain. Primer locations used to amplify the two isoforms in (C) are depicted as black arrows. NLS = nuclear localization sequence. (B) RNA-seq data indicates SRFfull is the predominant transcript in MEPMs independently of PDGF stimulation. (C) Reverse transcriptase PCR (rt-PCR) confirms expression of both SRF isoforms in MEPMs and the E13.5 palate. L = ladder. P2 = P2 E13.5 MEPMs (D) SRF is predominantly nuclear in E13.5 MEPMs regardless of PDGF stimulation. (E) Increased Elk1-SRF complex formation is observed following 30 minutes of PDGF or 30 minutes FGF treatment. Elk1 levels are not affected by PDGF or FGF treatment. (B) Elk1-SRF association requires both PI3K and ERK activation but is unaffected by MRTF activation. All biochemistry performed in E13.5 MEPMs. CD = cytochalasin D, PD = PD325901, LY = LY294002. U = untreated cells, P = 30 minutes 30 ng/mL PDGFAA, F = 30 minutes 50 ng/mL FGF1 + 1 μg/mL heparin.
Supplemental Figure 5, related to Figure 6. Correlation of PDGF and FGF target genes with published ChIP-Seq datasets. (A–C) ChIP-seq datasets for (A) Jun (another IEG induced in MEPMs), (B) Pax5 (not expressed in MEPMs), or (C) p300 (E13.5 palate transcriptional coactivator dataset) show no enrichment for target genes, in contrast to SRF ChIP-Seq peaks. (D TCF scores for PDGF, FGF, and shared target genes are similar. (E) PDGF target genes show greater MRTF scores while genes with high TCF scores tend to be shared targets of PDGF and FGF signaling (red = PDGF, purple = shared, blue = FGF). Many FGF genes show low TCF and MRTF scores, possibly suggesting cofactor independent SRF functions. MRTF and TCF ChIP-seq data (Esnault et al. 2014) was generated in 3T3 cells. (F) Table of p-values comparing MRTF and TCF scores for PDGF and FGF target genes. While MRTF scores are significantly increased in PDGF target genes compared to FGF target genes, TCF scores are distributed in an essentially identical manner.
Supplemental Figure 6, related to Figure 7. PDGF-SRF cytoskeletal target genes are important for craniofacial development and bound by SRF in E13.5 MEPMs. (A) PDGF specific SRF target genes have diverse roles in cytoskeletal organization and cell migration as well as high correlation with MRTF dependent transcription. Acta1, Acta2, and Actb are actin subunits, and mutations in ActB have been connected to craniofacial syndromes in patients (Riviere et al. 2012). Myh9 is part of the actomyosin contractile apparatus that has been linked to non-syndromic cleft lip and palate (NSCL/P) (Chiquet et al. 2009). Both Flna and Vcl are key components of the focal adhesion complex and both genes, as well as Ctgf, cause craniofacial phenotypes in mice (Robertson et al. 2003, West et al. 2006, Xu et al. 1998, Ivkovic et al. 2003). Finally, Tagln is a classic SRF target gene conserved across many different contexts (Li et al. 1997). (B) Endogenous SRF ChIP PCR shows binding at six out of eight selected target gene promoters in E13.5 MEPMs, reflecting the relevance of these genes in the midface. (C) SRF ChIP in E13.5 MEPMs reveals strong SRF binding at the Fos promoter in response to both PDGF and FGF signaling. The Arid5b promoter (an IEG induced by RTK signaling without any consensus SRF binding sequences or ChIP-seq binding events) shows low occupancy across all conditions, serving as a negative control. (D) Fos induction in MEPMs is unaffected by Latrunculin B treatment. ** p<0.05. Data plotted as mean ± SEM.
Supplemental Table 1, related to Figure 2. Summary of Srf neural crest conditional craniofacial phenotypes and interactions with Pdgfra mutants.
Wnt1-Cre; Srffl/fl mutants exhibit fully penetrant clefting and are not recovered postnatally. Wnt1-Cre; Srf+/fl; Pdgfra+/fl mutants have a partially penetrant clefting phenotype and are recovered below Mendelian ratios postnatally. Wnt1-Cre; Srffl/fl; Pdgfra+/fl compound mutants show even more severe phenotypes and are recovered below Mendelian ratios at all examined stages (E11.5 onwards). Further, we recover live Wnt1-Cre; Srffl/fl mutant embryos below Mendelian ratios at E13.5, consistent with one previous report (Wickramasinghe et al. 2008), while another study recovers some mutants as late as E16.5 (Newbern et al. 2008). These differences may be due in part to genetic background, as our mice were backcrossed 3 generations to 129S4 prior to phenotype analysis.
Supplemental Table 2, related to Figure 1. Cufflinks FPKM expression of 1 hr PDGF treated, 1 hr FGF treated, and serum starved E13.5 MEPMs (attached Excel spreadsheet). FPKM values and confidence intervals (generated by the Cufflinks pipeline) for all genes from the serum starved, 1 hr PDGF treated, and 1 hr FGF treated RNA-seq datasets.
Supplemental Table 3, related to Figure 6. Integration of SRF ChIP-seq with PDGF and FGF RNA-seq datasets (attached Excel spreadsheet).
Four sets of data are included: (1) list of significantly upregulated genes following 1 hr PDGF or 1 hr FGF treatment. (2) integration with ENCODE C2C12 SRF ChIP-seq, (3) integration with 3T3 SRF ChIP-seq, and (4) summary of RTK-SRF target genes. For each mapped SRF ChIP-seq peak, the first two columns reflect the following: (1) Gene name, (2) Distance of peak to TSS for the C2C12 SRF ChIP-seq dataset (Columns F and G) (ENCODE 2012). The next two columns (Column J and K) provide the same information for FGF target genes. The SRF ChIP-seq data from 3T3 cells (Esnault et al. 2014) for PDGF (columns N and O) and FGF (columns R and S) are similarly organized. Finally, Columns U-AF summarize the RTK responsive SRF target genes identified in each ChIP-seq dataset as well as the final merged list used in the main text. Note that there are fewer target gene names than ChIP-seq peaks because multiple SRF ChIP-seq peaks can be located near the same target gene TSS (genes with multiple peaks are highlighted in pink).
Supplemental Table 4, related to Figure 6 and 7. Integration of MRTF and TCF ChIP-seq scores with PDGF and FGF RNA-seq datasets (attached Excel spreadsheet). All SRF ChIP-seq peaks found in both the ENCODE and 3T3 cell datasets are provided, along with the MRTF and TCF scores for these peaks (Esnault et al. 2014). Mean scores and statistical testing are also displayed for each pair of gene sets.
Acknowledgments
We are very grateful to Fenglei He for performing the initial RNA-seq experiment. We thank Jason Newbern at the University of North Carolina for discussion on SRF mutant phenotypes. Srffl/fl mice were a gift from Dr. Xin Sun at the University of Wisconsin-Madison. Excellent genotyping and technical support were provided by Tony Chen, Aryel Heller, and Anne Levine. Live imaging was performed in the Microscopy Shared Resource Facility at the Icahn School of Medicine at Mount Sinai, and we thank Rumana Huq and Lauren O’Rourke for assistance. RNA-seq experiments were performed at the Mount Sinai Genomics Core, and we thank Omar Jabado, Yumi Kasai, Milind Mahajan, and Avi Ma’ayan for advice and discussions. We are grateful to the Developmental Studies Hybridoma Bank for reagents. ENCODE ChIP-seq datasets were downloaded for SRF (Wold lab), Jun (Snyder lab) and Pax5 (Hardison lab). FaceBase datasets were downloaded for E13.5 palate p300 ChIP-Seq and E13.5 palate RNA-seq (Visel lab). We thank Robert Krauss, Susan Parkhurst, and the members of the Soriano laboratory for helpful discussion and critical comments on this manuscript. This work was supported by NIH/NIDCR grants R01DE022363 and R01DE022778 to P.S. and NIH/NIDCR Ruth L. Kirschstein NRSA Individual Predoctoral Fellowship F31DE023456 to H.N.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MS, Gammill LS, Bronner-Fraser M. Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dynamics. 2008;237:1021–33. doi: 10.1002/dvdy.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Krause SM, Kretz O, Philippar U, Lemberger T, Casanova E, Wiebel FF, Schwarz H, Frotscher M, Schutz G, Nordheim A. Neuronal migration in the murine rostral migratory stream requires serum response factor. PNAS. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Kumagai Y, Sakurai A, Komatsu N, Fujita Y, Shionyu C, Matsuda M. Stochastic ERK Activation Induced by Noise and Cell-to-Cell Propagation Regulates Cell Density-Dependent Proliferation. Mol Cell. 2013;52:529–540. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Arsenian S, Weinhold B, Oelgeschläger M, Rüther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO Journal. 1998;17:6289–99. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Bio. 2011;12:104–17. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Belaguli NS, Zhou W, Trinh TH, Majesky MW, Schwartz RJ. Dominant negative murine serum response factor: alternative splicing within the activation domain inhibits transactivation of serum response factor binding targets. Mol Cell Bio. 1999;19:4582–91. doi: 10.1128/mcb.19.7.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentires-Alj M, Kontaridis MI, Neel BG. Stops along the RAS pathway in human genetic disease. Nature Medicine. 2006;12:283–285. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- Bush JO, Soriano P. Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes & Dev. 2010;24:2068–80. doi: 10.1101/gad.1963210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcia A, Gai G, Di Gregorio E, Talarico F, Naretto VG, Migone N, Pepe E, Grosso E, Brusco A. Bilaterally cleft lip and bilateral thumb polydactyly with triphalangeal component in a patient with two de novo deletions of HSA 4q32 and 4q34 involving PDGFC, GRIA2, and FBXO8 genes. American Journal of Med Gen. 2013;161:2656–62. doi: 10.1002/ajmg.a.36146. [DOI] [PubMed] [Google Scholar]
- Camoretti-Mercado B, Liu HW, Halayko J, Forsythe SM, Kyle JW, Li B, Fu Y, McConville J, Kogut P, Vieira JE, et al. Physiological control of smooth muscle-specific gene expression through regulated nuclear translocation of serum response factor. J Bio Chem. 2000;275:30387–93. doi: 10.1074/jbc.M000840200. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–9. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chen EY, Huilei X, Gordonov S, Lim MP, Perkins MH, Ma’ayan A. Expression 2 Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2012;28:105–11. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SJ, Marazita ML, Hart PS, Sulima PP, Field LL, McHenry TG, Govil M, Cooper ME, Letra A, Menezes R, et al. The PDGF-C regulatory region SNP rs28999109 decreases promoter transcriptional activity and is associated with CL/P. Eur Journal of Hum Gen. 2009;17:774–784. doi: 10.1038/ejhg.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IBN, Muha V, Klingseisen A, Leptin M, Müller HAJ. Fibroblast growth factor signalling controls successive cell behaviours during mesoderm layer formation in Drosophila. Development. 2011;138:2705–15. doi: 10.1242/dev.060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran BH, Zullo J, Verma IM, Stiles CD. Expression of a c-fos gene and of an fos-related gene is stimulated by platelet-derived growth factor. Science. 1984;226:1080–1082. doi: 10.1126/science.6093261. [DOI] [PubMed] [Google Scholar]
- Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. 1993;117:409–29. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biology. 1998;8:1323–6. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Ding H, Wu X, Boström H, Kim I, Wong N, Tsoi B, O’Rourke M, Koh GY, Soriano P, Betsholtz C, et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nat Genetics. 2004;36:1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes & Dev. 2014;28:943–58. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchevers HC, Vincent C, Le Douarin NM, Couly GF. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–68. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- Fambrough D, McClure K, Kazlauskas A, Lander ES. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–41. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- Fantauzzo KA, Soriano P. PI3K-mediated PDGFRα signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes & Dev. 2014;28:1005–1017. doi: 10.1101/gad.238709.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fera E, O’Neil C, Lee W, Li S, Pickering JG. Fibroblast growth factor-2 and remodeled type I collagen control membrane protrusion in human vascular smooth muscle cells: biphasic activation of Rac1. J Bio Chem. 2004;279:35573–82. doi: 10.1074/jbc.M400711200. [DOI] [PubMed] [Google Scholar]
- Franco CA, Mericskay M, Parlakian A, Gary-Bobo G, Gao-Li J, Paulin D, Li Z. Serum response factor is required for sprouting angiogenesis and vascular integrity. Dev Cell. 2008;15:448–61. doi: 10.1016/j.devcel.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Herzog D, Sumara G, Büchmann-Møller S, Civenni G, Wu X, Chrostek-Grashoff A, Suter U, et al. Stage-specific control of neural crest stem cell proliferation by the small rho GTPases Cdc42 and Rac1. Cell Stem Cell. 2009;4:236–47. doi: 10.1016/j.stem.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Gerber A, Esnault C, Aubert G, Treisman R, Pralong F, Schibler U. Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell. 2013;152:492–503. doi: 10.1016/j.cell.2012.12.027. [DOI] [PubMed] [Google Scholar]
- Gineitis D, Treisman R. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J Bio Chem. 2001;276:24531–9. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary Divergence of Platelet-Derived Growth Factor Alpha Receptor Signaling Mechanisms. Mol Cell Bio. 2003;23:4013–25. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Soriano P. A critical role for PDGFRα signaling in medial nasal process development. PLoS Genetics. 2013;9:e1003851. doi: 10.1371/journal.pgen.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497:507–11. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch RV, Soriano P. Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development. 2006;133:663–73. doi: 10.1242/dev.02242. [DOI] [PubMed] [Google Scholar]
- Iyer D, Chang D, Marx J, Wei L, Olson EN, Parmacek MS, Balasubramanyam A, Schwartz RJ. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. PNAS. 2006;103:4516–21. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer R, Hamilton TG, Hoch R, Soriano P. An allelic series at the PDGFαR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell. 2002;2:103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- Lau LF, Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells coordinate regulation with c-fos or c-myc. PNAS. 1987;84:1182–6. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, Bush JO. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Bio. 2013;379:229–34. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat Cell Bio. 2011;13:203–14. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nature Cell Bio. 2009;11:257–68. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou A, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Newbern J, Zhong J, Wickramasinghe SR, Li X, Wu Y, Samuels I, Cherosky N, Karlo JC, O’Loughlin B, Wikenheiser J, et al. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. PNAS. 2008;105:17115–17120. doi: 10.1073/pnas.0805239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Bio. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlakian A, Tuil D, Hamard G, Hentzen D, Concordet J, Paulin D, Li Z, Daegelen D. Targeted Inactivation of Serum Response Factor in the Developing Heart Results in Myocardial Defects and Embryonic Lethality. Mol Cell Bio. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes GCT, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes & Dev. 2006;20:1545–56. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends in Cell Biology. 2006;16:588–96. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- Rattanasopha S, Tongkobpetch S, Srichomthong C, Siriwan P, Suphapeetiporn K, Shotelersuk V. PDGFRa mutations in humans with isolated cleft palate. Eur J Hum Gen. 2012;20:1058–62. doi: 10.1038/ejhg.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera VM, Miranti CK, Misra RP, Ginty DD, Chen RH, Blenis J, Greenberg ME. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol Cell Bio. 1993;13:6260–6273. doi: 10.1128/mcb.13.10.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahl J, Raymond CS, Soriano P. PDGF signaling specificity is mediated through multiple immediate early genes. Nat Genetics. 2007;39:52–60. doi: 10.1038/ng1922. [DOI] [PubMed] [Google Scholar]
- Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J Cell Bio. 2002;156:737–50. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;2700:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stritt C, Stern S, Harting K, Manke T, Sinske D, Schwarz H, Vingron M, Nordheim A, et al. Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat Neuroscience. 2009;12:418–27. doi: 10.1038/nn.2280. [DOI] [PubMed] [Google Scholar]
- Sullivan AL, Benner C, Heinz S, Xie L, Miano JM, Glass CK, Huang W. Serum Response Factor Utilizes Distinct Mechanisms To Regulate Cytoskeletal Gene Expression in Macrophages Serum Response Factor Utilizes Distinct Promoter- and Enhancer-Based Mechanisms To Regulate Cytoskeletal Gene Expression. Mol Cell Bio. 2011;31:861–875. doi: 10.1128/MCB.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Boyd K, Xu W, Ma J, Jackson CW, Fu A, Shillingford JM, Robinson GW, Hennighausen L, Hitzler JK, et al. Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator of mammary gland function. Mol Cell Bio. 2006;26:5809–26. doi: 10.1128/MCB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130:507–518. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Dev Bio. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Thomas PS, Kim J, Nunez S, Glogauer M, Kaartinen V. Neural crest cell-specific deletion of Rac1 results in defective cell-matrix interactions and severe craniofacial and cardiovascular malformations. Dev Bio. 2010;340:613–25. doi: 10.1016/j.ydbio.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse S, Seedorf K, Paterson H, Marshall CJ, Cohen P, Ullrich A. EGF triggers neuronal differentiation of PC12 cells that overexpress the EGF receptor. Curr Biology. 1994;4:694–701. doi: 10.1016/s0960-9822(00)00154-8. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO Journal. 1987;6:2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opinion Cell Bio. 1996;8:205–15. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes & Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–52. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Wang C, Chang JYF, Yang C, Huang Y, Liu J, You P, Li X. Type 1 fibroblast growth factor receptor in cranial neural crest cell-derived mesenchyme is required for palatogenesis. J Bio Chem. 2013;288:22174–83. doi: 10.1074/jbc.M113.463620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang D, Hockemeyer D, Mcanally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe SR, Alvania RS, Ramanan N, Wood JN, Mandai K, Ginty D. Serum response factor mediates NGF-dependent target innervation by embryonic DRG sensory neurons. Neuron. 2008;58:532–45. doi: 10.1016/j.neuron.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1, related to Figure 1. SRF phosphorylation and PDGFRα and SRF coexpression in the developing mouse face. (A) PDGFAA treatment results in SRF phosphorylation, as CIP treatment of SRF immunoprecipitated lysates followed by anti-SRF Western blot results in loss of the upper band in the SRF doublet. (B) SRF protein is broadly expressed throughout the E11.5 neural crest derived craniofacial mesenchyme, including the MNP, LNP, and MxP. At high magnification, strong SRF (red) co-expression was noted with PDGFRα (green) in the (B′) MNP as well as the (B″) MxP (which eventually gives rise to the secondary palate), as shown in sections from PDGFRαGFP/+ embryos. Weaker co-expression is also visible in the (B‴) LNP. (C) Pdgfra and (C′) Srf mRNA are both expressed in the anterior E13.5 palate along with (D, D′) SRF protein (green). Note PDGFRα expression is strictly mesenchymal but SRF expression extends to both the mesenchyme and epithelium.
MNP = medial nasal process, LNP = lateral nasal process. MxP = maxillary process
IF scale bar (B, D): 100 μm
ISH scale bar (C): 200 μm
Supplemental Figure 2, related to Figure 1 and 2. FGF signaling activates Srf, but Fgfr1 conditional mutants do not interact genetically with Srf conditional mutants. (A) Timecourse of Srf mRNA expression following FGF1 treatment reveals a peak of Srf expression at 60 minutes (n=3, plotted as mean ± SEM, * p < 0.05). (B) FGF mediated SRF induction requires Erk signaling but not PI3K or MRTF activity. PD = PD325901, LY = LY294002, LB = Latrunculin B, CD = cytochalasin D. (C) Wnt1-Cre; Srf+/fl; Fgfr1+/fl double heterozygotes are overtly normal. Cells treated with 50 ng/mL FGF1 + 1 μg/mL heparin for desired duration.
Supplemental Figure 3, related to Figure 4 Wnt1-Cre2; Srffl/fl mice exhibit overt facial clefting similar to Wnt1-Cre; Srffl/fl mice. (A) Wnt1-Cre2; Srffl/fl mutant embryos show overt facial clefting at E11.5. Both Wnt1-Cre and Wnt1-Cre2 mediated deletion of Srf results in fully penetrant facial clefting phenotypes and embryonic lethality by E13.5. No Wnt1-Cre Srffl/fl or Wnt1-Cre2; Srffl/fl mutants are recovered postnatally. (B) FPCs were generated from E11.5 embryos by dissecting the indicating regions (red), comprising the medial nasal process (MNP), lateral nasal process (LNP), and maxillary process (MxP). Wnt1-Cre2; Srffl/fl FPCs express very low Srf mRNA levels but show robust expression of both Pdgfra and Fgfr1 compared to control embryos. (n=3, plotted as mean ± SEM) (C) Immunofluorescence staining shows reduced levels of SRF protein in Srf mutant FPCs. (D) Static images taken at 2 hours and 24 hours post-wounding in both control Wnt1-Cre2; Srf+/fl and mutant Wnt1-Cre2; Srffl/fl FPCs show increased closure with 10% FBS, 0.1% FBS, and PDGF in control compared to mutant cells. (E) Trajectories for 10 representative cells in both control Wnt1-Cre2; Srf+/fl and mutant Wnt1-Cre2; Srffl/fl FPCs shows decreased motility in mutant FPCs. Interestingly, a minority of SRF deficient cells (red arrowhead) exhibit normal trajectories.
Supplemental Figure 4, related to Figure 5. SRF is not alternatively spliced and does not undergo nuclear translocation following PDGF stimulation. (A) Schematic representation of the SRF locus and protein domains depicts the two major isoforms identified in our RNA-seq data: full length SRF (SRFfull) and a dominant negative form of SRF (SRFΔex5) which lacks the transactivation domain. Primer locations used to amplify the two isoforms in (C) are depicted as black arrows. NLS = nuclear localization sequence. (B) RNA-seq data indicates SRFfull is the predominant transcript in MEPMs independently of PDGF stimulation. (C) Reverse transcriptase PCR (rt-PCR) confirms expression of both SRF isoforms in MEPMs and the E13.5 palate. L = ladder. P2 = P2 E13.5 MEPMs (D) SRF is predominantly nuclear in E13.5 MEPMs regardless of PDGF stimulation. (E) Increased Elk1-SRF complex formation is observed following 30 minutes of PDGF or 30 minutes FGF treatment. Elk1 levels are not affected by PDGF or FGF treatment. (B) Elk1-SRF association requires both PI3K and ERK activation but is unaffected by MRTF activation. All biochemistry performed in E13.5 MEPMs. CD = cytochalasin D, PD = PD325901, LY = LY294002. U = untreated cells, P = 30 minutes 30 ng/mL PDGFAA, F = 30 minutes 50 ng/mL FGF1 + 1 μg/mL heparin.
Supplemental Figure 5, related to Figure 6. Correlation of PDGF and FGF target genes with published ChIP-Seq datasets. (A–C) ChIP-seq datasets for (A) Jun (another IEG induced in MEPMs), (B) Pax5 (not expressed in MEPMs), or (C) p300 (E13.5 palate transcriptional coactivator dataset) show no enrichment for target genes, in contrast to SRF ChIP-Seq peaks. (D TCF scores for PDGF, FGF, and shared target genes are similar. (E) PDGF target genes show greater MRTF scores while genes with high TCF scores tend to be shared targets of PDGF and FGF signaling (red = PDGF, purple = shared, blue = FGF). Many FGF genes show low TCF and MRTF scores, possibly suggesting cofactor independent SRF functions. MRTF and TCF ChIP-seq data (Esnault et al. 2014) was generated in 3T3 cells. (F) Table of p-values comparing MRTF and TCF scores for PDGF and FGF target genes. While MRTF scores are significantly increased in PDGF target genes compared to FGF target genes, TCF scores are distributed in an essentially identical manner.
Supplemental Figure 6, related to Figure 7. PDGF-SRF cytoskeletal target genes are important for craniofacial development and bound by SRF in E13.5 MEPMs. (A) PDGF specific SRF target genes have diverse roles in cytoskeletal organization and cell migration as well as high correlation with MRTF dependent transcription. Acta1, Acta2, and Actb are actin subunits, and mutations in ActB have been connected to craniofacial syndromes in patients (Riviere et al. 2012). Myh9 is part of the actomyosin contractile apparatus that has been linked to non-syndromic cleft lip and palate (NSCL/P) (Chiquet et al. 2009). Both Flna and Vcl are key components of the focal adhesion complex and both genes, as well as Ctgf, cause craniofacial phenotypes in mice (Robertson et al. 2003, West et al. 2006, Xu et al. 1998, Ivkovic et al. 2003). Finally, Tagln is a classic SRF target gene conserved across many different contexts (Li et al. 1997). (B) Endogenous SRF ChIP PCR shows binding at six out of eight selected target gene promoters in E13.5 MEPMs, reflecting the relevance of these genes in the midface. (C) SRF ChIP in E13.5 MEPMs reveals strong SRF binding at the Fos promoter in response to both PDGF and FGF signaling. The Arid5b promoter (an IEG induced by RTK signaling without any consensus SRF binding sequences or ChIP-seq binding events) shows low occupancy across all conditions, serving as a negative control. (D) Fos induction in MEPMs is unaffected by Latrunculin B treatment. ** p<0.05. Data plotted as mean ± SEM.
Supplemental Table 1, related to Figure 2. Summary of Srf neural crest conditional craniofacial phenotypes and interactions with Pdgfra mutants.
Wnt1-Cre; Srffl/fl mutants exhibit fully penetrant clefting and are not recovered postnatally. Wnt1-Cre; Srf+/fl; Pdgfra+/fl mutants have a partially penetrant clefting phenotype and are recovered below Mendelian ratios postnatally. Wnt1-Cre; Srffl/fl; Pdgfra+/fl compound mutants show even more severe phenotypes and are recovered below Mendelian ratios at all examined stages (E11.5 onwards). Further, we recover live Wnt1-Cre; Srffl/fl mutant embryos below Mendelian ratios at E13.5, consistent with one previous report (Wickramasinghe et al. 2008), while another study recovers some mutants as late as E16.5 (Newbern et al. 2008). These differences may be due in part to genetic background, as our mice were backcrossed 3 generations to 129S4 prior to phenotype analysis.
Supplemental Table 2, related to Figure 1. Cufflinks FPKM expression of 1 hr PDGF treated, 1 hr FGF treated, and serum starved E13.5 MEPMs (attached Excel spreadsheet). FPKM values and confidence intervals (generated by the Cufflinks pipeline) for all genes from the serum starved, 1 hr PDGF treated, and 1 hr FGF treated RNA-seq datasets.
Supplemental Table 3, related to Figure 6. Integration of SRF ChIP-seq with PDGF and FGF RNA-seq datasets (attached Excel spreadsheet).
Four sets of data are included: (1) list of significantly upregulated genes following 1 hr PDGF or 1 hr FGF treatment. (2) integration with ENCODE C2C12 SRF ChIP-seq, (3) integration with 3T3 SRF ChIP-seq, and (4) summary of RTK-SRF target genes. For each mapped SRF ChIP-seq peak, the first two columns reflect the following: (1) Gene name, (2) Distance of peak to TSS for the C2C12 SRF ChIP-seq dataset (Columns F and G) (ENCODE 2012). The next two columns (Column J and K) provide the same information for FGF target genes. The SRF ChIP-seq data from 3T3 cells (Esnault et al. 2014) for PDGF (columns N and O) and FGF (columns R and S) are similarly organized. Finally, Columns U-AF summarize the RTK responsive SRF target genes identified in each ChIP-seq dataset as well as the final merged list used in the main text. Note that there are fewer target gene names than ChIP-seq peaks because multiple SRF ChIP-seq peaks can be located near the same target gene TSS (genes with multiple peaks are highlighted in pink).
Supplemental Table 4, related to Figure 6 and 7. Integration of MRTF and TCF ChIP-seq scores with PDGF and FGF RNA-seq datasets (attached Excel spreadsheet). All SRF ChIP-seq peaks found in both the ENCODE and 3T3 cell datasets are provided, along with the MRTF and TCF scores for these peaks (Esnault et al. 2014). Mean scores and statistical testing are also displayed for each pair of gene sets.