Abstract
Disruptions in procollagen synthesis, trafficking and secretion by cells occur in multiple connective tissue diseases. Traditionally, these disruptions are studied by pulse-chase labeling with radioisotopes. However, significant DNA damage, excessive accumulation of reactive oxygen species and formation of other free radicals have been well documented in the literature at typical radioisotope concentrations used for pulse-chase experiments. Therefore, it is important to keep in mind that the resulting cell stress response might affect interpretation of the data, particularly with respect to abnormal function of procollagen-producing cells. Here, we describe an alternative method of pulse-chase procollagen labeling with azidohomoalanine, a noncanonical amino acid that replaces methionine in newly synthesized protein chains and can be detected via highly selective click chemistry reactions. At least in fibroblast culture, this approach is more efficient than traditional radioisotopes and has fewer, if any unintended effects on cell function. To illustrate its applications, we demonstrate delayed procollagen folding and secretion by cells from an osteogenesis imperfecta patient with a Cys substitution for Gly766 in the triple helical region of the α1(I) chain of type I procollagen.
Keywords: Azidohomoalanine, collagen, osteogenesis imperfecta, metabolic labeling, pulse-chase
Introduction
Collagens are the most highly abundant proteins in vertebrates distinguished by one or more characteristic triple helical domains. These domains are formed by three amino acid chains that have an obligatory glycine (Gly) in every third position. A significant fraction of X and Y positions in the Gly-X-Y repeats are occupied by proline (Pro) and hydroxyproline (Hyp). Like Gly, both Pro and Hyp are required for maintaining the structural integrity and stability of the triple helix. Procollagen precursors of collagens are synthesized by different types of cells, in some of which collagens account for about a third of all proteins produced by the cell. Secreted mature collagens form structural scaffolds of bone, cartilage, and other tissues as well as glue together different components of extracellular matrix (ECM) throughout the body.
Abnormal collagen biosynthesis is involved in a variety of disorders, ranging from relatively rare skeletal dysplasias (1-3) and vascular syndromes (4, 5) to such common ailments as fibrosis (6) and cancer (7, 8). Protein biosynthesis pathways and their role in pathology are often studied by pulse-chase measurements, in which labeled amino acids are introduced into cell culture media and incorporated by cells into newly synthesized chains (9, 10). After the labeling “pulse”, synthesis, folding, trafficking, secretion, and tissue integration of the labeled chains is followed by a “chase” in the media that contains only normal, unlabeled amino acids.
The most common approach to pulse-chase measurements utilizes amino acids containing radioactive isotopes. Labeling with 35S-Met is particularly popular since it is easily detected and highly active, and methionine is an essential amino acid that is not synthesized by human cells de novo (11, 12). Collagen is also frequently labeled with 3H- or 14C-Pro (13, 14). Because of much higher proline content relative to most other proteins, 3H- or 14C-Pro labeled collagen chains may be visualized by gel electrophoresis without purification. Another approach is labeling with non-radioactive, stable isotopes, but detection of such isotopes is more difficult, requires expensive instruments and is challenging for gel electrophoresis measurements (15-18).
A particularly appealing alternative to radioisotopes is non-canonical amino acids that are incorporated by cells into proteins instead of regular amino acids (19). For instance, azidohomolanine (Aha) is a methionine analogue (Fig. 1a) that is efficiently incorporated into aminoacyl-tRNA and proteins instead of methionine (20, 21). Unlike inorganic azide ions, the azide group of Aha is stable and nontoxic (22-24). It can be efficiently conjugated with fluorescent dyes via highly specific “click chemistry” reactions, even within live cells (22, 25). Previously published studies did not reveal any significant effects of this labeling on translation initiation, chain synthesis or protein folding (21, 25, 26).
Figure 1.
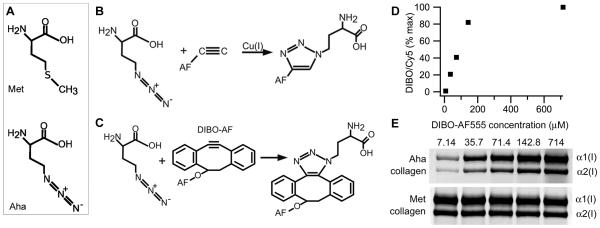
Procollagen labeling with azidohomoalanine (Aha). (A) Aha and methionine (Met) structures. (B) Cu-catalyzed and (C) Cu-free conjugation of Aha with Alexa Fluor (AF) fluorescent dyes. (D) Aha conjugation efficiency with DIBO-AF555 at different concentrations of the dye calculated from AF555/Cy5 fluorescence intensity ratio in the same gel band (panel E). The conjugation efficiency at 714 uM DIBO-AF555 is assumed to be 100%. (E) Gel electrophoresis of pepsin-treated procollagen (collagen) after Aha conjugation with DIBO-AF555 and Lys conjugation with Cy5. Aha-labeled collagen is revealed by fluorescence scanning at 532 nm excitation and 570±10 nm emission wavelength (AF555) while total collagen (Met collagen) is revealed by scanning of the same gel bands at 635 nm excitation and ≥ 665 nm emission (Cy5).
In the present study, we found collagen pulse-chase labeling with Aha to be more economical, efficient and convenient compared to radioisotopes. For instance, it allows analysis of gel electrophoresis results without a one-two week delay for capturing the gel image on an x-ray film or imaging plate for autoradiography. Moreover, we also found no appreciable non-target effects of Aha on the cells, in contrast to significant DNA damage, cell cycle changes and growth arrest reported in studies of radioisotope labeling (27-33).
Given our focus on nonradioactive labeling as well as safety and environmental considerations, we did not perform parallel pulse-chase measurements with radioisotopes. We believe that radioisotopes might be useful for some studies but should not be viewed as a reference standard for other pulse-chase approaches. On the contrary, we argue that well-established and well-published radioisotope effects on cellular function might require one to be more cautious in interpreting the results of radioisotope-based experiments.
Here, we analyze optimal conditions and potential non-target effects of collagen labeling with Aha. To illustrate this assay, we describe procollagen folding kinetics in fibroblasts from an osteogenesis imperfecta (OI) patient with a Gly766 to Cys substitution in the triple helical region of the α1(I) collagen chain. Like in an earlier report for other Gly substitutions (34), we observed a delay in folding of procollagen molecules containing the mutant chain. Slower procollagen folding and resulting misfolding might cause bone pathology by affecting the function of osteoblasts (3, 35).
Materials and methods
Cell culture
Normal control dermal human fibroblasts were generously provided by Dr. Joan Marini, National Institute of Child Health and Human Development, National Institutes of Health. Human dermal fibroblasts containing a type I collagen α1-chain-Gly766→Cys substitution were generously provided by Dr. Peter Byers, University of Washington School of Medicine. Fibroblasts were cultured at 37°C, 5% CO2 for less than 15 passages; 0.05% Trypsin-EDTA (Invitrogen) was utilized for cell passage.
Growth medium, Dulbecco’s modified Eagle’s medium (DMEM) with GlutaMAX™ (Invitrogen) and 10% fetal bovine serum (Cell Grow or Sigma) was used for general cell culture. (b) Labeling medium, Met and Cys free DMEM and 250 μM ascorbic acid 2-phosphate (Sigma) was used for labeling with AHA. (c) Chase medium, Met and Cys free DMEM, 250 μM ascorbic acid 2-phosphate, and 10 mM Met was used in pulse-chase experiments.
Labeling with azidohomoalanine and procollagen isolation
Confluent cells were treated with 250 μM ascorbic acid 2-phosphate (Sigma) in the growth medium for 1 day, depleted of Met in the labeling medium, and incubated with different concentrations of azidohomoalanine (Aha, IRIS, Germany) in the labeling medium. In pulse-chase experiments, the cells were subsequently incubated in the chase medium. Media procollagen was precipitated with 176 mg/mL ammonium sulfate. To measure the amount of secreted procollagen, internal collagen standard was added to 0.8 μg/mL final concentration before the precipitation. The internal standard (rat-tail-tendon collagen cleaved with MMP-1 and fluorescently labeled with Alexa Fluor 488) allows accurate measurement of the initial collagen concentration and provides a control for the efficiency of subsequent fluorescent labeling; its ratio to full-length collagen and procollagen remains constant within our purification procedure (8). Cell procollagen was extracted by washing the cells with PBS for 10 minutes at 4 °C on a rocker, followed by lysis with cell extraction buffer (1% NP40, 50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 mM benzamidine, 10 mM N-ethylmaleimide (NEM), pH 8.0) and precipitation with 176 mg/mL ammonium sulfate. Internal collagen standard was added (0.15 μg/mL final concentration) as needed before the precipitation. Note that NEM covalently modifies cysteine residues, but normal type I collagen chains do not contain such residues and we have never observed any effect of NEM on purification or gel electrophoresis of mutant collagen chains with Gly to Cys substitutions.
Fluorescent labeling and gel electrophoresis
Collagen was purified from procollagen precipitates by resuspension and overnight incubation in 0.5 M acetic acid, 0.1 mg/mL pepsin at 4 °C followed by precipitation with 0.7 M NaCl (final concentration). Click-IT Alexa Fluor 555 dibenzocyclooctyne (DIBO-AF555, Invitrogen) and monoreactive N-hydroxysuccinimide ester of Cy5 (Cy5, GE Healthcare) were suspended in anhydrous dimethylformamide, aliquoted, lyophilized, and stored with desiccant at 4 °C (Cy5) or −20 °C (DIBO-AF555) protected from light. For AHA labeling, collagen resuspended in 0.2 M phosphate, 150 mM NaCl, pH 8.0 was rapidly mixed 10:1 with DIBO-AF555 resuspended in dimethylformamide at different concentrations, vortexed and incubated for 1 h at room temperature on a shaker. Labeling of collagen Lys with Cy5 was performed as previously described (36). In double labeling experiments, the reaction of AHA with DIBO-AF555 was followed by the reaction of Lys with Cy5 in the same buffer.
Fluorescently labeled samples were mixed with lithium dodecyl sulfate sample buffer (Invitrogen), denatured for 10 min at 55-60 °C, and separated on precast 3-8% Tris-acetate mini gels (Invitrogen). The dye front was run off the gel to reduce fluorescence background. Gel images were captured in an FLA5000 fluorescence scanner (Fuji Medical Systems, Stamford, CT) and analyzed with Multigauge 3.0 software supplied with the scanner. The total amount of secreted and intracellular collagen was determined from the known concentration of the internal standard and measured ratio of Cy5 fluorescence intensities in the collagen and internal standard bands as previously described (8).
Procollagen folding experiments
Fibroblasts were plated in 22-cm2 dishes, grown to confluence, stimulated with 250 uM ascorbic acid 2-phosphate for 1 day, depleted of methionine for 30 min, pulse labeled with 500 μM AHA for 10 min in labeling media, and chased in 10 mM methionine in chase media for different time intervals from 0 to 120 min. All media were pre-warmed to 37°C and cell culture dishes were maintained on a heat block set to 38°C to ensure that the cells were at 37°C during all media changes. After the chase, a dish was incubated at 22°C for 10 s, cell lysis buffer concentrate was added directly to the media for a final concentration of 1% NP40, 200 μg/mL chymotrypsin, 80 μg/mL trypsin, 2 mM EGTA (all from Sigma). The dish was shaken for 10 s, incubated for 50 s, and shaken again for 10 s. At 75 s after the lysis buffer addition, the dish was tilted slightly to allow lysate to pool. At 90 s, the lysate was collected into a clean 2.0 mL microcentrifuge tube. At 120 s, protease inhibitors (5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 mM benzamidine, 10 mM NEM) were added to stop the enzymatic digestion. The lysate was vortexed, placed on ice, and mixed with internal collagen standard (0.15 μg/mL final concentration) and Tris-HCl (100 mM final concentration, pH 7.4). Collagen was precipitated with 176 mg/mL ammonium sulfate, treated with 0.1 mg/mL pepsin in 0.5 M acetic acid, reprecipitated with 0.7 M NaCl, fluorescently labeled with DIBO-AF555 and Cy5, and analyzed by gel electrophoresis.
Differential scanning calorimetry (DSC)
Procollagen precipitated with ammonium sulfate was either resuspended and dialyzed in PBS at 4°C or treated with 0.1 mg/mL pepsin in 0.5 M acetic acid overnight at 4°C, followed by 0.7 M NaCl precipitation, resuspension and dialysis in 2 mM HCl. The resulting ~0.1 mg/ml solutions of procollagen and collagen were scanned at 0.125°C/min heating rate in a Nano III DSC instrument (Calorimetry Sciences Corporation).
Quantitative real time PCR (qPCR)
RNA was isolated from fibroblasts with an RNeasy kit (Qiagen), reverse transcribed with SuperScript III First Strand Synthesis Supermix and random hexamers as primers (Invitrogen), and analyzed in a 7500 Fast Real Time PCR system (Applied Biosystems) with Taqman gene expression assays (DDIT3 (CHOP), Hs00358796_g1; HSPA5 (BIP), Hs00607129_gH; BCL2 (BCL2), Hs00608023_m1; CRYAB (crystallin αB), Hs00157107_m1; COL1A1 (procollagen α1(I)), Hs00164004_m1; Applied Biosystems). The same CT threshold value was used for all samples. Relative mRNA quantity was calculated from ΔΔCT values by utilizing HPRT1 (HPRT), Hs99999909_m1 and B2M (B2M), Hs99999907_m1 (Applied Byosystems) as endogenous controls and assuming 100% PCR efficiency (37).
Results
Azidohomoalanine conjugation with fluorescent dyes
Our attempts to utilize traditional, copper-catalyzed click chemistry for conjugation of Aha incorporated into α1(I) and α2(I) chains of type I collagen with alkyne derivatives of fluorescent dyes (Fig. 1b) were largely unsuccessful due to Cu-induced collagen precipitation. In buffers that inhibit this precipitation, we observed only low-efficiency, inconsistent Aha conjugation (data not shown).
In contrast, Aha conjugation with commercially available dibenzocyclooctyne (DIBO) derivatives of Alexa Fluor dyes (DIBO-AF) provided more efficient and consistent labeling of collagen chains (Fig. 1c). Labeling of collagen with DIBO derivative of Alexa Fluor 555 (DIBO-AF555) reached saturation around 150 μM (Fig. 1d,e). For our experiments, we chose to use 71.4 μM DIBO-AF555, where the conjugation efficiency was approximately 50% to reduce reagent used yet still get efficient labeling.
To determine the conjugation efficiency, we also labeled lysine (Lys) with Cy5 (N-hydroxysuccinimide ester) within the same sample and measured the AF555/Cy5 ratio of fluorescence intensities in the same gel electrophoresis bands of α1(I) and α2(I) chains. To correct for possible sample-to-sample variation in Lys labeling, MMP1-cleavage fragments of rat-tail-tendon collagen prelabeled with Alexa Fluor 488 were added to each sample before labeling with Cy5. These fragments provided an internal standard for Lys labeling efficiency, gel loading, and calculation of absolute collagen concentration in the sample (8).
Optimization of Aha incorporation into collagen
To optimize Met replacement with Aha, we varied the length of Met depletion, length of Aha pulse, and Aha concentration during the pulse and measured the DIBO-AF555/Cy5 ratio in gel bands of α1(I) and α2(I) chains. We found that 500 μM Aha concentration was sufficient for achieving near maximum Aha incorporation into procollagen newly secreted by fibroblasts (Fig. 2a). A 30 min incubation of the cells in Met and Cys free media prior to the addition of Aha was necessary and sufficient for maximum Aha incorporation (Fig. 2b). The saturating concentration of Aha was achieved after 4 hours in secreted procollagen (Fig. 2c) and after 2 hours in intracellular procollagen (Fig. 2d).
Figure 2.
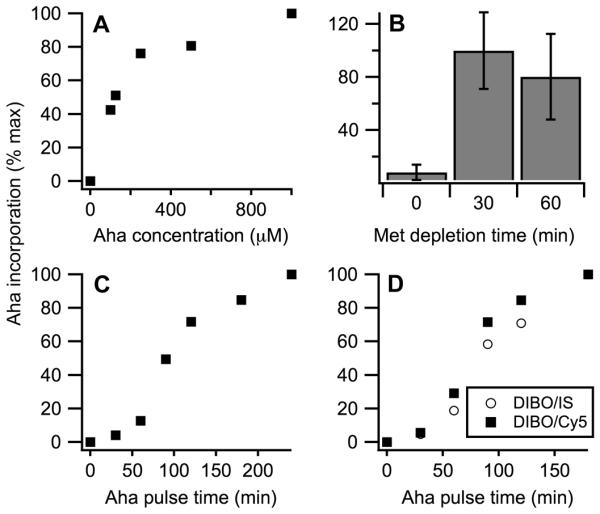
Optimization of Aha incorporation into procollagen. Aha incorporation is measured from DIBO-AF555/Cy5 fluorescence intensity ratio in secreted (A-C) and intracellular (D) procollagen as described in Fig. 1D. (A) Aha incorporation after 30 min Met depletion followed by 4 h pulses of different Aha concentrations. (B) Aha incorporation after Met depletions of different lengths followed by 4 h 500 μM Aha pulses. Error bars represent standard deviation. (C) Aha incorporation after 30 min Met depletions followed by 500 μM Aha pulses of different lengths. (D) Aha incorporation into intracellular procollagen after 30 min Met depletions followed by 500 μM Aha pulses of different lengths measured from DIBO-AF555/Cy5 (squares) and from DIBO-AF555/IS-AF488 (circles) fluorescence intensity ratios. The latter ratio normalizes the amount of Aha conjugated with DIBO-AF555 by the amount of internal standard (IS) conjugated with AF488, which is added to the cell lysis buffer. The two methods of evaluating Aha incorporation produce consistent results.
Effects of Aha labeling on fibroblast function and procollagen biosynthesis
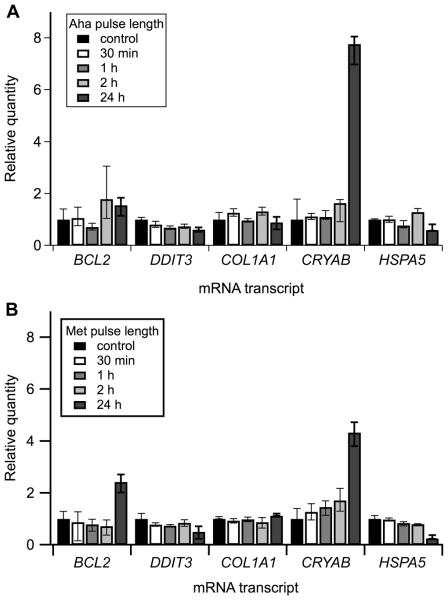
To test effects of this labeling procedure on cell function and procollagen synthesis, we compared: (i) fibroblast cultures in which 30 min Met/Cys depletion was followed by a 500 μM Aha pulse, (ii) cultures in which the same depletion was followed by a 500 μM Met pulse, and (iii) control cultures without the depletion and Met or Aha pulses. We found no effects of Met depletion and short Aha and Met pulses on transcript levels of key cell stress proteins. We observed decreased BIP and increased crystallin transcription after 24 h pulses, indicating slow activation of some cellular response (Fig. 3a,b). The response was similar for Met and Aha and likely associated with low serum concentration in the media (commonly used in pulse-chase studies to promote utilization of labeled amino acids by cells, rather than amino acids from serum proteins). However, the latter response should not pose a significant problem for typical pulse-chase experiments that do not extend beyond 4-8 h.
Figure 3.
Quantitative real-time PCR analysis of Aha effects on different cell stress markers. (A) 500 μM Aha pulses of increasing length were compared with (B) 500 μM Met pulses under identical cell culture conditions. Expression of BCL2, DDIT3 (CHOP), COL1A1 (procollagen α1(I)), CRYAB (crystallin αB), and HSPA5 (BiP) transcript levels were measured relative to cells in growth medium (control) and normalized to HPRT1 and B2M as endogenous controls. All error bars represent standard deviation.
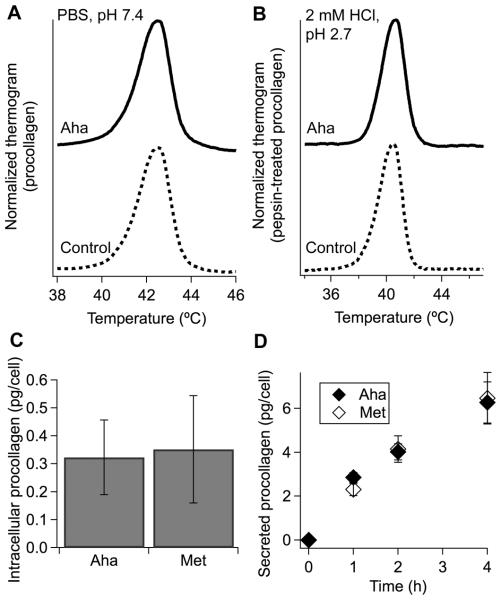
We did not observe any detectable effects of Aha labeling on procollagen biosynthesis either. Procollagen transcription and secretion rates as well as the amount of intracellular procollagen remained the same as in control cells for at least 4h (Fig. 4c,d). Normal electrophoretic mobility and thermal denaturation thermograms of collagen and procollagen indicated that Aha incorporation did not disrupt posttranslational modification, stability or structure of the triple helix (Figure 1e, 4a, b).
Figure 4.
Effects of Aha incorporation on procollagen stability, synthesis, and secretion. (A,B) Denaturation thermograms of secreted protein after ammonium sulfate isolation of procollagen (A) and subsequent purification of pepsin-treated collagen (B). (C) Amount of procollagen inside each cell after 4 h stimulation with 250 μM ascorbic acid 2-phosphate in growth medium followed by 30 min Met depletion and a 4 h pulse of 500 μM Aha or Met. (D) Amount of procollagen secreted by each cell after the same treatment, except for varying Aha/Met pulse length.
Analysis of procollagen misfolding and retention in OI fibroblasts by pulse-chase Aha labeling
To test Aha labeling in the context of practically important experiments, we analyzed procollagen folding and secretion in fibroblasts from an OI patient with a Cys substitution for Gly766 in the α1(I) chain (38). Analysis of collagen secretion represents a typical pulse-chase experiment utilized in studies of collagen metabolism disorders (14). Measurement of procollagen folding kinetics represents one of the most challenging pulse-chase experiments. Since newly synthesized procollagen is folded and secreted by fibroblasts within ~30-60 min, measurement of the folding kinetics requires ~10 min or shorter pulse labeling and therefore high sensitivity of detecting the labeled molecules (39).
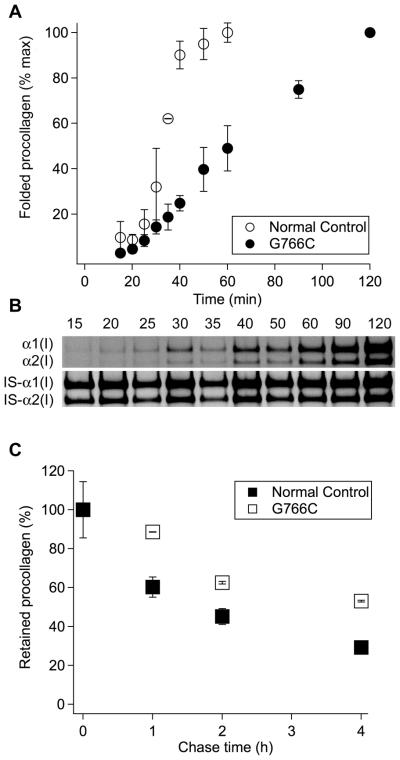
We observed significantly slower folding of procollagen molecules in OI compared to normal control fibroblasts (Fig. 5a,b), consistent with folding of procollagen with other Gly substitutions studied by pulse-chase labeling with 35S-Met (34). We quantified the kinetics of formation of cleavage-resistant folded triple helices by gel electrophoresis, after other proteins and unfolded procollagen chains in cell lysates were degraded by chymotrypsin and trypsin (34). We detected only full-length helices but not degradation products of partially folded molecules, in which the C-to-N-terminal triple helix propagation paused at the mutation site.
Figure 5.
Folding and intracellular retention of procollagen with an α1(I)-G766C substitution. (A,B) Measurement of procollagen folding kinetics by pulse-chase labeling with Aha. The fraction of fully folded procollagen (A) is calculated relative to the latest time point from DIBO-AF555/IS-AF488 fluorescence intensity ratios of gel electrophoresis bands (B); IS-α1 and IS-α2 are long fragments of α1(I) and α2(I) chains of rat-tail-tendon collagen cleaved with MMP1 used as internal standard (IS). (C). Kinetics of procollagen clearance from cells measured by pulse-chase labeling with Aha from DIBO-AF555/IS-AF488 fluorescence intensity ratios of gel electrophoresis bands of intracellular procollagen after different chase time. The average DIBO-AF555/IS-AF488 ratio at zero chase time is taken as 100%. All error bars represent standard deviation in triplicate experiments.
We also observed much slower clearance of Aha labeled procollagen from OI compared to normal control fibroblasts. Figure 5c shows the relative content of Aha in triple helices of intracellular procollagen during the chase, which was evaluated from DIBO-AF555/IS-AF488 fluorescence intensity ratio of DIBO-AF555 labeled collagen to AF488 labeled internal standard (IS). These helices were purified by pepsin treatment and salt fractionation from cell lysates free of cell debris and ECM. By directly measuring their Aha content, our assay accurately quantifies the clearance kinetics of labeled procollagen from the cell.
Discussion
Effects of pulse-chase labeling with radioisotopes on cell function
Pulse-chase labeling of procollagen is most commonly performed with 25-500 μCi/ml 3H-proline (14, 40), 0.5-5 μCi/ml 14C-proline (13, 40), or 10-200 μCi/mL 35S-methionine (34, 41). However, recent studies indicate that these low-energy β-emitters cause significant cell stress and malfunction even at low concentrations and short exposure (28, 29, 32, 33, 42). Significant fragmentation of DNA was observed after 2h at 10 μCi/ml 35S-methionine and 2-20 μCi/ml 3H-thymidine in the cell culture media (30). Radioisotope incorporation into multiple molecules and subsequent radioactive decay inside the cell was found to cause persistent DNA fragmentation long after replacing the media, suggesting that even short pulse labeling might have severe consequences (28, 30). The observed response to radioisotopes was similar in different cell types. Radioisotope labeling at or below typical concentrations was reported to elevate the level of p53, induce formation of reactive oxygen species (ROS), inhibit cell cycle progression, cause growth arrest, and increase apoptosis (28-30, 33, 42). Gene expression profiling revealed dramatic changes in multiple cell stress response proteins (31).
The observations of significant DNA damage, cell cycle changes, and growth arrest after 3H and 35S exposure indicate that other cellular functions are likely to be altered as well. Radioisotope effects might be particularly severe and difficult to account for in pulse-chase labeling studies of procollagen secretion by cells from patients with procollagen biosynthesis abnormalities. Such experiments require at least 2-4 h equilibration in the radioactive media followed by 2-4 h chase and therefore at least 4-8 h cumulative exposure to radioactive isotopes outside and inside the cells. During this time, DNA damage and ROS might significantly alter procollagen biosynthesis, e.g. because they affect Wnt/β-catenin, TGF-β, BMP and other key signaling pathways (43, 44). Retention of abnormal procollagen by the cells might worsen the radioactive damage and its downstream effects by enhancing radioisotope accumulation, particularly in the case of 3H-Pro or 14C-Pro labeling. Combined with differences in the general metabolic activity and susceptibility to radioactive damage, these effects might significantly complicate the comparison between patient and normal control cells.
The response of cells to the DNA damage, ROS and other highly reactive free radicals may therefore affect the interpretation of pulse-chase radioisotope labeling experiments. While these experiments may provide useful information about procollagen chain association, folding and secretion, full understanding of their results requires characterization of potential radioactive damage effects on the cells. Since such damage might depend on specific experimental conditions, its consequences are difficult to predict a priori. One potential alternative is labeling with non-radioactive isotopes, which may be detected by a variety of spectroscopic techniques (16, 18, 45). Another alternative, which we are pursuing in the present study, is labeling with noncanonical amino acids.
Labeling with Aha as an alternative approach
At least for procollagen, replacement of methionine with azidohomoalanine provides a particularly useful labeling approach. It is sufficiently sensitive, utilizes commercially available reagents, reduces environmentally toxic waste, eliminates exposure to radioactivity, and lowers the cost of experiments. Detection of Aha-labeled molecules by conjugation with fluorescent dyes is based on a reaction not found in living systems and is highly specific, eliminating background labeling (22, 24). It shortens the time for visualizing labeled proteins on gels. Instead of several days or weeks typically required for x-ray film or imaging plate exposure in autoradiography, fluorescence scanning of gels takes minutes. A pre-labeled internal standard and conjugation of Aha and Lys with different fluorescent dyes allow data analysis with fewer or no assumptions compared to traditional methods that implicitly assume similar procollagen labeling efficiency and extraction yield in patient and control cells, which may not be the case.
In contrast to radioisotopes, Aha does not affect cell viability (21) and has fewer, if any, unintended effects on cell function. In fibroblasts, we found no changes in expression of BCL2, BIP, and CHOP after replacing Met with Aha (Fig. 3), suggesting minimal or no cell stress. We also detected no changes in collagen gene or protein expression (Fig. 3, 4c,d), supporting previous observations of normal protein synthesis rate (20, 25) and therefore normal translation initiation at Met codon. Approximately 400 fold slower incorporation of Aha into aminoacyl-tRNA by methionyl-tRNA synthetase does not appear to hinder protein synthesis either (46).
Normal procollagen synthesis rate, concentration in the cell, and lifetime in the cell as well as unaltered triple helix stability, all indicate minimal or no effect of the Met to Aha substitution on procollagen folding, structure or trafficking through the cell. Normal expression of the key regulator of unfolded protein response, BIP, suggests normal folding, structure and trafficking of other proteins as well, consistent with previously reported studies of Aha effects (21, 25, 26). Because Met residues within the triple helix are exposed to the solvent, it is not unexpected that their replacement with Aha has no significant consequences for the triple helix folding or structure. More surprisingly, this substitution appears to have no effect on folding and structure of even the globular C-propeptide and other globular proteins (26), in which Met is buried inside the protein core.
Apparently, Aha is not just a safer, cheaper, and more convenient alternative to radioisotopes for pulse-chase studies of procollagen biosynthesis, but it also has fewer unintended consequences and its results are easier to interpret. On a cautionary note, however, our and previously published studies do not exclude unintended consequences of Aha labeling for proteins and cell functions that have not been examined so far. For instance, our study revealed no effects of Aha on procollagen biosynthesis or function of fibroblasts. Previously published studies revealed no effects of Aha on other proteins and other types of cells (20, 21, 23, 25, 26). Yet, this evidence is not sufficient to exclude all potential effects of Aha. Further characterization of potential unintended consequences might be necessary for other applications of Aha labeling or for making conclusions that extend beyond procollagen synthesis, folding, and trafficking in fibroblasts. At the same time, it is important to keep in mind that such a characterization is even more critical for radioisotope labeling, since the latter does lead to multiple known problems discussed above.
Procollagen Misfolding in OI Fibroblasts
Pulse-chase labeling with Aha enabled us to study abnormal folding and clearance of procollagen with Cys substitution for Gly766 in the α1(I) chain (Fig. 5), in cultured dermal fibroblasts from an OI patient. Consistent with a previous, radioisotope-based study of fibroblasts from OI patients with other Gly substitutions (34) and the established role of obligatory Gly residues (1), we observed significantly slower triple helix folding in OI compared to normal control cells (Fig. 5a). Our assay did not detect a folding intermediate paused at the mutation site. Note that in (34), such an intermediate was detected only for a Gly94 substitution at the N-terminal end of the triple helix but not for Gly substitutions located closer to the C-terminal end. In the latter mutations, the folding intermediates might not have sufficiently long lifetimes to be detected by the trypsin/chymotrypsin digestion assay or the corresponding collagen digestion products might not co-purify with full-length triple helices.
Slower folding likely contributes to slower clearance of Aha-labeled procollagen molecules from OI fibroblasts (Fig. 5c). However, less than 50% decrease in Aha-labeled molecules inside the cells after 4 h chase is difficult to explain just by the delay in their folding. The dramatic increase in the lifetime of a labeled molecule inside the cell points to a delay in trafficking through or export from the cell. In particular, misfolded molecules with Gly substitutions might be selectively retained and targeted for intracellular degradation (14, 41, 47-49). Cell stress response to such misfolding and accumulation of mutant procollagen molecules in osteoblasts might contribute to bone pathology in OI (3, 35).
In conclusion, the goal of the present manuscript is to establish pulse-chase labeling with Aha as a promising technique for studies of procollagen biosynthesis. In the context of OI studies, the experiments described above represent just a first step in developing this technique. For instance, direct visualization of Aha-labeled molecules by conjugation with fluorescent dyes, which can be done even in live cells (25), may be exploited for much more detailed analysis of procollagen synthesis, trafficking and degradation. Since understanding how OI cells handle mutant procollagen might provide important clues to OI pathophysiology and even novel approaches to treatment, we are currently developing assays and strategies for such studies. We hope that the present report will stimulate the interest of other researchers to this approach, thereby helping to unravel molecular mechanisms of a variety of collagen metabolism disorders.
Acknowledgements
The authors thank Dr. Dan L. Sackett for insightful comments and suggestions, Ms. Linda Powers for technical assistance, as well as Drs. Peter H. Byers and Joan C. Marini for generously providing cells used in this study.
Funding
This work was funded by the Intramural Research Program, NICHD, NIH.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Shapiro JR, Byers PH, Glorieux FH, Sponseller PD, editors. Osteogenesis imperfecta: a translational approach to brittle bone disease. Academic Press; Amsterdam: 2014. [Google Scholar]
- 2.Arnold WV, Fertala A. Skeletal diseases caused by mutations that affect collagen structure and function. Int J Biochem Cell B. 2013;45:1556–67. doi: 10.1016/j.biocel.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Bateman JF, Boot-Handford RP, Lamande SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat Rev Genet. 2009;10:173–83. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 4.De Paepe A, Malfait F. The Ehlers-Danlos syndrome, a disorder with many faces. Clin Genet. 2012;82:1–11. doi: 10.1111/j.1399-0004.2012.01858.x. [DOI] [PubMed] [Google Scholar]
- 5.Byers PH, Murray ML. Ehlers-Danlos syndrome: A showcase of conditions that lead to understanding matrix biology. Matrix Biol. 2014;33:10–5. doi: 10.1016/j.matbio.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 8.Makareeva E, Han SJ, Vera JC, Sackett DL, Holmbeck K, Phillips CL, et al. Carcinomas Contain a Matrix Metalloproteinase-Resistant Isoform of Type I Collagen Exerting Selective Support to Invasion. Cancer Res. 2010;70:4366–74. doi: 10.1158/0008-5472.CAN-09-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansens A, Braakman I. Pulse-chase labeling techniques for the analysis of protein maturation and degradation. Methods Mol Biol. 2003;232:133–45. doi: 10.1385/1-59259-394-1:133. [DOI] [PubMed] [Google Scholar]
- 10.Pollard J. Radioisotopic Labeling of Proteins for Polyacrylamide Gel Electrophoresis. In: Walker J, editor. The Protein Protocols Handbook. Humana Press; 1996. pp. 121–6. [Google Scholar]
- 11.Bonifacino JS. Metabolic labeling with amino acids. Current Protocols in Molecular Biology. 2001;44:10.8.1–8. doi: 10.1002/0471142727.mb1018s44. VI. [DOI] [PubMed] [Google Scholar]
- 12.Coligan JE, Gates FT, Kimball ES, Maloy WL. Radiochemical Sequence-Analysis of Biosynthetically Labeled Proteins. Methods Enzymol. 1983;91:413–34. doi: 10.1016/s0076-6879(83)91039-x. [DOI] [PubMed] [Google Scholar]
- 13.Bateman JF, Mascara T, Chan D, Cole WG. Abnormal Type-I Collagen-Metabolism by Cultured Fibroblasts in Lethal Perinatal Osteogenesis Imperfecta. Biochem J. 1984;217:103–15. doi: 10.1042/bj2170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barsh GS, Byers PH. Reduced Secretion of Structurally Abnormal Type-I Procollagen in a Form of Osteogenesis Imperfecta. Proc Natl Acad Sci U S A. 1981;78:5142–6. doi: 10.1073/pnas.78.8.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–60. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 16.Van Hoof D, Pinkse MWH, Oostwaard DWV, Mummery CL, Heck AJR, Krijgsveld J. An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat Methods. 2007;4:677–8. doi: 10.1038/nmeth0907-677. [DOI] [PubMed] [Google Scholar]
- 17.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–94. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 18.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Bio. 2006;7:952–8. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 19.Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr Opin Chem Biol. 2010;14:774–80. doi: 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dieterich D, Lee J, Link A, Graumann J, Tirrell D, Schuman E. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–40. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 21.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103:9482–7. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prescher J, Bertozzi C. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 23.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, et al. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:16793–7. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin RJ. The medicinal chemistry of the azido group. Prog Med Chem. 1994;31:121–232. doi: 10.1016/s0079-6468(08)70020-1. [DOI] [PubMed] [Google Scholar]
- 25.Dieterich D, Hodas J, Gouzer G, Shadrin I, Ngo J, Triller A, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taskent-Sezgin H, Chung J, Banerjee P, Nagarajan S, Dyer R, Carrico I, et al. Azidohomoalanine: A Conformationally Sensitive IR Probe of Protein Folding, Protein Structure, and Electrostatics. Angew Chem. 2010;49:7473–5. doi: 10.1002/anie.201003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dover R, Jayaram Y, Patel K, Chinery R. p53 expression in cultured cells following radioisotope labelling. J Cell Sci. 1994;107:1181–4. doi: 10.1242/jcs.107.5.1181. [DOI] [PubMed] [Google Scholar]
- 28.Yeargin J, Haas M. Elevated Levels of Wild-Type P53 Induced by Radiolabeling of Cells Leads to Apoptosis or Sustained Growth Arrest. Curr Biol. 1995;5:423–31. doi: 10.1016/s0960-9822(95)00083-2. [DOI] [PubMed] [Google Scholar]
- 29.Hu VW, Heikka DS. Radiolabeling revisited: metabolic labeling with (35)S-methionine inhibits cell cycle progression, proliferation, and survival. FASEB J. 2000;14:448–54. doi: 10.1096/fasebj.14.3.448. [DOI] [PubMed] [Google Scholar]
- 30.Hu VW, Heikka DS, Dieffenbach PB, Ha L. Metabolic radiolabeling: experimental tool or Trojan horse? (35)S-Methionine induces DNA fragmentation and p53-dependent ROS production. FASEB J. 2001;15:1562–8. doi: 10.1096/fj.01-0102com. [DOI] [PubMed] [Google Scholar]
- 31.Marko NF, Dieffenbach PB, Yan G, Ceryak S, Howell RW, Mccaffrey TA, et al. Does metabolic radiolabeling stimulate the stress response? Gene expression profiling reveals differential cellular responses to internal beta vs. external gamma radiation. FASEB J. 2003;17:1470–86. doi: 10.1096/fj.02-1194com. [DOI] [PubMed] [Google Scholar]
- 32.Solary E, Bertrand R, Jenkins J, Pommier Y. Radiolabeling of DNA Can Induce Its Fragmentation in Hl-60 Human Promyelocytic Leukemic-Cells. Exp Cell Res. 1992;203:495–8. doi: 10.1016/0014-4827(92)90027-6. [DOI] [PubMed] [Google Scholar]
- 33.Yanokura M, Takase K, Yamamoto K, Teraoka H. Cell death and cell-cycle arrest induced by incorporation of [H-3]thymidine into human haemopoietic cell lines. Int J Radiat Biol. 2000;76:295–303. doi: 10.1080/095530000138637. [DOI] [PubMed] [Google Scholar]
- 34.Raghunath M, Bruckner P, Steinmann B. Delayed triple helix formation of mutant collagen from patients with osteogenesis imperfecta. J Mol Biol. 1994;236:940–9. doi: 10.1006/jmbi.1994.1199. [DOI] [PubMed] [Google Scholar]
- 35.Makareeva E, Aviles NA, Leikin S. Chaperoning osteogenesis: new protein-folding disease paradigms. Trends Cell Biol. 2011;21:168–76. doi: 10.1016/j.tcb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makareeva E, Cabral WA, Marini JC, Leikin S. Molecular mechanism of alpha 1(I)-osteogenesis Imperfecta/Ehlers-Danlos syndrome - Unfolding of an N-anchor domain at the N-terminal end of the type I collagen triple helix. J Biol Chem. 2006;281:6463–70. doi: 10.1074/jbc.M511830200. [DOI] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, Milgrom S, et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209–21. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruckner P, Eikenberry EF. Formation of the triple helix of type I procollagen in cellulo. Temperature-dependent kinetics support a model based on cis in equilibrium trans isomerization of peptide bonds. Eur J Biochem. 1984;140:391–5. doi: 10.1111/j.1432-1033.1984.tb08114.x. [DOI] [PubMed] [Google Scholar]
- 40.Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007;39:359–65. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forlino A, Kuznetsova NV, Marini JC, Leikin S. Selective retention and degradation of molecules with a single mutant alpha1(I) chain in the Brtl IV mouse model of OI. Matrix Biol. 2007;26:604–14. doi: 10.1016/j.matbio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Dover R, Jayaram Y, Patel K, Chinery R. P53 Expression in Cultured-Cells Following Radioisotope Labeling. J Cell Sci. 1994;107:1181–4. doi: 10.1242/jcs.107.5.1181. [DOI] [PubMed] [Google Scholar]
- 43.Lander H. An essential role for free radicals and derived species in signal transduction. FASEB J. 1997;11:118–24. [PubMed] [Google Scholar]
- 44.Chau JF, Jia D, Wang Z, Liu Z, Hu Y, Zhang X, et al. A crucial role for bone morphogenetic protein-Smad1 signalling in the DNA damage response. Nat Commun. 2012;3:836. doi: 10.1038/ncomms1832. [DOI] [PubMed] [Google Scholar]
- 45.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 46.Kiick K, Saxon E, Tirrell D, Bertozzi C. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bateman JF, Chan D, Mascara T, Rogers JG, Cole WG. Collagen defects in lethal perinatal osteogenesis imperfecta. Biochem J. 1986;240:699–708. doi: 10.1042/bj2400699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel BE, Minor RR, Freund M, Prockop DJ. A point mutation in a type I procollagen gene converts glycine 748 of the alpha 1 chain to cysteine and destabilizes the triple helix in a lethal variant of osteogenesis imperfecta. J Biol Chem. 1987;262:14737–44. [PubMed] [Google Scholar]
- 49.Ishida Y, Yamamoto A, Kitamura A, Lamande SR, Yoshimori T, Bateman JF, et al. Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell. 2009;20:2744–54. doi: 10.1091/mbc.E08-11-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]