Abstract
Neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and frontotemporal dementia have several important features in common. They are progressive, they affect a relatively inaccessible organ, and we have no disease-modifying therapies for them. For these brain-based diseases, current diagnosis and evaluation of disease severity rely almost entirely on clinical examination, which may only be a rough approximation of disease state. Thus, the development of biomarkers – objective, relatively easily measured and precise indicators of pathogenic processes – could improve patient care and accelerate therapeutic discovery. Yet existing, rigorously tested neurodegenerative disease biomarkers are few, and even fewer biomarkers have translated into clinical use. To find new biomarkers for these diseases, an unbiased, high-throughput screening approach may be needed. In this review, I will describe the potential utility of such an approach to biomarker discovery, using Parkinson’s disease as a case example.
Introduction
The two most prevalent neurodegenerative diseases are Alzheimer’s Disease (AD) and Parkinson’s Disease (PD). As of 2010, >35 million people worldwide suffered from dementia, with the vast majority due to AD (Wimo and Prince, 2010). Similarly, as of 2005, >4 million people worldwide suffered from PD (Dorsey et al., 2007). Moreover, risk for both of these neurodegenerative diseases increases with age, with both of these diseases projected to double in numbers over the next two decades (Dorsey et al., 2007; Wimo and Prince, 2010). As a consequence, the economic burden associated with these incurable, neurodegenerative diseases is enormous and continues to grow (Dorsey et al., 2013; Kowal et al., 2013; Wimo and Prince, 2010).
It is increasingly recognized that to tackle this looming crisis, we need better tools, including tools for the early recognition and precise measurement of these diseases (Marek et al., 2008; Mueller et al., 2005; Perrin et al., 2009; Sherer, 2011). Thus, several large efforts to develop biomarkers -- objective, proxy indicators of pathophysiological state or therapeutic response -- have been recently launched in AD (Weiner et al., 2013) and in PD (Marek et al., 2011).
Launching an effort neither dictates the methodology nor ensures success, however, and a high-throughput, unbiased screening approach may be needed to successfully find and develop biomarkers for neurodegenerative diseases. To provide concrete examples that may illustrate more broadly-applicable ideas, this review will focus on the development of PD biomarkers. Specifically, I will point out areas of need for PD biomarkers, discuss existing biomarkers in PD, and make a case for an unbiased screening approach to the development of new biomarkers. I will then discuss various methods that could be applied in this type of approach, highlighting successes in other fields and evidence for their potential in PD. Finally, I will suggest concrete measures that may accelerate the pace of biomarker discovery in PD and beyond.
Parkinson’s Disease
Parkinson’s disease (PD) is a progressive neurodegenerative disease first described clinically by James Parkinson nearly 200 years ago (Parkinson, 1817). The defining motor features of PD -- bradykinesia accompanied by various other features such as resting tremor, hypertonia, or postural instability -- cause considerable morbidity (Hughes 1992). In addition, both the personal and societal tolls of cognitive impairment and dementia due to PD have been increasingly recognized (Pressley et al., 2003). Indeed, over 80% of patients with longstanding PD will develop dementia (Buter et al., 2008; Hely et al., 2008; Mayeux et al., 1992). Altogether, the United States national economic burden of PD is estimated to have exceeded $14 billion in 2010 (Kowal et al., 2013).
Approximately one hundred years after the first clinical description of PD, a characteristic cytoplasmic eosinophilic inclusion body was demonstrated in neuropathological studies of PD patient brains by Frederick Lewy (Lewy, 1912), and this pathognomonic inclusion body subsequently came to bear his name (Trétiakoff, 1919). In the 1990’s, Lewy bodies were reported to consist largely of the protein alpha-synuclein (Spillantini et al., 1997), and pathological forms of this protein are now strongly implicated in the development of PD (Desplats et al., 2009; Luk et al., 2012; Polymeropoulos et al., 1997; Singleton et al., 2003). While some Mendelian genetic causes, as well as some common genetic variant risk factors, for PD are known (reviewed in Trinh and Farrer, 2013), for the most part, PD remains a sporadic, idiopathic disease, diagnosed during life on clinical grounds.
At present, the gold standard for PD diagnosis is the neuropathological finding of dopaminergic neurodegeneration accompanied by the presence of alpha-synuclein-containing Lewy bodies (Dickson et al., 2009). However, clinical diagnosis during life agrees with neuropathological diagnosis at autopsy only 70–80% of the time (Hughes et al., 1992). In PD, like in AD, amyotrophic lateral sclerosis, frontotemporal dementia, and other neurodegenerative diseases, no disease-modifying therapies are available, despite nearly two decades of failed trials (Olanow et al., 2008; Rascol et al., 2011a).
The intractability of PD to attempts at disease-modifying therapy is likely multifactorial. One factor, though, that extends to our current conception of all the adult-onset neurodegenerative diseases, may be the advanced stage of pathophysiology at the time of clinical diagnosis (Berg et al., 2014). Specifically, it is estimated that at the time of clinical PD diagnosis, ~50% of substantia nigra dopaminergic neurons may already be lost (Fearnley and Lees, 1991). Moreover, in recent years, a number of prodromal features for PD have been recognized – two that have received much attention are hyposmia (impairment in one’s sense of smell) and REM behavior disorder (inability to suppress movements during dreaming) (Berg et al., 2014). For example, individuals suffering from hyposmia may have a fivefold increased risk of developing PD (Ross et al., 2008), and ~40% of RBD patients may develop PD or related neurodegenerative diseases over 10 years (Postuma et al., 2009; Schenck et al., 1996). With the advent of these data has come the recognition that there is a prodrome indicative of onset of a pathophysiological cascade of events, and that this prodrome may predate formal diagnosis of a neurodegenerative disease by years or even decades (Berg et al., 2014; Braak and Del Tredici, 2008).
Biomarkers in neurodegenerative conditions: Definitions and needs
As defined by the Biomarkers Definitions Working Group convened by the National Institutes of Health (NIH), a biomarker is “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” (Biomarkers Definitions Working Group., 2001). A key point in this definition includes the emphasis on objective measurement – this stands in contrast to the clinical context, in which many aspects of assessment may be to various extents subjective. An additional inherent assumption is that the surrogate biomarker indicator will be relatively easy to measure compared to the biological or pathogenic process itself.
In PD, both the objectivity and the ease inherent in biomarkers are attractive features. PD is a brain-based disease diagnosed and followed primarily on clinical grounds, with significant day-to-day and even hour-to-hour fluctuations in clinical presentation. As a consequence, PD presents challenges in both the establishment of diagnosis and the assessment of disease severity that would benefit from objective corroborative data.
As previously mentioned, neuropathological diagnosis is presently the gold standard for the determination of PD diagnosis (Dickson et al., 2009). However, for obvious reasons, in actual practice the diagnosis is made on clinical grounds. Clinical diagnosis is approximately 80% accurate in patients followed longitudinally with moderate symptoms (Hughes et al., 1992). In best-case scenarios, where the diagnosis is made by movement disorders specialists applying strict criteria, the accuracy may rise to 90% (Hughes et al., 2001). However, this accuracy may also fall substantially, to approximately 65%, in earlier stages of PD (Rajput et al., 1991). Because in PD there likely exists a long prodromal phase in which pathophysiological events are already in motion, a situation arises in which it is precisely in those patients in whom clinical diagnosis is difficult that there exists the greatest opportunity for therapeutic intervention.
A PD diagnostic biomarker could be used to corroborate or confirm clinical diagnosis. In addition, in the case of very robust markers, diagnostic biomarkers could be used to screen individuals for enrollment in clinical trials. Notably, such a diagnostic-biomarker-screened approach to clinical trial enrollment has recently been pioneered for clinical trials in AD (Kozauer and Katz, 2013), using two proteins – tau and amyloid beta – measured in cerebrospinal fluid (CSF). A PD diagnostic biomarker that could be used in the earliest stages of disease would be particularly valuable.
Aside from biomarkers that could classify patients easily into PD versus other diagnostic groups, biomarkers providing an objective measurement for the assessment of PD severity would also be valuable in the clinical care of existing PD patients. These biomarkers of disease severity might prove particularly useful in a clinical trial context, even serving as potential surrogate endpoints. To date, PD clinical trials have relied on clinical endpoints such as timing of the need to start levodopa (e.g. Parkinson Study Group PRECEPT Investigators, 2007) or change in the clinical Unified Parkinson’s Disease Rating Scale (e.g. Rascol et al., 2011b) to determine efficacy. After two decades of largely unsuccessful clinical trials (Olanow et al., 2008; Rascol et al., 2011a), it may be worth re-examining not just the therapeutic mechanisms that have been targeted, but also the ways that efficacy has been measured. That is, without fine-scaled, precise measures of disease severity, subtler benefits may have escaped detection. This biomarker goal is admittedly ambitious, and experience to date in AD has proved disappointing, but the ramifications of discovering and validating such a surrogate endpoint biomarker in any of the neurodegenerative diseases would be profound (Greenberg et al., 2013).
A third area in which biomarkers may be of particular utility in PD is in prognostication for various motor and non-motor outcomes. A frequent question from the newly-diagnosed PD patient is one about prognosis and expected disease course. Unfortunately, while population-level data suggest that certain demographic features (e.g. older age, associated comorbidities) may predict a more rapid rate of progression (Suchowersky et al., 2006), or certain motor phenotypes (e.g. lack of tremor) may associate with faster rates of decline (Marras et al., 2002), these data are not particularly informative for prognostic purposes on an individual scale. Prognostic biomarkers – analogous to a measure such as cholesterol level in assessing risk for cardiovascular events, or tumor estrogen receptor status in assessing prognosis in breast cancer – would therefore also address a significant unmet need in PD.
Biomarker discussions often separate markers into two conceptual categories, biomarkers of state and trait. Biomarkers of state are envisioned as indicators of current disease presence and severity, and biomarkers of trait as indicators of risk for disease or potential for various future outcomes. The first two classes of PD biomarkers discussed above – diagnostic biomarkers and biomarkers of disease severity – might fall into the “state” category, while prognostic biomarkers might fall into the “trait” category. That said, as we understand more about pathophysiology in PD, the lines begin to blur. For example, how might one classify a marker that accurately identifies normal-appearing individuals who, if they were to live to the age of 85, would develop clinically manifest PD 90% of the time? It would depend, presumably, on what “state” one is interested in – manifest PD or the presence of occult PD pathophysiology. – Despite defiance of easy classification, though, development of precisely such a pre-symptomatic diagnostic biomarker might be one of the most pressing needs in PD biomarker research. Indeed, at a meeting convened by the NIH-National Institute of Neurological Disorders and Stroke (NIH-NINDS) in January 2014 to set priorities for PD research, the development of means to identify prodromal PD subjects was the highest priority clinical recommendation (NIH-NINDS, 2014).
In summary, there is a significant need for PD biomarkers – objective, relatively easily measured indicators of PD pathogenic processes. Three specific scenarios in which biomarkers would be especially useful are: (1) establishing early and/or pre-symptomatic diagnosis, (2) following disease progression, and (3) assessing motor and non-motor prognosis.
National and international efforts to develop PD biomarkers
The recognition of a significant need for PD biomarkers has been accompanied by the development of important multi-site cohorts and biorepositories to aid in their development. Two important efforts with complementary roles are the Parkinson’s Disease Biomarker Program (PDBP) launched by the National Institute for Neurological Disorders and Stroke (NINDS) in late 2012, and the Parkinson’s Progression Marker Initative (PPMI) launched by the Michael J. Fox Foundation in 2010. Both efforts will follow their cohorts longitudinally for at least five years, with serial visits in which clinical data and biosamples are obtained.
The PDBP is an NINDS effort aimed at novel biomarker discovery. To accomplish this objective, the PDBP has three main arms: (1) support of research efforts to develop PD biomarkers, (2) creation of a biorepository of PD and control samples for use in early biomarker discovery efforts, and (3) maintenance of an easily-accessible database for the housing and sharing of sample data (Parkinson’s Disease Biomarkers Program, 2014). As of August 2014, >900 samples of DNA; >600 baseline samples of plasma, serum, and RNA; and >200 baseline samples of CSF have been collected and are available in the biorepository. For the biofluids and RNA, subsequent follow-up samples are available for some individuals as well. Clinical data with an emphasis on Common Data Elements (NINDS Common Data Elements, 2014) for these individuals is housed in the corresponding database, known as the Data Management Resource (The Lancet Neurology, 2013). Data and samples are available, with the former nearly universally accessible, and the latter also highly accessible through a specimen request process. The over-arching goal of the PDBP is to facilitate early-stage, high-risk PD biomarker discovery efforts.
Like the PDBP, PPMI also aims to facilitate the development of PD biomarkers (Parkinson’s Progression Markers Initiative, 2014; Marek et al., 2011). However, three important distinctions give these two efforts complementary, rather than overlapping, roles. First, for inclusion in the PPMI cohort, PD patients (400 PD subjects, to accompany 200 normal controls, with all 600 individuals already enrolled) must not be on PD medication at the time of enrollment; since the PDBP PD subjects do not have this restriction, PPMI subjects tend to be at an earlier stage of PD than PDBP. Second, PPMI subjects are recruited from 32 clinical sites in 12 countries; in contrast, PDBP subjects are recruited primarily from 5 clinical sites in the United States. Thus, PPMI is a more heterogeneous cohort than PDBP. Finally, the over-arching goal of PPMI is to serve as a replication cohort for PD biomarkers discovered in other cohorts, such as those recruited from individual academic centers or earlier-stage efforts such as the PDBP.
Because both PDBP and PPMI are new efforts, it remains to be seen how the biorepositories will be used. In keeping with the confirmatory goal of PPMI, pilot projects using PPMI samples have focused primarily on the replication of several previously-reported biomarker candidates (e.g. CSF tau and amyloid beta, Kang et al., 2013). Within the biomarker research discovery arm of PDBP, projects evaluating candidate PD biomarkers – such as specific magnetic resonance imaging (MRI) measures – exist alongside unbiased screening efforts to find new biomarkers with techniques such as bottom-up shotgun mass-spectrometry, RNA sequencing, and screening via an aptamer-based platform (Parkinson’s Disease Biomarkers Program, 2014). An overview of these various unbiased screening techniques will be provided in a later section of this review. One anticipates that novel biomarkers emerging from these discovery efforts – as well as early discovery efforts proposed from outside the PDBP consortium using other discovery biorepositories such as the BioFIND cohort (BioFIND Clinical Study, 2014) – will make use of the PDBP and PPMI biorepository samples, respectively, in a pipeline from early testing to international replication.
A review of existing biomarkers in PD
A need for biomarkers exists in PD, and large-scale biorepositories have been developed to facilitate their development. What, then, is the status of existing PD biomarkers?
Recent, comprehensive reviews of this topic exist (Brooks, 2010; Chahine et al., 2014; Henchcliffe et al., 2011; Magdalinou et al., 2014; Mollenhauer et al., 2014; Parnetti et al., 2013; Sherer, 2011; Svenningsson et al., 2012; Wang et al., 2013), so this review will focus only on those biomarkers that have been the most extensively studied in large cohorts and/or by multiple research groups. These biomarkers may be grouped into (1) biochemical biomarkers, (2) biomarkers based on brain imaging, and (3) biomarkers based on other modalities. While a complete review of biomarkers in other neurodegenerative diseases is beyond the scope of this article, I note that this general biomarker categorization scheme may be extended to all the neurodegenerative diseases. I will also discuss one mature biochemical biomarker and one mature imaging-based biomarker that are in use in AD, as they may serve as benchmarks for where biomarkers have progressed within neurodegenerative diseases as a whole.
Biochemical biomarkers in PD
Biochemical biomarkers are proteins, metabolites, or other entities that can be quantitated in tissues or biofluids from PD patients. To date, PD biochemical biomarkers have been measured primarily in the CSF or blood of subjects.
Alpha-synuclein (aSyn), the main component of the characteristic Lewy body inclusions of PD, is one of the most-studied biochemical biomarkers in PD. Specifically, various species of the aSyn protein – monomeric vs. oligomeric vs. fibrillar forms, total vs. phosphorylated forms – have been evaluated in human CSF, blood, saliva, and other biofluids/tissues (Malek et al., 2014; Schmid et al., 2013). Of these, the most mature area of research lies in studies of CSF total aSyn.
Following an early demonstration that aSyn could be detected in human CSF (Borghi et al., 2000), CSF total aSyn levels have been studied by many different groups, primarily as a potential diagnostic biomarker in PD. While some groups have reported significant differences between PD patients and normal controls, with PD patients exhibiting lower levels (Hong et al., 2010; Mollenhauer et al., 2008; Tokuda et al., 2006), others have not observed this difference (Foulds et al., 2012; Park et al., 2011). It is possible that these differing findings could result from differences in assay sensitivity, cohort, or other factors. However, even in those groups reporting a difference, total CSF aSyn levels in PD patients and normal controls demonstrate considerable overlap (Mollenhauer et al., 2011; Shi et al., 2011), precluding practical use as a diagnostic biomarker. In addition, total CSF aSyn does not appear to distinguish PD patients from individuals with other neurodegenerative diseases (Foulds et al., 2012; Tateno et al., 2012). Finally, total CSF aSyn levels do not consistently correlate with PD severity (Foulds et al., 2012; Mollenhauer et al., 2011; Shi et al., 2011; Tokuda et al., 2006; Tokuda et al., 2010; Wang et al., 2012). Thus, despite considerable effort, evidence to date suggests that total CSF aSyn may have significant limitations as either a diagnostic biomarker in PD or as a biomarker for disease severity in PD (reviewed in Parnetti et al., 2013).
Less studied than CSF total aSyn but potentially promising are measures of oligomeric species of CSF aSyn. To date, three different groups have reported elevations in oligomeric CSF aSyn in PD relative to controls (Park et al., 2011; Sierks et al., 2011; Tokuda et al., 2010). At present, however, small samples sizes in all of these studies preclude definitive conclusions. Also promising is a report in a large cohort of subjects with PD and other neurodegenerative diseases that CSF phosphorylated aSyn may be elevated in PD (Wang et al., 2012), although this finding awaits confirmation by others.
In addition to alpha-synuclein, CSF amyloid beta and tau, and serum urate have received attention as potential PD biomarkers.
Amyloid beta – specifically, the disease-implicated form of amyloid beta known as Aβ42 – and tau – have emerged as robust CSF biomarkers for AD. In AD, CSF Aβ42 appears to decrease with increasing amyloid plaque burden in the brain (Fagan et al., 2007; Jack Jr and Holtzman, 2013; Mattsson et al., 2009; Shaw et al., 2009), while CSF total tau and CSF phosphorylated tau appear to increase with progressive neurodegeneration (Jack Jr and Holtzman, 2013; Shaw et al., 2009; Tapiola et al., 2009). Indeed, CSF Aβ42 and CSF tau as biomarkers for AD are arguably the current gold standard for biochemical biomarkers in any of the neurodegenerative diseases.
As coincident AD pathology is frequently found in the brains of PD patients (Irwin et al., 2013), and many questions about how one measures and interprets CSF Aβ42 and CSF tau have already been answered in AD, it is hardly surprising that the potential for CSF Aβ42 and tau levels as biomarkers in PD has been explored as well. While data on CSF tau as a biomarker for PD are conflicting (reviewed in Parnetti et al., 2013), decreased levels of CSF Aβ42 appear to characterize PD or particular subgroups of PD, as well as AD. That is, some authors have found CSF Aβ42 levels to be decreased in PD compared to controls (Alves et al., 2010; Shi et al., 2011; Zhang et al., 2008), while others have found CSF Aβ42 to be lower in PD patients with dementia vs. PD patients without dementia (Compta et al., 2009; Montine et al., 2010). In our cohort, we have found no differences in CSF Aβ42 levels comparing diagnostic groups; however, we have found decreased CSF Aβ42 in PD patients with faster cognitive decline (Siderowf et al., 2010). In all cases, observed effect sizes were small, with significant overlap among groups being compared. Taken together, these data suggest that CSF Aβ42 may be, at best, partially informative as a biomarker for cognition in PD.
Finally, serum and plasma levels of the purine metabolites urate and uric acid have been examined as biomarkers of PD risk. Beginning with an observation in the Honolulu Heart Study in 1996, increased levels of serum or plasma urate have been found to associate with decreased risk for the development of PD in several large epidemiological cohort studies (Davis et al., 1996; de Lau et al., 2005; Weisskopf et al., 2007). In addition, within PD, increased serum urate levels also correlate with slower rates of motor progression (Schwarzschild et al., 2008), with stronger effects in men.
In summary, CSF total aSyn levels, CSF Aβ42, CSF tau, and serum or plasma urate have been studied extensively as potential biochemical biomarkers in PD. Decreased CSF Aβ42 may associate with poorer cognition in PD, while decreased serum or plasma urate may associate with increased risk for PD. Data for CSF tau are negative or conflicting, and conflicting results also suggest that CSF total aSyn has limitations as a diagnostic or prognostic biomarker for PD. Because of the biological importance of aSyn in the pathophysiology of PD, however, CSF aSyn may still hold promise as a biomarker for target engagement, should aSyn-directed therapeutic strategies emerge. Modified forms of CSF aSyn may also have promise as biomarkers, although additional corroboration is needed.
Biomarkers for PD based on brain imaging
It is well established that the dopaminergic neurodegeneration central to PD results in the decreased binding of ligands by the dopamine transporter (DAT) protein, which is found on the presynaptic terminal of dopaminergic neurons (Uhl, 2003). This reduction in DAT binding may be imaged using various radiolabeled DAT ligands, captured by techniques such as positron emission tomography (PET) or single-photon emission computed tomography (SPECT) (Sherer, 2011). In 2011, a specific DAT imaging protocol – known as the DaTSCAN – was approved by the US FDA for confirmation of PD clinical diagnosis; the same protocol has been in clinical use in Europe since 2000 (Berardelli et al., 2013; Sherer, 2011). Thus, some have argued that dopaminergic system neuroimaging techniques such as DaTSCAN are a gold standard biomarker for PD diagnosis (Sherer, 2011). Indeed, DAT imaging is now a screening step in the enrollment of the large, international PPMI cohort (Marek et al., 2011). Moreover, DAT imaging is performed in two cohorts of asymptomatic subjects at increased risk for development of PD – the Parkinson’s Associated Risk Study (Siderowf et al., 2012), and the newly-enrolling prodromal arm of PPMI, sometimes referred to as Pre-PPMI (Parkinson’s Progression Markers Initiative, 2014). The goal of these investigations is to understand whether DAT imaging may be informative as a biomarker in pre-symptomatic stages of PD pathogenesis as well.
Despite these promising features, dopaminergic system imaging measures such as DAT imaging have limitations (Bajaj et al., 2013). First, because other diseases may also exhibit dopaminergic dysfunction, DAT imaging cannot definitively diagnose PD (Ravina et al., 2005). Second, various aspects of DAT imaging – the need for a radioligand, the variance associated with age and sex, the potential for interference from medications such as methylphenidate, modafinil, benztropine (Booij and Kemp, 2008; Cummings et al., 2011; Volkow et al., 2005), the inability to reliably discriminate PD from other parkinsonian syndromes (Cummings et al., 2011) – may in practice limit its use in clinical settings.
Aside from dopaminergic system imaging, other brain imaging-based techniques have been proposed as biomarkers for PD. These have been reviewed in detail elsewhere (Brooks, 2010; Sherer, 2011), but as one example, high-resolution diffusion-tensor imaging (DTI) has been reported to demonstrate decreased fractional anisotropy in the substantia nigra of early-stage PD patients compared to controls (Vaillancourt et al., 2009). Of note, this finding corroborated earlier reports of structural differences in the substantia nigra in PD, detected on less technologically advanced MRI platforms (Hutchinson and Raff, 2000). However, to date, most MRI-based studies have been performed in a small number of individuals (reviewed in Brooks, 2010), limiting ability to interpret and generalize findings.
In a look to the future, considerable effort by both industry-led and foundation-sponsored projects is currently underway to develop aSyn imaging tools in human subjects (Shah and Catafau, 2014). As a corollary, it is worth noting that radioactive tracers specific for amyloid beta have been developed and are in use in AD.
Biomarkers for PD based on other modalities
Aside from biochemical and brain-imaging based markers for PD, other strategies have been pursued in biomarker discovery and may yield promising future leads as well. While the purpose of this article is not to extensively review all existing candidate biomarkers in PD, a few examples may be useful, as similar approaches are being used across the neurodegenerative diseases. First, imaging has been performed outside of the brain as a potential biomarker in PD. Specifically, imaging of cardiac sympathetic innervation (Druschky et al., 2000) and imaging of the retina (Hajee et al., 2009) have been reported to demonstrate differences in PD patients compared to normal controls. Second, alternative modalities such as gait assessment are being evaluated (Lord et al., 2013). Finally, approaches examining tissues other than biofluids (Beach et al., 2010), such as biopsy of the submandibular gland (Adler et al., 2014) or colon (Lebouvier et al., 2010) for assessment for Lewy body pathology, are areas of active exploration for their potential as PD biomarkers.
Detection of amyloid beta by imaging or CSF analysis in AD
It would be remiss not to mention that in AD, the road from discovery of biochemical and imaging-based biomarkers to translation of these biomarkers to clinical research use has been walked before. Specifically, since the advent of CSF studies of amyloid beta species, along with the development over a decade ago of the Pittsburgh B (PiB) compound, which can be used in PET scans to detect amyloid beta (Klunk et al., 2004), the concept of detecting the core proteinopathy in neurodegenerative diseases by imaging or other modalities has gained substantial traction. Indeed, detection of high amyloid beta burden through imaging by PiB or other radioactive tracers has even been incorporated into 2011 recommended diagnostic criteria for AD and research recommendations for studies of preclinical AD (Albert et al., 2011; Jack Jr et al., 2011; McKhann et al., 2011; Sperling et al., 2011). As an alternative to the detection of elevated amyloid beta by imaging, thought leaders in AD have also recommended the use of CSF Aβ42 measures, which, as summarized previously, decrease as amyloid plaque burden increases in the brain (Molinuevo et al., 2014).
It is notable that the translation from research finding to diagnostic criteria for these biochemical and imaging-based amyloid beta biomarkers in AD took more than two decades. In the process of their discovery, replication, and validation, many important principles have emerged that can inform, and hopefully accelerate, the timeline for future biomarker development in AD, PD, and other neurodegenerative diseases. For example, the importance of controlling potential sources of pre-analytical variability to the best extent possible cannot be overstated, and best practices for how to do this have been put forward by multiple groups in the AD field (O’Bryant et al., 2014; Vanderstichele et al., 2012). Second, the development of a few robust tests (e.g. PiB imaging, Alzbio3 biochemical biomarker assay) has been key to understanding whether multiple groups see the same thing, assigning fixed “cutoff” values, etc. (Kang et al., 2012). Third, the widespread adoption of a specific set of biomarkers, along with the same robust tests for ascertaining their values, has been instrumental in the translation of amyloid beta imaging and CSF Aβ42 measures into clinical research use (Kang et al., 2013).
Summary of existing PD biomarkers
Potential biochemical biomarkers, biomarkers based on brain imaging, and biomarkers based on other modalities have been reported in PD. However, with the exception of dopaminergic system imaging (primarily DAT imaging), and a handful of biochemical markers (CSF total aSyn, CSF Aβ42, CSF tau, and serum or plasma urate) most potential biomarkers have yet to be assessed in large cohorts, or replicated in multiple groups.
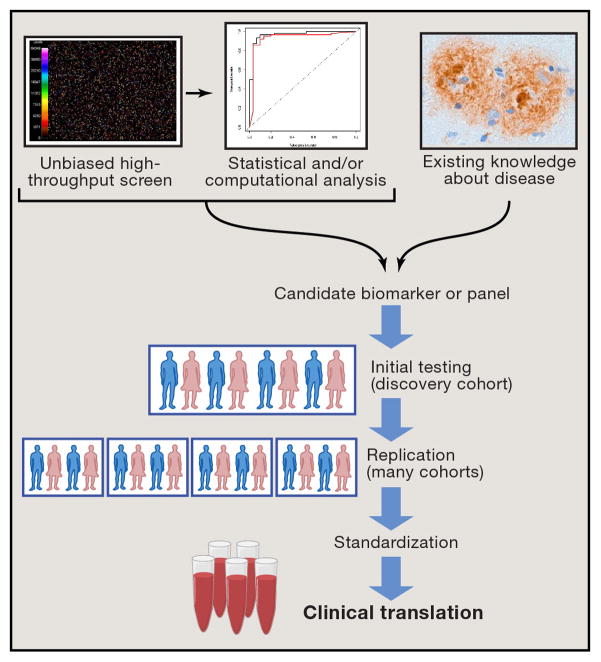
If we return to the pressing needs for biomarkers discussed in the previous section and ask which of these needs are adequately met by existing, extensively studied biomarkers, it becomes clear that we must continue to find and develop new biomarkers in PD (see Table 1). Two different strategies exist for accomplishing this goal (see Figure 1).
Table 1.
Areas of need for biomarkers in Parkinson’s disease are presented along with existing potential biomarkers.
| Need | Potential Marker | Ease of obtaining measure | Ease of interpreting result |
|---|---|---|---|
| Confirming PD diagnosis | DAT imaging | Difficult | Easy |
| Diagnosing pre-symptomatic PD | None | ||
| Following disease progression | None | ||
| Serving as a surrogate endpoint | None | ||
| Assessing motor prognosis | Serum urate | Easy | Moderately difficult |
| Assessing non-motor prognosis | CSF Aβ42 | Moderately difficult | Difficult |
Figure 1.
Strategies for biomarker development
Candidate vs. unbiased screening approaches
The existing biomarkers for PD discussed in the previous section were found by a candidate marker approach. That is, based on existing knowledge about the pathophysiology of PD or other neurodegenerative diseases, specific proteins, imaging modalities, or other readouts were tested for their association with PD. This approach can be quite successful, leading to the discovery and development of CSF tau and Aβ42 as confirmatory diagnostic biomarkers in AD, for example. However, in the case of PD as well as all the neurodegenerative diseases, the plausible list of candidates is nearly exhausted, with large gaps remaining in our biomarker armamentarium.
An alternative approach is to screen many possible biomarkers simultaneously without a priori assumptions about their potential for conveying important biological information. Such an unbiased screening approach requires the ability to perform massively parallel screening of hundreds, thousands, or more potential markers -- a technological hurdle that has only been surmountable in recent years. In addition, such an approach requires a substantial understanding of bioinformatics to interpret the large datasets generated. However, this type of approach has been extremely successful in other arenas (e.g. the discovery of genetic risk factors for disease). Moreover, there is also evidence for early success in the arena of PD biomarker discovery.
Techniques for unbiased screening
Permissive technology has been key to the emergence of unbiased screening approaches in many areas of science. In the area of biomarker development, these technologies have been employed in the discovery of protein biomarkers as well as non-protein biomarkers.
Unbiased screening techniques for protein biomarkers
Perhaps the most prototypical biomarker is a protein that can be measured in an easily-accessible biofluid such as blood, CSF, or urine. Proteins are attractive from a laboratory testing perspective because they can be measured by a number of techniques, and they are relatively stable and abundant.
When one considers the techniques that are available to measure many proteins, or parts of proteins, at once, there are two major types: mass spectrometry-based methods and antibody-based methods (Solier and Langen, 2014). In addition, an emerging technology has also been employed by several groups in the biomarker-development field.
Mass spectrometry (MS) is an analytical chemistry technique based on the ionization of a sample being studied. As a consequence of ionization, molecules within the sample are broken up and can be separated according to mass and charge. From the resulting pattern, information can be gained to identify the chemical nature of the sample tested. MS has been used in a number of ways for potential biomarker discovery, including matrix-assisted laser desorption/ionization, followed by time-of-flight MS (MALDI-TOF); liquid chromatography, followed by tandem MS (LC-MS/MS); and two-dimensional gel-based techniques. The specifics of each technique have been reviewed elsewhere (Duncan and Hunsucker, 2005). From the perspective of unbiased biomarker development, the important advantage is that because in each case the procedure is applied to the whole sample, and data are collected on the whole sample, MS-based techniques are by definition unbiased. That said, in practice, MS-based techniques do have limitations (reviewed in Boja and Rodriguez, 2012). First, a highly abundant protein/peptide in the sample of origin may dominate the data gathered, thus limiting the ability to find differentiating biomarkers that are present in lower amounts. Second, most MS-based techniques require stringency in sample preparation, and all MS-based techniques face significant challenges in terms of conversion of raw mass spectral data into interpretable readouts. Third, with a few notable exceptions in microbiology (Seng et al., 2009), drug monitoring (Vogeser and Seger, 2008) and newborn screening for inborn errors of metabolism (la Marca, 2014), MS-based techniques have seen little translation into the clinical realm, despite the fact that they have been available for decades.
In contrast, since the advent of an immunoassay for insulin in 1959 (Yalow and Berson, 1959), antibody-based methods have made up the vast majority of protein tests currently used in clinical laboratory testing. These types of assays rely on the recognition of a specific protein epitope -- or antigen -- by an antibody. Variations exist in terms of how each partner -- antigen and antibody -- are presented, and specific techniques have been reviewed elsewhere (Ellington et al., 2010; Solier and Langen, 2014). From the perspective of biomarker development, antibody-based methods, as a whole, offer as their major advantage ease of use and clinical translation. The primary limitation of antibody-based methods for unbiased screening, however, is a practical ceiling on the number of proteins one could reasonably assess simultaneously. These limitations arise both because of the reliance on the ability of a peptide/protein to trigger an immune response (and therefore antibody formation) and because of interference effects from multiplexing a large number of protein assays (reviewed in Ellington et al., 2010). At present, within the realm of antibody-based methods, bead-based multiplex immunoassay formats are frequently used to screen a large number of proteins from a single sample (Schwenk et al., 2007). We (Chen-Plotkin et al., 2011; Qiang et al., 2013) and others (Gurbel et al., 2008) have found, however, that this ceiling in protein number hovers at approximately 100 measured out of the >250,000 estimated proteins (What is Proteomics? 2014) in the human proteome.
MS-based methods offer the potential for massive coverage of the proteome at the expense of ease of use and clinical translation, while antibody-based methods offer the potential for ease of use at the expense of adequate proteomic coverage. In an era of technological development, though, it is fitting to end this section with a look at one emerging platform that may offer an “in-between” solution for unbiased protein screening.
In nature, antibodies are not the only entities that can specifically recognize proteins -- many proteins such as transcription factors and splicing factors bind specifically to stretches of nucleic acid, offering the possibility that this type of protein-nucleic acid interaction could be exploited to develop a highly-multiplexed protein screening panel. Thus, in 1990, the selection of oligonucleotides targeting specific proteins -- called aptamers -- was first described (Ellington and Szostak, 1990; Tuerk and Gold, 1990). In the intervening decades, aptamers have been used in a number of research contexts, and in the last few years, a modified aptamer-based platform for screening >1000 proteins from a small sample has entered commercial use (Lollo et al., 2014), developed by the company Somalogic. While it remains to be seen whether this method will produce consistent results across time and different sites, early efforts (Baird et al., 2012; Mehan et al., 2014) have piloted the use of this aptamer-based platform for biomarker discovery.
Unbiased screening for non-protein biomarkers
While prototypical, proteins are by no means the only potential type of biomarkers one could identify by unbiased screening. In recent years, efforts have emerged to develop biomarkers for neurodegenerative disease by metabolomic, lipidomic, and mRNA/microRNA expression profiling.
Metabolomics refers to the systematic detection and quantitation of a large number of low-molecular weight products (often defined as <1kD) of metabolic processes in living organisms (Goodacre et al., 2004; Holmes et al., 2008b). Often, the detection and quantitation methods used in metabolomics are based on MS or nuclear magnetic resonance (NMR) techniques, although other methods have been applied as well (reviewed in Holmes et al., 2008b). Technical limitations depend on the specific metabolomic methodology applied, but in general, limitations mirror those of MS-based techniques – in particular, the difficulty in converting raw data into interpretable, robust readouts. A subfield of metabolomics which has seen considerable activity is the field of lipidomics, which refers to the systemic detection and quantitation of those small-molecular metabolites which are lipids (Han and Gross, 2005).
Because metabolic processes are dynamic, metabolomics has the potential to detect differences in groups that emanate from a physiologic or pathophysiologic process directly. Thus, metabolomic approaches have been used successfully to delineate the metabolic consequences of specific in vivo perturbations, such as gene alterations (Raamsdonk et al., 2001) or dietary differences (Holmes et al., 2008a). For some human illnesses in which the primary pathophysiologic process is known (or, in fact, defining of the condition), the resulting metabolomic signature of this process has been proposed as a biomarker. For example, in cardiovascular disease, a characteristic metabolomic profile detectable in serum has been reported to distinguish individuals with vs. without coronary artery disease with high accuracy (Brindle et al., 2002).
In the area of neurodegenerative disease, metabolomic approaches have been applied to the development of biomarkers for PD. For instance, a liquid chromatography electrocoulometric array approach has been described which can completely separate the plasma metabolite profiles of PD vs. normal controls by a partial-least-square discriminant analysis approach (Bogdanov et al., 2008), although it is probable that for clinical translation to occur, significant simplification of both the methodology and the data analysis stream will be needed.
In addition to small-molecule products of metabolic processes, mRNAs or microRNAs (miRNAs, small non-coding RNAs often serving as master regulators of many genes) are other moieties that may be suitable for large-scale unbiased screening for biomarker development. Global profiling of these two potential RNA-based biomarker classes is aided by the development of many array-based and, more recently, high-throughput sequencing-based methods for fast and accurate RNA quantitation. Robust tools for the rapid acquisition and preservation of RNA from human subjects are also available, thus making many of the technical hurdles associated with biomarker development potentially surmountable for RNA-based biomarkers (Bartels and Tsongalis, 2009). It remains to be seen, however, whether robust and reproducible signals can be found within this class of potential biomarkers (Nair et al., 2014).
Successful applications of an unbiased screening approach
Success in other fields
To establish successful leads by unbiased screening is not dissimilar to finding the proverbial needle (or several needles) in a haystack. Thus, for an unbiased screening approach to succeed, several conditions must be met. First, potentially useful biomarkers must be present within the pool of candidates screened (i.e. one must be looking in the right haystack). Second, rapid and reliable methods must be available for screening (i.e. one must be able to look at all the straws in the haystack quickly). In several areas outside of neurodegenerative biomarker development, the confluence of these conditions has already led to prominent successes.
The most obvious area in which an unbiased screening approach has to date been successful has been in the discovery of genetic risk factors for various traits, including human diseases. While various combinations of screening method and choice of “haystack” have been tried, the dominant mode of investigation is to screen hundreds of thousands of single nucleotide polymorphisms (SNPs) for association with the trait in question, through the use of large arrays. This type of study, the prototypical genomewide association study (GWAS), has yielded thousands of genetic loci associated with thousands of traits/diseases (Welter et al., 2014), since its first successful application in 2005 (Edwards et al., 2005; Haines et al., 2005; Klein et al., 2005). Notably, the results of the first GWAS, associating genotypes at the complement factor H gene with the eye neurodegenerative disease age-related macular degeneration, have since led to pharmacogenetic insights into therapeutic drug dosing in this disease. Some may argue that gains such as these afforded by GWAS are modest (Goldstein, 2009), and in almost all cases the difficult work of translating scientific insights gained by GWAS into tangible benefit to disease therapy still remains. However, the gains to our understanding of genetic trait architecture afforded by the advent of GWAS are clear, and there is also little question that many genetic loci now associated by GWAS with various diseases could not have been found by a candidate approach (Stranger et al., 2011).
A second area in which unbiased screening has led to many discoveries, some with material biomedical benefits already, is in the area of cancer biomarkers. While this area has been reviewed elsewhere (Sawyers, 2008; Tian et al., 2012), one prominent example may illustrate the contribution of biomarkers discovered by unbiased screening to the clinical diagnosis and treatment of breast cancer.
In 2000, investigators used gene expression profiling of just over 8000 genes to derive four main subtypes of breast cancer based on their molecular signature (Perou et al., 2000). This classification scheme has been largely upheld by the advent of more sophisticated tools for molecular profiling (Cancer Genome Atlas Network, 2012); moreover, molecular-signature-based breast cancer classification has also been used for prognostic purposes (van’t Veer et al., 2002). From the viewpoint of clinical translation, the concept of molecular profiling of breast cancer tumors by array-based methods subsequently led to the development of small panels of genes used for prognostic purposes (Paik et al., 2004). Some of these small-panel platforms have been commercialized (Ross et al., 2008), with the OncotypeDx platform, available since 2004, as a prototype that has been shown to influence treatment decisions (Carlson and Roth, 2013; Partin and Mamounas, 2011) by estimating an individual patient’s risk of recurrence.
Early efforts in neurodegenerative disease biomarker development through unbiased discovery approaches
While the field is still in its infancy, early efforts to discover and develop biomarkers through unbiased screening have been reported in both AD and PD, with mixed results. Several high-profile reports have failed to replicate, leading to a skepticism that is probably healthy for the field; however, several PD biomarker candidates nominated by unbiased screening have been successfully replicated. I highlight here a few examples that may, depending on their outcome, serve as cautionary tales or as indicators of better days to come.
In AD, an early effort to develop biomarkers used a screen of 120 proteins from a multiplex immunoassay panel to nominate 18 plasma proteins for the differentiation of AD plasma samples from those of normal controls (Ray et al., 2007). An attempt to commercialize this protein panel was begun, but subsequent efforts by other groups to replicate these results were unsuccessful (Björkqvist et al., 2012).
More recently, a lipidomic approach has been applied to the development of biomarkers for Alzheimer’s disease (AD), with a panel of ten plasma lipids reported to differentiate individuals who would convert to AD or amnestic mild cognitive impairment (MCI) from normal, non-converting controls (Mapstone et al., 2014). It remains to be seen whether this finding can be replicated by the originating group or other groups.
In PD, a blood-based mRNA expression profiling approach has been used to develop an eight-gene panel to differentiate PD from normal controls (Scherzer et al., 2007). Despite excellent performance in the training set used for development of the panel, classification accuracy in a separate test set demonstrated considerably poorer performance (Scherzer et al., 2007). Subsequent efforts to differentiate PD from normal controls based on mRNA expression profiling have been reported using various tissues (Diao et al., 2012; Molochnikov et al., 2012), with one blood-based mRNA profiling paper describing a five-gene diagnostic panel that overlapped with the prior eight-gene panel by one gene (Molochnikov et al., 2012), and one analysis of three different mRNA profiling datasets (from human PD brain, human PD blood, and rotenone-treated immortalized cells) finding exactly one gene (SRRM2) differentially expressed in all three datasets (Shehadeh et al., 2010). While it is encouraging to see some degree of overlap in results from multiple mRNA expression profiling efforts to develop PD biomarkers, a more robust overlap and overt replication would be needed to really move forward from a practical standpoint. Indeed, it is very difficult to establish whether there was substantial replication of findings from the initial study by subsequent studies, possibly because of methodological differences.
Over the last few years, our group has worked to develop biomarkers for PD through unbiased screening. To optimize potential for downstream clinical translation, we have screened proteins in the blood plasma. In 2010, we reported the results of a ~100-protein multiplex immunoassay screen for plasma proteins correlating with cognition in PD. We found that plasma epidermal growth factor (EGF) levels correlated with cognitive performance in PD, with lower levels seen in PD patients with poorer cognition cross-sectionally and lower levels also appearing to predict future cognitive decline in a small, longitudinally-followed PD cohort (Chen-Plotkin et al., 2011). Both the cross-sectional and predictive findings of our study were subsequently replicated by an independent research group, investigating an early, unmedicated PD cohort (Pellecchia et al., 2013). More recently, we have evaluated the potential for plasma EGF to serve as a biomarker for cognition in PD in the international PPMI cohort. Unfortunately, EGF levels were strongly correlated with clinical site of origin, suggesting that sensitivity to sample handling differences may pose a significant technical hurdle to the widespread use of EGF as a biomarker (unpublished observations).
In contrast, we have also reported the results of a ~100-protein multiplex immunoassay screen for plasma proteins associating with age at PD onset. Plasma ApoA1 emerged as a top candidate biomarker, with higher ApoA1 levels correlating with older age at PD onset, less severe motor symptoms in PD, and increased DAT putaminal binding in asymptomatic individuals at high risk for PD (Qiang et al., 2013), all suggesting a protective effect. Subsequent to our initial report, we have demonstrated that the association between higher ApoA1 plasma levels and older age at PD onset is independent of the platform used for measurement of ApoA1 and replicates in multiple cohorts (Qiang et al., 2013; Swanson et al., 2014), including the international PPMI cohort.
It is notable that whereas CSF Aβ42 was developed through a candidate marker approach over decades, ApoA1 emerged from an unbiased screen with less than 2 years’ time from initial report to successful replication across platforms and cohorts. The speed of this progress may illustrate both the potential of an unbiased approach for the discovery of PD biomarkers and the importance of emerging national and international cohorts for sample sharing.
Unbiased screening: Promise and pitfalls
Why have so many unbiased screening efforts to develop biomarkers failed to replicate, and what pitfalls may we try to avoid in the future? By examining certain features that differentiate the successes from the failures, several recommendations emerge for charting a path forwards.
First and foremost, there is need for replication, early and often. This is true in the development of any biomarker, but it is even more important if the biomarker emerges from unbiased screening, because there is no a priori reason to know or suspect that a particular candidate is better than any other candidate. Thus, to avoid many false positives and wasted effort, the field should hold as a standard for publication that biomarkers need to be replicated in an independent group of samples from the ones in which they were discovered. This has become a de facto standard in most genetics publications.
In most cases, the initial replication, if contained within the first report as suggested above, will be performed by the investigator who discovers the biomarker, often in separate samples from the same clinical site. However, it will be important for moving a biomarker forwards in a translational pipeline to see that it replicates in samples from other clinical sites and ideally, in multi-site cohorts like PDBP and PPMI.
Two pragmatic points arise here. First, the large and complex datasets arising from unbiased screening approaches lend themselves to many different analytical approaches, sometimes with quite different opinions on which analytical approaches best get at the biology within the dataset. Some standardization of bioinformatics analysis will likely be needed, although it may come at the expense of innovation in analysis. Because the field is still very young, this stifling of innovation is undesirable at present. Instead, it should be incumbent on replication efforts, even if reporting other analytical methods and results, to at least establish whether their result substantially corroborates prior reports. Moreover, the field as a whole would be greatly aided by deposition of data in a public biomarkers database allowing for meta-analyses of multiple datasets, as is routine at this point for mRNA and genetic studies. Second, from a scientific cultural standpoint, it is vastly easier to publish a discovery effort than a replication effort. Thus, incentives are strongly weighted towards discovery and novelty, making it even more difficult to evaluate whether similar scientific experiments yield largely similar results. In the past, this problem was partially addressed by the fact that further development of tests and therapeutics from initial academic discovery settings would be carried forward primarily in the industrial sector, with less of an emphasis on the need for publication than in academia. In this regard, it is probable that some of the reason for the historical “valley of death” between promising academic research leads and eventual clinical translation has to do with the emphasis on novelty, at the expense of replication, in academia vs. the emphasis on deliverables in industry. As more of this translational work may be moving into academic partnerships (Institute of Medicine (US) Forum on Neuroscience and Nervous System Disorders, 2008), however, we need to consider how to create new incentive structures for the painstaking work of replication, if academic research labs are actually to have a role in the development of usable biomarkers.
Because unbiased screening efforts need to start with a great number of candidates (hundreds, thousands, or more) screened, a second recommendation is that a high level of statistical rigor be applied to screening results from the outset. This is most likely to lead to true positives. In essence, this has been one of the major lessons from a decade of GWAS efforts.
A third recommendation is that, while novel technical platforms may be essential to the ability to screen many candidates at once, it is important to quickly establish whether technical aspects of the biomarker assay and the biomarker itself are robust. With respect to assay development, the careful and laborious efforts that have resulted in the establishment of robust assays for CSF tau and CSF Aβ42 are instructive (reviewed in Hampel et al., 2010). In addition, the development of a small biomarker panel of mRNAs for breast cancer prognostication was predicated on the establishment of robust methods for measuring mRNAs (both by microarray and by quantitative PCR) over decades.
Fourth, the CSF tau/Aβ42 assay required partnership between researchers in academia and industry to eventually translate into clinical and clinical research use (Hampel et al., 2010). It is likely that this will also be the case for successful biomarker development by candidate or unbiased methods in the future as well. The public-private structure of efforts such as PPMI and the Alzheimer’s Disease Neuroimaging Initiative (Alzheimer’s Disease Neuroimaging Initiative, 2013), with resultant intermingling of researchers from multiple different sectors, may thus be a major asset.
Fifth, another kind of partnership will be needed to most efficiently exploit an unbiased screening approach. Specifically, unbiased screening efforts usually lead to large, and sometimes massive, datasets that require computational analysis and statistical expertise. However, leads that emerge from these unbiased screens need to be empirically tested in more samples, on alternate technical platforms, to assess their reliability. In the most effective instances, there should be an open dialogue between those analyzing the big datasets and those testing leads empirically, to delineate the best set of analysis assumptions and to strike the best balance between statistical certainty and real-world requirements (e.g. that a potential biomarker be robust to handling variability, or that a potential biomarker be tractable with existing tools). As a consequence, this type of approach to biomarker development will require “dry” expertise in computational and statistical methods, “wet” expertise in bench-based techniques, and clinical expertise to access and select human samples for testing. Traditionally, these types of expertise have been gained through different types of training. Yet recognition of a need to cross these boundaries has been in place for nearly a decade (Zerhouni, 2005). In the case of unbiased biomarker development, we need effective and close partnerships between different types of experts, or we need to train hybrid experts.
Conclusions
Neurodegenerative diseases affect the brain, an organ that is impractical to sample, and existing clinical measures are subjective approximations of disease severity only. Thus, the development of biomarkers – objective proxy measures of disease or disease-relevant responses – may be essential to the discovery of much-needed therapeutics.
To date, most efforts to develop biomarkers in these diseases have pursued a candidate marker approach – one decides a priori that a particular protein or other measure has relevance to disease and then tests whether this is true. With a few exceptions, this approach has not yielded reliable biomarkers that are needed, specifically (1) biomarkers for diagnostic confirmation or pre-symptomatic diagnosis, (2) biomarkers for objective measurement of disease severity, and (3) biomarkers for assessing motor and non-motor prognosis. As a consequence, we need to consider alternative approaches.
One such alternative approach is to begin with an unbiased screen of hundreds or thousands of potential candidates. This type of approach was not always possible; it is predicated on the development of permissive technologies for high-throughput screening. While this type of approach has its own challenges – chiefly in the form of requiring new types of partnership or hybrid expertise, and a culture that provides incentive for replication it has also been successful in the discovery of genetic risk factors for many diseases (including AD and PD) and in the discovery of clinically useful biomarkers in cancer. The clear successes of unbiased screening approaches in other areas, as well as early examples in PD biomarker discovery where unbiased screening leads have been successfully replicated, speak to the promise of this type of approach. For diseases already affecting ~40 million people worldwide, with no disease-modifying therapy available, it could not come soon enough.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Adler CH, Dugger BN, Hinni ML, Lott DG, Driver-Dunckley E, Hidalgo J, Henry-Watson J, Serrano G, Sue LI, Nagel T, et al. Submandibular gland needle biopsy for the diagnosis of Parkinson disease. Neurology. 2014;82:858–864. doi: 10.1212/WNL.0000000000000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Petersen RC. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G, Bronnick K, Aarsland D, Blennow K, Zetterberg H, Ballard C, Kurz MW, Andreasson U, Tysnes OB, Larsen JP, Mulugeta E. CSF amyloid-beta and tau proteins, and cognitive performance, in early and untreated Parkinson’s disease: the Norwegian ParkWest study. J Neurol Neurosurg Psychiatry. 2010;81:1080–1086. doi: 10.1136/jnnp.2009.199950. [DOI] [PubMed] [Google Scholar]
- Alzheimer’s Disease Neuroimaging Initiative. 2013 Retrieved from http://www.adni-info.org.
- Baird GS, Nelson SK, Keeney TR, Stewart A, Williams S, Kraemer S, Peskind ER, Montine TJ. Age-dependent changes in the cerebrospinal fluid proteome by slow off-rate modified aptamer array. The American journal of pathology. 2012;180:446–456. doi: 10.1016/j.ajpath.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj N, Hauser RA, Grachev ID. Clinical utility of dopamine transporter single photon emission CT (DaT-SPECT) with (123I) ioflupane in diagnosis of parkinsonian syndromes. J Neurol Neurosurg Psychiatry. 2013;84:1288–1295. doi: 10.1136/jnnp-2012-304436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- Beach TG, Adler CH, Sue LI, Vedders L, Lue L, White CL, III, Akiyama H, Caviness JN, Shill HA, Sabbagh MN. Multi-organ distribution of phosphorylated α-synuclein histopathology in subjects with Lewy body disorders. Acta Neuropathol. 2010;119:689–702. doi: 10.1007/s00401-010-0664-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardelli A, Wenning G, Antonini A, Berg D, Bloem B, Bonifati V, Brooks D, Burn D, Colosimo C, Fanciulli A. EFNS/MDS-ES recommendations for the diagnosis of Parkinson’s disease. European Journal of Neurology. 2013;20:16–34. doi: 10.1111/ene.12022. [DOI] [PubMed] [Google Scholar]
- Berg D, Postuma RB, Bloem B, Chan P, Dubois B, Gasser T, Goetz CG, Halliday GM, Hardy J, Lang AE. Time to redefine PD? Introductory statement of the MDS Task Force on the definition of Parkinson’s disease. Movement Disorders. 2014;29:454–462. doi: 10.1002/mds.25844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioFIND Clinical Study. 2014 Retrieved from https://www.michaeljfox.org/page.html?biofind-clinical-study.
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- Björkqvist M, Ohlsson M, Minthon L, Hansson O. Evaluation of a previously suggested plasma biomarker panel to identify Alzheimer’s disease. PloS one. 2012;7:e29868. doi: 10.1371/journal.pone.0029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Matson WR, Wang L, Matson T, Saunders-Pullman R, Bressman SS, Flint Beal M. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain. 2008;131:389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- Boja ES, Rodriguez H. Mass spectrometry-based targeted quantitative proteomics: Achieving sensitive and reproducible detection of proteins. Proteomics. 2012;12:1093–1110. doi: 10.1002/pmic.201100387. [DOI] [PubMed] [Google Scholar]
- Booij J, Kemp P. Dopamine transporter imaging with [123I] FP-CIT SPECT: potential effects of drugs. European journal of nuclear medicine and molecular imaging. 2008;35:424–438. doi: 10.1007/s00259-007-0621-0. [DOI] [PubMed] [Google Scholar]
- Borghi R, Marchese R, Negro A, Marinelli L, Forloni G, Zaccheo D, Abbruzzese G, Tabaton M. Full length α-synuclein is present in cerebrospinal fluid from Parkinson’s disease and normal subjects. Neurosci Lett. 2000;287:65–67. doi: 10.1016/s0304-3940(00)01153-8. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. Invited Article: Nervous system pathology in sporadic Parkinson disease. Neurology. 2008;70:1916–1925. doi: 10.1212/01.wnl.0000312279.49272.9f. [DOI] [PubMed] [Google Scholar]
- Brooks DJ. Imaging approaches to Parkinson disease. J Nucl Med. 2010;51:596–609. doi: 10.2967/jnumed.108.059998. [DOI] [PubMed] [Google Scholar]
- Buter TC, van den Hout A, Matthews FE, Larsen JP, Brayne C, Aarsland D. Dementia and survival in Parkinson disease: a 12-year population study. Neurology. 2008;70:1017–1022. doi: 10.1212/01.wnl.0000306632.43729.24. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JJ, Roth JA. The impact of the Oncotype Dx breast cancer assay in clinical practice: a systematic review and meta-analysis. Breast Cancer Res Treat. 2013;141:13–22. doi: 10.1007/s10549-013-2666-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahine LM, Stern MB, Chen-Plotkin A. Blood-based biomarkers for Parkinson’s disease. Parkinsonism Relat Disord. 2014;20:S99–S103. doi: 10.1016/S1353-8020(13)70025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Hu WT, Siderowf A, Weintraub D, Goldmann Gross R, Hurtig HI, Xie SX, Arnold SE, Grossman M, Clark CM. Plasma epidermal growth factor levels predict cognitive decline in Parkinson disease. Ann Neurol. 2011;69:655–663. doi: 10.1002/ana.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compta Y, Martí MJ, Ibarretxe-Bilbao N, Junqué C, Valldeoriola F, Muñoz E, Ezquerra M, Ríos J, Tolosa E. Cerebrospinal tau, phospho-tau, and beta-amyloid and neuropsychological functions in Parkinson’s disease. Movement Disorders. 2009;24:2203–2210. doi: 10.1002/mds.22594. [DOI] [PubMed] [Google Scholar]
- Cummings JL, Henchcliffe C, Schaier S, Simuni T, Waxman A, Kemp P. The role of dopaminergic imaging in patients with symptoms of dopaminergic system neurodegeneration. Brain. 2011;134:3146–3166. doi: 10.1093/brain/awr177. [DOI] [PubMed] [Google Scholar]
- Davis JW, Grandinetti A, Waslien CI, Ross GW, White LR, Morens DM. Observations on serum uric acid levels and the risk of idiopathic Parkinson’s disease. Am J Epidemiol. 1996;144:480–484. doi: 10.1093/oxfordjournals.aje.a008954. [DOI] [PubMed] [Google Scholar]
- de Lau LM, Koudstaal PJ, Hofman A, Breteler M. Serum uric acid levels and the risk of Parkinson disease. Ann Neurol. 2005;58:797–800. doi: 10.1002/ana.20663. [DOI] [PubMed] [Google Scholar]
- Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S A. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao H, Li X, Hu S, Liu Y. Gene expression profiling combined with bioinformatics analysis identify biomarkers for Parkinson disease. PloS one. 2012;7:e52319. doi: 10.1371/journal.pone.0052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, Braak H, Duda JE, Duyckaerts C, Gasser T, Halliday GM, Hardy J, Leverenz JB, Del Tredici K, Wszolek ZK. Neuropathological assessment of Parkinson’s disease: refining the diagnostic criteria. The Lancet Neurology. 2009;8:1150–1157. doi: 10.1016/S1474-4422(09)70238-8. [DOI] [PubMed] [Google Scholar]
- Dorsey ER, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, Marshall FJ, Ravina BM, Schifitto G, Siderowf A, Tanner CM. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology. 2007;68:384–386. doi: 10.1212/01.wnl.0000247740.47667.03. [DOI] [PubMed] [Google Scholar]
- Dorsey ER, George BP, Leff B, Willis AW. The coming crisis: obtaining care for the growing burden of neurodegenerative conditions. Neurology. 2013;80:1989–1996. doi: 10.1212/WNL.0b013e318293e2ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druschky A, Hilz M, Platsch G, Radespiel-Tröger M, Druschky K, Kuwert T, Neundörfer B. Differentiation of Parkinson’s disease and multiple system atrophy in early disease stages by means of I-123-MIBG SPECT. J Neurol Sci. 2000;175:3–12. doi: 10.1016/s0022-510x(00)00279-3. [DOI] [PubMed] [Google Scholar]
- Duncan MW, Hunsucker SW. Proteomics as a tool for clinically relevant biomarker discovery and validation. Exp Biol Med (Maywood) 2005;230:808–817. doi: 10.1177/153537020523001105. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin Chem. 2010;56:186–193. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/β-amyloid42 ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- Fearnley JM, Lees AJ. Ageing and Parkinson’s disease: substantia nigra regional selectivity. Brain. 1991;114 (Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- Foulds P, Yokota O, Thurston A, Davidson Y, Ahmed Z, Holton J, Thompson J, Akiyama H, Arai T, Hasegawa M. Post mortem cerebrospinal fluid α-synuclein levels are raised in multiple system atrophy and distinguish this from the other α-synucleinopathies, Parkinson’s disease and Dementia with Lewy bodies. Neurobiol Dis. 2012;45:188–195. doi: 10.1016/j.nbd.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Greenberg BD, Carrillo MC, Ryan JM, Gold M, Gallagher K, Grundman M, Berman RM, Ashwood T, Siemers ER. Improving Alzheimer’s disease phase II clinical trials. Alzheimer’s & Dementia. 2013;9:39–49. doi: 10.1016/j.jalz.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Gurbel PA, Kreutz RP, Bliden KP, DiChiara J, Tantry US. Biomarker analysis by fluorokine multianalyte profiling distinguishes patients requiring intervention from patients with long-term quiescent coronary artery disease: a potential approach to identify atherosclerotic disease progression. Am Heart J. 2008;155:56–61. doi: 10.1016/j.ahj.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Hajee ME, March WF, Lazzaro DR, Wolintz AH, Shrier EM, Glazman S, Bodis-Wollner IG. Inner retinal layer thinning in Parkinson disease. Arch Ophthalmol. 2009;127:737–741. doi: 10.1001/archophthalmol.2009.106. [DOI] [PubMed] [Google Scholar]
- Hampel H, Frank R, Broich K, Teipel SJ, Katz RG, Hardy J, Herholz K, Bokde AL, Jessen F, Hoessler YC. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nature Reviews Drug Discovery. 2010;9:560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- Hely MA, Reid WG, Adena MA, Halliday GM, Morris JG. The Sydney multicenter study of Parkinson’s disease: the inevitability of dementia at 20 years. Movement Disorders. 2008;23:837–844. doi: 10.1002/mds.21956. [DOI] [PubMed] [Google Scholar]
- Henchcliffe C, Dodel R, Beal MF. Biomarkers of Parkinson’s disease and Dementia with Lewy bodies. Prog Neurobiol. 2011;95:601–613. doi: 10.1016/j.pneurobio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- Hong Z, Shi M, Chung KA, Quinn JF, Peskind ER, Galasko D, Jankovic J, Zabetian CP, Leverenz JB, Baird G, et al. DJ-1 and alpha-synuclein in human cerebrospinal fluid as biomarkers of Parkinson’s disease. Brain. 2010;133:713–726. doi: 10.1093/brain/awq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55:181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AJ, Daniel SE, Lees AJ. Improved accuracy of clinical diagnosis of Lewy body Parkinson’s disease. Neurology. 2001;57:1497–1499. doi: 10.1212/wnl.57.8.1497. [DOI] [PubMed] [Google Scholar]
- Hutchinson M, Raff U. Structural changes of the substantia nigra in Parkinson’s disease as revealed by MR imaging. AJNR Am J Neuroradiol. 2000;21:697–701. [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine (US) Forum on Neuroscience and Nervous System Disorders. Paper presented at Neuroscience Biomarkers and Biosignatures: Converging Technologies, Emerging Partnerships, Workshop Summary. Washington, D.C: [PubMed] [Google Scholar]
- Irwin DJ, Lee VM, Trojanowski JQ. Parkinson’s disease dementia: convergence of [alpha]-synuclein, tau and amyloid-[beta] pathologies. Nature Reviews Neuroscience. 2013;14:626–636. doi: 10.1038/nrn3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Albert MS, Knopman DS, McKhann GM, Sperling RA, Carrillo MC, Thies B, Phelps CH. Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia. 2011;7:257–262. doi: 10.1016/j.jalz.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Holtzman DM. Biomarker modeling of Alzheimer’s disease. Neuron. 2013;80:1347–1358. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Irwin DJ, Chen-Plotkin AS, Siderowf A, Caspell C, Coffey CS, Waligórska T, Taylor P, Pan S, Frasier M. Association of cerebrospinal fluid β-amyloid 1–42, T-tau, P-tau181, and α-synuclein levels with clinical features of drug-naive patients with early Parkinson disease. JAMA neurology. 2013;70:1277–1287. doi: 10.1001/jamaneurol.2013.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Irwin DJ, Chen-Plotkin AS, Siderowf A, Caspell C, Coffey CS, Waligórska T, Taylor P, Pan S, Frasier M. Association of cerebrospinal fluid β-amyloid 1–42, T-tau, P-tau181, and α-synuclein levels with clinical features of drug-naive patients with early Parkinson disease. JAMA neurology. 2013;70:1277–1287. doi: 10.1001/jamaneurol.2013.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Vanderstichele H, Trojanowski JQ, Shaw LM. Simultaneous analysis of cerebrospinal fluid biomarkers using microsphere-based xMAP multiplex technology for early detection of Alzheimer’s disease. Methods. 2012;56:484–493. doi: 10.1016/j.ymeth.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergström M, Savitcheva I, Huang G, Estrada S. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- Kowal SL, Dall TM, Chakrabarti R, Storm MV, Jain A. The current and projected economic burden of Parkinson’s disease in the United States. Movement Disorders. 2013;28:311–318. doi: 10.1002/mds.25292. [DOI] [PubMed] [Google Scholar]
- Kozauer N, Katz R. Regulatory innovation and drug development for early-stage Alzheimer’s disease. N Engl J Med. 2013;368:1169–1171. doi: 10.1056/NEJMp1302513. [DOI] [PubMed] [Google Scholar]
- la Marca G. Mass spectrometry in clinical chemistry: the case of newborn screening. J Pharm Biomed Anal. 2014 doi: 10.1016/j.jpba.2014.03.047. [DOI] [PubMed] [Google Scholar]
- Lebouvier T, Neunlist M, des Varannes SB, Coron E, Drouard A, N’Guyen J, Chaumette T, Tasselli M, Paillusson S, Flamand M. Colonic biopsies to assess the neuropathology of Parkinson’s disease and its relationship with symptoms. PloS one. 2010;5:e12728. doi: 10.1371/journal.pone.0012728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewy F. Paralysis agitans. I Pathologische anatomie. Handbuch der neurologie. 1912;3:920–933. [Google Scholar]
- Lollo B, Steele F, Gold L. Beyond antibodies: New affinity reagents to unlock the proteome. Proteomics. 2014;14:638–644. doi: 10.1002/pmic.201300187. [DOI] [PubMed] [Google Scholar]
- Lord S, Galna B, Rochester L. Moving forward on gait measurement: toward a more refined approach. Movement Disorders. 2013;28:1534–1543. doi: 10.1002/mds.25545. [DOI] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, Lee VM. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdalinou N, Lees AJ, Zetterberg H. Cerebrospinal fluid biomarkers in parkinsonian conditions: an update and future directions. J Neurol Neurosurg Psychiatry. 2014 doi: 10.1136/jnnp-2013-307539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek N, Swallow D, Grosset K, Anichtchik O, Spillantini M, Grosset D. Alpha-synuclein in peripheral tissues and body fluids as a biomarker for Parkinson’s disease a systematic review. Acta Neurol Scand. 2014 doi: 10.1111/ane.12247. [DOI] [PubMed] [Google Scholar]
- Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley JM. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014 doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S. The parkinson progression marker initiative (PPMI) Prog Neurobiol. 2011;95:629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S. The parkinson progression marker initiative (PPMI) Prog Neurobiol. 2011;95:629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek K, Jennings D, Tamagnan G, Seibyl J. Biomarkers for Parkison’s disease: tools to assess Parkinson’s disease onset and progression. Ann Neurol. 2008;64:S111–S121. doi: 10.1002/ana.21602. [DOI] [PubMed] [Google Scholar]
- Marras C, Rochon P, Lang AE. Predicting motor decline and disability in Parkinson disease: a systematic review. Arch Neurol. 2002;59:1724–1728. doi: 10.1001/archneur.59.11.1724. [DOI] [PubMed] [Google Scholar]
- Mattsson N, Zetterberg H, Hansson O, Andreasen N, Parnetti L, Jonsson M, Herukka S, van der Flier, Wiesje M, Blankenstein MA, Ewers M. CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. JAMA. 2009;302:385–393. doi: 10.1001/jama.2009.1064. [DOI] [PubMed] [Google Scholar]
- Mayeux R, Denaro J, Hemenegildo N, Marder K, Tang M, Cote LJ, Stern Y. A population-based investigation of Parkinson’s disease with and without dementia: relationship to age and gender. Arch Neurol. 1992;49:492–497. doi: 10.1001/archneur.1992.00530290076015. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehan MR, Williams SA, Siegfried JM, Bigbee WL, Weissfeld JL, Wilson DO, Pass HI, Rom WN, Muley T, Meister M. Validation of a blood protein signature for non-small cell lung cancer. Clinical proteomics. 2014;11:32. doi: 10.1186/1559-0275-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinuevo JL, Blennow K, Dubois B, Engelborghs S, Lewczuk P, Perret-Liaudet A, Teunissen CE, Parnetti L. The clinical use of cerebrospinal fluid biomarker testing for Alzheimer’s disease diagnosis: A consensus paper from the Alzheimer’s Biomarkers Standardization Initiative. Alzheimer’s & Dementia. 2014 doi: 10.1016/j.jalz.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Mollenhauer B, Cullen V, Kahn I, Krastins B, Outeiro TF, Pepivani I, Ng J, Schulz-Schaeffer W, Kretzschmar HA, McLean PJ. Direct quantification of CSF α-synuclein by ELISA and first cross-sectional study in patients with neurodegeneration. Exp Neurol. 2008;213:315–325. doi: 10.1016/j.expneurol.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Mollenhauer B, Locascio JJ, Schulz-Schaeffer W, Sixel-Döring F, Trenkwalder C, Schlossmacher MG. α-Synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: a cohort study. The Lancet Neurology. 2011;10:230–240. doi: 10.1016/S1474-4422(11)70014-X. [DOI] [PubMed] [Google Scholar]
- Mollenhauer B, Rochester L, Chen-Plotkin A, Brooks D. What can biomarkers tell us about cognition in Parkinson’s disease? Movement Disorders. 2014;29:622–633. doi: 10.1002/mds.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molochnikov L, Rabey JM, Dobronevsky E, Bonucelli U, Ceravolo R, Frosini D, Grunblatt E, Riederer P, Jacob C, Aharon-Peretz J. A molecular signature in blood identifies early Parkinson’s disease. Mol Neurodegener. 2012;7:26. doi: 10.1186/1750-1326-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montine TJ, Shi M, Quinn JF, Peskind ER, Craft S, Ginghina C, Chung KA, Kim H, Galasko DR, Jankovic J. CSF Aβ42 and tau in Parkinson’s disease with cognitive impairment. Movement disorders. 2010;25:2682–2685. doi: 10.1002/mds.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–877. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair VS, Pritchard CC, Tewari M, Ioannidis JP. Design and Analysis for Studying microRNAs in Human Disease: A Primer on -Omic Technologies. Am J Epidemiol. 2014;180:140–152. doi: 10.1093/aje/kwu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH-NINDS. Paper presented at Parkinson’s Disease 2014: Advancing Research, Improving Lives; Washington, D.C. Jan 6, 2014. [Google Scholar]
- NINDS Common Data Elements. 2014 Retrieved from www.commondataelements.ninds.nih.gov/PD.aspx.
- O’Bryant SE, Gupta V, Henriksen K, Edwards M, Jeromin A, Lista S, Bazenet C, Soares H, Lovestone S, Hampel H. Guidelines for the standardization of preanalytic variables for blood-based biomarker studies in Alzheimer’s disease research. Alzheimer’s & Dementia. 2014 doi: 10.1016/j.jalz.2014.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanow CW, Kieburtz K, Schapira AH. Why have we failed to achieve neuroprotection in Parkinson’s disease? Ann Neurol. 2008;64:S101–S110. doi: 10.1002/ana.21461. [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Park MJ, Cheon S, Bae H, Kim S, Kim JW. Elevated levels of α-synuclein oligomer in the cerebrospinal fluid of drug-naïve patients with Parkinson’s disease. Journal of Clinical Neurology. 2011;7:215–222. doi: 10.3988/jcn.2011.7.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson Study Group PRECEPT Investigators. Mixed lineage kinase inhibitor CEP-1347 fails to delay disability in early Parkinson disease. Neurology. 2007;69:1480–1490. doi: 10.1212/01.wnl.0000277648.63931.c0. [DOI] [PubMed] [Google Scholar]
- Parkinson J. An essay on the shaking palsy. Whittingham and Rowland; 1817. [Google Scholar]
- Parkinson’s Disease Biomarkers Program. 2014 Retrieved from https://pdbp.ninds.nih.gov.
- Parkinson’s Progression Markers Initiative. 2014 Retrieved from www.ppmi-info.org.
- Parnetti L, Castrioto A, Chiasserini D, Persichetti E, Tambasco N, El-Agnaf O, Calabresi P. Cerebrospinal fluid biomarkers in Parkinson disease. Nature Reviews Neurology. 2013;9:131–140. doi: 10.1038/nrneurol.2013.10. [DOI] [PubMed] [Google Scholar]
- Partin JF, Mamounas EP. Impact of the 21-gene recurrence score assay compared with standard clinicopathologic guidelines in adjuvant therapy selection for node-negative, estrogen receptor-positive breast cancer. Annals of surgical oncology. 2011;18:3399–3406. doi: 10.1245/s10434-011-1698-z. [DOI] [PubMed] [Google Scholar]
- Pellecchia MT, Santangelo G, Picillo M, Pivonello R, Longo K, Pivonello C, Vitale C, Amboni M, De Rosa A, Moccia M. Serum epidermal growth factor predicts cognitive functions in early, drug-naive Parkinson’s disease patients. J Neurol. 2013;260:438–444. doi: 10.1007/s00415-012-6648-6. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature. 2009;461:916–922. doi: 10.1038/nature08538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Postuma RB, Gagnon JF, Vendette M, Fantini ML, Massicotte-Marquez J, Montplaisir J. Quantifying the risk of neurodegenerative disease in idiopathic REM sleep behavior disorder. Neurology. 2009;72:1296–1300. doi: 10.1212/01.wnl.0000340980.19702.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressley JC, Louis ED, Tang MX, Cote L, Cohen PD, Glied S, Mayeux R. The impact of comorbid disease and injuries on resource use and expenditures in parkinsonism. Neurology. 2003;60:87–93. doi: 10.1212/wnl.60.1.87. [DOI] [PubMed] [Google Scholar]
- Qiang JK, Wong YC, Siderowf A, Hurtig HI, Xie SX, Lee VM, Trojanowski JQ, Yearout DB, Leverenz J, Montine TJ. Plasma apolipoprotein A1 as a biomarker for Parkinson disease. Ann Neurol. 2013;74:119–127. doi: 10.1002/ana.23872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- Rajput AH, Rozdilsky B, Rajput A. Accuracy of clinical diagnosis in parkinsonism--a prospective study. Can J Neurol Sci. 1991;18:275–278. doi: 10.1017/s0317167100031814. [DOI] [PubMed] [Google Scholar]
- Rascol O, Fitzer-Attas CJ, Hauser R, Jankovic J, Lang A, Langston JW, Melamed E, Poewe W, Stocchi F, Tolosa E. A double-blind, delayed-start trial of rasagiline in Parkinson’s disease (the ADAGIO study): prespecified and post-hoc analyses of the need for additional therapies, changes in UPDRS scores, and non-motor outcomes. The Lancet Neurology. 2011;10:415–423. doi: 10.1016/S1474-4422(11)70073-4. [DOI] [PubMed] [Google Scholar]
- Rascol O, Lozano A, Stern M, Poewe W. Milestones in Parkinson’s disease therapeutics. Movement disorders. 2011;26:1072–1082. doi: 10.1002/mds.23714. [DOI] [PubMed] [Google Scholar]
- Ravina B, Eidelberg D, Ahlskog JE, Albin RL, Brooks DJ, Carbon M, Dhawan V, Feigin A, Fahn S, Guttman M, et al. The role of radiotracer imaging in Parkinson disease. Neurology. 2005;64:208–215. doi: 10.1212/01.WNL.0000149403.14458.7F. [DOI] [PubMed] [Google Scholar]
- Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, Friedman LF, Galasko DR, Jutel M, Karydas A. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- Ross GW, Petrovitch H, Abbott RD, Tanner CM, Popper J, Masaki K, Launer L, White LR. Association of olfactory dysfunction with risk for future Parkinson’s disease. Ann Neurol. 2008;63:167–173. doi: 10.1002/ana.21291. [DOI] [PubMed] [Google Scholar]
- Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13:477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- Schenck CH, Bundlie SR, Mahowald MW. Delayed emergence of a parkinsonian disorder in 38% of 29 older men initially diagnosed with idiopathic rapid eye movement sleep behaviour disorder. Neurology. 1996;46:388–393. doi: 10.1212/wnl.46.2.388. [DOI] [PubMed] [Google Scholar]
- Scherzer CR, Eklund AC, Morse LJ, Liao Z, Locascio JJ, Fefer D, Schwarzschild MA, Schlossmacher MG, Hauser MA, Vance JM, et al. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc Natl Acad Sci U S A. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid AW, Fauvet B, Moniatte M, Lashuel HA. Alpha-synuclein post-translational modifications as potential biomarkers for Parkinson disease and other synucleinopathies. Mol Cell Proteomics. 2013;12:3543–3558. doi: 10.1074/mcp.R113.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzschild MA, Schwid SR, Marek K, Watts A, Lang AE, Oakes D, Shoulson I, Ascherio A. Serum urate as a predictor of clinical and radiographic progression in Parkinson disease. Arch Neurol. 2008;65:716–723. doi: 10.1001/archneur.2008.65.6.nct70003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk JM, Lindberg J, Sundberg M, Uhlen M, Nilsson P. Determination of binding specificities in highly multiplexed bead-based assays for antibody proteomics. Mol Cell Proteomics. 2007;6:125–132. doi: 10.1074/mcp.T600035-MCP200. [DOI] [PubMed] [Google Scholar]
- Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- Shah M, Catafau AM. Molecular Imaging Insights into Neurodegeneration: Focus on Tau PET Radiotracers. J Nucl Med. 2014;55:871–874. doi: 10.2967/jnumed.113.136069. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehadeh LA, Yu K, Wang L, Guevara A, Singer C, Vance J, Papapetropoulos S. SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson’s disease. PLoS One. 2010;5:e9104. doi: 10.1371/journal.pone.0009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer TB. Biomarkers for Parkinson’s disease. Sci Transl Med. 2011;3:79ps14. doi: 10.1126/scitranslmed.3002488. [DOI] [PubMed] [Google Scholar]
- Shi M, Bradner J, Hancock AM, Chung KA, Quinn JF, Peskind ER, Galasko D, Jankovic J, Zabetian CP, Kim HM. Cerebrospinal fluid biomarkers for Parkinson disease diagnosis and progression. Ann Neurol. 2011;69:570–580. doi: 10.1002/ana.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siderowf A, Jennings D, Eberly S, Oakes D, Hawkins KA, Ascherio A, Stern MB, Marek K. Impaired olfaction and other prodromal features in the Parkinson At-Risk Syndrome study. Movement Disorders. 2012;27:406–412. doi: 10.1002/mds.24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siderowf A, Xie SX, Hurtig H, Weintraub D, Duda J, Chen-Plotkin A, Shaw LM, Van Deerlin V, Trojanowski JQ, Clark C. CSF amyloid {beta} 1–42 predicts cognitive decline in Parkinson disease. Neurology. 2010;75:1055–1061. doi: 10.1212/WNL.0b013e3181f39a78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierks MR, Chatterjee G, McGraw C, Kasturirangan S, Schulz P, Prasad S. CSF levels of oligomeric alpha-synuclein and beta-amyloid as biomarkers for neurodegenerative disease. Integrative Biology. 2011;3:1188–1196. doi: 10.1039/c1ib00018g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Solier C, Langen H. Antibody-based proteomics and biomarker research—Current status and limitations. Proteomics. 2014;14:774–783. doi: 10.1002/pmic.201300334. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Jr, Kaye J, Montine TJ. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchowersky O, Reich S, Perlmutter J, Zesiewicz T, Gronseth G, Weiner WJ Quality Standards Subcommittee of the American Academy of Neurology. Practice Parameter: diagnosis and prognosis of new onset Parkinson disease (an evidence-based review): report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology. 2006;66:968–975. doi: 10.1212/01.wnl.0000215437.80053.d0. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, Westman E, Ballard C, Aarsland D. Cognitive impairment in patients with Parkinson’s disease: diagnosis, biomarkers, and treatment. The Lancet Neurology. 2012;11:697–707. doi: 10.1016/S1474-4422(12)70152-7. [DOI] [PubMed] [Google Scholar]
- Swanson CR, Li K, Unger TL, Gallagher MD, Van Deerlin VM, Agarwal P, Leverenz J, Roberts J, Samii A, Goldmann Gross R, et al. Lower plasma ApoA1 levels are found in Parkinson’s disease and associate with APOA1 genotype. Movement disorders. 2014 doi: 10.1002/mds.26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapiola T, Alafuzoff I, Herukka S, Parkkinen L, Hartikainen P, Soininen H, Pirttilä T. Cerebrospinal fluid β-amyloid 42 and tau proteins as biomarkers of Alzheimer-type pathologic changes in the brain. Arch Neurol. 2009;66:382–389. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- Tateno F, Sakakibara R, Kawai T, Kishi M, Murano T. Alpha-synuclein in the cerebrospinal fluid differentiates synucleinopathies (Parkinson Disease, dementia with Lewy bodies, multiple system atrophy) from Alzheimer disease. Alzheimer Dis Assoc Disord. 2012;26:213–216. doi: 10.1097/WAD.0b013e31823899cc. [DOI] [PubMed] [Google Scholar]
- The Lancet Neurology. NINDS boosts research for biomarkers in Parkinson’s disease. Lancet Neurol. 2013;12 doi: 10.1016/S1474-4422(13)70029-2. 217–4422(13)70029–2. [DOI] [PubMed] [Google Scholar]
- Tian Q, Price ND, Hood L. Systems cancer medicine: towards realization of predictive, preventive, personalized and participatory (P4) medicine. J Intern Med. 2012;271:111–121. doi: 10.1111/j.1365-2796.2011.02498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda T, Salem SA, Allsop D, Mizuno T, Nakagawa M, Qureshi MM, Locascio JJ, Schlossmacher MG, El-Agnaf O. Decreased α-synuclein in cerebrospinal fluid of aged individuals and subjects with Parkinson’s disease. Biochem Biophys Res Commun. 2006;349:162–166. doi: 10.1016/j.bbrc.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Tokuda T, Qureshi MM, Ardah MT, Varghese S, Shehab SA, Kasai T, Ishigami N, Tamaoka A, Nakagawa M, El-Agnaf OM. Detection of elevated levels of alpha-synuclein oligomers in CSF from patients with Parkinson disease. Neurology. 2010;75:1766–1772. doi: 10.1212/WNL.0b013e3181fd613b. [DOI] [PubMed] [Google Scholar]
- Trétiakoff C. Contribution a l’etude l’anatomie pathologique du locus Niger de soemmering: avec quelques déductions relatives à la pathogénie des troubles du tonus musculaire et de la maladie de Parkinson. 1919. [Google Scholar]
- Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nature Reviews Neurology. 2013;9:445–454. doi: 10.1038/nrneurol.2013.132. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Uhl GR. Dopamine transporter: basic science and human variation of a key molecule for dopaminergic function, locomotion, and parkinsonism. Movement disorders. 2003;18:S71–S80. doi: 10.1002/mds.10578. [DOI] [PubMed] [Google Scholar]
- Vaillancourt DE, Spraker MB, Prodoehl J, Abraham I, Corcos DM, Zhou XJ, Comella CL, Little DM. High-resolution diffusion tensor imaging in the substantia nigra of de novo Parkinson disease. Neurology. 2009;72:1378–1384. doi: 10.1212/01.wnl.0000340982.01727.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderstichele H, Bibl M, Engelborghs S, Le Bastard N, Lewczuk P, Molinuevo JL, Parnetti L, Perret-Liaudet A, Shaw LM, Teunissen C. Standardization of preanalytical aspects of cerebrospinal fluid biomarker testing for Alzheimer’s disease diagnosis: a consensus paper from the Alzheimer’s Biomarkers Standardization Initiative. Alzheimer’s & Dementia. 2012;8:65–73. doi: 10.1016/j.jalz.2011.07.004. [DOI] [PubMed] [Google Scholar]
- van’t Veer LJ, Dai H, Van De Vijver, Marc J, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Vogeser M, Seger C. A decade of HPLC MS/MS in the routine clinical laboratory—goals for further developments. Clin Biochem. 2008;41:649–662. doi: 10.1016/j.clinbiochem.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang G, Fowler JS, Ding Y. Imaging the effects of methylphenidate on brain dopamine: new model on its therapeutic actions for attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1410–1415. doi: 10.1016/j.biopsych.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Wang J, Hoekstra JG, Zuo C, Cook TJ, Zhang J. Biomarkers of Parkinson’s disease: current status and future perspectives. Drug Discov Today. 2013;18:155–162. doi: 10.1016/j.drudis.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi M, Chung KA, Zabetian CP, Leverenz JB, Berg D, Srulijes K, Trojanowski JQ, Lee VM, Siderowf AD, et al. Phosphorylated alpha-synuclein in Parkinson’s disease. Sci Transl Med. 2012;4:121ra20. doi: 10.1126/scitranslmed.3002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer’s & Dementia. 2013;9:e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf MG, O’Reilly E, Chen H, Schwarzschild MA, Ascherio A. Plasma urate and risk of Parkinson’s disease. Am J Epidemiol. 2007;166:561–567. doi: 10.1093/aje/kwm127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- What is Proteomics? 2014 Retrieved from http://proteomics.cancer.gov/whatisproteomics.
- Wimo A, Prince M. World Alzheimer Report 2010 The Global Economic Impact of Dementia. Alzheimer’s Disease International 2010 [Google Scholar]
- Yalow RS, Berson SA. Assay of plasma insulin in human subjects by immunological methods. 1959 doi: 10.1038/1841648b0. [DOI] [PubMed] [Google Scholar]
- Zerhouni EA. Translational and clinical science—time for a new vision. N Engl J Med. 2005;353:1621–1623. doi: 10.1056/NEJMsb053723. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sokal I, Peskind ER, Quinn JF, Jankovic J, Kenney C, Chung KA, Millard SP, Nutt JG, Montine TJ. CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am J Clin Pathol. 2008;129:526–529. doi: 10.1309/W01Y0B808EMEH12L. [DOI] [PMC free article] [PubMed] [Google Scholar]