Abstract
Aquaculture production has nearly tripled in the last two decades, bringing with it a significant increase in the use of antibiotics. Using liquid chromatography/tandem mass spectrometry (LC-MS/MS), the presence of 47 antibiotics was investigated in U.S. purchased shrimp, salmon, catfish, trout, tilapia, and swai originating from 11 different countries. All samples (n= 27) complied with U.S. FDA regulations and five antibiotics were detected above the limits of detection: oxytetracycline (in wild shrimp, 7.7 ng/g of fresh weight; farmed tilapia, 2.7; farmed salmon, 8.6; farmed trout with spinal deformities, 3.9), 4-epioxytetracycline (farmed salmon, 4.1), sulfadimethoxine (farmed shrimp, 0.3), ormetoprim (farmed salmon, 0.5), and virginiamycin (farmed salmon marketed as antibiotic-free, 5.2). A literature review showed that sub-regulatory levels of antibiotics, as found here, can promote resistance development and publications linking aquaculture to this have increased more than 8-fold from 1991–2013. Although this study was limited in size and employed sample pooling, it represents the largest reconnaissance of antibiotics in U.S. seafood to date, providing data on previously unmonitored antibiotics and on farmed trout with spinal deformities. Results indicate low levels of antibiotic residues and general compliance with U.S. regulations. The potential for development of microbial drug resistance was identified as a key concern and research priority.
Keywords: Aquaculture, Oxytetracycline, Antibiotics, Antibiotic Resistance, Seafood
1. Introduction
It is estimated that within the next few years, aquaculture will account for almost 40% of total global seafood production by weight, up from 4% in 1970 [1, 2]. This increase to a projected worldwide production of 83 million metric tons in 2013 has been due to a heightened demand for seafood, improved aquaculture techniques, emergence as a key cash crop in certain regions of the world, and recognition as a cheaper way to obtain high-quality protein [2, 3]. However, as production surges, many aquaculture facilities resort to antibiotics to combat diseases in an environment that creates ample opportunities for bacterial pathogens to thrive [4]. Antibiotics are also commonly used as a prophylactic, sometimes on a daily basis [5]. Although some promising alternatives such as short-chain fatty acids and bacteriophage therapy have been proposed, many are not ready for mass usage [5]. Developed vaccines show promise in reducing antibiotic usage [4], but are only available to treat certain diseases and are not as cost-effective as antibiotics. Thus, the usage of antibiotics in aquaculture remains high.
Consequences associated with the use of antibiotics in aquaculture include the spread of antibiotics into the environment [6, 7], residual concentrations left in seafood, high exposure by aquaculture facility personnel, and antibiotic resistance development [3, 4]. Another issue is the impact of antibiotics on the animals themselves, such as potential changes in genetic expression [8, 9] and physiological anomalies. These physiological anomalies include malformation of the spine reported in fish exposed to oxytetracycline [9, 10].
Many of the antibiotics used in aquaculture are also used in human medicine [11]. Amoxicillin and ampicillin are commonly prescribed for treating bacterial infections such as pneumonia and gastroenteritis [12]. As fish are a potential source of bacterial pathogens for humans, it is important to monitor the spread of antibiotic resistance amongst seafood [13]. Resistance to the most commonly applied antibiotics has been found in previous studies [3, 14, 15, 16], including several that are multi-drug resistant (MDR) to many classes of antibiotics important in treating human infections [16, 17, 18, 19]. Thus, detecting and monitoring antibiotic residues in seafood is critically important to reduce potential environmental and human health risks.
A large portion of aquaculture takes place in countries with few regulations and limited enforcement [20], creating the need to monitor imported seafood strictly for antibiotic residues and presence of pathogens. In this study, twenty-seven seafood samples were collected by the National Oceanic and Atmospheric Administration (NOAA) from stores in Arizona and California for analysis. Samples included five of the top ten most consumed seafood varieties in the U.S.: shrimp, tilapia, catfish, swai, and Atlantic salmon. Trout with visible deformed spines were also analyzed. Using liquid chromatography tandem mass spectrometry (LC-MS/MS), 47 antibiotics identified from literature as drugs of concern were analyzed for using two methods. We also conducted a meta-analysis of published data on antibiotics and resistance development to note trends in aquaculture over the last few decades.
2. Materials and Methods
2.1 Samples and Preparation
A collaborating NOAA consumer safety officer obtained samples (n= 27) from retail grocery stores in Arizona and California (in southwest U.S.) over a period of three months from June to August in 2012 (Table 1). Samples originated from 11 different countries. Each sample was sold as a pre-packed unit or bought from store counter displays, meaning that each sample sometimes included multiple fish. Negative controls consisted of catfish donated from Louisiana State University that were never exposed to antibiotics. Normal and deformed rainbow trout (n=3 for each) were obtained to survey the potential link between antibiotic exposure and spinal deformities. Atlantic salmon marketed as “antibiotic-free” was also obtained from a local health food store.
Table 1.
Aquaculture information and demographics on samples used in this study.
| GENERAL INFORMATION FOR THE U.S. | THIS STUDY | ||||||
|---|---|---|---|---|---|---|---|
| Seafood Type | 2011 Ranka | 2012 Imports & Valueb | 2011 Production & Valuec | Composite Sample #d | Origin # of Samplese | Fillet (F) or Whole (W) | Packagedf |
| Shrimp | 1 | 531,840 $4,440M |
148,000 $6M |
1. Farmed Shrimp | Ind-2; Tha-1; Ban-1; Vie-1 | W | Y |
| 2. Wild-caught Shrimp | Mex-1 | W | N | ||||
| Tilapia | 5 | 227,440 $970M |
10,000 $54M |
3. Farmed Tilapia | Pan-1; Chi-2 | F | Y |
| Catfish | 7 | 107,690 $370M |
163,000 $395M |
4. Farmed Catfish | U.S.-2 | W | N |
| 5. AB-Free Farmed Catfishg | U.S. LSU-3 | W | N | ||||
| Trout | N/A | 9,310 $70M |
15,300 $53M |
6. Farmed Trout w/ D Spine | U.S.-3 | W | N |
| 7. Farmed Trout w/ Normal Spine | U.S.-3 | W | N | ||||
| Salmon | 3 | 120,640 $720 |
373,000 $720 |
8. Farmed International Atlantic Salmon | Can-2 Chl-1 | F | Y |
| 9. Farmed AB-Free Atlantic Salmonh | Sco-1 | ||||||
| 10. Farmed US Atlantic Salmon | U.S.-1 | ||||||
| Swai | 6 | N/Ai | N/Ai | 11. Farmed Swai | Vie-2 | F | Y |
Rank in most consumed seafood. Data from National Fisheries Institute [46];
Units: metric tons and millions of U.S. dollars. Fresh and frozen seafood imported for human consumption. Data from National Oceanic and Atmospheric Administration (NOAA) for the 50 U.S. states, District of Columbia, Puerto Rico, and the U.S. Virgin Islands [47]. Numbers have been rounded;
Units: metric tons and millions U.S. dollars. Commercial U.S. landings and aquaculture. Data from NOAA [47]. Numbers have been rounded. 2012 US aquaculture data were unavailable, thus limiting reported values to 2011 data;
11 total composites were made;
Ind= Indonesia, Tha= Thailand, Ban= Bangladesh, Vie= Vietnam, Mex= Mexico, Pan= Panama, Chi= China, U.S.= United States, LSU= Louisiana State University, Can= Canada, Chl= Chile, Sco= Scotland;
Pre-packaged seafood was provided in factory-sealed plastic packages;
Catfish bred from eggs for research purposes never exposed to antibiotics were provided by Dr. Javier Santander of Arizona State University and from Louisiana State University;
Salmon sold as “antibiotic-free” salmon;
Swai is also marketed as pangasius, channel catfish, catfish, basa, and tra, among other names. Thus, import data were not available, due to this inconsistency in labeling.
Whole fish were filleted and only edible parts were used for analysis. Shrimp (n=6), tilapia (n=3), catfish (n=5), rainbow trout (n=6), Atlantic salmon (n=5), and swai (n=2) were stored at minus 20°C prior to processing by homogenization, using a commercial meat grinder (STX Turbo Force 3000 Series Electric Meat Grinder, Lincoln, Nebraska). Between processing of individual samples, the grinder was cleaned with warm water and soap, and then rinsed separately with acetone, ethanol, and distilled water three times each. Composite samples were prepared by pooling equal amounts of individual samples to result in 11 composite samples: farmed shrimp, wild-caught shrimp, farmed tilapia, farmed catfish, antibiotic-free catfish, farmed rainbow trout of normal habitus, farmed rainbow trout with deformed spine, farmed international Atlantic salmon, farmed antibiotic-free Atlantic salmon, farmed U.S. Atlantic salmon, and farmed swai (Table 1).
2.2 Sample Analysis
Samples pre-processed as described above were frozen and shipped to a commercial laboratory (AXYS Analytical Services Ltd., Sydney, British Columbia, Canada). Approximately 2.5 grams fresh weight (wet weight) of homogenized seafood was subsampled and spiked with isotope-labeled surrogates. Samples were then extracted by bath sonication with 15 mL acetonitrile that was acidified to pH 2 using 0.14 M NaH2PO4/ 85% H3PO (1.93 g NaH2PO4 · H2O, 99 mL reagent water, 1 mL 85% H3PO4). The extract was then treated with 500 mg of solid ethylenediaminetetraacetic acid (EDTA). Resultant extracts were then filtered and cleaned using solid phase extraction (Waters Oasis HLB SPE cartridges 20 cc/1g LP; Hartford, CT). For each sample, 30 mL of extract was diluted to 200 mL total volume with ultra pure water. Prior to sample loading, the cartridges were conditioned using 20 mL of methanol, 6 mL ultra pure water, and 6 mL pH 2 water. The cartridges were then washed with 10 mL of ultra pure water and subsequently dried under a vacuum. Analytes were eluted using 12 mL methanol, and the eluate concentrated under vacuum to a volume of 4 mL prior to analysis. The full 2.5 g of sample was extracted and contained in the final 4 mL extract.
Samples were analyzed by positive electrospray ionization on a triple quadrupole LC-MS/MS in multiple reaction monitoring (MRM) mode using a Waters Micromass Quattro Ultima LC-MS/MS System paired with a Waters LC 2795. Chromatography was conducted using reverse-phased C18 column (Waters, Milford, MA). A total of 60 pharmaceuticals were analyzed according to the AXYS Method MLA-075, a modification of the USEPA Method 1694 as described previously [21]. Out of the 60 analytes screened for, 47 were antibiotics and are the focus of this paper (Table 2 and SI table S1). Two methods were used on the same extract (injection volume: 10 μL) to analyze for tetracyclines and non-tetracyclines, respectively. The tetracyclines method, totaling 30 min in duration, had solvent A consisting of an equal mixture of acetonitrile and methanol with 0.5 mM oxalic acid and 0.5% (v/v) formic acid; solvent B consisted of HPLC-grade water containing 0.5 mM oxalic acid and 0.5% (v/v) formic acid. The starting mixture was 10% solvent A (flow rate 0.2 mL/min), increased to 90% A by minute 20 at a flow rate of 0.23 mL/min. The non-tetracyclines method had a run time of 33 min, using as solvent A HPLC-grade water with 0.1% formic acid and 0.1% ammonium formate, and as solvent B a mixture of equal amounts of acetonitrile and methanol. The starting mixture was 95% solvent A (flow rate 0.15 mL/min), increased to 100% solvent B by minute 23 at a flow rate 0.3 mL/min. For the 10 of the 60 total compounds for which a respective stable-isotope labeled analog was available, the concentration was determined using the isotope dilution technique [22]. For the remaining 50 compounds where a labeled analog was not available, the concentration was determined using an alternate isotope-labeled internal standard (see supplemental information).
Table 2.
Antibiotics analyzed, recovery percentages, method detection limits, and concentrations detected in seafood samples in units of ng/g fresh weight.
| Antibiotic Class | Compound, Recovery %, (MDL*), Concentration If Detected | |
|---|---|---|
| DETECTED | NOT DETECTED | |
| Tetracyclines | Oxytetracycline, 100, (2.4), 7.72, 2.73 3.96, 8.68 4-Epioxytetracycline, 112.5, (3.9), 4.18 |
Anhydrochlortetracycline, 46.8, (7.4); Anhydrotetracycline, 137.5, (6.0); Chlortetracycline, 130.5, (9.2); Demeclocycline, 97.7, (6.0); Doxycycline, 117, (2.4); 4-Epianhydrochlortetracycline, 15.9, (24.1); 4-Epianhydrotetracycline, 104.1, (6.2); 4-Epichlortetracycline, 104, (9.1); 4-Epitetracycline, 130.5, (4.2); Isochlortetracycline, 87.2, (2.4); Minocycline, 109.5, (25.5); Tetracycline, 135, (3.5) |
| Sulfonamides | Sulfadimethoxine, 79.5, (0.2), 0.31 | Sulfachloropyridazine, 83, (0.6); Sulfadiazine, 102.3, (0.6); Sulfamerazine, 111, (0.2); Sulfamethazine, 109, (0.4); Sulfamethizole, 85.5, (0.9); Sulfamethoxazole, 112.4, (0.2); Sulfanilamide, 56.5, (6.0); Sulfathiazole, 138, (0.6) |
| Macrolides | Virginiamycin, 89.5, (4.2), 5.29 | Azithromycin, 97.7, (0.7); Clarithromycin, 96.4, (0.6); Erythromycin-H2O, 117, (0.9); Lincomycin, 129.5, (1.2); Roxithromycin, 75.1, (0.1); Tylosin, 72.1, (2.4); |
| Quinolones | - | Ciprofloxacin, 99.6, (2.); Clinafloxacin, 119, (2.6); Enrofloxacin, 119, (1.2); Flumequine, 104.7, (0.6); Lomefloxacin, 72.7, (1.2); Norfloxacin, 114, (6.); Ofloxacin, 81.8, (0.6); Oxolinic Acid, 54.8, (0.3); Sarafloxacin, 65.7, (0.6) |
| Penicillins | - | Cloxacillin, 86, (1.2); Oxacillin, 87.7, (1.2); Penicillin G, 28.3, (1.2); Penicillin V, 120.5, (1.2) |
| Cephalosporin | - | Cefotaxime, 65.1, (9.9) |
| Other | Ormetoprim, 93.1, (0.4), 0.510 | Carbadox, 24.7, (0.6); Trimethoprim, 91.5, (0.6) |
Superscripts of detected concentrations indicate sample number; see Table 1 for additional sample information.
Highest method detection limit (MDL) for each analyte is reported. See Table S2 in the Supplemental Information for all MDLs.
Precision between intraday samples and duplicates was expressed as relative percent difference (RPD), which was calculated using the following expression as reported previously [23]:
| (Eq. 1) |
where Csample and Cduplicate are the concentrations detected in the original sample and in its duplicate, respectively.
2.3 Quality Assurance and Control
Several tests were performed before and during sample analysis to ensure system and laboratory performance. Initial calibration was performed using labeled surrogates, recovery standards and authentic targets to encompass the working concentration range. Retention times of native and labeled compounds had to be within 0.4 minutes of the respective retention time established during the previous calibration. A mid-level solution was analyzed every 12 hours or every 20 samples, whichever occurred first. All calibration curves consisted of at least 5 consecutive calibration levels. Native compounds with labeled surrogate standards had to elute within 0.1 minutes of the associated labeled surrogates in order to be authenticated. Method blanks and matrix spikes to evaluate recovery rates were also conducted, and duplicates were also analyzed for 5% of test samples within each batch on the same day (containing 7 or more test samples). Method detection limits (MDLs) were determined as specified by EPA Federal Regulation 40 CFR Part 136, Appendix B.
2.4 Meta-Analysis of the Peer-reviewed Literature for Antibiotic Resistance Articles
A literature search of the Web of Knowledge was performed for studies published between 2003 and November 2013 using the search terms “antibiotic resistance AND aquaculture” and “antibiotic resistance AND seafood” to identify relevant strains of bacteria isolated from seafood shown to contain antibiotic resistant microorganisms. Only microbial strains isolated from finned fish or shrimp were included to make it relevant to this study and only seafood for human consumption was included; strains further had to show resistance to one or more specific antibiotics (as opposed to mere classes of antibiotics). Resistance to only four antibiotic classes, tetracyclines, sulfonamides, penicillins, and quinolones, was examined because these are the top drug classes customarily screened for in our study.
The same search words were used to identify connections between antibiotic resistance and aquacultural practices (i.e., sediment, water pollution, resistant strains found on aquaculture facilities or seafood). Articles focusing on non-antibiotic pathogen reduction methods and/or ornamental fish were excluded. No publication-year limit was employed.
2.5 Calculation of Theoretical Maximum Concentrations in Individual Samples Used in Composites
This study employed a composite sampling approach. Samples were pooled to create 11 composites from 27 individual samples. Theoretical maximum concentrations in individual samples processed were calculated using the conservative formula:
| (Eq. 2) |
where Ccomposite is the concentration determined experimentally in the pool of samples, n is the number of samples contributing to the pool, and Cindividual sample is the calculated theoretical maximum concentration of the analyte in individual samples contributing to the pool. Each composite sample was constructed from a different number of individual samples, depending on the species. See Table 1 for a complete list.
3. Results and Discussion
3.1 Method Performance
As this paper focuses on antibiotics, further discussion will only pertain to the 47 antibiotic analytes that were screened for. Method detection limits for the various antibiotics ranged from 0.1 ng/g (roxithromycin/sulfadimethoxine) to 25.5 ng/g (minocycline) fw of seafood (Table 2; Supplementary Information: Table S2). Recoveries of the 47 antibiotics ranged from 15.9% (4-epianhydrochlortetracycline) to 138% (sulfathiazole), with the majority (35 out of 47) placing in the preferred range of 70 to 130% (Table 2). No laboratory contamination was observed in method blanks. Method performance in this study was favorable and comparable to previously reported results [23, 24].
3.2 Occurrence of Antibiotics in Seafood
Seven out of eleven composite samples were found to have detectible quantities of antibiotics, including oxytetracycline, 4-epioxytetracycline, sulfadimethoxine, ormetoprim, and virginiamycin (Table 2). The most commonly detected antibiotic was oxytetracycline, which is the number one used antibiotic in aquaculture, with 12 of the top 15 aquaculture-producing countries reporting usage [3]. It was detected at a concentration of 8.6 ng/g fw, along with its 4-epimer at 4.1 ng/g fw, in farmed international Atlantic salmon comprised of samples from Chile and Canada (Fig 1), which are among the top four salmon-producing countries [1]. As the 4-epimer is a known degradation product of oxytetracycline [25] it is likely that a higher oxytetracycline concentration was originally in these samples. Tetracyclines are regulated in the U.S. as a sum of all parent antibiotics and their 4-epimers [26]. The resultant combined concentration in farmed international Atlantic salmon of 12.6 ng/g was still under the maximum permitted concentration of 2 μg/g in finfish (Table 3).
Figure 1.
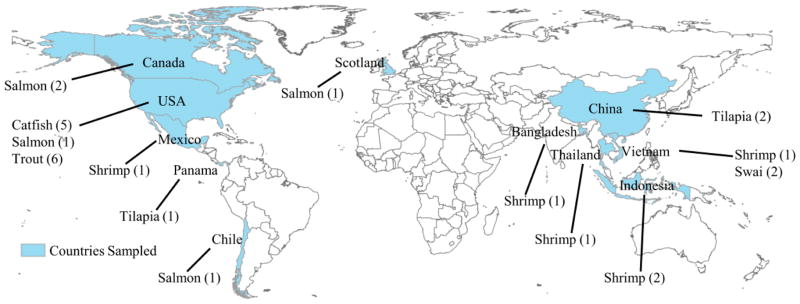
Map showing countries from which seafood samples orginated (n= number of samples).
Table 3.
Maximum Residue Limits (MRLs) of antibiotics allowed for the USA, EU, Chile, and CODEX (μg/g fresh weight). For antibiotics lacking regulatory guidelines in seafood, values are given for other food animal varieties when available.
| Antibiotic | USAa | European Unionb | Chilec | CODEXd |
|---|---|---|---|---|
| Carbadox | 0.03e | - | - | - |
| Cloxacillin | 0.01g | 0.3m | - | - |
| Doxycycline | 2f | 0.1i | - | - |
| Enrofloxacin | 0.1h | 0.1n | 0 | - |
| Tetracyclinesr | 2f | 0.1o | - | - |
| Erythromycin-H2O | 0.1g | 0.2m | 0.2m | 0.1q |
| Lincomycin | 0.1i | 0.1m | - | 0.2q |
| Ormetoprim | 0.1j | - | - | - |
| Oxytetracycline | 2f | 0.1o | 0.2m | 0.2m |
| Penicillin G | 0k | 0.05m | - | 0.05i |
| Penicillin V | 0k | - | - | - |
| Sulfadimethoxine | 0.1j | 0.1 (sum of sulfonamides) | - | - |
| Sulfamerazine | 0l | - | - | |
| Sulfathiazole | 0.1i | - | - | |
| Tetracycline | 2f | 0.1o | - | 0.2p |
| Tylosin | 0.2g | 0.1m | - | 0.1g |
| Virginiamycin | 0.1i | - | - | - |
FDA USDA CFR 21 [26];
EU commission regulation no. 37/2010, Dec. 2009 [48];
Values for salmon only [36];
Codex Alimentarius Commenssion (CAC), 2009 [49];
Swine liver;
Sum of tetracyclines in finfish;
Cattle;
Cattle liver;
Swine;
Salmonids and catfish;
Different forms of penicillin are not differentiated. Chicken;
Trout;
All fish;
Sum of ciprofloxacin and enrofloxacin;
Sum of 4-epimer plus parent drug;
Sum of parent drugs;
Poultry;
Includes 4-epianhydrotetracycline, 4-epianhydrotetracycline, 4-epichlortetracycline, 4-epioxytetracycline 4-epitetracycline, demeclocycline, isochlortetracycline, minocycline. Currently unregulated/information not available for: anhydrochlortetracycline, anhydrotetracycline, azithromycin, cefotaxime, clarithromycin, clinafloxacin, omefloxacin, norfloxacin ofloxacin, and roxithromycin. Currently, no MRLs have been set in U.S. for ciprofloxacin, flumequine, oxacillin, oxolinic acid, sarafloxacin, and trimethoprim.
The unexpected detection of oxytetracycline at a concentration of 7.7 ng/g fw in wild-caught shrimp imported from Mexico may be due to several reasons. Unintentional or intentional mislabeling of the product and cross-contamination of seafood during handling, processing and packaging are possible. Uptake of the drug from coastal waters and sediments impacted by inputs of raw and treated wastewater [27] also could explain the observed detection but ultimately the origin of contamination remains unknown.
Oxytetracycline was also detected at concentrations of 2.7 and 3.9 ng/g fw, respectively, in farmed tilapia and in farmed rainbow trout with visibly deformed spines (Fig 2A). Oxytetracycline was not detected above the detection limit of 2.4 ng/g in trout without visible spinal deformities (supplemental information T2). Detection of the latter corroborates earlier reports that this antibiotic may cause spinal deformities in certain species [10]; however, due to the limited number of individual samples available (n =3), the present study was underpowered and cannot ascertain causation. As trout is a major market in the U.S., with over 700 trout-rearing farms [28], further work with a larger sample size is needed to elucidate the connection between oxytetracycline dosing and spinal deformities in trout and other fish species. Among the large group of sulfonamides, only sulfadimethoxine was detected and only in a single seafood variety, in farmed shrimp at 0.3 ng/g fw. Sulfadimethoxine reportedly is used by 4 of the top 15 aquaculture-producing countries [3]. Yet, although screened for previously [29, 30] and several detection methods have been developed [31, 32], the result reported here constitutes the first detection of this drug in shrimp. There is no U.S. MRL set for this drug in shrimp, although it is regulated in salmonids and catfish at a level of 0.1 μg/g fw (Table 3).
Figure 2. Farmed trout with visible spinal deformities and applicable U.S. and E.U. MRLs in composite and individual samples.
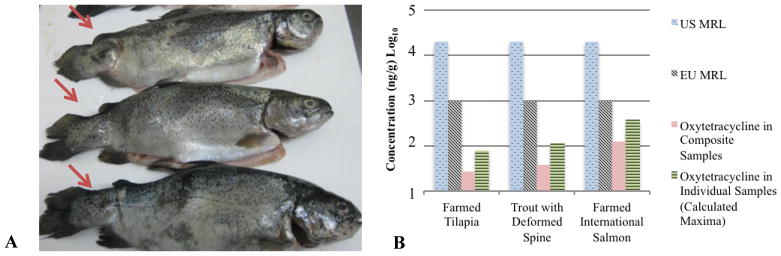
Panel A shows an image of spinal deformities in trout analyzed in this work. Arrows indicate abnormal spinal curvatures (Photo credit: Don McBride, NOAA, 2012). Panel B shows a comparison of oxytetracycline concentrations determined in this study to maximum residue limits (MRLs) allowed in the United States (U.S.) and the European Union (EU) [26, 47]. Concentrations of oxytetracycline and 4-epioxytetracycline in farmed international salmon were added together for this data point, as regulation maximum total tetracyclines.
Ormetoprim, an antibiotic commonly used with sulfonamides, was detected at a concentration of 0.5 ng/g fw in farmed Atlantic salmon from the U.S. This concentration is about 200 times less than the regulatory limit of 0.1 μg/g.
Contrary to the label stating culturing without antibiotics, virginiamycin was found at a concentration of 5.2 ng/g fw in farmed Atlantic salmon. The apparent presence of virginiamycin indicates that either the labeling was inaccurate or contamination of the seafood occurred. Although the detected concentration was much lower than the regulatory limit of 0.1 μg/g (Table 3, this finding is still important, as it indicates that the “antibiotic-free” label does not always accurately represent whether antibiotics are absent or present.
The occurrence of antibiotics in seafood above method detection limits in the low ng/g range attained here appears to be the exception rather than the norm. Five antibiotics were detected at low ng/g concentrations in this survey. The present study is the first to consider the top consumed seafoods in the U.S. as well as the first to survey a large number of antibiotics. The majority of these antibiotics have never been screened for in our food supply. This study also represents samples from 11 countries (Fig 1), 8 of which are among the top 15 aquaculture-producing countries [3]. Results of this study of modest sample size suggest that seafood, regardless of whether wild-caught, farmed, imported, or domestically produced, is typically compliant with U.S. chemical regulations. However, the results need further confirmation, ideally by studies featuring a large sample size.
3.3 Antibiotic Resistance Development in Seafood
Although the concentrations reported here are less than the FDA allowed maxima, these sub-therapeutic drug concentrations can often select for and enrich resistant bacteria [33]. There has been a notable increase in resistant microbial strains associated with the antibiotics and seafoods examined in this study. Out of 179 Escherichia coli strains isolated from commercial seafood in a study by Ryu et al., 55 strains were found to be resistant to tetracycline [14]. Another 34 strains were found to hold intermediate resistance to tetracycline, which can be affected and selected for by sub-therapeutic antibiotic concentrations. Nawaz et al. also reported isolation of MDR Klebsiella spp. bacteria from imported shrimp obtained from grocery stores [34]. The identification of these strains may be interpreted as being the result of extensive human use and misuse of antibiotics in the clinic, community, agriculture, and in animal husbandry such as aquaculture [33]. The top antibiotics used by heavy aquaculture producers include the following: oxytetracycline, oxolinic acid, chloramphenicol, erythromycin, furazolidone, trimethoprim, sulfadiazine, ampicillin, florfenicol, flumequine, and sulfadimethoxine [3]. All of these antibiotics are included on the WHO list of critically/highly important antibiotics for human health [11, 34, 35]. Multiple studies in the last three decades have revealed resistance to many of these antibiotics, the majority of which were screened for in this study (Fig 3A). The fact that seafood examined for bacteria has resulted in isolates belonging to pathogenic genera causing infections in humans (e.g. Salmonella, Vibrio, Escherichia) [7, 14, 16] increases the likelihood of resistance spread from aquaculture to people. This poses a risk to consumers as well as employees coming into contact the with seafood from production to store delivery.
Figure 3. Published studies reporting resistant bacteria isolated from aquaculture and seafood.
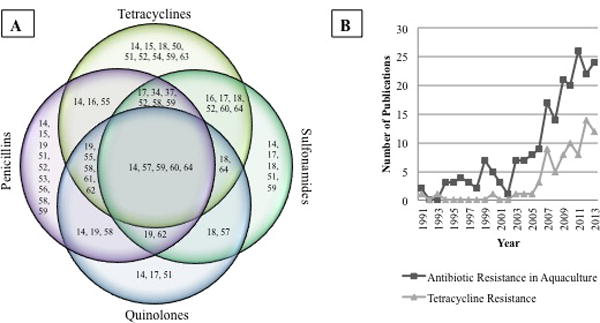
Panel A shows select studies from 2003–2013 reporting the presence of bacteria resistant to 4 groups of antibiotics found on seafood available for human consumption. Numbers correspond to references. For example, reference 60 showed isolates that were resistant to antibiotics in all four classes. This reference also showed a separate isolate that showed resistance to just tetracyclines and sulfonamides. Panel B shows the number of publications featuring antibiotic resistance development in aquaculture and seafood (dark gray) and number of publications featuring resistance to the antibiotic class of tetracyclines (light gray).
Indeed, literature volume statistics summarized in Fig 3 show that the topic of resistance to many antibiotics screened here is a major area of concern for the aquaculture community. The number of publications linking resistance to seafood has increased by 800% between the 1990s and today (Fig 3B). The majority of papers report the ineffectiveness of tetracycline and oxytetracycline as one of the most commonly seen resistances. The observed publication trend also acknowledges an increased awareness of the fact that exponential growth has taken place in the aquaculture industry in the past few decades. This trend also suggests an association between the heavy usage of oxytetracycline (the number one used antibiotic in aquaculture) and resistance development.
Some bacterial strains identified in our literature review were found to be completely or intermediately resistant to certain antibiotics [14, 36]. Furthermore, the transfer of plasmids among bacteria on seafood has been reported [37]. Strains were found to have minimal inhibitory concentrations (MIC) far lower than the MIC requirement for the “resistant” classification, indicating that very low concentrations of antibiotics can select for resistance. One study found that only about half of the isolates from their aquaculture samples had MICs above the “resistant” concentration of 128 μg/mL; some isolates exhibited MICs as low as 0.25 μg/mL, over 500 times less than the classification of resistance-promoting concentration [38]. In Chile, the reported dose of oxytetracycline through feed is 100–120 μg per g fish per day, administered for 14–21 days, depending on the disease [39]. In China, the preventative dose for the fluoroquinolone compound oxolinic acid is 10–20 μg per g fish per day for 4–7 days [40]. These concentrations currently in use are known to exert selective pressure. Since many of these antibiotics also are used in human medicine, selective pressure may promote the occurrence of resistant strains of potential human health concern. Overall, the information compiled in Fig 3 shows that the development and occurrence of drug resistant bacteria in seafood is an issue that is both timely and of notable importance. Thus, to ensure the safety of the food supply in the U.S. and abroad, the monitoring of seafood has to focus on both the residues of aquacultural drugs themselves and the drug resistance in pathogens these antibiotics can trigger.
3.4 Study Limitations
This study employed composite sampling. This approach is well suited for the economical screening of a large number of analytes and for accurately determining average concentrations therein [41, 42]. This method of sampling was chosen here because the purpose of this study was to conduct a large-scale screening of many analytes. However, this methodology is inappropriate for determining the full range of concentrations (i.e., minima and maxima) as well as detection frequencies. Accordingly, theoretical maximum concentrations of oxytetracycline and sulfadimethoxine were calculated for individual samples and the resultant values represent conservative estimates that are likely higher than the true concentration. The oxytetracycline values of 8.1, 11.7, and 37.8 ng/g calculated, respectively, in farmed tilapia, farmed trout with spinal deformities, and farmed international salmon are well below the U.S. limit of 2,000 ng/g (Fig 2B). Note that the concentration of 37.8 ng/g calculated for salmon includes both oxytetracycline and 4-epioxytetracycline; it is provided in this form because tetracyclines are regulated as a sum of drugs of this class. Values calculated for sulfadimethoxine (1.7 ng/g for each country’s sample) is also significantly under U.S. regulatory limits.
Another limitation is that sampling was done only in Arizona and California. The obtained results may not necessarily apply to other states and alternate sources (i.e., countries) of commercial seafood. Many wild-caught seafood varieties were not available for this survey because the vast majority of seafood for consumption in the U.S. is only readily available from aquaculture operations. Also, as we obtained fresh seafood in the form most consumers choose, samples were either whole animals or fillets and either pre-packaged or loose, which means that variation in handling and processing by the producer may affect antibiotic preservation and degradation in the tissue. This variation, as well as antibiotic sources that do not originate from aquaculture, could also have contaminated the seafood and affected our data.
Samples were collected in June-August, 2012 and analyzed in November 2012, following storage for 3–5 months at −20°C. A previous study, examining the effect of sample storage at −18°C, showed that tetracyclines, sulfonamides, quinolones, macrolides, and aminoglycosides are stable and remain intact structurally and quantitatively, as demonstrated using a porcine muscle matrix [43]. However, penicillins were observed to attenuate, by about 30% and 20%, respectively, for ampicillin and cloxacillin over the course of 3-6 months [43]. Hence, the concentrations of penicillins at the time of purchase in samples of seafood analyzed here may have been higher than the values of less than <1.2 to <1.6 ng/g fw reported here.
Our sample size of 27 is of a magnitude similar to other studies that utilized composite sampling to investigate poorly characterized potential human exposure sources [44, 45] The goal of the present work was not necessarily to identify specific antibiotics in individual samples, but rather to conduct a large-scale screening of U.S. seafood to assess whether there is a need for more aggressive monitoring. Whereas the present dataset cannot prove the safety or danger of imported seafoods, it provides an incremental, yet significant step forward in assessing the safety of the U.S. seafood supply. Data made available here suggest that there is no immediate threat to human health from trace levels of the analytes surveyed in this work. However, additional studies using a larger sample size would be beneficial to confirm the findings and conclusions of the results obtained here.
Our literature review considered only a subset of papers based on the inclusion criteria stated. A less stringent search would have resulted in an even larger body of literature supporting the conclusion reached here that the promotion of antibiotic resistance constitutes a major health concern in aquaculture.
5. Conclusions
This study surveyed the concentrations of 47 antibiotics in 6 different seafood varieties originating in 11 countries purchased exclusively from the southwestern U.S. All samples studied demonstrated compliance under current federal regulations, suggesting that they are chemically safe to consume. This conclusion could be drawn from the analysis of pooled samples, an approach that did not permit to determine the actual concentration in each individual sample entering the survey, however. Five antibiotics were found at detectable levels and estimated concentrations were relatively low (0.3–8.6 ng/g fw). However, the development and spread of antibiotic resistance is a public health priority that is divorced from the regulatory limits designed to prevent adverse outcomes from human ingestion of drugs. Antibiotics present at levels well below regulatory limits still can promote the emergence of (multi-) drug resistant microorganisms. Future studies are warranted to fully understand the connection between aquacultural use of antibiotics, development of drug resistance, human exposure to resistant pathogens, and ensuing morbidity and mortality in seafood consumers. The trend in the last 3 decades of notable increases in the number of resistant and multi-drug resistant strains identified in seafood is of concern. Monitoring studies such as the present work are one of multiple steps required to understand and manage potential risks posed by use of antibiotics in aquaculture and in society at large. The present study was limited in sample size and employed sample pooling. It is desirable to perform additional surveys to confirm the findings and preliminary conclusions reported here.
Supplementary Material
Highlights.
5 out of 47 antibiotics were detected in shrimp, salmon, tilapia and trout.
Oxytetracycline is the most commonly detected antibiotic compound.
Publications reporting antibiotic resistance in aquaculture have increased 8-fold over 3 decades.
We report a low risk of drug exposure from consumption of U.S. seafoods
We recommend vigilance toward stemming microbial risks.
Acknowledgments
The authors thank Don McBride of NOAA for acquiring the seafood samples and Dr. Javier Santander of the Biodesign Institute’s Center for Infectious Diseases and Vaccinology for providing antibiotic-free catfish. We also thank Isaac Roll and Dr. Arjun Venkatesan of the Center for Environmental Security for editing and for providing the GIS map, respectively. We acknowledge the services provided by AXYS Analytical Services Ltd, British Columbia, Canada. This project was supported in part by the Johns Hopkins Center for a Livable Future and by Award Numbers R01ES015445 and RO1ES020889 of the National Institute of Environmental Health Sciences (NIEHS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIEHS or the National Institutes of Health (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Food and Agricultural Organization, FAO. FishStateJ-2, Version 2.1.0. Food and Agriculture Organization of the United Nations; 2013. Downloaded November 1, 2013 from http://www.fao.org/fishery/statistics/software/en. [Google Scholar]
- 2.Cole DW, Cole R, Gaydos SJ, Gray J, Hyland G, Jacques ML, Powell-Dunford N, Sawhney C, Au WW. Aquaculture: Environmental, toxicological, and health issues. Int J Hyg Environ Health. 2009;212:369–377. doi: 10.1016/j.ijheh.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Sapkota A, Sapkota AR, Kucharski M, Burke J, McKenzie S, Walker P, Lawrence R. Aquaculture practices and potential human health risks: current knowledge and future priorities. Environ Int. 2008;34:1215–1226. doi: 10.1016/j.envint.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol. 2006;8:1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x. [DOI] [PubMed] [Google Scholar]
- 5.Defoirdt T, Sorgeloos P, Bossier P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol. 2011;14:251–258. doi: 10.1016/j.mib.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Christensen AM, Ingerslev F, Baun A. Ecotoxicity of mixtures of antibiotics used in aquacultures. Environ Toxicol Chem. 2006;25:2208–2215. doi: 10.1897/05-415r.1. [DOI] [PubMed] [Google Scholar]
- 7.Baker-Austin C, McArthur JV, Tuckfield RC, Najarro M, Lindell AH, Gooch J, Stepanauskas R. Antibiotic resistance in the shellfish pathogen Vibrio parahaemolyticus isolated from the coastal water and sediment of Georgia and South Carolina. USA J Food Protect. 2008;71:2552–2558. doi: 10.4315/0362-028x-71.12.2552. [DOI] [PubMed] [Google Scholar]
- 8.Barros-Becker F, Romero J, Pulgar A, Feijoo CG. Persistent oxytetracycline exposure induces an inflammatory process that improves regenerative capacity in zebrafish larvae. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lunden T, Miettinen S, Lonnstrom LG, Lilius EM, Bylund G. Influence of oxytetracycline and oxolinic acid on the immune response of rainbow trout (Oncorhynchus mykiss) Fish and Shellfish Immunology. 1998;8(3):217–230. [Google Scholar]
- 10.Toften H, Jobling M. Development of spinal deformities in Atlantic salmon and Arctic charr fed diets supplemented with oxytetracycline. J Fish Biol. 1996;49:668–677. [Google Scholar]
- 11.Heuer OE, Kruse H, Grave K, Collignon P, Karunasagar I, Angulo FJ. Human health consequences of use of antimicrobial agents in aquaculture. Clin Infect Dis. 2009;49:1248–1253. doi: 10.1086/605667. [DOI] [PubMed] [Google Scholar]
- 12.Struthers JK, Westran R. Clinical Bacteriology. CRC Press; FL, USA: 2003. p. 152.p. 166. [Google Scholar]
- 13.Novotny L, Dvorska L, Lorencova A, Beran V, Pavlik I. Fish: a potential source of bacterial pathogens for human beings. Vet Med-Czech. 2004;49:343–358. [Google Scholar]
- 14.Ryu SH, Park SG, Choi SM, Hwang YO, Ham HJ, Kim SU, Lee YK, Kim MS, Park GY, Kim KS, Chae YZ. Antimicrobial resistance and resistance genes in Escherichia coli strains isolated from commercial fish and seafood. Int J Food Microbiol. 2012;152:14–18. doi: 10.1016/j.ijfoodmicro.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Nawaz M, Khan AA, Khan S, Sung K, Kerdahi K, Steel R. Molecular Characterization of tetracycline-resistant genes and integrons from avirulent strains of Escherichia coli isolated from catfish. Foodborne Pathog Dis. 2009;6:553–559. doi: 10.1089/fpd.2008.0204. [DOI] [PubMed] [Google Scholar]
- 16.Ponce E, Khan AA, Cheng CM, Summage-West C, Cerniglia CE. Prevalence and characterization of Salmonella enterica serovar Weltevreden from imported seafood. Food Microbiol. 2008;25:29–35. doi: 10.1016/j.fm.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhao SH, Datta AR, Ayers S, Friedman S, Walker RD, White DG. Antimicrobial-resistant Salmonella serovars isolated from imported foods. Int J Food Microbiol. 2003;84:87–92. doi: 10.1016/s0168-1605(02)00402-6. [DOI] [PubMed] [Google Scholar]
- 18.Labella A, Gennari M, Ghidini V, Trento I, Manfrin A, Borrego JJ, Lleo MM. High incidence of antibiotic multi-resistant bacteria in coastal areas dedicated to fish farming. Mar Pollut Bull. 2013;70:197–203. doi: 10.1016/j.marpolbul.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 19.Chiu TH, Kao LY, Chen ML. Antibiotic resistance and molecular typing of Photobacterium damselae subsp damselae, isolated from seafood. J Appl Microbiol. 2013;114:1184–1192. doi: 10.1111/jam.12104. [DOI] [PubMed] [Google Scholar]
- 20.Pruden A, Joakim Larsson GD, Amezquita A, Collignon P, Brandt KK, Graham DW, Lazorchak JM, Suzuki S, Silley P, Snape JR, Topp E, Zhang T, Zhu YG. Management options for reducing the release of antibiotics and antibiotic resistance genes for the environment. Environ Health Perspect. 2013;121:878–885. doi: 10.1289/ehp.1206446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chari BP, Halden RU. Validation of mega composite sampling and nationwide mass inventories for 26 previously unmonitored contaminants in archived biosolids from the U.S National Biosolids Repository. Water Res. 2012;46:4814–4824. doi: 10.1016/j.watres.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halden RU, Paull DH. Co-occurrence of triclocarban and triclosan in U.S. water resources. Environ Sci Technol. 2005;39:1420–1426. doi: 10.1021/es049071e. [DOI] [PubMed] [Google Scholar]
- 23.McClellan K, Halden RU. Pharmaceuticals and personal care products in archives U.S. biosolids from the 2001 EPA national sewage sludge survey. Water Res. 2010;44:658–668. doi: 10.1016/j.watres.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love RC, Halden RU, Davis MF, Nachman KE. Feather meal: a previously unrecognized Route for reentry into the food supply of multiple pharmaceuticals and personal care products (PPCPs) Environ Sci Technol. 2012;46:3795–3802. doi: 10.1021/es203970e. [DOI] [PubMed] [Google Scholar]
- 25.Loke ML, Jespersen S, Vreeken R, Halling-Sorensen B, Tjornelund J. Determination of oxytetracycline and its degradation products by high-performance liquid chromatography-tandem mass spectrometry in manure-containing anaerobic test systems. J Chromatogr B. 2003;783:11–23. doi: 10.1016/s1570-0232(02)00468-3. [DOI] [PubMed] [Google Scholar]
- 26.Code of Federal Regulations Title 21. U.S. Food and Drug Administration; Silver Spring, MD: [Last Accessed November 14, 2013]. FDA CFR Title 21. from http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfCFR/CFRSearch.cfm?CFRPart=556. [Google Scholar]
- 27.Kim SC, Carlson K. Temporal and spatial trends in the occurrence of human and veterinary antibiotics in aqueous and river sediment matrices. Environ Sci Technol. 2007;41:50–57. doi: 10.1021/es060737+. [DOI] [PubMed] [Google Scholar]
- 28.Agricultural Marketing Research Center. Trout Profile. Iowa State University; Ames, IA: [Last accessed November 14, 2013]. from http://www.agmrc.org/commoditiesproducts/aquaculture/trout-profile/ [Google Scholar]
- 29.Won SY, Lee CH, Chang HS, Kim SO, Lee SH, Kim DS. Monitoring of 14 sulfonamide antibiotic residues in marine products using HPLC-PDA and LC-MS/MS. Food Control. 2011;22:1101–1107. [Google Scholar]
- 30.Tittlemier SA, Van de Riet J, Burns G, Potter R, Murphy C, Rourke W, Pearce H, Dufresne G. Analysis of veterinary drug residues in fish and shrimp composites collected during the Canadian Total Diet Study, 1993–2004. Food Addit Contam. 2007;24:14–20. doi: 10.1080/02652030600932937. [DOI] [PubMed] [Google Scholar]
- 31.Gehring TA, Griffin B, Williams R, Geiseker C, Rushing LG, Siitonen PH. Multiresidue determination of sulfonamides in edible catfish, shrimp and salmon tissues by high-performance liquid chromatography with postcolumn derivatization and fluorescence detection. J Chromatogr B. 2006;840:132–138. doi: 10.1016/j.jchromb.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 32.Villar-Pulido M, Gilbert-Lopez B, Garcia-Reyes JF, Martos NR, Molina-Diaz A. Multiclass detection and quantitation of antibiotics and veterinary drugs in shrimps by fast liquid chromatography time-of-flight mass spectrometry. Talanta. 2011;85:1419–1427. doi: 10.1016/j.talanta.2011.06.036. [DOI] [PubMed] [Google Scholar]
- 33.Andersson DI, Hughes D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist Update. 2012;15:162–172. doi: 10.1016/j.drup.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Nawaz M, Khan SA, Tran Q, Sung K, Khan AA, Adamu I, Steele RS. Isolation and characterization of multidrug-resistant Klebsiella spp. Isolated from shrimp imported from Thailand. Int J of Food Microbiol. 2012;155:179–184. doi: 10.1016/j.ijfoodmicro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization (WHO) Critically important antibacterial agents for human medicine; for risk management strategies of non-human use. Report of a WHO working group consultation; Canberra, Australia. 15–18 February 2005; Geneva: WHO; 2005. [Google Scholar]
- 36.World Health Organization (WHO) Critically important antimicrobials for human medicine: categorization for the development of risk management strategies to contain antimicrobial resistance due to non-human use. Report of the Second WHO expert Meeting; Copenhagen, Denmark. 29–31 May 2007; Geneva: WHO; 2007. [Google Scholar]
- 37.Ferrini AM, Mannoni V, Suffredini E, Cozzi L, Croci L. Evaluation of antibacterial resistance in Vibrio strains isolated from imported seafood and Italian aquaculture settings. Food Anal Method. 2008;1:164–170. [Google Scholar]
- 38.Guglielmetti E, Korhonen JM, Heikkinen J, Morelli L, von Wright A. Transfer of plasmid-mediated resistance to tetracycline in pathogenic bacteria from fish and aquaculture environments. FEMS Microbiol Lett. 2009;293:28–34. doi: 10.1111/j.1574-6968.2009.01512.x. [DOI] [PubMed] [Google Scholar]
- 39.Akinbowale OL, Peng H, Barton MD. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J Appl Microbiol. 2006;100:1103–1113. doi: 10.1111/j.1365-2672.2006.02812.x. [DOI] [PubMed] [Google Scholar]
- 40.Bravo S. Environmental impacts and management of veterinary medicines in aquaculture: the case of salmon aquaculture in Chile. In: Bondad-Reantaso MG, Arthur JR, Subasinghe RP, editors. Improving biosecurity through prudent and responsible use of veterinary medicines in aquatic food production. Rome: FAO; pp. 51–67.pp. 207 FAO Fisheries and Aquaculture Technical Paper No. 547. [Google Scholar]
- 41.Yuan X, Chen W. Use of veterinary medicines in Chinese aquaculture: current status. In: Bondad-Reantaso MG, Arthur JR, Subasinghe RP, editors. Improving biosecurity through prudent and responsible use of veterinary medicines in aquatic food production. Rome: FAO; pp. 51–67.pp. 207 FAO Fisheries and Aquaculture Technical Paper No. 547. [Google Scholar]
- 42.Baron PA, Love DC, Nachman KE. Pharmaceuticals and personal care products in chicken meat and other food animal products: a market-basket pilot study. Sci Total Environ. 2014;490:296–300. doi: 10.1016/j.scitotenv.2014.04.112. [DOI] [PubMed] [Google Scholar]
- 43.Berendsen BJA, Elbers IJW, Stolker AAM. Determination of the stability of antibiotics in matrix and reference solutions using a straightforward procedure applying mass spectrometric detection. Food Addit Contam A. 2011;28:1657–1666. doi: 10.1080/19440049.2011.604045. [DOI] [PubMed] [Google Scholar]
- 44.Kim SR, Halden RU, Buckley TJ. Volatile organic compounds in human milk: methods and measurements. Environ Sci Technol. 2007;41:1662–1667. doi: 10.1021/es062362y. [DOI] [PubMed] [Google Scholar]
- 45.Kim SR, Halden RU, Buckley TJ. Polycyclic aromatic hydrocarbons in human milk of nonsmoking U.S. women. Environ Sci Technol. 2008;42:2663–2667. doi: 10.1021/es702275x. [DOI] [PubMed] [Google Scholar]
- 46.National Fisheries Institute. [Last accessed November 14, 2013];Top 10 Consumed Seafoods. from http://www.aboutseafood.com/about/about-seafood/top-10-consumed-seafoods.
- 47.National Oceanic and Atmospheric Administration (NOAA) Current Fishery Statistics No. 2012-2 Imports and exports of fishery products annual summary, 2012. U.S. Department of Commerce; [Last accessed November 21, 2013]. from http://www.st.nmfs.noaa.gov/commercial-fisheries/fus/fus12/index. [Google Scholar]
- 48.European Union Commission Regulation No 37/2010 of 22 December 2009 on pharmacologically active substances and their classification regarding maximum residue limits in foodstuffs of animal origin. [Last accessed November 20, 2013]; from http://ec.europa.eu/health/files/mrl/mrl_20101212_consol.pdf.
- 49.Codex Alimentarius Commenssion (CAC) [Last accessed November 19, 2013];Maximum residue limits for veterinary drugs in foods. 2009 from www.codexalimentarius.net/vetdrugs/data/MRL2_e_2009.pdf.
- 50.Fallah AA, Saei-Dehkordi SS, Mahzounieh M. Occurrence and antibiotic resistance profiles of Listeria monocytogenes isolated from seafood products and market and processing environments in Iran. Food Control. 2013;34:630–636. [Google Scholar]
- 51.Ansari M, Rahimi E, Raissy M. Antibiotic susceptibility and resistance of Aeromonas spp. isolated from fish. Afr J of Microbiol Res. 2011;5:5772–5775. [Google Scholar]
- 52.Khan AA, Ponce E, Nawaz MS, Cheng CM, Khan JA, West CS. Identification and Characterization of Class 1 Integron Resistance Gene Cassettes among Salmonella Strains Isolated from Imported Seafood. Appl and Environ Microb. 2009;75:1192–1196. doi: 10.1128/AEM.02054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar R, Lalitha KV. Prevalence and molecular characterization of Vibrio cholerae O1, non-O1 and non-O139 in tropical seafood in Cochin, India. Foodborne Pathog Dis. 2013;10:278–283. doi: 10.1089/fpd.2012.1310. [DOI] [PubMed] [Google Scholar]
- 54.Budiati T, Rusul G, Wan-Abdullah WN, Arip YM, Ahmad R, Thong KL. Prevalence, antibiotic resistance and plasmid profiling of Salmonella in catfish (Clarias gariepinus) and tilapia (Tilapia mossambica) obtained from wet markets and ponds in Malaysia. Aquaculture. 2013;372:127–132. [Google Scholar]
- 55.Raissy M, Moumeni M, Ansari M, Rahimi E. antibiotic resistance pattern of some Vibrio strains isolated from seafood. Iran J Fish Sci. 2012;11:618–626. [Google Scholar]
- 56.Deekshit VK, Kumar BK, Kai P, Srikumar S, Karunasagar I, Karunasagar I. Detection of class 1 integrons in Salmonella Weltevreden and silent antibiotic resistance genes in some seafood-associated nontyphoidal isolates of Salmonella in south-west coast of India. J Appl Microbiol. 2012;112:1113–1122. doi: 10.1111/j.1365-2672.2012.05290.x. [DOI] [PubMed] [Google Scholar]
- 57.Yan H, Li L, Alam J, Shinoda S, Miyoshi S, Shi L. Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int J Food Microbiol. 2010;143:230–234. doi: 10.1016/j.ijfoodmicro.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 58.Kakatkar AS, Pansare LS, Gautam RK, Shashidhar R, Karani M, Bandekar JR. Molecular characterization of antibiotic resistant Salmonella isolates from Indian foods. Food Res Int. 2011;44:3272–3275. [Google Scholar]
- 59.Liu F, Guan W, Alam MJ, Shen Z, Zhang S, Li L, Shinoda S, Shi L. Pulsed-field gel electrophoresis typing of multidrug-resistant Vibrio parahaemolyticus isolated from various sources of seafood. J Health Sci. 2009;55:783–789. [Google Scholar]
- 60.Kumar R, Surendran PK, Thampuran N. Foodborne Pathog Dis. 2009;6:621–625. doi: 10.1089/fpd.2008.0252. [DOI] [PubMed] [Google Scholar]
- 61.Adeyemi A, Enyinnia V, Nwanze R, Smith S, Omonigbehin E. Antimicrobial susceptibility of potentially pathogenic halophilic Vibrio species isolated from seafood in Lagos, Nigeria. Afr J Biotechnol. 2008;7:3788–3791. [Google Scholar]
- 62.Thayumanavan T, Vivekanandhan G, Savithamani K, Subashkumar R, Lakshmanaperumalsamy P. Incidence of haemolysin-positive and drug-resistant Aeromonas hydrophila in freshly caught finfish and prawn collected from major commercial fishes of coastal South India. FEMS Immunol Med Microbiol. 2003;36:41–45. doi: 10.1016/S0928-8244(03)00037-3. [DOI] [PubMed] [Google Scholar]
- 63.Kim SR, Nonaka L, Suzuki S. Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol Lett. 2004;237:147–156. doi: 10.1016/j.femsle.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 64.Sarter S, Nguyen HNK, Hung LT, Lazard J, Didier D. Antibiotic resistance in Gram-negative bacteria isolated from farmed catfish. Food Control. 2006;18:1391–1396. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


