Abstract
The trillions of beneficial commensal microorganisms that normally reside in the gastrointestinal tract have emerged as a critical source of environmentally-derived stimuli that can impact health and disease. However, the underlying cellular and molecular mechanisms that recognize commensal bacteria-derived signals and regulate mammalian homeostasis are just beginning to be defined. Highly coordinated epigenomic modifications allow mammals to alter the transcriptional program of a cell in response to environmental cues. These modifications may play a key role in regulating the dynamic relationship between mammals and their microbiota. Here we review recent advances in understanding of the interplay between the microbiota and mammalian epigenomic pathways, and highlight emerging findings that implicate a central role for histone deacetylases (HDACs) in orchestrating host-microbiota interactions.
The microbiota in human health and disease
It is now clear that multiple human diseases, including asthma, allergy, diabetes, obesity, autism, cancer and inflammatory bowel disease (IBD) develop as a result of complex mammalian gene-environment interactions [1–6]. In addition to diet and drugs, there is increasing evidence that signals derived from the microbiota that normally colonize the mammalian body can act as environmental triggers that influence the balance between health and disease [5–9]. Extensive studies spanning the last several years have demonstrated that the mammalian host has formed a symbiotic relationship with these commensal bacteria. The majority of the commensal bacteria reside within the intestine where they directly interact with a single layer of intestinal epithelial cells (IECs) and influence underlying immune cell populations.
As reviewed extensively recently elsewhere, the intestinal microbiota play a critical role in regulating the immune system [10–13] (Maloy et al, this issue) and nutrient metabolism [14–16]. In addition, a wide range of chronic immune-mediated diseases, such as IBD, diabetes and allergy, have been associated with dysregulation of the host-microbiota relationship and alterations in the diversity of intestinal commensal bacteria [5, 9, 17, 18]. While it is evident that regulation of the host-commensal relationship is essential to mammalian health, the host mechanisms involved in integrating signals derived from commensal bacteria are just beginning to be elucidated. Understanding the cellular and molecular pathways that regulate host-commensal interactions could aid in the development of novel therapeutics to prevent or limit several human diseases associated with changes in the microbiota. Here, we review recent findings that implicate a central role for epigenomic mechanisms in orchestrating the host-microbiota relationship.
Environmental sensing through epigenomics
Eukaryotic cells package their DNA around histone proteins to form a higher order structure termed chromatin. The repetitive element within chromatin, called the nucleosome, is composed of DNA tightly wound around a histone octamer and histone H1 functions as a linker between nucleosomes that permits further condensation of the chromatin structure [19]. The condensed chromatin structure is considered to be generally repressive of gene expression, as this condensed state physically limits access of transcriptional machinery to the genome [20]. Epigenetics involves the study of the molecular processes that permit changes in gene expression without a change in the genetic code and encompasses ATP-dependent chromatin remodeling, regulation by non-coding RNAs, and covalent nucleosomal modifications (Glossary) [21].
Covalent nucleosomal modifications as well as ATP-dependent remodeling enzymes enable chromatin flexibility in response to specific cellular signals. Thus, the chromatin structure can undergo local condensation or relaxation to regulate various processes such as DNA replication, repair, or transcription [21]. The most well characterized covalent epigenetic modifications are DNA methylation and histone modifications, each of which can influence gene expression without altering the associated DNA sequence. Histone N-terminus tails extend from the nucleosomal core and provide a template for various covalent modifications such as acetylation, phosphorylation, methylation, SUMOylation, and ubiquitination. These modifications establish a “histone code” which directs specific recruitment of transcriptional machinery and cofactors, resulting in differential effects on gene expression [22, 23].
While covalent DNA and histone modifications are most commonly discussed as epigenetic phenomena, the term epigenetics often suggests heritability, although multigenerational analyses are often not performed. More recently, a broader concept of the epigenome has been adopted to refer to the combination of histone and DNA modifications and associated proteins that package the genome and guide transcription [21]. Epigenomic mechanisms alter the transcriptional response to environmental cues and thus represent a critical mechanism that can link host genetic predisposition and environmental triggers in the pathogenesis of disease. Therefore, this phenomena has been implicated in the development of most chronic conditions with complex multifactorial etiologies, including cancer, diabetes, allergy, atherosclerosis, and IBD [1, 24–26]. Epigenomic modifications are maintained by the balanced activity of various epigenomic-modifying enzymes, such as DNA methyltransferases (DNMTs), histone acetyltransferases (HATs)/histone deacetylases (HDACs), and histone methyltransferases (HMTs)/histone demethylases (HDMs), that provide a potentially significant interface by which environmental signals can dynamically interact with the genome.
Regulation of epigenomic modifications and enzymes by the microbiota
A complex relationship exists between the intestinal microbiota, the intestinal epithelium and the immune system [27, 28], and dynamic transcriptional regulation is essential in regulating host-commensal bacteria interactions. IECs function as a fundamental cell lineage that resides at the interface between the mammalian host and commensal bacteria. These cells form a physical barrier, sense bacterial-derived signals, and secrete antimicrobial peptides and cytokines/chemokines that, in turn, regulate the microbiota and immune cell homeostasis [29, 30]. Further, epithelial permeability, proliferation, and expression of antimicrobial proteins are regulated by cytokines produced by immune cells [31]. The microbiota, itself, can influence immune cell homeostasis and is critical in the development/maturation of both the innate and adaptive system [32, 33]. In turn, the innate and adaptive immune systems are key components that shape diversity and localization of the microbiota [34].
While there is growing appreciation that bacterial pathogens may modulate host epigenomics [35], studies comparing conventionally-housed and germ-free mice have just begun to uncover potential links between the presence of commensal bacteria and host epigenomic pathways. IECs directly interact with the microbiota and function as essential non-hematopoietic cellular mediators of innate immune responses in the intestine. Pattern-recognition receptors (PRRs) in IECs, including Toll-like receptors (TLRs), recognize and integrate signals from microbial-associated motifs to direct intestinal barrier function and immunoregulatory responses [29, 30, 36]. Examination of epigenomic modifications in IECs revealed that DNA methylation, a generally repressive modification, was lower in the TLR4 gene from large intestinal IECs of germ-free mice compared to IECs from conventionally housed mice, supporting the hypothesis that commensal bacteria may induce tolerance in IECs by repressing TLR4 gene expression through DNA methylation [37].
Similar to other tissues and cells, the epigenome clearly mediates immune cell development [26–29]. Recently, direct links between the presence of commensal bacteria and methylation-dependent pathways have been implicated in regulation of immune cells. Germ-free mice were found to exhibit accumulations of invariant natural killer T (iNKT) cells in the colon and lung compared to conventional controls, resulting in increased susceptibility to mucosal pathology [38]. Remarkably, colonization of neonatal germ-free mice, but not adult germ-free mice, with a conventional microbiota decreased methylation levels of the Cxcl16 gene, which corresponded with decreased Cxcl16 expression, decreased mucosal iNKT accumulation, and protection from mucosal pathology in murine models of IBD and allergic asthma, thus suggesting that neonatal contact with commensal bacteria may establish protection from immune-mediated diseases via DNA methylation[38]. Evaluation of the DNA-methylation adaptor protein, Uhrf1, in regulatory T cells (Tregs) later demonstrated that this factor is upregulated in response to the intestinal microbiota and that Uhrf1 expression is required for DNA methylation of specific genes and proper colonic Treg proliferation and function [39]. Evaluation of the effects of commensal bacteria-derived metabolites on Treg homeostasis is discussed further below.
Comparison of mononuclear phagocytes from conventionally-housed mice and germ-free mice demonstrated that the presence of commensal-bacterial derived signals corresponded with a relative increase in histone H3 trimethylation, a mark of transcriptionally active genes, at or around the transcriptional start sites of multiple inflammatory genes including Ifnb1 and Il-6. This elevation in histone H3 methylation was suggested as a potential mechanism underlying how commensal bacterial-dependent induction of permissive chromatin modifications may enable basal inflammatory gene expression needed for effective priming of cytotoxic lymphocytes such as NK cells and, subsequently, improved immunity to intracellular pathogens [40].
In addition to DNA and histone methylation, an emerging body of work over the last year has brought regulation of histone acetylation by HDACs to the forefront as a critical factor in epigenomic regulation that mediates the interplay between mammalian host cells and the intestinal microbiota. Acetylation of histone tails by HATs is believed to disrupt the DNA-histone interaction, causing local relaxation of the chromatin and permitting access for transcription machinery [41]. Furthermore, histone acetylated-lysines are the preferred substrate for bromodomain containing proteins that include several coactivators with HAT activity, which propagate increased acetylation and transcriptional activation [42–44]. Conversely, removal of the acetyl groups by HDACs generally promotes tighter DNA-histone associations and represses transcriptional activity.
HDACs and the intestinal epithelium
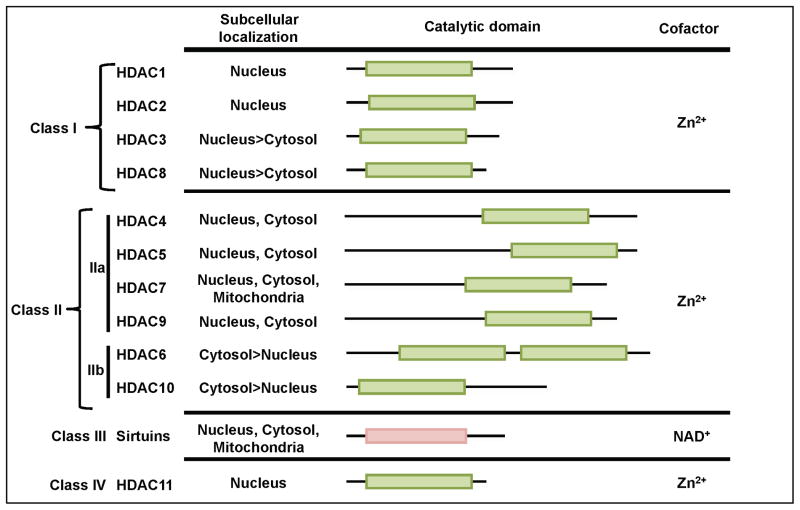
There are 18 known HDACs that are classified into four groups based on their homology to yeast HDACs and subcellular location[45]. The class I, II, and IV HDACs require zinc for their enzymatic deacetylase activities, whereas class III HDACs (sirtuins) depend on nicotine adenine dinucleotide as a cofactor [46] (Figure 1). As their name indicates, these enzymes regulate transcription through histone deacetylation, but HDACs may also deacetylate non-histone targets and possess enzyme-independent effects [47–49]. HDACs are often found in large complexes that are recruited to the chromatin through interactions with transcription factors. Further, their activity and recruitment to the genome can be altered by endogenous hormones and metabolites, dietary compounds and bacterial-derived products, such as lipopolysaccharide (LPS) [45, 50–54]. In vivo studies suggest that the specificity of HDACs in regulating distinct gene programs differs with cell identity, available associating proteins, and the cell signaling environment [45].
Figure 1. Histone Deacetylase (HDAC) Classification.
Isoforms of HDACs are divided into four classes based on the sequence similarity of their catalytic domain to yeast homologues. The distribution of the conserved catalytic domain and dominant region of subcellular localization are indicated.
Expression of the class I HDACs, HDAC1, HDAC2, and HDAC3, was originally characterized in IECs in relation to their role in intestinal development and cancer [55, 56]. Recent work has found that HDAC expression in IECs mediates commensal bacteria-dependent regulation of intestinal homeostasis [57] (Figure 2). Specifically, loss of IEC-intrinsic HDAC3 expression led to alterations in histone acetylation, decreased anti-microbial gene expression, impaired survival of the Paneth cell IEC lineage, and decreased intestinal barrier function. However, generation of germ-free HDAC3-deficient mice revealed that elimination of the microbiota restored Paneth cell homeostasis and intestinal barrier function in HDAC3 knockout mice to levels observed in wildtype germ-free mice, suggesting that HDAC3 integrates signals from the microbiota to regulate the intestinal barrier when commensal bacteria are in the environment. Further, HDAC3 expression levels in the colon have been shown to be induced in the presence of commensal bacteria, and HDAC3 and histone deacetylation have been implicated in IL-10 mediated inhibition of IL-12 p40 expression in colonic macrophages [58]. The underlying microbiota-dependent mechanisms that orchestrate HDAC3-directed histone or non-histone deacetylation remain to be defined.
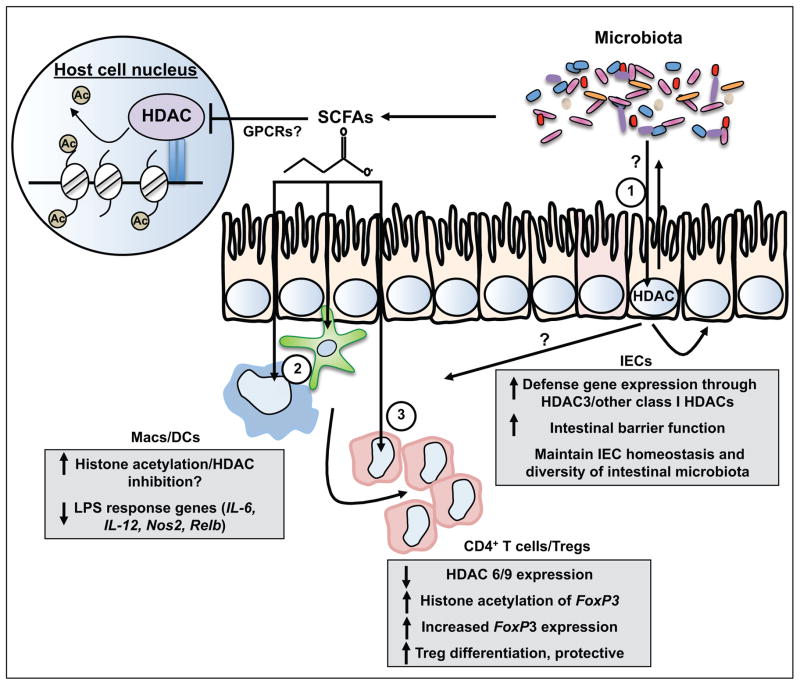
Figure 2. Epigenomic regulation of microbiota-dependent intestinal homeostasis via HDACs.
A series of recent studies identify that HDACs, a family of epigenomic-modifying enzymes that remove acetyl groups from lysine residues on histone tails, mediate dynamic regulation between the microbiota and multiple cell lineages including (1) IECs [57], (2) monocytes [68, 69], and (3) CD4+ T cells/ Tregs [65, 69, 70]. Crosstalk between the microbiota and HDAC-dependent transcriptional networks in IECs and/or commensal bacterial-derived SCFA inhibition of HDACs in innate and adaptive immune cell populations could significantly impact intestinal immune homeostasis, barrier function, and susceptibility to damage and inflammation. Depending on the cellular target and HDAC isoform, manipulation of these pathways could either protect or promote intestinal inflammation, disrupt the diversity of commensal bacteria in the intestine, and potentially result in epigenomic modifications that influence cell fate and antigen tolerance. HDAC: Histone deacetylase; IEC: intestinal epithelial cell; Treg: regulatory T cell; DC: dendritic cell; Mac: Macrophage, SCFA: short chain fatty acid.
Expression of other class I HDACs, such as HDAC1/2, have also recently been found to be essential in regulation of IEC homeostasis and epithelial barrier function [59]. However, in contrast to IEC-specific HDAC3 or double HDAC1/2 knockout mice, IEC-specific deletion of HDAC2 alone protected mice from experimental colitis [60], suggesting differential roles for specific HDACs and sensitivity to alterations in levels of HDAC activity in IECs. Whether expression of HDAC1/2 or other classes of HDACs mediate commensal bacteria-dependent regulation of IEC-intrinsic gene expression and intestinal homeostasis remains to be determined.
Commensal bacteria-derived metabolites regulate HDACs in hematopoietic cells
The microbiota contributes diet-dependent products, such as lipids, amino acids, vitamins and short-chain fatty acids (SCFAs), and diet-independent products, such as lipopolysaccharide and peptidoglycan, to the intestinal microenvironment [15]. These commensal bacterial-derived byproducts have the potential to modify the epigenome of host cells and in turn alter the cell’s development and function. Several recent studies have examined commensal bacteria-derived SCFAs that are produced through bacterial fermentation of dietary carbohydrates in the colon and can be incorporated by IECs or diffuse across the epithelium into the underlying intestinal lamina propria [61–63]. SCFAs have been shown to activate G-protein-coupled-receptors (GPCRs), such as GPR41 and GPR43 [64–67], although recent reports have suggested GPCR-independent regulation by SCFAs [68–70]. The most abundant SCFAs in the intestinal lumen are butyrate, propionate, and acetate. Germ-free mice exhibit significantly decreased levels of all three of these SCFAs in comparison to conventionally-housed mice [71, 72], indicating that the microbiota is essential for their synthesis. SCFAs alone have been found to regulate the development and function of several immune cell lineages [15, 65, 69, 70]. Further, commensal bacteria such as Clostridia and Bifidobacteria can produce SCFAs in the intestinal lumen, and these microbes have also been found to regulate host defense responses [63, 73, 74].
Previous work has determined that SCFAs can potently inhibit HDAC activity in vitro [53, 63, 75, 76]; therefore HDACs in intestinal immune cells have been highlighted as potentially important targets of microbiota-derived SCFAs. A combination of recent publications have uncovered a model in which SCFAs derived from commensal bacteria exert anti-inflammatory effects in the colon in part by stimulating histone acetylation of the FoxP3 locus in naïve CD4+ T cells, increasing FoxP3 expression, and promoting the differentiation of Tregs [65, 69, 70] (Figure 2). Furusawa et al. utilized chromatin immunoprecipitation sequencing (ChIP-seq) for histone H3 acetylation to directly analyze the effects of butyrate on the epigenome at specific genomic loci in naïve CD4+ T cells under Treg polarizing-conditions [70]. Butyrate induced upregulation of histone H3 acetylation at both the promoter and intragenic enhancer elements (conserved noncoding sequence) of the FoxP3 locus. H3 acetylation at these loci positively correlated with FoxP3 expression, and increased at one intragenic site during the course of Treg differentiation, providing strong evidence that the effects of butyrate on FoxP3 expression and Treg differentiation are dependent on regulation of histone H3 acetylation at these loci. In addition to regulating histone acetylation, Arpaia et al. demonstrated that butyrate also results in increased acetylation of the Foxp3 protein, suggesting that acetylated Foxp3 is more stable and exhibits enhanced function [69].
Although thorough examination into how or which HDACs are directly mediating SCFA-dependent effects on Treg homeostasis is ongoing, butyrate and propionate, both SCFAs with HDAC-inhibitory activity, promoted Treg differentiation in the periphery, whereas acetate, a SCFA that lacks significant HDAC activity but is a potent ligand of GPR43, did not induce the same Treg effect, supporting that HDACs mediate effects of specific commensal-derived SCFAs on Tregs [69, 70]. Smith et al. suggested a mechanism by which SCFAs decreased expression of the class II HDACs, HDAC6 and/or HDAC9 in colonic Tregs. Consistent with these SCFA findings, synthetic HDAC inhibitors were previously found to limit colitis through expansion of Foxp3+ Tregs in association with differences in HDAC9 expression [77–79]. Targeting of HDAC6 and HDAC9 through deletion and inhibitor studies improved Treg suppressive function, however regulation of acetylation of the FoxP3 transcription factor, rather than histone H3 within the FoxP3 gene, was characterized as a potential mechanism in these reports [78, 80].
The effects of commensal bacteria-derived SCFAs on histone acetylation in myeloid cell lineages have also recently been examined. For instance, in addition to confirming direct effects of butyrate in promoting CD4+ Treg responses, Arpaia et al. suggest that butyrate-dependent HDAC inhibition in DCs may result in indirect promotion of colonic Treg differentiation [69]. DCs demonstrated increased global H3 acetylation levels in response to butyrate, indirectly implicating HDACs as targets of butyrate in DCs. Treatment of DCs with butyrate also resulted in decreased expression of LPS response genes, including IL-12, IL-6, and Relb [68, 69]. Similarly, Chang et al. demonstrated that treatment of macrophages with butyrate increased global histone acetylation and decreased expression of LPS-induced pro-inflammatory cytokines IL-6 and IL-12, independent of GPCRs and TLR signaling [68, 69]. (Figure 2). Inhibition of HDAC activity or loss of HDAC expression has been repeatedly been found to result in decreased enrichment of pro-inflammatory gene expression profiles [54, 57, 81, 82]. Investigation is ongoing into whether this decreased transcription of pro-inflammatory genes represents a direct, but counterintuitive, increase in histone acetylation at inflammatory genes, increased expression/recruitment of a transcriptional repressor, and/or decreased expression/recruitment of a transcriptional activator, possibly through altered acetylation of non-histone targets [54, 81, 83].
While butyrate increases levels of histone acetylation in immune cells, it remains unclear how this relates to physiologic concentrations of SCFAs in the colon and whether commensal-derived SCFAs increase histone acetylation primarily through regulation of HDAC expression versus direct inhibition of its enzymatic activity. Further, while loss of HDAC3 expression in IECs impairs microbiota-dependent intestinal barrier function, inhibition of HDACs by commensal bacteria-derived SCFAs in Tregs generally protects from pathologic intestinal inflammation, possibly underlying why multiple outcomes and mechanisms have been suggested for butyrate treatment during colitis [68, 84–87]. These seemingly opposing effects of HDACs in different intestinal cell populations warrant a more thorough examination of the differential effects of SCFAs on specific HDAC isoforms in different host cells, in the context of protective and pathologic immunity [66, 68, 84, 85, 87].
Complex interplay between microbiota and HDACs in the intestinal microenvironment
In addition to mediating microbiota-derived signals, it is likely that the interplay between the microbiota and epigenomic pathways involves complex feedback signals that influence one another, directly or indirectly (Figure 2). Deletion of HDAC3 from IECs in conventionally-housed mice (HDAC3ΔIEC mice) resulted in significant alterations in the composition of commensal bacteria, but this intestinal dysbiosis alone was not sufficient to transfer the IEC dysregulation observed in HDAC3ΔIEC mice to wildtype germ-free mice. Therefore, in addition to mediating commensal bacteria-dependent regulation of intestinal homeostasis, downstream responses of HDAC3-dependent regulation in IECs maintain normal diversity of the intestinal microbiota [57]. As discussed earlier, recent studies have underscored a critical role for naïve CD4+ T cells, Tregs and other immune cell populations in sensing and responding to commensal bacterial-derived SCFAs, in part through HDACs [65, 69, 70], however it remains unknown how pathways downstream of SCFAs or exogenous HDAC inhibitors feedback on commensal bacterial populations to regulate diversity of the microbiota.
Additional layers of complexity that influence cellular levels and targets of lysine acetylation will likely impact the dynamic crosstalk between the microbiota and epigenomic regulation by HDACs. Histone acetylation by HATs requires availability of the substrate donor, acetyl-coA, so interactions of HDACs with histones and subsequent deacetylation may vary based on the cellular levels of acetyl-CoA. There is increasing evidence that cellular metabolism of acetyl-coA is altered in response to environmental signals such as nutrient availability [50, 88, 89]. Therefore, conditions of cell stress or increased metabolism could influence the response of HDACs to microbiota-derived signals. Further, introduction of a conventional microbiota to germ-free mice led to acetylation of lysine residues on numerous non-histone proteins in the liver and colon [90]. Therefore, in addition to regulating histone acetylation, commensal bacterial-derived signals appear to also impact acetylation of non-histone substrates. Future studies are needed to determine the contributions of specific HATs or HDACs in regulating microbiota-dependent acetylation of non-histone proteins.
HDACs can be targeted by a large class of inhibitors that are currently being utilized or examined for the treatment of various types of cancer, as well as inflammatory and degenerative conditions [45, 91, 92]. Currently, two HDAC inhibitors, vorinostat (suberoylanilide hydroxamic acid) and depsipeptide (romidepsin) are approved for treatment of refractory cutaneous T-cell lymphoma and more recently, depsipeptide was approved for treatment of peripheral T-cell lymphoma [93]. The therapeutic potential of HDAC inhibitors is promising and numerous compounds that either target multiple HDACs or specific isoforms are being evaluated clinically. However, as the mechanisms underlying their clinical effects are not fully understood, studies directed towards better understanding their specificity and mode of action are ongoing [93, 94].
Concluding Remarks
Although the recent advances discussed here offer promising steps toward recognizing the importance of epigenomics in mediating the host-microbiota relationship, understanding the mechanisms and extent to which epigenomic regulation orchestrates this relationship is still in its infancy (Box 1). Determining the critical pathways and modes of action by which cell-specific epigenomic-modifying enzymes differentially respond to commensal bacteria-derived signals and regulate the epigenome will be essential. Future investigation is also needed into the clinical implications of utilizing epigenomic-targeting drugs, such as DNMT and HDAC inhibitors, on the microbiota and associated microbiota-dependent immune and metabolic health. Further, epigenomics may provide an underlying mechanism for how long-term and multigenerational effects of the microbiota are inherited. In order to better understand and, eventually, intervene therapeutically at this level of regulation, more thorough examination into the cellular heritability of microbiota-dependent epigenomic modifications must be conducted.
Box 1. Important Areas of future research.
Identifying critical pathways by which cell lineage-specific epigenomic-modifying enzymes differentially respond to commensal bacteria-derived signals and regulate the epigenome.
Characterizing how alterations in the levels of microbiota-derived SCFAs are sensed in vivo and the mode of action by which SCFAs regulate HDACs in hematopoietic and non-hematopoietic cells.
Exploring the clinical implications of utilizing epigenomic-targeting drugs, such as DNMT and HDAC inhibitors, on the microbiota and microbiota-associated immune and metabolic homeostasis.
Examining the cellular heritability of microbiota-dependent epigenomic modifications.
Epigenomic pathways likely play a central role in regulating susceptibility to several human diseases that are influenced by both genetic and microbe-derived factors. However, progress towards understanding how these pathways can be manipulated to improve host-microbiota interactions and treat microbiota-influenced diseases will require continued cross-disciplinary basic science and translational investigation into this emerging field.
Highlights.
Epigenomic modifications enable environmental cues to alter transcriptional programs in mammalian cells.
Signals derived from the microbiota in the intestinal microenvironment likely regulate the epigenome.
Recent advances suggest that histone deacetylases are key epigenomic-modifying enzymes in the mammalian host that mediate interactions with the microbiota.
Acknowledgments
The authors thank members of the Alenghat and Artis laboratories for discussions and critical reading of the manuscript. This work is supported by the National Institutes of Health (DK093784 to T.A.; AI061570, AI095608, AI087990, AI074878, AI095466, AI106697, AI102942, AI097333 to D.A.), the Crohns and Colitis Foundation of America (T.A. and D.A.), the Burroughs Wellcome Fund Career Award for Medical Scientists (T.A.), and the Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award (D.A.).
Glossary
- Chromatin
The DNA-protein complex that is packaged into the nucleus of a cell. Chromatin is a higher order, dynamic structure
- Nucleosome
The basic unit of chromatin that is composed of approximately 147 base pairs of DNA wrapped around a single histone octamer
- Epigenetics
Changes in gene expression that are heritable through cell divisions and potentially generations, but do not result from alterations in the genetic code
- Epigenome
The combination of chromatin modifications and associated proteins that collectively package the genome and direct transcription
- Epigenomic-modifying enzymes
Enzymes that alter the epigenome by catalyzing the covalent addition or removal of specific small molecules
- Histone acetyltransferases (HATs)
An epigenomic-modifying enzyme that transfers an acetyl group from acetyl CoA to lysine residues on histones or non-histone proteins. Bromodomains that are present in various proteins preferentially interact with acetyl-lysines generated by HAT activity
- Histone deacetylases (HDACs)
An epigenomic-modifying enzyme that removes acetyl groups from acetylated lysines on histones or non-histone proteins. HATs and HDACs are also now called lysine acetyltransferases (KATs) and lysine deacetylases (KDACs) to acknowledge their ability to deacetylate lysine residues on non-histone targets
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Renz H, et al. Gene-environment interactions in chronic inflammatory disease. Nat Immunol. 2011;12:273–277. doi: 10.1038/ni0411-273. [DOI] [PubMed] [Google Scholar]
- 2.Slomko H, et al. Minireview: Epigenetics of obesity and diabetes in humans. Endocrinology. 2012;153:1025–1030. doi: 10.1210/en.2011-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee AB, Zhang Z. Allergic asthma: influence of genetic and environmental factors. J Biol Chem. 2011;286:32883–32889. doi: 10.1074/jbc.R110.197046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strober W, et al. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kau AL, et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaser A, et al. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamada N, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 10.Spasova DS, Surh CD. Blowing on embers: commensal microbiota and our immune system. Frontiers in immunology. 2014;5:318. doi: 10.3389/fimmu.2014.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duerkop BA, et al. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Kabat AM, et al. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014 doi: 10.1016/j.it.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 15.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musso G, et al. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 17.McLoughlin RM, Mills KH. Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. J Allergy Clin Immunol. 2011;127:1097–1107. doi: 10.1016/j.jaci.2011.02.012. quiz 1108–1099. [DOI] [PubMed] [Google Scholar]
- 18.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 21.Arrowsmith CH, et al. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 22.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 23.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 24.Begin P, Nadeau KC. Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin Immunol. 2014;10:27. doi: 10.1186/1710-1492-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 26.Handel AE, et al. Epigenetics: molecular mechanisms and implications for disease. Trends Mol Med. 2010;16:7–16. doi: 10.1016/j.molmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Hooper LV, et al. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 29.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roda G, et al. Intestinal epithelial cells in inflammatory bowel diseases. World J Gastroenterol. 2010;16:4264–4271. doi: 10.3748/wjg.v16.i34.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahan S, et al. Epithelia: lymphocyte interactions in the gut. Immunol Rev. 2007;215:243–253. doi: 10.1111/j.1600-065X.2006.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khosravi A, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimitriu PA, et al. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environmental microbiology reports. 2013;5:200–210. doi: 10.1111/j.1758-2229.2012.00393.x. [DOI] [PubMed] [Google Scholar]
- 35.Bierne H, et al. Epigenetics and bacterial infections. Cold Spring Harbor perspectives in medicine. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rakoff-Nahoum S, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, et al. Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J Biol Chem. 2011;286:35755–35762. doi: 10.1074/jbc.M111.271007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obata Y, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15:571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 40.Ganal SC, et al. Priming of Natural Killer Cells by Nonmucosal Mononuclear Phagocytes Requires Instructive Signals from Commensal Microbiota. Immunity. 2012 doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, et al. HATs on and beyond chromatin. Curr Opin Cell Biol. 2001;13:218–224. doi: 10.1016/s0955-0674(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 42.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 43.Owen DJ, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. Embo J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haberland M, et al. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauve AA, et al. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 47.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 48.You SH, et al. The interaction between nuclear receptor corepressor and histone deacetylase 3 regulates both positive and negative thyroid hormone action in vivo. Mol Endocrinol. 2010;24:1359–1367. doi: 10.1210/me.2009-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donohoe DR, Bultman SJ. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J Cell Physiol. 2012;227:3169–3177. doi: 10.1002/jcp.24054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 52.Kim GW, et al. Dietary, metabolic, and potentially environmental modulation of the lysine acetylation machinery. Int J Cell Biol. 2010;2010:632739. doi: 10.1155/2010/632739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–369. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012;109:E2865–2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson AJ, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 56.Tou L, et al. Regulation of mammalian epithelial differentiation and intestine development by class I histone deacetylases. Mol Cell Biol. 2004;24:3132–3139. doi: 10.1128/MCB.24.8.3132-3139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alenghat T, et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature. 2013;504:153–157. doi: 10.1038/nature12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi T, et al. IL-10 regulates Il12b expression via histone deacetylation: implications for intestinal macrophage homeostasis. J Immunol. 2012;189:1792–1799. doi: 10.4049/jimmunol.1200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turgeon N, et al. HDAC1 and HDAC2 restrain the intestinal inflammatory response by regulating intestinal epithelial cell differentiation. PLoS One. 2013;8:e73785. doi: 10.1371/journal.pone.0073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turgeon N, et al. The acetylome regulators Hdac1 and Hdac2 differently modulate intestinal epithelial cell dependent homeostatic responses in experimental colitis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G594–605. doi: 10.1152/ajpgi.00393.2013. [DOI] [PubMed] [Google Scholar]
- 61.Cummings JH, et al. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vinolo MA, et al. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 64.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim MH, et al. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. e391–310. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 67.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 68.Chang PV, et al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 71.Burlingham WJ, et al. IL-17-dependent cellular immunity to collagen type V predisposes to obliterative bronchiolitis in human lung transplants. Journal of Clinical Investigation. 2007;117:3498–3506. doi: 10.1172/JCI28031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 73.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 74.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Candido EP, et al. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 76.Waldecker M, et al. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 77.de Zoeten EF, et al. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010;138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tao R, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 79.Glauben R, et al. Histone deacetylases: novel targets for prevention of colitis-associated cancer in mice. Gut. 2008;57:613–622. doi: 10.1136/gut.2007.134650. [DOI] [PubMed] [Google Scholar]
- 80.Beier UH, et al. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci Signal. 2012;5:ra45. doi: 10.1126/scisignal.2002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roger T, et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- 82.Jeong Y, et al. Histone deacetylase isoforms regulate innate immune responses by deacetylating mitogen-activated protein kinase phosphatase-1. J Leukoc Biol. 2014;95:651–659. doi: 10.1189/jlb.1013565. [DOI] [PubMed] [Google Scholar]
- 83.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 84.Berndt BE, et al. Butyrate increases IL-23 production by stimulated dendritic cells. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1384–1392. doi: 10.1152/ajpgi.00540.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamer HM, et al. Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin Nutr. 2010;29:738–744. doi: 10.1016/j.clnu.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Diehl GE, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tarrerias AL, et al. Short-chain fatty acid enemas fail to decrease colonic hypersensitivity and inflammation in TNBS-induced colonic inflammation in rats. Pain. 2002;100:91–97. doi: 10.1016/s0304-3959(02)00234-8. [DOI] [PubMed] [Google Scholar]
- 88.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40:323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simon GM, et al. Quantitative assessment of the impact of the gut microbiota on lysine epsilon-acetylation of host proteins using gnotobiotic mice. Proc Natl Acad Sci U S A. 2012;109:11133–11138. doi: 10.1073/pnas.1208669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang L. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases. J Cell Physiol. 2006;209:611–616. doi: 10.1002/jcp.20781. [DOI] [PubMed] [Google Scholar]
- 92.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014 doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 93.Ververis K, et al. Histone deacetylase inhibitors (HDACIs): multitargeted anticancer agents. Biologics. 2013;7:47–60. doi: 10.2147/BTT.S29965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bode KA, Dalpke AH. HDAC inhibitors block innate immunity. Blood. 2011;117:1102–1103. doi: 10.1182/blood-2010-11-315820. [DOI] [PubMed] [Google Scholar]